ABSTRACT
The present study employed mass sequencing of small RNA libraries to identify the repertoire of small noncoding RNAs expressed in normal CD4+ T cells compared to cells transformed with human T-cell leukemia virus type 1 (HTLV-1), the causative agent of adult T-cell leukemia/lymphoma (ATLL). The results revealed distinct patterns of microRNA expression in HTLV-1-infected CD4+ T-cell lines with respect to their normal counterparts. In addition, a search for virus-encoded microRNAs yielded 2 sequences that originated from the plus strand of the HTLV-1 genome. Several sequences derived from tRNAs were expressed at substantial levels in both uninfected and infected cells. One of the most abundant tRNA fragments (tRF-3019) was derived from the 3′ end of tRNA-proline. tRF-3019 exhibited perfect sequence complementarity to the primer binding site of HTLV-1. The results of an in vitro reverse transcriptase assay verified that tRF-3019 was capable of priming HTLV-1 reverse transcriptase. Both tRNA-proline and tRF-3019 were detected in virus particles isolated from HTLV-1-infected cells. These findings suggest that tRF-3019 may play an important role in priming HTLV-1 reverse transcription and could thus represent a novel target to control HTLV-1 infection.
IMPORTANCE Small noncoding RNAs, a growing family of regulatory RNAs that includes microRNAs and tRNA fragments, have recently emerged as key players in many biological processes, including viral infection and cancer. In the present study, we employed mass sequencing to identify the repertoire of small noncoding RNAs in normal T cells compared to T cells transformed with human T-cell leukemia virus type 1 (HTLV-1), a retrovirus that causes adult T-cell leukemia/lymphoma. The results revealed a distinct pattern of microRNA expression in HTLV-1-infected cells and a tRNA fragment (tRF-3019) that was packaged into virions and capable of priming HTLV-1 reverse transcription, a key event in the retroviral life cycle. These findings indicate tRF-3019 could represent a novel target for therapies aimed at controlling HTLV-1 infection.
INTRODUCTION
Adult T-cell leukemia/lymphoma (ATLL) is an aggressive neoplasm of mature CD4+ cells that is etiologically linked to infection with human T-cell leukemia virus type 1 (HTLV-1). About 15 to 25 million people are infected with HTLV-1 worldwide, with infection most prevalent in southwestern Japan and the Caribbean basin. The virus is transmitted through blood, semen, and breast milk. While most infected individuals remain asymptomatic, about 3% eventually develop ATLL after decades of clinical latency. HTLV-1 also causes tropical spastic paraparesis/HTLV-associated myelopathy (TSP/HAM), a progressive demyelinating disease that targets mainly the thoracic spinal cord; similar to ATLL, TSP/HAM arises in about 3% of infected individuals, but after a latency period of years rather than decades (for reviews of HTLV-1 pathogenesis, see references 1 and 2).
HTLV-1 was the first human retrovirus to be identified and is the only one with a direct etiological link to cancer. HTLV-1 is classified as a “complex” retrovirus, as its genome contains extra open reading frames (ORFs), in addition to the gag, pol, pro, and env genes common to all retroviruses (reviewed in reference 3). The extra ORFs in HTLV-1 code for a transcriptional transactivator named Tax, a posttranscriptional regulatory protein named Rex, and four accessory proteins named HBZ, p30, p13, and p12/p8 (4).
HTLV-1 is found mainly in CD4+ T cells in vivo. Infection of peripheral blood mononuclear cells (PBMC) with HTLV-1 yields interleukin 2 (IL-2)-dependent immortalized T cells, some of which progress to a fully transformed phenotype with IL-2-independent growth. The immortalizing potential of HTLV-1 is attributable primarily to the viral protein Tax. In addition to transactivating the viral promoter, Tax affects the expression and function of cellular genes controlling signal transduction, cell growth, apoptosis, and chromosomal stability (5) and is able to cause leukemia when expressed as a transgene in mice (6). HBZ also likely contributes to the oncogenic properties of HTLV-1; it is mitogenic for T cells (7) and is able to induce leukemia in transgenic mice (8). Other accessory proteins may affect viral transmission and persistence (9–11).
In the present study, we investigated the expression repertoire of small noncoding RNAs, in particular microRNAs (miRNAs) and tRNA fragments (tRFs), in HTLV-1-infected cells. MicroRNAs negatively regulate gene expression at the posttranscriptional level by base pairing to specific target mRNAs in RNA-induced silencing complexes (RISC) containing Argonaute proteins. Perfect base pairing leads to degradation of the mRNA in an RNA interference (RNAi)-like manner, while imperfect base pairing (the more frequent interaction) results in a block of translation (12). Posttranscriptional regulation of gene expression by microRNAs is of critical importance in normal cell physiology, and aberrant expression of microRNAs is emerging as a key component of a wide range of pathologies, including solid and hematological tumors (13).
Expression studies based on quantitative reverse transcription (RT)-PCR and microarray analysis identified a number of cellular microRNAs that are downregulated or upregulated in HTLV-1-infected cell lines and ATLL cells (14–17). Viruses may also produce microRNAs as a means of regulating the expression of viral or host genes (18). Recent studies of bovine leukemia virus (BLV), a retrovirus in the same subfamily as HTLV-1, revealed the expression of a cluster of viral microRNAs (19, 20). A computational analysis of the HTLV-1 genome identified 11 sequences with potential to form stem-loop structures that could yield viral microRNAs (21).
tRFs are produced from the 3′ ends of tRNA precursors or from the 5′ ends or the 3′ ends of mature tRNAs and are designated tRF-1, tRF-5, and tRF-3, respectively (22). tRFs have an average length of 19 nucleotides (nt) (23) and, similar to microRNAs, are produced by specific cleavage events rather than through degradation. tRF-1 sequences are cleaved from tRNA precursors by RNase Z, while Dicer was shown to be responsible for cleavage to produce a tRF-5 sequence (23) and a tRF-3 sequence (24). Although not much is known about the function of tRFs, some are able to form complexes with Argonaute proteins (23, 24), and one tRF-3 sequence repressed expression of specific mRNA targets through a microRNA-like mechanism (24).
We employed massive sequencing to identify the repertoire of microRNAs and tRFs expressed in HTLV-1-infected cells compared to normal CD4+ T cells. Comparison of the frequencies of known microRNAs in the libraries revealed 3 microRNAs that were differentially expressed in the infected cell lines compared to CD4 controls, 2 small RNAs that matched HTLV-1 sequences, and several abundant tRFs. We provide evidence that a tRF corresponding to the 3′ end of tRNA-proline (Pro) is incorporated into virus particles and can function as a primer for viral reverse transcriptase.
MATERIALS AND METHODS
Cell culture.
Cell lines C91PL and MT-2, chronically infected with HTLV-1 (25), were maintained in RPMI (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 2 mM glutamine (Invitrogen), 100 units/ml penicillin, and 20 units/ml streptomycin (complete RPMI). PBMC were isolated from buffy coat fractions obtained from healthy plasma donors attending the Transfusion Unit of Padua City Hospital by centrifugation through Ficoll-Hypaque (GE Healthcare). Half of the PBMC sample was immediately processed using the MACS CD4+ T cell Isolation Kit II (Miltenyi Biotec), and the resulting CD4 cells were harvested for total RNA (see below). The other half of the PBMC preparation was placed in complete RPMI (1 × 106 cells/ml) supplemented with 100 μg/ml phytohemagglutinin (PHA) (Sigma-Aldrich) and cultured for 48 h. The culture was then supplemented with 50 U/ml IL-2 (Proleukin-Chiron), incubated for an additional 48 h, and harvested for isolation of CD4 cells with the MACS kit. Flow cytometry analysis revealed that the isolated cell preparations contained more than 99% and 94% CD4 cells in the unstimulated and stimulated preparations, respectively. Total RNA was isolated using TRIzol (Invitrogen). The quality of the RNA was assessed by electrophoresis using the RNA 6000 Nano Assay LabChip Kit and Agilent 2100 Bioanalyzer, and the RNA concentration was measured using a Nanodrop spectrophotometer.
Generation of small RNA libraries.
Small RNA libraries were generated according to the method of Lau et al. (26) as follows. A 10-μg aliquot of each total RNA preparation was spiked with a 32P-labeled 23-nt RNA tracer and then subjected to polyacrylamide gel electrophoresis (PAGE) through a 15% denaturing gel, along with a 32P-labeled aliquot of Decade RNA ladder (Ambion); the presence of the RNA tracer permitted visualization of each modification and purification step after exposure of the gel to a phosphorimaging screen (Storm; GE Healthcare). Species migrating in the ∼18- to 25-nucleotide size range were excised from the gel, eluted, and ethanol precipitated. The RNA was then modified by addition of a 17-nt oligonucleotide linker (miRNA Cloning Linker 1; IDT) at the 3′ end with RNA ligase (GE Healthcare), PAGE purified through a 12% denaturing gel, modified with a second 17-nt oligonucleotide linker at the 5′ end, and PAGE purified again through a 10% denaturing gel. The resulting modified RNA was reverse transcribed and PCR amplified in preparation for sequencing according to a protocol provided by G. Hannon (Cold Spring Harbor Laboratory), as described previously (27). The resulting samples were subjected to 454 massive sequencing using a Roche Life Sciences platform. The total numbers of sequence reads were 7,709 (freshly isolated CD4), 7,818 (stimulated CD4), 7,603 (C91PL), and 6,801 (MT-2). The library prepared from freshly isolated CD4+ cells was described previously (27). Lists of sequence reads are available upon request.
Identification of microRNAs and tRFs by bioinformatics analysis.
The sequence reads were trimmed from matches with sequencing primers or linker sequences and subjected to nonredundancy analysis with the NCBI nrdb program of the BLAST suite (28).
The sequences were compared with the mature microRNA subset of the mirBase database, version 18 (http://www.mirbase.org/) using the Smith-Waterman search program from the FastA sequence analysis suite (29), allowing a maximum of one mismatch between the sequence read and the reference mature miRNA. Read counts annotated with the mature microRNA names were tested for differential expression with the Bioconductor edgeR package (30). Reads that did not match mature microRNAs were subjected to a further comparison with mirBase precursor sequences. Sequences that did not map to known microRNA precursors were compared with the downloaded FastA sequences from the Genomic tRNA database (http://gtrnadb.ucsc.edu/) using the SHRiMP program (http://compbio.cs.toronto.edu/shrimp/). tRF-3019 and other tRFs were identified by searching sequence lists with Excel tools. The small RNA sequences that did not correspond to known microRNAs or tRFs were subjected to novel small RNA prediction with the mirDeep2 pipeline (31), but no suitable novel candidates were found. The analyses were integrated with ad hoc written perl scripts.
To identify potential viral microRNAs, the sequence sets were compared to the HTLV-1 genome sequence ATK (GenBank accession no. J02029 and DDBJ accession no. M33896). Sequences with ≥90% identity with HTLV-1 and ≤2 gaps and ≤2 mismatches were further analyzed as described in Results below.
RT assay using tRF-3019. (i) Preparation of RNA template.
A DNA fragment corresponding to nt 721 to 822 of HTLV-1 ATK was PCR amplified using the HTLV-1 molecular clone ACH (32) as a template and primers U5-s and Gag-as (see Table S1 in the supplemental material). A 20-nt tail was added to the 5′ end of the product with a second round of PCR using primers Tail-U5-s and Gag-as. The 129-nt fragment was cloned into vector pSG5E, which is a modified version of pSG5 (Stratagene) containing the polylinker of pBluescript (Stratagene) 3′ to the T7 promoter. The resulting plasmid (pSG-U5-PBS) was linearized 3′ to the insert and in vitro transcribed with T7 RNA polymerase (Invitrogen). After a DNase 1 digestion to eliminate the plasmid, the mixture was extracted with phenol-chloroform and ethanol precipitated to recover the RNA. The resulting pellet was resuspended in distilled H2O and stored at −80°C.
(ii) Preparation of virus particle lysates containing HTLV-1 reverse transcriptase.
Confluent cultures of C91PL cells were centrifuged at low speed to remove the cells. The supernatant was passed through a 0.45-μm filter (Sartorius) and centrifuged at 24,000 rpm in an SW28 rotor for 2 h. The pelleted material was resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 0.5% Nonidet-P40; 10 μl per 10 ml centrifuged supernatant) and stored at −80°C.
(iii) RT assay.
The RT assay was based on a published method (33, 34). For each RT assay, a 100-ng aliquot of in vitro-transcribed RNA was combined with 10 pmol of either tRF-3019 RNA, miR-150-5p RNA (negative control), tRF-3019 DNA (positive control), or water instead of primer in a 10.5-μl volume; annealed at 70°C for 10 min; and then cooled on ice. The mixtures were brought to a final volume of 20 μl containing 1 mM each deoxynucleoside triphosphate (dNTP), 10 U RNase inhibitor, RT buffer (25 mM Tris-HCl, pH 8.3, 5 mM MgCl2, 50 mM KCl, 2 mM dithiothreitol [DTT]), and 2 μl virion lysate and incubated at 37°C for 1 h, followed by 95°C for 5 min. A 2.5-μl aliquot of the cDNA product was amplified in a final volume of 25 μl containing 1× Taq Gold PCR buffer, 2 mM MgCl2, 200 μM dNTPs, 5 pmol each of primers Tail-s and U5-as, and 0.5 U AmpliTaq Gold DNA polymerase (Life Technologies). The PCR method consisted of a denaturation step at 94°C for 1 min, followed by 25 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 45 s. The products were analyzed on a 6% polyacrylamide gel and stained with ethidium bromide. Images were obtained using a Bio-Rad Gel Doc XRS system.
RT-PCR to detect tRFs, tRNAs, and gag/pol RNA.
FBS may contain exosomes carrying small RNAs; to avoid this possible source of contamination, complete RPMI containing 20% FBS was centrifuged at 24,000 rpm for 4 h using a Beckman-Coulter SW28 rotor to pellet any exosomes. The supernatant medium was then passed through a 0.2-μm filter and brought to 10% FBS by adding an equal volume of RPMI containing antibiotics and glutamine. C91PL cells were cultured to confluence in the exosome-depleted medium, and virus particles were recovered as described above. RNA was isolated from the pelleted particles and the producer C91PL cells using TRIzol LS (Life Technologies). Aliquots of the RNA were subjected to denaturing PAGE through a 15% polyacrylamide gel to separate species in the size range of full-length tRNAs from the small RNAs, with tRNAs visible in the cellular RNA sample and 5 pmol synthetic miR-150-5p serving as size markers for the 2 fractions. The gel was stained with ethidium bromide, and the regions containing tRNAs and small RNAs (about 15 to 30 nt) were excised, crushed, and incubated with gentle mixing in elution buffer (300 mM sodium acetate, pH 5.2, 1 mM EDTA) overnight at 4°C. RNA was ethanol precipitated and resuspended in distilled H2O (10 μl per 19 ml of original culture supernatant) (see Fig. 6, where the fractions are labeled tRNA and tRF).
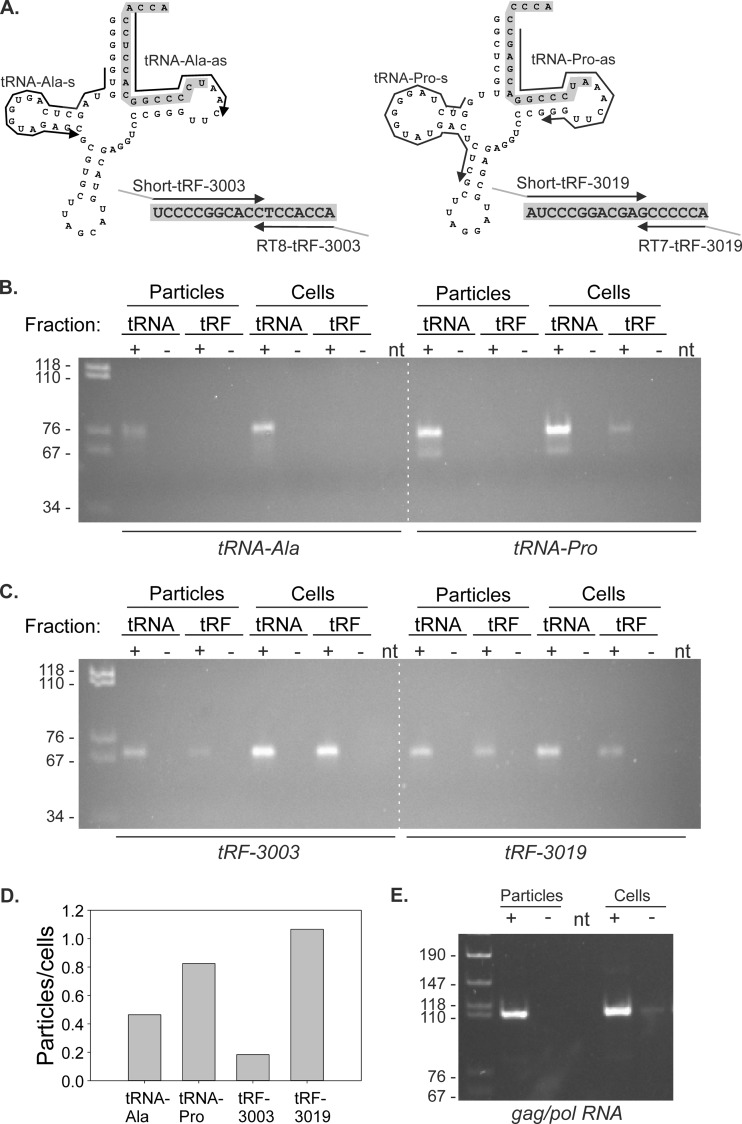
FIG 6.
RT-PCR to detect tRNAs, tRFs, and gag/pol RNA in virus particles and C91PL cells. (A) As described in Materials and Methods, RNA from virus particles and producer C91PL cells was subjected to denaturing PAGE; regions of the gel containing tRNA and tRFs were excised, and RNA was recovered by passive elution and ethanol precipitation. The resulting fractions were subjected to RT-PCR to detect tRNA-Ala, tRNA-Pro, and their tRF-3 sequences, tRF-3003 and tRF-3019, respectively. (B and C) Images of the RT-PCR products after separation on 6% polyacrylamide gels. The intensities of RT-PCR bands obtained for tRNAs and tRFs (measured in tRNA and tRF fractions, respectively) were measured using a Bio-Rad Gel Doc XRS imager. (D) Plot of ratios of band intensities obtained for virus particles versus cells. The calculated ratios were as follows: tRNA-Ala, particles/cells = 0.46; tRNA-Pro, particles/cells = 0.82; tRF-3003, particles/cells = 0.18; tRF-3019, particles/cells = 1.07. (E) Results of RT-PCR performed on RNA from the virus particles and producer cells to detect HTLV-1 genomic gag/pol RNA. RT-PCR was carried out using primers U5-s and Gag-as as described in Materials and Methods. The dashed white lines were added to panels B and C to aid in their alignment. The first lane on each gel contained MspI-digested pBluescript as a size marker; band sizes in basepairs are indicated on the left. The plus and minus signs above the lanes indicate RT reactions carried out in the presence (+) or absence (−) of reverse transcriptase. RNA template was omitted from the RT reaction in lanes labeled nt.
The primer sets used to amplify tRF-3019, tRF-3003, and the tRNAs from which they are derived (tRNA-Pro and tRN-Ala, respectively) are listed in Table S1 in the supplemental material (also see Fig. 6A). RT-PCR to detect tRF-3019 and tRF-3003 was based on a protocol for detecting microRNAs (35). Size-fractionated RNA (2 μl) was annealed with 2 pmol primer RT7-tRF-3019 or RT8-tRF-3003 at 70°C for 10 min in a 7.5-μl volume. The mixture was brought to 10 μl with the addition of 1 mM dNTP, 1× RT buffer, and 5 U avian myeloblastosis virus (AMV) reverse transcriptase (Finnzymes) and reverse transcribed at 40°C for 1 h. Two-microliter aliquots of the resulting cDNAs were PCR amplified in a final volume of 25 μl containing 1× Taq Gold PCR buffer, 1.5 mM MgCl2, 200 μM dNTP, 0.1 pmol of primer Short-tRF-3019 or Short-tRF-3003, 2.5 pmol each of primers PCR-tRF-s and PCR-tRF-as (specific for tails added by the RT primer and Short-tRF primer), and 0.5 U AmpliTaq Gold DNA polymerase. The PCR method consisted of a denaturation step at 95°C for 10 min and 5 cycles of denaturation at 95°C for 30 s, annealing at 40°C for 45 s, and extension at 72°C for 30 s, followed by 22 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 45 s, and extension at 72°C for 30 s.
To detect tRNA-Pro and tRNA-Ala, 1 μl of size-fractionated RNA was reverse transcribed in a 10-μl reaction mixture at 53°C for 50 min using the antisense primer and Superscript III (Life Technologies). The resulting cDNA (2.5 μl) was PCR amplified using sense and antisense primers and AmpliTaq Gold, with a denaturation step at 95°C for 8 min, followed by cycles of denaturation at 95°C for 40 s, annealing at 60°C for 40 s, and extension at 72°C for 40 s (30 cycles for tRNA-Ala and 26 cycles for tRNA-Pro). To detect gag/pol RNA, 0.5 μl of RNA from virus particles or 200 ng total RNA from producer C91PL cells was reverse transcribed using primer Gag-as and Superscript III and then PCR amplified using primers U5-s and Gag-as (see Table S1 in the supplemental material) and AmpliTaq Gold as described above for 30 cycles with an annealing temperature of 59°C and a final extension step for 5 min at 72°C. The PCR products were separated on 6% polyacrylamide gels.
RESULTS
Generation, sequencing, annotation, and quantification of small RNA libraries.
Total RNA was isolated from the HTLV-1-infected cell lines C91PL and MT-2 (25), which have a CD4+ phenotype, and from control normal unstimulated and in vitro-stimulated CD4+ T cells. RNA species of ∼18 to 25 nt were separated by polyacrylamide gel electrophoresis and modified with tags at the 5′ and 3′ ends according to the method of Lau et al. (26) and sequenced using a Roche 454 Life Sciences platform. Sequence reads were mapped against mirBase release 18 to identify the populations of mature known microRNAs in the investigated cell lines, and the edgeR Bioconductor statistical package (see Materials and Methods) was employed to identify statistically significant differences in the frequencies of known microRNAs.
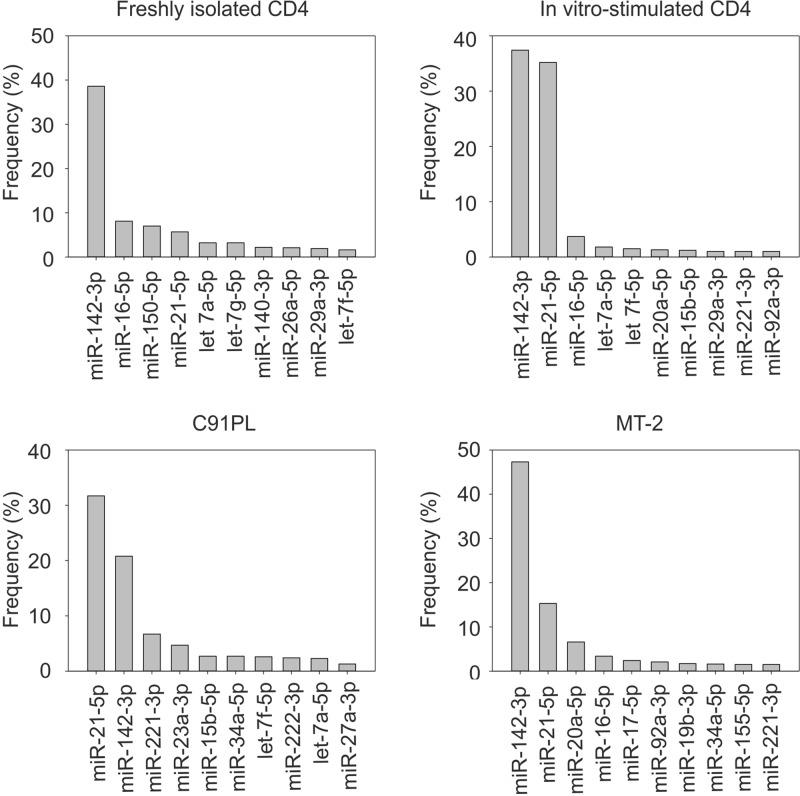
The graphs in Fig. 1 illustrate the 10 most abundant known microRNAs identified in the libraries. miR-142-3p, a marker of hematopoietic cells, stood out as the most abundant microRNA in freshly isolated CD4+ T cells and in the infected cell line MT-2, with a frequency more than triple that of the other microRNAs. miR-21, a microRNA known to be linked to T-cell activation and transformation (36), became nearly as frequent as miR-142-3p upon in vitro stimulation of CD4+ cells and was also abundant in the two infected cell lines.
FIG 1.
Relative abundances of microRNAs identified by sequencing. Shown are the 10 most abundant microRNAs in normal CD4 T cells and HTLV-1-infected cell lines. Frequencies were calculated by dividing the number of sequence reads for each microRNA by the total number of sequence reads for all known microRNAs.
To identify the microRNAs connected with HTLV-1 infection, we calculated differences in the frequencies of microRNAs in infected cell lines versus resting and stimulated CD4+ T cells. Three microRNAs were differentially expressed in both infected cell lines compared to control CD4 cells (indicated in boldface in Table 1: miR-34a-5p was upregulated, and miR-150-5p and miR-146b-5p were both downregulated). An analysis performed using the miRDeep2 software did not yield any putative new microRNA candidates among the sequence reads detected in this study.
TABLE 1.
Differentially expressed microRNAsa
| Cells | MicroRNA | Log2 FC | P value |
|---|---|---|---|
| MT-2 vs. CD4 | miR-34a-5p | 6.15 | 0.00015 |
| miR-4448 | 4.89 | 0.00366 | |
| miR-7-5p | 4.35 | 0.01564 | |
| miR-150-5p | −10.08 | 0.00327 | |
| miR-30c-5p | −8.05 | 0.01329 | |
| miR-146b-5p | −7.52 | 0.03318 | |
| miR-29c-3p | −6.87 | 0.03802 | |
| C91PL vs. CD4 | miR-34a-5p | 6.66 | 0.00001 |
| miR-92b-3p | 3.82 | 0.01313 | |
| miR-23a-3p | 3.15 | 0.01166 | |
| miR-150-5p | −10.61 | 0.00127 | |
| miR-342-5p | −7.02 | 0.02719 | |
| miR-26a-5p | −6.84 | 0.00776 | |
| miR-20b-5p | −6.74 | 0.03088 | |
| miR-146b-5p | −4.76 | 0.02268 | |
| miR-19b-3p | −3.83 | 0.03015 | |
| miR-16b-5p | −3.65 | 0.02867 |
MicroRNAs with statistically significant differences in expression are indicated in boldface. FC, fold change.
Small RNAs expressed by HTLV-1.
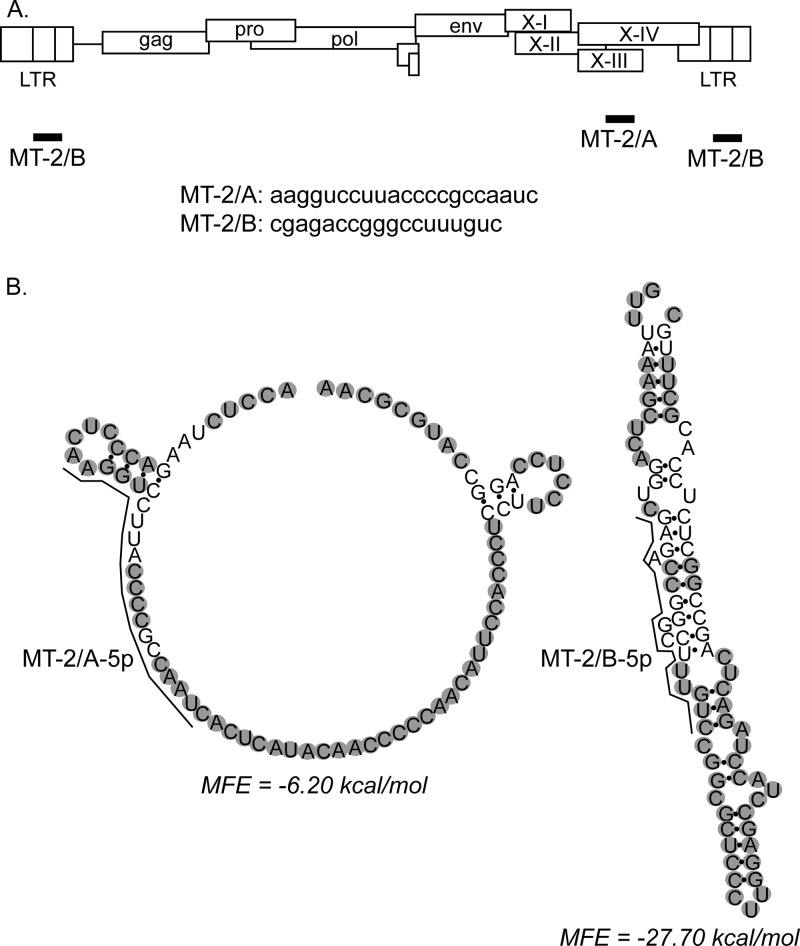
With the aim of identifying viral microRNAs, the reads obtained from MT-2 and C91PL cells were aligned to the HTLV-1 genome. This analysis yielded 25 sequences with ≥90% identity with HTLV-1 and ≤2 gaps and ≤2 mismatches (data not shown). Two sequences shown in Fig. 2A perfectly matched the primary plus-strand HTLV-1 transcript. Both sequences were present only in the MT-2 library. Sequence MT-2/A was positioned in exon 3, between the stop codons for p30/p13 and Rex. Sequence MT-2/B was located in the R region, in a position within stem-loop D of the Rex response element (RXRE) (37). This segment of the HTLV-1 genome was predicted to form a microRNA precursor hairpin (pre-miRNA)-like structure and thus to have the potential to give rise to a viral microRNA (21). Figure 2B shows the predicted secondary structures of genomic regions containing MT-2/A and MT-2/B with 5′ and 3′ flanking sequences to simulate their position in the 5′ portion of a pre-miRNA. Results showed that sequence MT-2/A is likely to be present mostly in an unstructured region, while sequence MT-2/B has a high probability to be positioned in a stem.
FIG 2.
Small RNAs expressed by HTLV-1. (A) Positions and nucleotide sequences of the two small RNA species identified in MT-2 cells. MT-2/A corresponded to nt 7582 to 7602 in exon 3 of the HTLV-1 ATK sequence. MT-2/B corresponded to nt 513 to 530 of the 5′ R region and nt 8792 to 8808 of the 3′ R region of ATK. (B) Secondary structure predicted by RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi; University of Vienna) for HTLV-1 sequences containing MT-2/A and MT-2/B with 15 nt added at the 5′ end and 50 nt at the 3′ end to simulate a pre-miR. The optimal secondary structures and their minimum free energy (MFE) values are indicated. Nucleotides in grey circles have a high probability of occupying the indicated paired or unpaired position and were identified with red or orange circles in the original RNAfold diagram.
tRFs expressed in HTLV-1-infected cells.
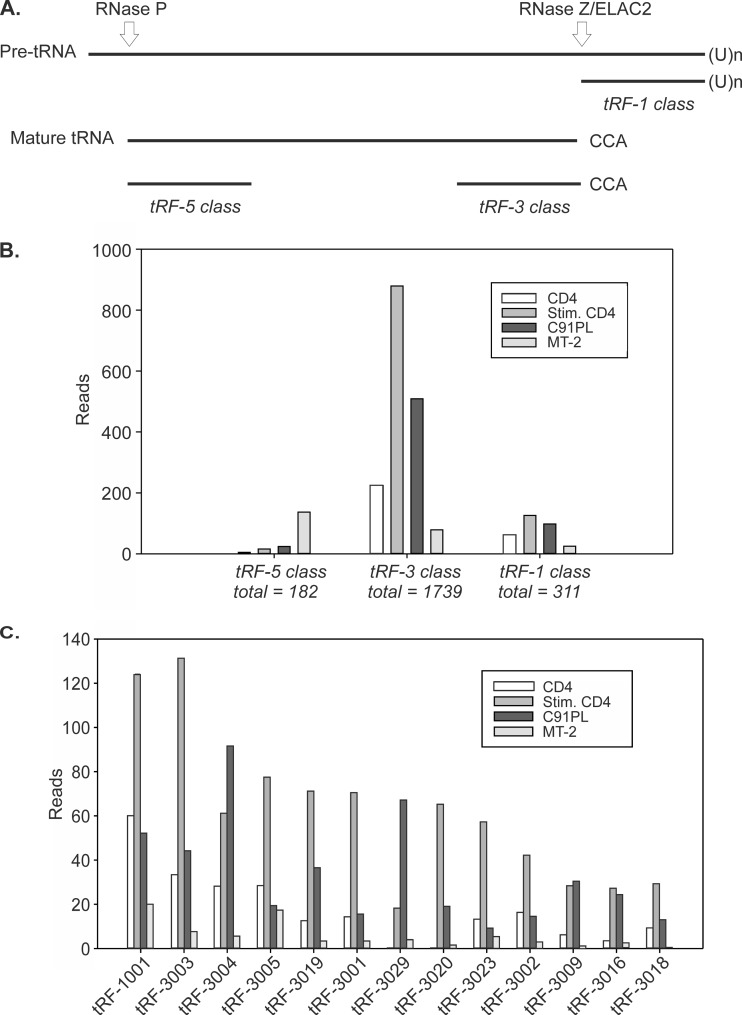
We next tested the sequences identified in the libraries for perfect matches to the 135 tRFs reported by Lee et al. in a study of prostate cancer cell lines (22). Table S2 in the supplemental material lists the number of sequence reads for each tRF, as well as isoforms showing variations at the 5′ or 3′ end. Overall, in both normal and HTLV-1-infected CD4 cells, fragments processed from the 3′ ends of mature tRNAs (tRF-3) were considerably more abundant than tRFs produced from the 3′ ends of tRNA precursors (tRF-1) or from the 5′ ends of mature tRNAs (tRF-5) (Fig. 3B).
FIG 3.
Relative abundances of tRFs. (A) The 3 classes of tRFs aligned to the tRNA precursor. (B) Total numbers of sequence reads with perfect matches to each of the tRF classes, together with 5′ and 3′ isoforms (see Table S2 in the supplemental material). (C) Sequence reads for tRFs with a total of at least 50 sequence reads summed among the 4 libraries.
Figure 3C shows the most abundant tRFs identified in the libraries. Among the 22 previously described tRF-1 sequences, tRF-1001 was the most abundant. tRF-1001, as well as the other tRFs, were upregulated in normal CD4+ cells upon mitogenic stimulation. tRF-3004 and tRF-3029 were more abundant in C91PL cells than in stimulated CD4+ controls, and MT-2 cells yielded few tRF sequences compared to the other 3 cell types.
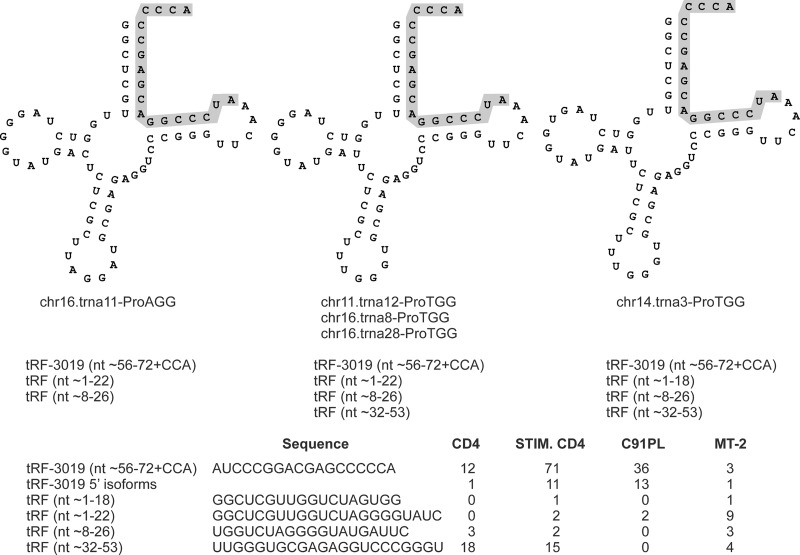
The tRF-3 class also includes tRF-3019 (22). This tRF corresponds to the 3′ end of tRNA-Pro, the tRNA considered to serve as the primer for HTLV-1 reverse transcriptase (38). tRF-3019 was the fifth most abundant tRF identified in our libraries and was most abundant in stimulated CD4+ cells. A BLAST search for tRNA genes able to produce tRF-3019 yielded 21 tRNA-Pro genes located on chromosomes 1, 5, 6, 11, 14, 16, and 17 (see Table S3 in the supplemental material). The four libraries contained several tRF-3019 isoforms with additional nucleotides at the 5′ end that perfectly matched the human genome but were not complementary to the viral genome (Fig. 4). The libraries also contained a small number of reads corresponding to fragments derived from other portions of tRNA-Pro.
FIG 4.
tRFs processed from tRNA-Pro. (Top) Three examples of the 21 tRNA-Pro molecules that are able to produce tRF-3019 (shaded). The diagrams were obtained from the University of California, Santa Cruz, tRNA database and modified by adding the 3′ CCA triplet that is present on mature tRNAs and tRF-3 sequences (Fig. 3). (Bottom) Sequences of the tRFs and number of reads identified in each library.
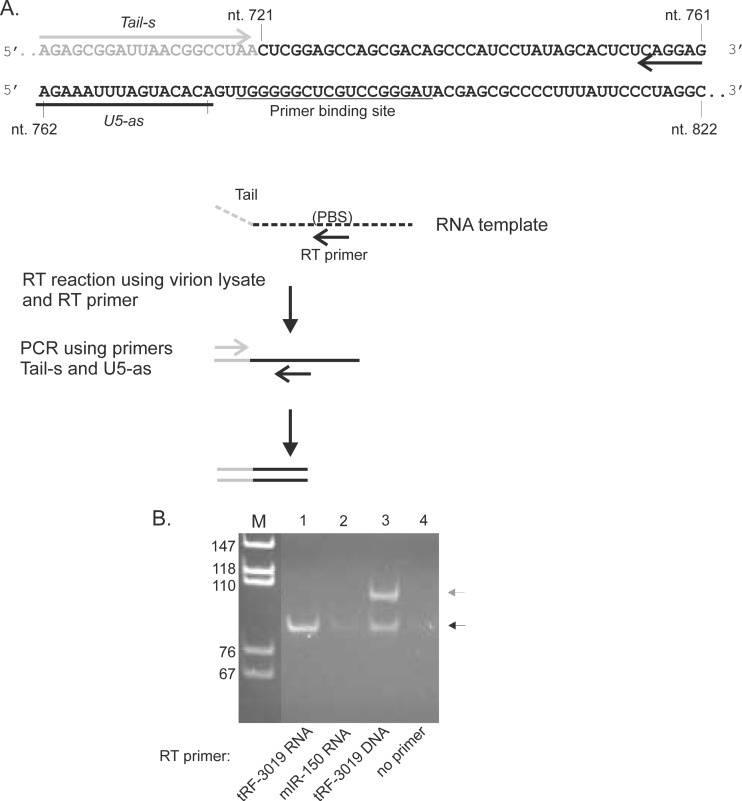
tRF-3019 functions as a primer for HTLV-1 reverse transcriptase.
It is noteworthy that only the portion of tRNA-Pro corresponding to tRF-3019 is complementary to the HTLV-1 primer binding site (PBS) (Fig. 5A), suggesting that the tRF would be fully sufficient as a primer for reverse transcription. We thus tested the primer activity of tRF-3019 in an in vitro reverse transcriptase assay carried out using a synthetic RNA template and the reverse transcriptase contained in HTLV-1 particles recovered from the culture supernatant of C91PL cells (Fig. 5A). The RT assay mixtures contained either no primer, synthetic tRF-3019 RNA, or tRF-3019 DNA as a positive control or miR-150-5p RNA as a negative control. The PCR mixture contained a sense primer specific for a tail sequence present in the synthetic RNA template and an antisense primer positioned immediately 5′ to the PBS. Figure 5B shows the results of the assays. The RT assay performed using tRF-3019 RNA yielded the expected 87-bp PCR product, thus confirming that tRF-3019 can function as a primer for HTLV-1 reverse transcriptase. The assay carried out using a tRF-3019 DNA primer yielded the 87-bp product, along with a longer product indicated by the gray arrow in Fig. 5B. This second band corresponded in size to an amplicon produced with the tail primer and residual tRF-3019 DNA present in the cDNA (i.e., 107 bp). Interestingly, trace amounts of the 87-bp product were also detected in the assays carried out using C91PL RT and miR-150-5p or no primer. This amplicon may have originated from cDNA primed by tRNA-Pro or tRF-3019 present in the viral particle lysate that was used as a source of reverse transcriptase (see below).
FIG 5.
tRF-3019 acts as a primer for HTLV-1 reverse transcriptase. (A) Summary of the RT assay. The template consisted of an in vitro-transcribed RNA spanning HTLV-1 nt 721 to 822 modified by the addition of a 20-nt tail at the 5′ end. The template was incubated with HTLV-1 reverse transcriptase present in virus particles recovered from the culture supernatant of C91PL cells and either tRF-3019 RNA, miR-150-5p RNA (negative control), tRF-3019 DNA (positive control), or no primer. The products of the RT reactions were amplified by PCR using PCR primers Tail-s and U5-as and separated by PAGE in a 6% polyacrylamide gel, along with MspI-digested pBluescript as a size marker. (B) Composite of the ethidium bromide-stained gel. The black arrow indicates the position of the 87-bp PCR product expected using primers Tail-s and U5-as. The additional band in lane 3 indicated by the gray arrow was consistent with a product amplified by Tail-s and residual tRF-3019 DNA primer added to the RT assay. The primer sequences are reported in Table S1 in the supplemental material.
HTLV-1-infected cells release particles containing tRF-3019.
Having established that tRF-3019 is capable of priming HTLV-1 reverse transcription, we next tested for the presence of tRNA-Pro and tRF-3019 in virus particles recovered from supernatants of C91PL cultures. As a control, we also assayed for tRF-3003, the most abundant tRF-3 detected in the 4 libraries, along with its parent, tRNA-alanine (Ala). As described in Materials and Methods, RNA isolated from the virus particles and producer cells was subjected to denaturing PAGE to separate species in the tRF size range from full-length tRNAs. This was necessary, as the tailed RT-PCR primers utilized to detect the tRFs also amplified the 3′ ends of the full-length tRNAs (Fig. 6A). RT-PCR products were separated by PAGE; the intensities of the resulting bands were measured to estimate relative abundances of the tRNAs and tRFs in virus particles versus cells.
Figure 6B shows PAGE analyses of the RT-PCR products obtained for tRNA-Ala and tRNA-Pro. As expected, both tRNA-Ala and tRNA-Pro were readily detected in the C91PL cells. However, a plot of the ratios of the band intensities (Fig. 6D) revealed that tRNA-Pro was enriched in virus particles compared to tRNA-Ala. The RT-PCR products for both tRNAs were much more evident in the full-length tRNA fraction than in the tRF fraction, indicating that the denaturing PAGE purification step resulted in acceptable separation of the 2 size classes.
Figure 6C shows the results of RT-PCR carried out on the same samples using primer sets that amplified the tRFs present in the tRF fraction and the 3′ ends of the tRNAs in the tRNA fraction. Calculation of band intensity ratios (Fig. 6D) showed that tRF-3003 was much less abundant in the virus particles than in the cells, while tRF-3019 was detected at comparable levels in virus particles and cells. Therefore, we concluded that tRF-3019 was enriched in virus particles compared to tRF-3003.
These findings indicate that both tRNA-Pro and tRF-3019 are incorporated into particles released in the supernatant of HTLV-1-infected cells. It is necessary to point out that these particles may also contain exosomes, which may also package proteins and RNA. However, the observation that the particles were enriched for tRNA-Pro and tRF-3019 compared to tRNA-Ala and tRF-3003 supports a specific packaging process directed by interaction of the tRNA/tRF with the HTLV-1 PBS. As shown in Fig. 6E, RT-PCR assays on RNA isolated from the particles confirmed that they contain the HTLV-1 genomic gag/pol mRNA. Therefore, although we cannot exclude the presence of exosomes in the particle preparations, our findings demonstrate that these particles contain reverse transcriptase activity (Fig. 5) and the viral genome and are enriched for the PBS-specific tRNA-Pro and tRF-3019. Taken together, these results strongly suggest that tRF-3019 participates in HTLV-1 reverse transcription.
DISCUSSION
In the present study, we employed mass sequencing to identify the repertoire of small noncoding RNAs expressed in normal T cells compared to cells transformed with HTLV-1. The results revealed a distinct pattern of microRNA expression in HTLV-1-infected cells, two virus-encoded small RNAs, and a number of tRFs. Interestingly, the tRNA fragment tRF-3019 was detected in virus particles and was capable of priming HTLV-1 reverse transcription.
Three microRNAs were differentially expressed in both infected cell lines compared to control CD4+ cells: miR-34a-5p was upregulated, and miR-150-5p and miR-146b-5p were both downregulated. Their shared pattern of regulation in the two infected cell lines suggests that these microRNAs play important roles in HTLV-1 infection/transformation, rather than representing markers of T-cell activation, which are also present in HTLV-1-infected cells.
The observation that miR-150-5p expression is reduced in HTLV-1-infected cell lines is consistent with other studies (15, 16). miR-150-5p undergoes upregulation during T-cell development (27) but is diminished upon stimulation of normal murine CD4+ T cells (39). Expression of miR-150-5p is increased in several hematological tumors, including ATLL samples (15, 16), but is downregulated in the cutaneous CD4+ T-cell lymphoma Sézary syndrome (40), in NK/T-cell lymphomas (41), and in several other hematological malignancies (reviewed in reference 42). Forced expression of miR-150-5p in B-lymphoma cell lines (43), T-acute lymphoblastic leukemia (T-ALL) cell lines (27), and NK cell lines (41) produced antiproliferative and/or proapoptotic effects. Validated targets of miR-150-5p include the oncogenes c-Myb (44) and NOTCH-3 (27), as well as the HIV-1 3′ untranslated region (UTR) (45). It is noteworthy that the minus-strand HTLV-1 transcripts coding for HBZ contain 2 potential binding sites for miR-150-5p (46).
miR-146b-5p is gradually upregulated during T-cell development from the double-positive CD4+ CD8+ to the single-positive CD4+ or CD8+ stage (27). The sequence of miR-146b-5p is almost identical to that of miR-146a, which was identified as upregulated through the action of Tax in previous studies of HTLV-1-infected cell lines (14, 47). miR-146b-5p mRNA targets, therefore, likely overlap those identified for miR-146a, which include the Toll-like receptor signaling pathway proteins TRAF6 and IRAK1 (48), the apoptosis signaling protein FADD (49), and the chemokine receptor CXCR4 (50). miR-146b-5p is downregulated in ATLL (16, 17), Sézary syndrome (40), and several other hematological malignancies but is upregulated in mycosis fungoides (51) and pediatric acute myeloid leukemia (52; reviewed in reference 42).
miR-34a-5p is known to be upregulated by p53 in response to genotoxic and oncogenic stresses. miR-34a-5p targets genes affecting cell proliferation and survival, resulting in growth arrest, senescence, and apoptosis; its downregulation in several solid tumors suggests a tumor suppressor role (53). miR-34a-5p was found to be more abundant in memory versus naive CD4+ T cells (54) and is upregulated in Epstein-Barr virus-transformed B cells (55) during latency type III (56) and in hepatitis B virus-associated hepatocellular carcinoma (57) and might thus exert diverse effects depending on the cell context (58). The results of RT-PCR assays indicated strong upregulation of miR-34a-5p in primary samples from ATLL patients (D. M. D'Agostino, unpublished data).
Recent studies revealed that BLV, a complex oncogenic retrovirus related to HTLV-1, encodes a cluster of viral microRNAs (19, 20). Our deep-sequencing analysis also revealed two virus-encoded small RNA species (MT-2/A and MT-2/B). However, the fact that MT-2/A and MT-2/B were detected with only one sequence read each suggests that, in contrast to BLV, HTLV-1 may not rely on viral microRNAs as a mechanism of posttranscriptional regulation. Alternatively, the production of viral microRNAs might not be favored in cells that are chronically infected, such as MT-2 and C91PL. Therefore, before concluding that HTLV-1 does not produce microRNAs, it will be important to measure their levels of expression in the context of primary samples obtained from infected patients.
The greater representation of tRF-3 sequences than of tRF-1 and tR-5 classes in the libraries is in line with the preponderance of tRF-3 sequences found in prostate cancer cell lines (see Table S2 in reference 22) and in mature B cells (24). Previous functional studies of tRF-1001, which was abundantly expressed in our libraries, revealed its elevated expression in cancer cell lines compared to normal tissue samples and indicated that it is required for cell proliferation (22). Among the tRF-3 sequences abundantly expressed in the four libraries, functional data are available for tRF-3018 in the context of B cells (24). This tRF, named CU1276 in the B-cell study, was differentially expressed in different stages of B-cell maturation, with the greatest expression found in the germinal center (GC) stage while it was absent in GC-derived lymphoma cells. Functional studies of tRF-3018/CU1276 verified its ability to associate with Argonaute proteins and to repress expression of RPA1, a protein involved in DNA replication and repair (24).
The present study focused on tRF-3019, as it corresponds to the 3′ end of tRNA-Pro, which is generally considered to be the primer for HTLV-1 reverse transcriptase (38). tRF-3019 was capable of priming HTLV-1 reverse transcription (Fig. 5) and was enriched in virus particles (Fig. 6). Taken together, these observations support a role for tRF-3019 in the life cycle of HTLV-1.
As shown in Fig. 4, 12 of the 18 nucleotides of tRNA-Pro that are complementary to the HTLV-1 PBS are based paired in the mature tRNA. This positioning of the primer portion of the tRNA in a closed stem is a characteristic of all retroviral tRNA primers. These hydrogen bonds must be disrupted in order for the primer to bind to the PBS, which would not be necessary if a tRF is used as a primer.
The libraries examined in the present study contained a few sequence reads for tRF-3015, which represents the 3′ end of tRNA-lysine (Lys), the primer for HIV-1 (see Table S2 in the supplemental material). Schopman et al. (59) pointed out the possibility that tRFs may serve as primers for reverse transcriptase but also presented experimental evidence from studies of HIV-1 that did not support this proposal. Efficient HIV-1 reverse transcription requires interactions of tRNA-Lys with the PBS, as well as other regions of the viral genome. Of particular importance is an 8-nt sequence termed the primer activation signal (PAS) located in the U5 region that binds to the third stem-loop (T arm) of tRNA-Lys and promotes initiation of reverse transcription and elongation of the cDNA (reviewed in reference 60). Although all retroviruses are predicted to contain a PAS (61), the putative PAS in HTLV-1, which is positioned approximately 10 nucleotides 5′ to the PBS, has not yet been functionally characterized.
The secondary structure of the tRNA primer must also be disrupted to allow nucleotides in the T arm to interact with the PAS. In HIV-1, the NC protein plays an important role in unfolding tRNA-Lys to allow its binding to the HIV-1 PAS (62). Interestingly, a study of NC proteins from several retroviruses indicated that the HTLV-1 NC protein possesses comparatively weak nucleic acid chaperone activity (63). It is possible that another mechanism is responsible for unfolding of tRNA-Pro or that the PAS interaction is not important for HTLV-1.
Alternatively, tRF-3019 may serve as the major primer. In fact, our in vitro assay showed that tRF-3019 permits reverse transcription of a segment of HTLV-1 RNA containing the PBS and predicted PAS. The detailed picture of the interactions between HIV-1 RNA elements and its tRNA primer raises the possibility that tRFs representing the 3′ ends of primer tRNAs might support the initiation of reverse transcription, but not progressivity, with failure to proceed to the strand transfer step. In this case, tRF-3019 might inhibit the overall process of reverse transcription, thus acting as a restriction factor for HTLV-1 replication. Further studies will be necessary to test these hypotheses by comparing the abilities of tRF-3019 and tRNA-Pro to prime and support strand transfer.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paola Dalla Pria for technical assistance, Katia Basso and Beatrice Macchi for sharing protocols, Subhamoy Mukherjee for helping with bioinformatics analyses, and Margherita Ghisi, Riccardo Dalla-Favera, Romain Parent, and Luigi Chieco-Bianchi for discussions.
The research was supported by investigator grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC; nos. 4175 and 13378), an AIRC-Cariverona Regional grant, and the University of Padua (Ateneo grant no. CPDA124913/12). A. Grassi was supported by a fellowship from the Centro Lincei Interdisciplinare Beniamino Segre, Accademia Nazionale dei Lincei.
Footnotes
Published ahead of print 8 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02823-13.
REFERENCES
- 1.Franchini G, Nicot C, Johnson JM. 2003. Seizing of T cells by human T-cell leukemia/lymphoma virus type 1. Adv. Cancer Res. 89:69–132. 10.1016/S0065-230X(03)01003-0 [DOI] [PubMed] [Google Scholar]
- 2.Matsuoka M. 2003. Human T-cell leukemia virus type I and adult T-cell leukemia. Oncogene 22:5131–5140. 10.1038/sj.onc.1206551 [DOI] [PubMed] [Google Scholar]
- 3.Lairmore M, Franchini G. 2007. Human T-cell leukemia virus types 1 and 2, p 2071–2106 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 4.Lairmore MD, Anupam R, Bowden N, Haines R, Haynes RAH, Ratner L, Green P. 2011. Molecular determinants of human T-lymphotropic virus type 1 transmission and spread. Viruses 3:1131–1165. 10.3390/v3071131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuoka M, Jeang KT. 2011. Human T-cell leukemia virus type 1 (HTLV-1) and leukemic transformation: viral infectivity, Tax, HBZ and therapy. Oncogene 30:1379–1389. 10.1038/onc.2010.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasegawa H, Sawa H, Lewis MJ, Orba Y, Sheehy N, Yamamoto Y, Ichinohe T, Tsunetsugu-Yokota Y, Katano H, Takahashi H, Matsuda J, Sata T, Kurata T, Nagashima K, Hall WW. 2006. Thymus-derived leukemia-lymphoma in mice transgenic for the Tax gene of human T-lymphotropic virus type I. Nat. Med. 12:466–472. 10.1038/nm1389 [DOI] [PubMed] [Google Scholar]
- 7.Satou Y, Yasunaga J, Yoshida M, Matsuoka M. 2006. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. U. S. A. 103:720–725. 10.1073/pnas.0507631103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satou Y, Yasunaga J, Zhao T, Yoshida M, Miyazato P, Takai K, Shimizu K, Ohshima K, Green PL, Ohkura N, Yamaguchi T, Ono M, Sakaguchi S, Matsuoka M. 2011. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 7:e1001274. 10.1371/journal.ppat.1001274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Prooyen N, Andresen V, Gold H, Bialuk I, Pise-Masison C, Franchini G. 2010. Hijacking the T-cell communication network by the human T-cell leukemia/lymphoma virus type 1 (HTLV-1) p12 and p8 proteins. Mol. Aspects Med. 31:333–343. 10.1016/j.mam.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silic-Benussi M, Biasiotto R, Andresen V, Franchini G, D'Agostino DM, Ciminale V. 2010. HTLV-1 p13, a small protein with a busy agenda. Mol. Aspects Med. 31:350–358. 10.1016/j.mam.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukumoto R, Andresen V, Bialuk I, Cecchinato V, Walser JC, Valeri VW, Nauroth JM, Gessain A, Nicot C, Franchini G. 2009. In vivo genetic mutations define predominant functions of the human T-cell leukemia/lymphoma virus p12I protein. Blood 113:3726–3734. 10.1182/blood-2008-04-146928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huntzinger E, Izaurralde E. 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12:99–110. 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- 13.Iorio MV, Croce CM. 2012. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 4:143–159. 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichler K, Schneider G, Grassmann R. 2008. MicroRNA miR-146a and further oncogenesis-related cellular microRNAs are dysregulated in HTLV-1-transformed T lymphocytes. Retrovirology 5:100. 10.1186/1742-4690-5-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung ML, Yasunaga J, Bennasser Y, Dusetti N, Harris D, Ahmad N, Matsuoka M, Jeang KT. 2008. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 68:8976–8985. 10.1158/0008-5472.CAN-08-0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellon M, Lepelletier Y, Hermine O, Nicot C. 2009. Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia. Blood 113:4914–4917. 10.1182/blood-2008-11-189845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamagishi M, Nakano K, Miyake A, Yamochi T, Kagami Y, Tsutsumi A, Matsuda Y, Matsubara A, Muto S, Utsunomiya A, Yamaguchi K, Uchimaru K, Ogawa S, Watanabe T. 2012. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-κB pathway in adult T-cell leukemia and other cancers. Cancer Cell 21:121–135. 10.1016/j.ccr.2011.12.015 [DOI] [PubMed] [Google Scholar]
- 18.Kincaid RP, Sullivan CS. 2012. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 8:e1003018. 10.1371/journal.ppat.1003018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kincaid RP, Burke JM, Sullivan CS. 2012. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. U. S. A. 109:3077–3082. 10.1073/pnas.1116107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosewick N, Momont M, Durkin K, Takeda H, Caiment F, Cleuter Y, Vernin C, Mortreux F, Wattel E, Burny A, Georges M, Van den Broeke A. 2013. Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma. Proc. Natl. Acad. Sci. U. S. A. 110:2306–2311. 10.1073/pnas.1213842110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li SC, Shiau CK, Lin WC. 2008. Vir-Mir db: prediction of viral microRNA candidate hairpins. Nucleic Acids Res. 36:D184–D189. 10.1093/nar/gkm610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee YS, Shibata Y, Malhotra A, Dutta A. 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23:2639–2649. 10.1101/gad.1837609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15:2147–2160. 10.1261/rna.1738409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. 2013. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 110:1404–1409. 10.1073/pnas.1206761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popovic M, Lange-Wantzin G, Sarin PS, Mann D, Gallo RC. 1983. Transformation of human umbilical cord blood T cells by human T-cell leukemia/lymphoma virus. Proc. Natl. Acad. Sci. U. S. A. 80:5402–5406. 10.1073/pnas.80.17.5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau NC, Lim LP, Weinstein EG, Bartel DP. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858–862. 10.1126/science.1065062 [DOI] [PubMed] [Google Scholar]
- 27.Ghisi M, Corradin A, Basso K, Frasson C, Serafin V, Mukherjee S, Mussolin L, Ruggero K, Bonanno L, Guffanti A, De Bellis G, Gerosa G, Stellin G, D'Agostino DM, Basso G, Bronte V, Indraccolo S, Amadori A, Zanovello P. 2011. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood 117:7053–7062. 10.1182/blood-2010-12-326629 [DOI] [PubMed] [Google Scholar]
- 28.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson WR, Lipman DJ. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U. S. A. 85:2444–2448. 10.1073/pnas.85.8.2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 26:407–415. 10.1038/nbt1394 [DOI] [PubMed] [Google Scholar]
- 32.Kimata JT, Wong FH, Wang JJ, Ratner L. 1994. Construction and characterization of infectious human T-cell leukemia virus type 1 molecular clones. Virology 204:656–664. 10.1006/viro.1994.1581 [DOI] [PubMed] [Google Scholar]
- 33.Balestrieri E, Pizzimenti F, Ferlazzo A, Giofre SV, Iannazzo D, Piperno A, Romeo R, Chiacchio MA, Mastino A, Macchi B. 2011. Antiviral activity of seed extract from Citrus bergamia towards human retroviruses. Bioorg. Med. Chem. 19:2084–2089. 10.1016/j.bmc.2011.01.024 [DOI] [PubMed] [Google Scholar]
- 34.Frezza C, Balestrieri E, Marino-Merlo F, Mastino A, Macchi B. 2014. A novel, cell-free PCR-based assay for evaluating the inhibitory activity of antiretroviral compounds against HIV reverse transcriptase. J. Med. Virol. 86:1–7. 10.1002/jmv.23748 [DOI] [PubMed] [Google Scholar]
- 35.Sharbati-Tehrani S, Kutz-Lohroff B, Bergbauer R, Scholven J, Einspanier R. 2008. miR-Q: a novel quantitative RT-PCR approach for the expression profiling of small RNA molecules such as miRNAs in a complex sample. BMC Mol. Biol. 9:34. 10.1186/1471-2199-9-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumarswamy R, Volkmann I, Thum T. 2011. Regulation and function of miRNA-21 in health and disease. RNA Biol. 8:706–713. 10.4161/rna.8.5.16154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toyoshima H, Itoh M, Inoue J, Seiki M, Takaku F, Yoshida M. 1990. Secondary structure of the human T-cell leukemia virus type 1 rex-responsive element is essential for rex regulation of RNA processing and transport of unspliced RNAs. J. Virol. 64:2825–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seiki M, Hattori S, Yoshida M. 1982. Human T-cell leukemia virus: molecular cloning of the provirus DNA and the unique terminal structure. Proc. Natl. Acad. Sci. U. S. A. 79:6899–6902. 10.1073/pnas.79.22.6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. 2006. A role for Dicer in immune regulation. J. Exp. Med. 203:2519–2527. 10.1084/jem.20061692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ballabio E, Mitchell T, van Kester MS, Taylor S, Dunlop HM, Chi J, Tosi I, Vermeer MH, Tramonti D, Saunders NJ, Boultwood J, Wainscoat JS, Pezzella F, Whittaker SJ, Tensen CP, Hatton CS, Lawrie CH. 2010. MicroRNA expression in Sezary syndrome: identification, function, and diagnostic potential. Blood 116:1105–1113. 10.1182/blood-2009-12-256719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T, Shimizu N, Sawada K. 2011. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia 25:1324–1334. 10.1038/leu.2011.81 [DOI] [PubMed] [Google Scholar]
- 42.D'Agostino DM, Zanovello P, Watanabe T, Ciminale V. 2012. The microRNA regulatory network in normal- and HTLV-1-transformed T cells. Adv. Cancer Res. 113:45–83. 10.1016/B978-0-12-394280-7.00002-6 [DOI] [PubMed] [Google Scholar]
- 43.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 40:43–50. 10.1038/ng.2007.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsay RG, Gonda TJ. 2008. MYB function in normal and cancer cells. Nat. Rev. Cancer 8:523–534. 10.1038/nrc2439 [DOI] [PubMed] [Google Scholar]
- 45.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13:1241–1247. 10.1038/nm1639 [DOI] [PubMed] [Google Scholar]
- 46.Ruggero K, Corradin A, Zanovello P, Amadori A, Bronte V, Ciminale V, D'Agostino DM. 2010. Role of microRNAs in HTLV-1 infection and transformation. Mol. Aspects Med. 31:367–382. 10.1016/j.mam.2010.05.001 [DOI] [PubMed] [Google Scholar]
- 47.Tomita M, Tanaka Y, Mori N. 2012. MicroRNA miR-146a is induced by HTLV-1 Tax and increases the growth of HTLV-1-infected T-cells. Int. J. Cancer 130:2300–2309. 10.1002/ijc.25115 [DOI] [PubMed] [Google Scholar]
- 48.Taganov KD, Boldin MP, Chang KJ, Baltimore D. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 103:12481–12486. 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curtale G, Citarella F, Carissimi C, Goldoni M, Carucci N, Fulci V, Franceschini D, Meloni F, Barnaba V, Macino G. 2010. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood 115:265–273. 10.1182/blood-2009-06-225987 [DOI] [PubMed] [Google Scholar]
- 50.Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, Biffoni M, Nuzzolo ER, Billi M, Foa R, Brunetti E, Grignani F, Testa U, Peschle C. 2008. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat. Cell Biol. 10:788–801. 10.1038/ncb1741 [DOI] [PubMed] [Google Scholar]
- 51.van Kester MS, Ballabio E, Benner MF, Chen XH, Saunders NJ, van der Fits L, van Doorn R, Vermeer MH, Willemze R, Tensen CP, Lawrie CH. 2011. miRNA expression profiling of mycosis fungoides. Mol. Oncol. 5:273–280. 10.1016/j.molonc.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, Zhou H, Qu LH, Xu L, Chen YQ. 2009. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One 4:e7826. 10.1371/journal.pone.0007826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hermeking H. 2010. The miR-34 family in cancer and apoptosis. Cell Death Differ. 17:193–199. 10.1038/cdd.2009.56 [DOI] [PubMed] [Google Scholar]
- 54.Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJ, Birolo RS, Moro M, Crosti MC, Gruarin P, Maglie S, Marabita F, Mascheroni D, Parente V, Comelli M, Trabucchi E, De Francesco R, Geginat J, Abrignani S, Pagani M. 2011. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat. Immunol. 12:796–803. 10.1038/ni.2057 [DOI] [PubMed] [Google Scholar]
- 55.Mrazek J, Kreutmayer SB, Grasser FA, Polacek N, Huttenhofer A. 2007. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 35:e73. 10.1093/nar/gkm244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cameron JE, Fewell C, Yin Q, McBride J, Wang X, Lin Z, Flemington EK. 2008. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology 382:257–266. 10.1016/j.virol.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizuguchi Y, Mishima T, Yokomuro S, Arima Y, Kawahigashi Y, Shigehara K, Kanda T, Yoshida H, Uchida E, Tajiri T, Takizawa T. 2011. Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PLoS One 6:e15304. 10.1371/journal.pone.0015304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dutta KK, Zhong Y, Liu YT, Yamada T, Akatsuka S, Hu Q, Yoshihara M, Ohara H, Takehashi M, Shinohara T, Masutani H, Onuki J, Toyokuni S. 2007. Association of microRNA-34a overexpression with proliferation is cell type-dependent. Cancer Sci. 98:1845–1852. 10.1111/j.1349-7006.2007.00619.x [DOI] [PubMed] [Google Scholar]
- 59.Schopman NC, Heynen S, Haasnoot J, Berkhout B. 2010. A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. 7:573–576. 10.4161/rna.7.5.13141 [DOI] [PubMed] [Google Scholar]
- 60.Abbink TE, Berkhout B. 2008. HIV-1 reverse transcription initiation: a potential target for novel antivirals? Virus Res. 134:4–18. 10.1016/j.virusres.2007.12.009 [DOI] [PubMed] [Google Scholar]
- 61.Beerens N, Berkhout B. 2002. Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS. RNA 8:357–369. 10.1017/S1355838202028194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beerens N, Jepsen MD, Nechyporuk-Zloy V, Kruger AC, Darlix JL, Kjems J, Birkedal V. 2013. Role of the primer activation signal in tRNA annealing onto the HIV-1 genome studied by single-molecule FRET microscopy. RNA 19:517–526. 10.1261/rna.035733.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. 2008. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 82:10129–10142. 10.1128/JVI.01169-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.