Abstract
Epstein-Barr virus (EBV) latent antigen EBNA3C is implicated in B-cell immortalization and linked to several B-cell malignancies. Deregulation of H2AX can induce genomic instability with increased chromosomal aberrations, which ultimately leads to tumorigenesis. Here we demonstrated that EBNA3C can attenuate H2AX expression at the transcript and protein levels. A reduction of total H2AX levels was clearly observed upon infection of primary B cells with wild-type EBV but not with EBNA3C knockout recombinant EBV. H2AX also interacted with EBNA3C through its N-terminal domain (residues 1 to 100). Furthermore, H2AX mutated at Ser139 failed to interact with EBNA3C. Luciferase-based reporter assays also revealed that the binding domain of EBNA3C is sufficient for transcriptional inhibition of the H2AX promoter. EBNA3C also facilitated H2AX degradation through recruitment of components of the ubiquitin proteasome pathway. We further demonstrated that knockdown of H2AX in lymphoblastoid cell lines (LCLs) led to the upregulation of the Bub1 oncoprotein and downregulated expression of p53. Overall, our study provides additional insights into EBV-associated B-cell lymphomas, which are linked to the regulation of the DNA damage response system in infected cells. The importance of these insights are as follows: (i) EBNA3C downregulates H2AX expression at the protein and transcript levels in epithelial cells, B cells, and EBV-transformed LCLs, (ii) EBNA3C binds with wild-type H2AX but not with the Ser139 mutant of H2AX, (iii) the N terminus (residues 1 to 100) of EBNA3C is critical for binding to H2AX, (iv) localization of H2AX is predominantly nuclear in the presence of EBNA3C, and (v) H2AX knocked down in LCLs led to enhanced expression of Bub1 and downregulation of the tumor suppressor p53, which are both important for driving the oncogenic process.
INTRODUCTION
Epstein-Barr virus (EBV) is a human gammaherpesvirus associated with infectious mononucleosis, and it is estimated that >95% of adults are carriers of EBV throughout their lifetime (1, 2). The contributory role of EBV in driving the oncogenic process is continually being explored. EBV transforms latently infected primary B cells into constantly proliferating lymphoblastoid cell lines (LCLs) (3). EBV is also commonly involved in numerous malignancies, including Burkitt's lymphoma (BL), posttransplant lymphoproliferative disorders (PTLDs), nasopharyngeal carcinoma (NPC), HIV-associated lymphomas, some types of T-cell lymphomas, and gastric cancer (4, 5).
Transformation of human B cells into LCLs by EBV establishes a latent type of infection typically known as type III latency (6). Three major viral latency programs have been described, with deferential expression profiles of specific viral gene transcription (7). EBV latency patterns are characterized by the expression of different EBV nuclear antigens (EBNAs), including EBNA1, -2, -3A, -3B, and -3C; LP/5; latent membrane protein 1 (LMP1); LMP2A; and LMP2B (8). Importantly, these latent proteins are significantly expressed during the latency III program (9, 10). Previous studies showed that EBNA2, EBNA3A, EBNA3C, and LMP1 play critical roles in B-cell transformation (11, 12).
Previous studies showed that one of the essential EBV latent antigens, EBNA3C, is important for modulating B-cell activation. For example, the B-cell activation marker CD21 was upregulated in the presence of EBNA3C in Burkitt's lymphoma cell lines (13, 14). EBNA3C binds to RBP-Jk, an important regulator of the Notch signaling pathway, through an amino-terminal motif, and the acidic domains are responsible for nuclear translocation due to the presence of the nuclear localization signals (15). Recently, we reported that the p53 tumor suppressor is negatively regulated by EBNA3C at both the transcriptional and posttranscriptional levels (16). Critically, EBNA3C has also been shown to regulate the major cell cycle checkpoints (17). Recently, it was suggested that EBV has a potential role in inducing genomic instability and that viral proteins associated with the latency III program can regulate the DNA damage response (DDR) (18). In addition, previous studies from our laboratory demonstrated that EBNA3C binds to Chk2, a major effector of the DDR, which also deregulates the cell cycle of EBV-infected cells at the G2/M phase (19, 20).
EBV infection of primary B cells was shown to activate the DDR by inducing phosphorylation of H2AX at Ser139 (γ-H2AX) (20). H2AX is a histone variant that has a key regulatory function during induction of the DDR. Induction of γ-H2AX is a hallmark of the DDR, which recruits various DNA damage proteins, repair proteins, as well as cell cycle checkpoints (21). Recently, we found that H2AX phosphorylation is important for Kaposi's sarcoma-associated herpesvirus (KSHV)-induced oncogenesis, which is mediated through one of its major latent proteins, LANA (22). However, upon EBV infection, the mechanism by which cells trigger the DDR and proceed toward oncogenesis is still not clearly understood. Furthermore, it still has not been determined how the DDR progresses without repairing the damaged DNA to bypass cell cycle arrest or apoptosis (16, 23, 24). In this study, we now demonstrate that the EBV latent antigen EBNA3C deregulates total H2AX levels transcriptionally and posttranslationally through involvement of the ubiquitin-mediated proteasome degradation pathway. Additionally, our study also showed dramatic changes in expression patterns of the tumor suppressor p53 and the oncoprotein Bub1 in H2AX knockdown LCLs. These results provide further clues as to the biological relevance of H2AX deregulation in EBV-induced oncogenesis. Overall, our study suggests that EBNA3C can play an important role in EBV-mediated oncogenesis through downmodulation of H2AX.
MATERIALS AND METHODS
Ethics statement.
The University of Pennsylvania School of Medicine CFAR (Center for AIDS Research) Immunology Core provided us human peripheral blood mononuclear cells (PBMCs) from unidentified donors, with written consent, which was approved by the Institutional Review Board (IRB) based on Declaration of Helsinki recommendations.
Constructs and transfection.
The plasmid expressing full-length EBNA3C or its various truncated domains was previously mentioned (25, 26). A plasmid expressing wild-type H2AX was previously described (27). Constructs expressing Flag-tagged wild-type H2AX and its mutated version, H2AX Ser139 [H2AX (S-A)], were kindly provided by Alvaro N. A. Monteiro (H. Lee Moffitt Cancer Center, Tampa, FL) (28). The pGL3-H2AX wild-type reporter construct was generously provided by Toru Ouchi (Roswell Park Cancer Institute, Buffalo, NY) (29). All construct sequences were further confirmed by DNA sequencing at the core facility at the University of Pennsylvania. All transfection procedures were described previously (30).
Cell cultures and antibodies.
EBV-positive LCL1 and LCL2, BJAB clones stably expressing EBNA3C (BJAB#7 and BJAB#10), and the EBV-negative cell lines BJAB, DG75, and Ramos were cultured in RPMI 1640 medium and as described previously (22).
Rabbit anti-H2AX antibody was obtained from Bethyl Laboratories Inc. (Montgomery, TX). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (anti-mouse) was purchased from U.S. Biological Corp. (Swampscott, MA). Mouse antibodies reactive to the Myc epitope (9E10), Flag epitope (M2), and EBNA3C (A10) were described previously (27).
Induction and infection of recombinant EBV.
Bacterial artificial chromosome (BAC) green fluorescent protein (GFP)-EBV was generated as previously described (13). An EBNA3C null mutant (ΔEBNA3C BAC GFP-EBV) was generated by deleting the EBNA3C region between bp 98370 and 102891 from a wild-type BAC GFP-EBV plasmid (J. Lu, H. C. Jha, and E. S. Robertson, unpublished data). Southern blot analysis, PCR, and sequencing confirmed that the mutation was correct. Cells harboring BAC GFP-EBV and ΔEBNA3C GFP-EBV were induced with TPA (12-O-tetradecanoylphorbol) and butyric acid (BA) (3 mM; Sigma-Aldrich Corp., St. Louis, MO) for 4 to 5 days in Dulbecco's modified Eagle's medium (DMEM). The cell suspension was centrifuged at 1,800 rpm for 15 min and filtered through 0.45-μm cellulose acetone filters. The viral particles were concentrated by ultracentrifugation at 23,500 rpm at 4°C, and viral particles were stored at −80°C.
For infection studies, we followed procedures similar to those described previously (31). Ten million PBMCs were mixed with either the wild-type or ΔEBNA3C virus supplemented with RPMI 1640 medium containing 10% fetal bovine serum (FBS) and incubated overnight at 37°C. Cells were centrifuged at 2,000 rpm for 10 min. Pelleted cells were resuspended in 2 ml of cell culture medium supplemented with 10% FBS. GFP expression was visualized to monitor infection, and reverse transcription-PCR (RT-PCR) of EBNA1 confirmed infection.
RNA interference.
Short hairpin oligonucleotides directed against EBNA3C (sh-EBNA3C) were designed (Dharmacon Research, Chicago, IL). All short hairpin knockdown procedures and the sequence for the sh-EBNA3C clone were described previously (28). To clone sh-H2AX, we used strategies and sequences similar to those described previously (32). Cell cultures were selected with puromycin.
Immunoprecipitation and Western blotting.
Transiently transfected cells were harvested and washed with ice-cold phosphate-buffered saline (PBS), followed by lysis of cells using radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 7.5], 150 mM sodium chloride, 2 mM EDTA, 1% NP-40), and a protease inhibitor cocktail was added before lysis of the cells. The cell debris was removed by centrifugation at 21,000 × g for 12 min (4°C), and the supernatant was transferred into autoclaved microcentrifuge tubes. The lysates were then precleared by spinning with normal mouse serum and a 1:1 mixture of protein A- and protein G-conjugated Sepharose beads for 1.5 h at 4°C. The beads were spun at low speed, and the supernatant was transferred into a fresh microcentrifuge tube. The specific protein of interest was incubated with 1 μg of suitable interacting antibody overnight at 4°C in a rotating chamber. Immune complexes were collected by binding with protein A and G beads, pelleted, and washed with ice-cold RIPA buffer 3 times. Western blotting (WB) was performed as described previously (33).
RNA isolation and quantitative real-time PCR.
TRIzol reagent (Invitrogen Inc., Carlsbad, CA) was used to isolate total RNA according to the manufacturer's instruction. RNA to cDNA was prepared by using a Superscript II reverse transcription kit (Invitrogen Inc., Carlsbad, CA). The primers used for RT-PCR were as follows: 5′-GTTCCCAGTGGGCCGTGTA-3′ and 5′-CGGTGAGGTACTCCAGCACT-3′ for H2AX, 5′-AGAAGGGGAGCGTGTTTGT-3′ and 5′-GGCTCGTTTTGACGTCGGC-3′ for EBNA3C, 5′-CATTGAGTCGTCTCCCCTTTGGAAT-3′ and 5′-TCATAACAAGGTCCTTAATCGCATC-3′ for EBNA1, and 5′-TGCACCACCAACTGCTTAG-3′ and 5′-CATGCAGGGATGATGTTC-3′ for GAPDH. Quantitative RT-PCR was carried out by using a Step One Plus real-time PCR system (Applied BioSystems, Foster City, CA), and all experiments were performed in triplicates.
Reporter assay.
Ten million HEK293 cells were cotransfected with 10 μg of pGL3H2AX reporter plasmids and increasing concentrations of EBNA3C. All procedures were performed as described previously (22).
Ubiquitination assay.
HEK293 cells were transfected with the vector, Flag-EBNA3C, the hemagglutinin (HA)-tagged ubiquitin moiety (HA-Ub), and Myc-H2AX. Cells were treated with 20 μM the drug MG132 (Enzo Life Sciences Inc., Plymouth Meeting, PA). All procedures were essentially performed as described previously (34). Briefly, after 36 h posttransfection, cell lysates were taken and immunoprecipitated with specific antibodies. Immunoprecipitated samples were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. Levels of ubiquitination were evaluated by using an HA-specific antibody (12CA5). H2AX antibody was used for immunoprecipitation, and Western blots were performed by using the specific antiubiquitin antibody.
Immunofluorescence analysis.
Immunofluorescence (IF) analysis was carried out essentially as described previously (35). Briefly, U2OS cells layered onto coverslips were transiently transfected with the indicated plasmids by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The B-cell lines BJAB, BJAB#7, and BJAB#10 as well as LCL1 and LCL2 were air dried and fixed. All procedures were performed as described previously (22).
GST pulldown assay.
Escherichia coli BL21(DE3) was transformed (by the heat shock method) with glutathione S-transferase (GST) and GST-H2AX plasmid constructs. All procedures were performed as described previously (22).
Colony formation assay (CFA).
Ten million HEK293 cells were transfected with EBNA3C and H2AX mutant and wild-type plasmids by electroporation. At 36 h posttransfection, cells were selected with G418 in DMEM supplemented with 5 μg/ml puromycin. The selection medium was changed on alternate days. After 2 weeks, cells were fixed on the plates by using 3% paraformaldehyde (PFA) for 30 min, followed by staining with 0.1% crystal violet. The total intensity of colonies was calculated by scanning the dish using the LiCor Odyssey system (LiCor Biosciences, Salt Lake City, UT) and quantified.
RESULTS
H2AX expression is downregulated in the presence of EBNA3C during primary infection and latency.
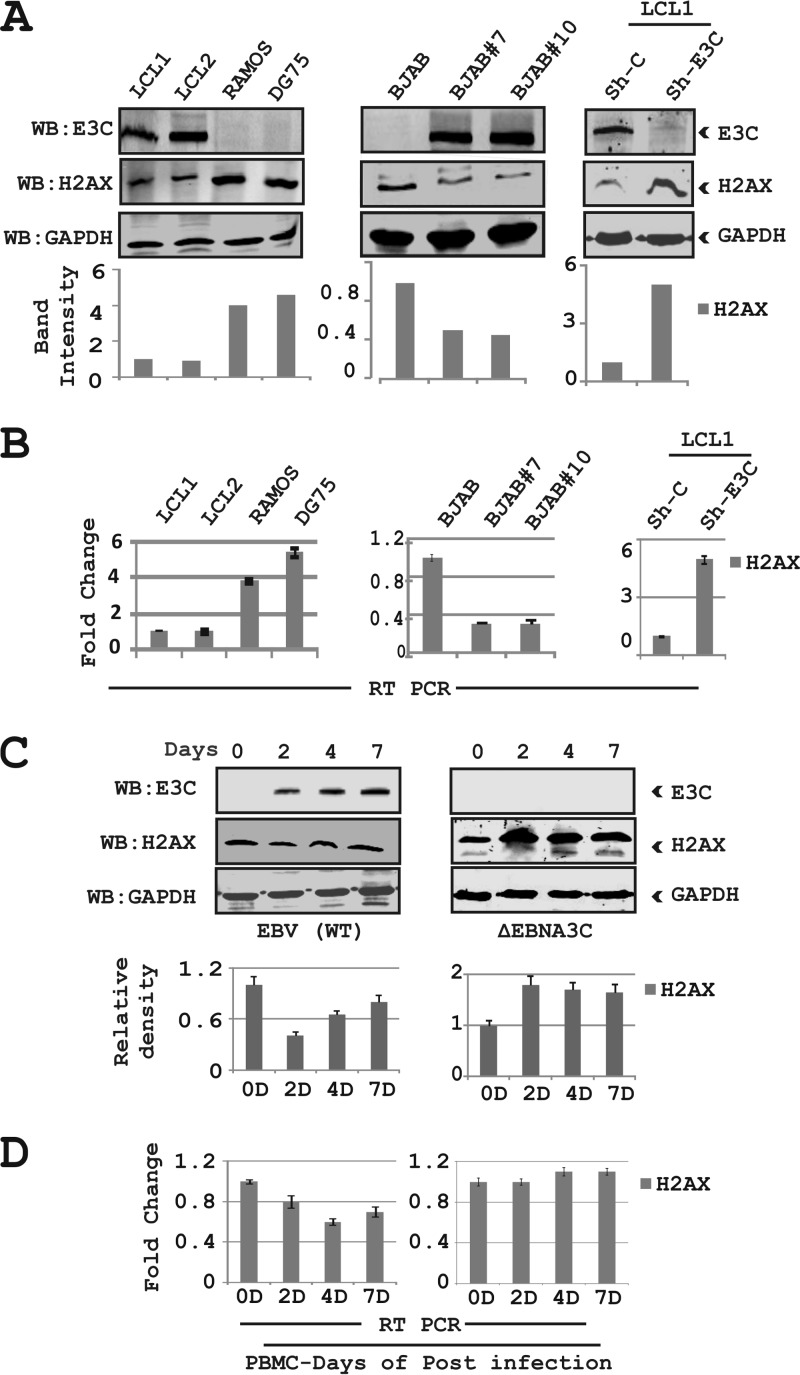
It was recently demonstrated that the EBV essential antigen EBNA3C contributes to attenuation of the DNA damage response (DDR) during latency and is particularly active during early infection of naive B cells in vitro by EBV (31). However, the mechanism of attenuation has not been elucidated. H2AX is ubiquitously expressed in mammalian cells, and the phosphorylation of H2AX at Ser139 is a well-known marker for the DDR (36). This is supported by previous studies which demonstrated that a reduction in H2AX levels is associated with higher genomic instability and tumor predisposition (18, 23, 26). To investigate the effects of EBNA3C on H2AX regulation, we evaluated H2AX levels in the presence of EBNA3C expression as well as in EBV-infected B cells. We first compared H2AX expression levels in EBV-transformed lymphoblastoid cell lines (LCL1 and LCL2) with those in EBV-negative Burkitt's lymphoma (BL), Ramos, and DG75 cells (Fig. 1A). The results showed that cellular H2AX levels were downregulated by approximately 3- to 4-fold in LCL1 and LCL2 compared to the EBV-negative cell lines DG75 and Ramos by Western blotting (Fig. 1A). To determine if EBNA3C alone can affect H2AX expression, we next investigated the levels of H2AX in BJAB cells stably expressing EBNA3C (BJAB#7 and BJAB#10) compared to a BJAB vector control cell line (Fig. 1A). Our results from Western blot analysis showed changes in H2AX levels with a drop of at least 2-fold in BJAB#7 and BJAB#10 cells compared to BJAB cells (Fig. 1A). In addition, when EBNA3C expression was stably knocked down using specific lentiviral constructs in LCLs, a dramatic enhancement of approximately 5-fold in H2AX levels was observed (Fig. 1A). These results strongly suggest that EBNA3C contributes directly to downregulation of H2AX in EBNA3C-expressing cells.
FIG 1.
EBNA3C downregulates total H2AX levels in B cells. (A, left) Endogenous expression of H2AX in EBV-positive cell lines (LCL1 and LCL2) and EBV-negative Burkitt's lymphoma cell lines (Ramos and DG75) was analyzed at the protein level. (Middle) Endogenous expression of H2AX in BJAB (EBV-negative) BJAB#7, and BJAB#10 (EBNA3C stably expressing) cells was analyzed by WB. (Right) LCL1 cells stably transduced for sh-control and sh-EBNA3C were harvested to determine the protein expression levels of H2AX. The relative density of H2AX was analyzed by normalization with GAPDH. (B) All groups of cells were extracted by using TRIzol reagent, and the RNA concentration was estimated by using a Nanodrop instrument (Eppendorf Inc., Hamburg, Germany). Transcript levels for the H2AX gene are represented by fold changes, and the experiments were performed in triplicates. (Left) Results of LCL1 and LCL2 compared to Ramos and DG75 cells. (Middle and right) Results of BJAB cells compared to BJAB#7 and BJAB#10 cells (middle) and LCL1 stably knocked down for EBNA3C and the control vector (right). (C and D) PBMCs were infected with wild-type (WT) EBV and ΔEBNA3C virus for up to 7 days. Cells were harvested, and expression of H2AX was monitored in a time-dependent manner for protein and transcript levels by performing WB analysis and real-time PCR. All panels show representative data of repeated experiments.
EBNA3C is a strong transcription factor capable of regulating transcription of both viral and cellular genes (16, 30, 32, 35). To determine if H2AX was directly modulated by EBNA3C at the transcript level, we evaluated H2AX transcripts in EBV-transformed and EBNA3C stable cell lines compared to EBV-negative B cells. The cell lines included two lymphoblastoid cell lines (LCL1 and LCL2), two BL cell lines stably expressing EBNA3C (BJAB#7 and BJAB#10), and LCL1 stably knocked down for EBNA3C by using a specific lentiviral construct along with its isogenic negative cell line (Fig. 1B). The results from real-time PCR analysis indicated that the H2AX levels were significantly reduced in both EBV-positive as well as EBNA3C-positive cell lines, whereas this was mostly reversed when EBNA3C was knocked down in an LCL background (Fig. 1B). These results indicated that EBNA3C can block H2AX expression under normal physiological conditions.
To provide another link to the physiological relevance of H2AX expression during primary infection, we performed an in vitro infection study using peripheral blood mononuclear cells (PBMCs) with recombinant viruses, including both wild-type virus as well as an EBNA3C-deleted EBV (ΔEBNA3C) (Fig. 1C). Ten million resting PBMCs were infected and grew over a period of 7 days post-EBV infection (Fig. 1C). The H2AX expression level was attenuated by day 2 postinfection, and this decrease was recovered at day 7 to just over 50% based on quantitation using a LiCor Odyssey system in the linear range (Fig. 1C). However, PBMCs infected with the ΔEBNA3C virus showed enhanced H2AX levels in postinfection cells to almost 2-fold, which was similar for up to 7 days (Fig. 1C). The Western blot results were also supported by subsequent real-time PCR results, where we observed an attenuation of H2AX at the transcript level in EBV-infected PBMCs. However, an increase in H2AX transcript levels was observed by using EBNA3C-deleted recombinant EBV (Fig. 1D). These results suggest that EBNA3C contributes to the reduction of total H2AX levels and may be important during the early stages of EBV-mediated B-cell infection, which leads to transformation.
Expression of EBNA3C leads to downregulation of H2AX in Burkitt's lymphoma and epithelial cells.
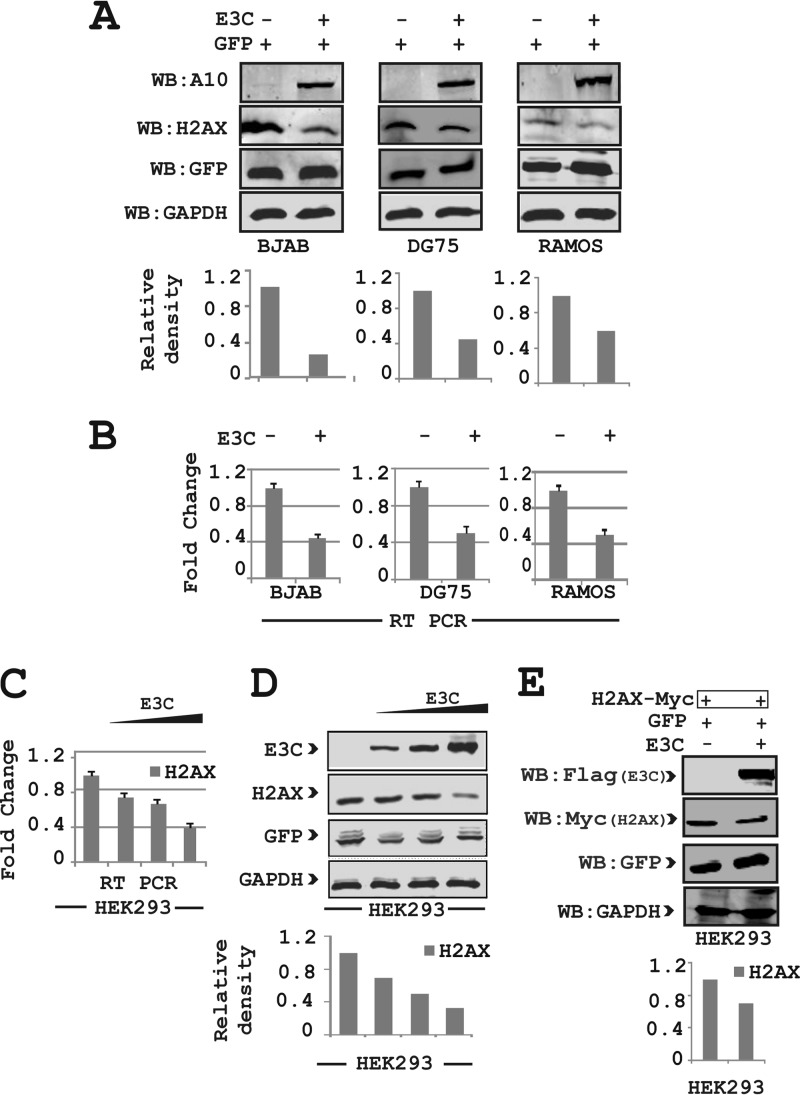
To determine the involvement of other viral oncoproteins in the effects of EBNA3C on H2AX expression, we performed expression analysis with both Burkitt's lymphoma (BL) and epithelial cells. First, an EBNA3C-expressing plasmid was cotransfected along with a GFP control vector into three virus-negative BL cell lines, namely, BJAB, DG75, and Ramos (Fig. 2A). A GFP expression vector was used to determine the overall transfection efficiency. Western blot results demonstrated that there was an approximately 2- to 4-fold reduction in H2AX protein levels in the presence of EBNA3C (Fig. 2A). In agreement with these Western blot results, H2AX transcript levels analyzed by using real-time PCR also showed a significant decrease of about 2- to 3-fold in the presence of EBNA3C compared to two EBV-negative BL cell lines (Fig. 2B). To determine whether the effect of EBNA3C on H2AX is cell type specific, we performed experiments similar to those described above for epithelial cells using HEK293 cells in a dose-dependent manner. As expected, H2AX levels were significantly attenuated with increasing expression of EBNA3C at the transcript and protein levels (Fig. 2C and D, respectively).
FIG 2.
EBNA3C attenuates H2AX levels in Burkitt's lymphoma and epithelial cells. (A) Approximately 30 million BJAB, DG75, and Ramos cells were harvested for WB analysis of H2AX, EBNA3C, GAPDH, and GFP levels after transient transfection of EBNA3C and GFP constructs. (B) Endogenous expression of H2AX levels in Burkitt's lymphoma cell lines transfected with EBNA3C was analyzed for transcript levels by real-time PCR. Approximately 15 million BJAB, DG75, and Ramos cells were cotransfected with the GFP-expressing vector with either EBNA3C or the control vector. Transcript levels of the H2AX gene are represented as fold changes. The experiments were performed in triplicate. (C) H2AX in HEK293 cells transfected with EBNA3C in a dose-dependent manner was analyzed at the transcript level by RT-PCR. Transcript levels of the H2AX gene are represented as fold changes from experiments performed in triplicate. (D) H2AX in HEK293 cells transfected with EBNA3C in a dose-dependent manner was analyzed at the protein level. (E) H2AX and EBNA3C constructs were transfected into HEK293 cells and analyzed for the detection of H2AX protein levels. HEK293 cells were transfected with GFP, EBNA3C-Flag, or the vector with Myc-H2AX. Data in all panels are representative of repeat experiments.
Importantly, coexpression of H2AX tagged with the Myc epitope and EBNA3C tagged with the Flag epitope from heterologous promoters also showed attenuation of H2AX at the protein level using the Myc-specific mouse monoclonal antibody (Fig. 2E). These results indicated that EBNA3C can also deregulate H2AX at the transcriptional level.
EBNA3C forms a complex with H2AX.
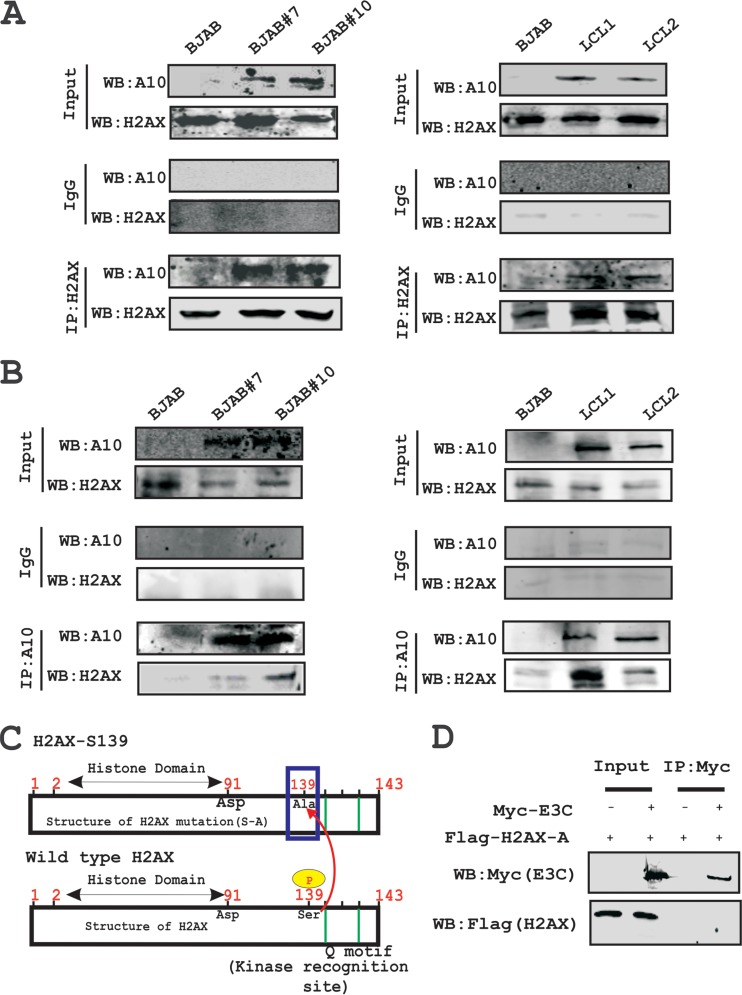
In an attempt to understand whether this attenuation of H2AX levels in the presence of EBNA3C expression is regulated through an interaction between these two molecules, we further investigated their binding activities in B cells. To support this, we performed immunoprecipitation assays with anti-H2AX antibody in BJAB, BJAB#7, BJAB#10, LCL1, and LCL2 cells (Fig. 3A). The results demonstrated that H2AX was stably associated with EBNA3C in both cells stably expressing EBNA3C (BJAB#7 and BJAB#10) as well as LCL1 and LCL2 (Fig. 3A). Furthermore, to support our interaction results, we performed a reverse immunoprecipitation assay, where we used the A10 mouse monoclonal antibody specific for EBNA3C and cell lines similar to those described above (Fig. 3B). These association studies provided strong evidence that EBNA3C interacted with H2AX in both EBV-transformed cells as well as EBNA3C-positive Burkitt's lymphoma cells (Fig. 3A).
FIG 3.
H2AX associates with EBNA3C in cells. (A) Complex formation between EBNA3C and H2AX was evaluated by using anti-H2AX antibody in Burkitt's lymphoma cells (left) and LCLs (right). (B) The association between EBNA3C and H2AX was evaluated by using an anti-EBNA3C (A10) antibody in Burkitt's lymphoma cells (left) and LCLs (right). (C) Schematic showing wild-type H2AX and a specific serine 139 mutation with alanine in the full-length protein. (D) The Flag-H2AX Ser139 [H2AX (S-A)] mutant was cotransfected with either Myc-EBNA3C or the control vector. WB was performed with anti-Myc and anti-Flag antibodies. IP, immunoprecipitation.
H2AX autophosphorylation plays a critical role in the interaction between H2AX and EBNA3C.
H2AX phosphorylation at Ser139 has previously been shown to be critical for maintenance of latency during multiple gammaherpesviruses infections, including mouse gammaherpesvirus 68 (MHV-68) and KSHV (22, 37). The interaction between EBNA3C and H2AX prompted us to further evaluate whether mutation at the Ser139 residue of H2AX was critical for complex formation with EBNA3C (Fig. 3C). To address this question, we performed an immunoprecipitation assay using HEK293 cells where Flag-tagged Ser139 H2AX (where the serine residue was replaced by an alanine residue) was coexpressed with Myc-tagged EBNA3C (Fig. 3D). Subsequently, immunoprecipitation was performed by using anti-Myc antibody to target Myc-tagged EBNA3C (Fig. 3D). The immunoprecipitated bands were visualized with anti-Flag antibody. Interestingly, no signal was detected for EBNA3C when H2AX was mutated at the Ser139 residue (Fig. 3D). This indicated that the Ser139 residue of H2AX was important for complex formation of H2AX with EBNA3C. Moreover, we found reduced expression levels of γ-H2AX in EBV-positive LCL1 and LCL2 compared to EBV-negative DG75 and Ramos cells (data not shown).
The first 100 residues of EBNA3C are responsible for the interaction of EBNA3C with H2AX.
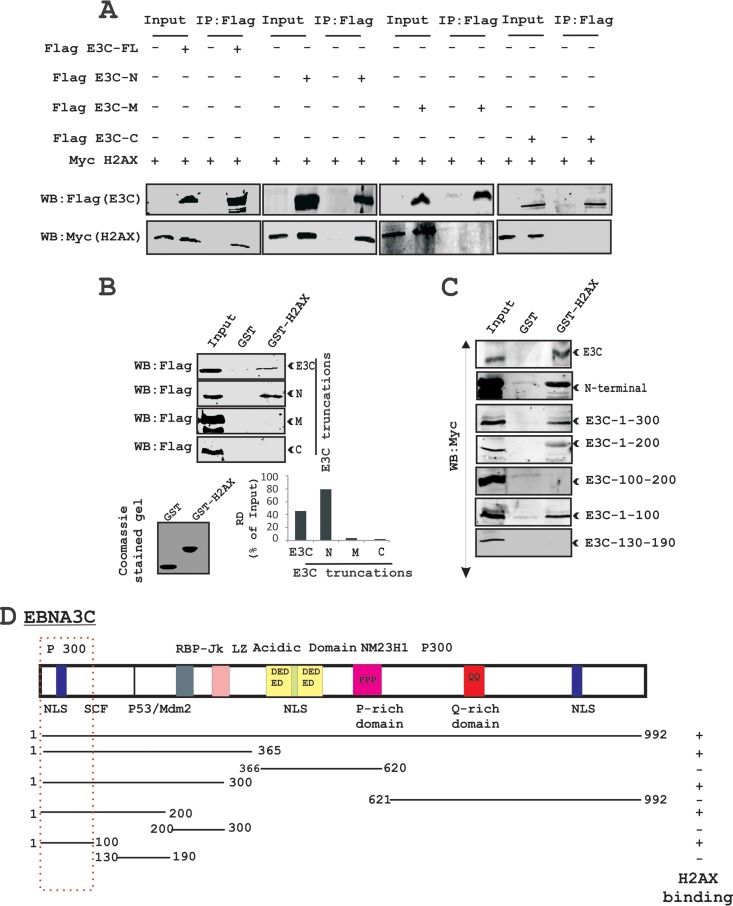
To identify the binding residues of EBNA3C important for the interaction with H2AX, we performed further similar immunoprecipitation experiments in a heterologous expression system using HEK293 cells. Cells were transfected with plasmids expressing Flag-tagged full-length EBNA3C (residues 1 to 992), the N-terminal region (residues 1 to 365), the middle (M) region (residues 366 to 620), and the C-terminal region (residues 621 to 992) along with a Myc-tagged H2AX expression vector (Fig. 4A). Immunoprecipitation assays were performed by using a Flag-specific mouse monoclonal antibody (M2). The results revealed that H2AX forms a complex with residues within the N-terminal region of EBNA3C but not with the middle or carboxyl-terminal domains (Fig. 4A).
FIG 4.
The first 100 amino acid residues of the EBNA3C protein are critical for complex formation with H2AX. (A) Complex formation between EBNA3C and H2AX in HEK293 cells was investigated by immunoprecipitation. Approximately 10 million HEK293 cells were transfected with either the control vector, full-length EBNA3C, or different truncations of EBNA3C (amino-terminal, middle, and carboxy-terminal regions) tagged with the Flag vector. (B) A GST pulldown assay was performed by using EBNA3C truncations (full-length EBNA3C [aa 1 to 992], N-terminal aa 1 to 365, middle domain aa 366 to 620, and C-terminal aa 621 to 992) fused to GST. Smaller truncations of the N-terminal region of EBNA3C (aa 1 to 300, 1 to 200, 100 to 200, 1 to 100, 1 to 50, and 130 to 190) were cloned into the pA3M vector and monitored for binding activity with H2AX (right). A Coomassie-stained gel is shown for the GST control and the GST-H2AX fusion protein. RD, relative density. (C) Schematic showing different EBNA3C domains associated with other reported proteins as well as its binding domain for H2AX.
To further confirm the above-described results of association studies, we performed GST pulldown experiments using bacterially purified GST-fused H2AX with HEK293 cell extracts transiently transfected with plasmids expressing full-length EBNA3C and the three major truncated domains of EBNA3C (N-terminal, M, and C-terminal domains) (Fig. 4B). The results from GST pulldown assays also confirmed a strong interaction of full-length EBNA3C and the N-terminal residues (amino acids [aa] 1 to 365) with H2AX (Fig. 4B, top). The amounts of control GST and the GST-H2AX fusion protein used in the GST pulldown experiment were similar, as shown by corresponding Coomassie brilliant blue-stained SDS-PAGE gels (Fig. 4B, bottom). Additionally, to narrow down the binding residues in the N-terminal domain of EBNA3C, we further performed GST pulldown assays using various truncated regions of the EBNA3C N-terminal region (Fig. 4C). A strong association between GST-fused H2AX and EBNA3C residues 1 to 300, 1 to 200, and 1 to 100 was clearly detected, while no association was found beyond the first 100 amino acid residues of EBNA3C (Fig. 4C). These results demonstrate that the EBNA3C residues 1 to 100 are responsible for the formation of a stable complex with H2AX in cells. The schematic in Fig. 4D represents the mapping of the H2AX interaction domain within EBNA3C and further outlines the studies to identify the interacting residues.
EBNA3C colocalizes with H2AX in EBV-transformed LCLs.
Our results from both immunoprecipitation as well as GST pulldown assays strongly supported the formation of a complex between EBNA3C and H2AX under physiological conditions using EBV-transformed B cells. This prompted us to investigate whether EBNA3C and H2AX can colocalize within similar cellular compartments. Immunofluorescence (IF) studies were performed by using B cells stably expressing EBNA3C as well as EBV-transformed LCLs and EBV-negative controls (Fig. 5). First, IF was carried out with BJAB control cells along with the isogenic EBNA3C-expressing cells (BJAB#7 and BJAB#10) (data not shown). The assay was carried out by using specific antibodies against EBNA3C (A10; mouse monoclonal) and H2AX (rabbit polyclonal) along with Texas Red mouse and fluorescein isothiocyanate (FITC) anti-rabbit secondary antibodies for visualization of the complexes. The results showed a strong colocalization between H2AX and EBNA3C in similar nuclear compartments (data not shown). Furthermore, an analysis of the fluorescence intensity of H2AX signals in EBNA3C-expressing BJAB cells demonstrated a significant reduction compared to control BJAB cells (data not shown). This further corroborated our biochemical studies discussed above, which showed reduced H2AX levels in the presence of either EBV or EBNA3C.
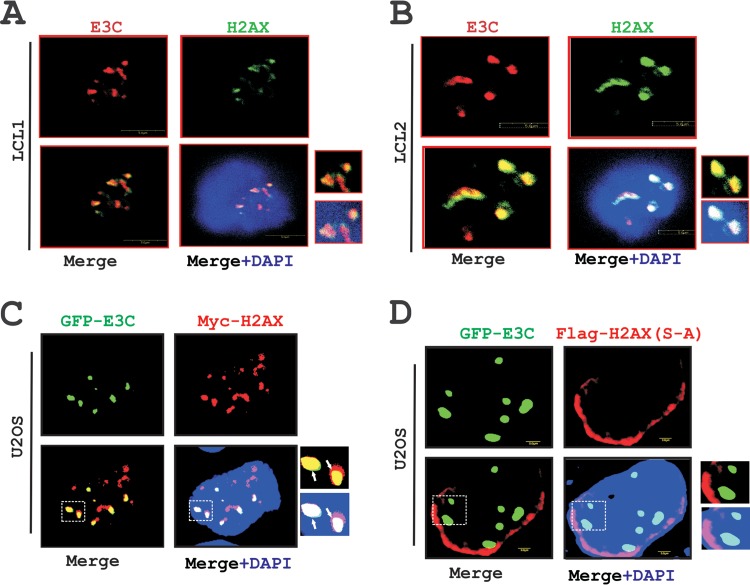
FIG 5.
EBNA3C colocalizes with H2AX in nuclear compartments. (A and B) In vitro EBV-transformed B-cell lines LCL1 and LCL2 were plated onto slides and subsequently air dried at room temperature. Cells were fixed by using 3% PFA and blocked with 5% skimmed milk. Protein expression were detected by using anti-mouse EBNA3C monoclonal antibody A10 (1:150) and anti-rabbit H2AX antibody (1:500) followed by anti-mouse Alexa Fluor 594 (red) for EBNA3C and anti-rabbit Alexa Fluor 488 (green) for H2AX. DAPI (4′,6-diamidino-2-phenylindole) was used for nuclear staining. (C) A total of 0.3 million epithelial U2OS cells were transfected with GFP-EBNA3C and Myc-H2AX plasmids by using Lipofectamine. H2AX was detected by using anti-Myc (9E10) antibody (1:200) followed by anti-rabbit Alexa Fluor 594 (red) secondary antibody. Colocalization of H2AX with GFP-EBNA3C was monitored by using an Olympus confocal microscope. (D) A total of 3 × 105 U2OS cells were seeded onto a coverslip and transfected with GFP-EBNA3C and mutant H2AX (S-A) plasmids by using Lipofectamine.
Importantly, IF analysis using LCL1 and LCL2 showed a similar colocalization pattern of H2AX and EBNA3C (Fig. 5A and B). To further validate this observation, we performed similar IF experiments using GFP-tagged EBNA3C and Myc-tagged H2AX in U2OS epithelial cells at 36 h posttransfection (Fig. 5C). Monitoring of the GFP signal of EBNA3C and a specific mouse monoclonal antibody against H2AX tagged with the Myc epitope clearly showed a punctuate pattern of colocalization between H2AX molecules and EBNA3C in similar nuclear compartments (Fig. 5C).
Our above-described binding results demonstrated that phosphorylation of H2AX at Ser139 is crucial for binding with EBNA3C. To further determine whether this residue was also important for colocalization, we next performed an IF study with GFP-tagged EBNA3C and a Flag-tagged mutated version of H2AX [H2AX (S-A)] in U2OS cells (Fig. 5D). At 36 h posttransfection, IF was performed by using the specific mouse monoclonal Flag antibody (M2) along with GFP fluorescence at a 488-nm excitation (Fig. 5D). Interestingly, the IF signals for H2AX (S-A) were observed mostly at the periphery of the nucleus, instead of the punctuate dots seen for the wild-type version (compare Fig. 5C and D). Consequently, little or no association of EBNA3C was observed in nuclear compartments with H2AX (S-A) (Fig. 5D). In agreement with our binding studies, the IF results once again showed that Ser139 of H2AX may play a critical role in the formation of a complex as well as colocalization with EBNA3C in similar nuclear compartments.
EBNA3C inhibits the transcription activity of the H2AX promoter.
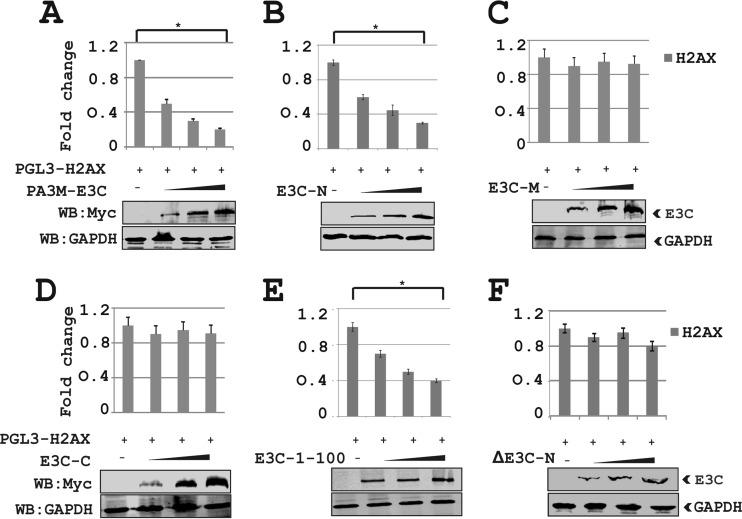
The above-described results showed that H2AX transcripts were downregulated in the presence of EBNA3C. To more specifically investigate this phenomenon, we performed luciferase-based reporter assays using the wild-type H2AX promoter linked to the luciferase reporter gene in the presence of EBNA3C (Fig. 6). First, the H2AX reporter plasmid H2AX-pGL3 was transiently transfected into HEK293 cells in the presence of the vector or full-length Myc-tagged EBNA3C (Fig. 6A). Cells were also transfected with the GFP control plasmid to monitor the transfection efficiency in this experiment. The results demonstrated that EBNA3C dramatically attenuated transcriptional activity from the H2AX promoter in a dose-dependent fashion in HEK293 cells (Fig. 6A). The levels of EBNA3C and GAPDH were analyzed by WB (Fig. 6A). Furthermore, to evaluate whether the binding domain of EBNA3C is necessary to control H2AX transcription, we performed similar reporter assays using various truncation domains of EBNA3C (Fig. 6B to F). The results showed that the N-terminal binding region (residues 1 to 365) but not the nonbinding domains (residues 366 to 992) of EBNA3C substantially attenuated transcription from the H2AX promoter (Fig. 6B to D). In agreement, the smaller N-terminal binding region of EBNA3C at residues 1 to 100 also showed a similar inhibitory trend of transcriptional regulation at the H2AX promoter (Fig. 6E). This indicated that the amino-terminal binding residues of EBNA3C are required as well as sufficient for transcriptional inhibition of the H2AX promoter. To further validate the above-described results, we transfected HEK293 cells with the N-terminally deleted region of EBNA3C (ΔN EBNA3C) along with the reporter and GFP expression plasmids (Fig. 6F). This result supported our above-described data showing that the N-terminal binding residues of EBNA3C are important for regulating H2AX transcription. All reporter experiments were carefully examined by WB analysis to monitor the expression levels of EBNA3C and GAPDH as an internal loading control (Fig. 6).
FIG 6.
The N-terminal binding residues of EBNA3C are sufficient for suppressing H2AX transcription. HEK293 cells were transfected with 10 μg of pGL3-H2AX and 1, 2, and 4 μg of EBNA3C truncations (EBNA3C aa 1 to 992, aa 1 to 365, aa 366 to 620, and aa 621 to 992). The N-terminally deleted region of EBNA3C (ΔEBNA3C-N) was also included. Data from the reporter assays are represented as relative luciferase units (RLU), with means and standard deviations represented by error bars. The fold activation was calculated by comparison of the promoter activity in the presence of the pGL3-H2AX promoter with the value of the pA3M vector alone.
EBNA3C can induce degradation of H2AX by recruiting the ubiquitin-proteasome system.
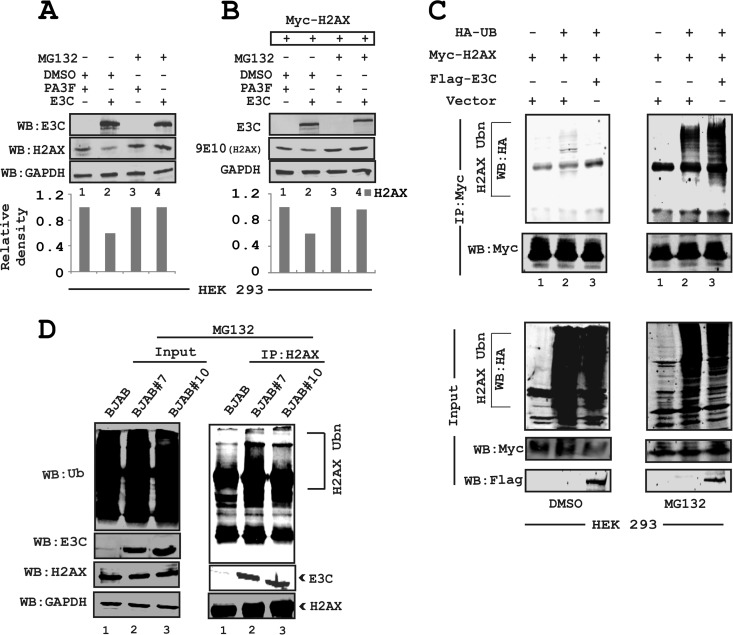
To establish latency, EBV has evolved strategies for targeted inactivation of cellular factors to escape from the DDR induced during early infection (38–41). However, the underlying mechanism is still not clear. Overall, our results described above strongly suggested that in addition to transcriptional regulation, EBNA3C can also attenuate H2AX at the protein level. Moreover, it has also been reported that the H2AX protein level can be regulated through polyubiquitination (23). Interestingly, EBNA3C has been previously shown to utilize a ubiquitin-mediated proteasome degradation strategy for targeting and degrading multiple tumor suppressor proteins to establish successful latent infection in infected B cells (31, 32). Along these lines, we wanted to determine whether downregulation of H2AX protein levels was associated with EBNA3C-mediated degradation through the ubiquitin-proteasome pathway. To explore this possibility, HEK293 cells were transfected with EBNA3C or the vector alone and subsequently treated with either the dimethyl sulfoxide (DMSO) control or MG132 over a period of 12 h at 48 h posttransfection (Fig. 7A). MG132 blocks the degradation of ubiquitin-conjugated proteins in mammalian cells by the 26S proteasome complex (42). The results demonstrated that the reduced levels of endogenous H2AX in the presence of EBNA3C were rescued by MG132 compared to control cells treated with DMSO (Fig. 7A, compare lanes 2 and 4). This observation was further validated by using transient expression of Myc-tagged H2AX expressed from a heterologous promoter in HEK293 cells (Fig. 7B). These results also demonstrated a rescue of H2AX protein levels from EBNA3C-mediated reduction in the presence of MG132 (Fig. 7B). These results indicate that EBNA3C can contribute to H2AX degradation through recruitment of the ubiquitin-proteasome machinery, the function of which was blocked by MG132 (Fig. 7A and B).
FIG 7.
EBNA3C facilitates H2AX degradation through the ubiquitin-mediated proteasomal pathway. (A) HEK293 cells were transfected with plasmids and treated with either MG132 (20 μg/ml) or DMSO as a control for 30 h posttransfection to inhibit proteasome activity. (B) HEK293 cells were transfected with plasmids expressing EBNA3C-Flag or Myc-H2AX. (C) HEK293 cells were transfected with various combinations of expression vectors, as shown. (D) In vivo ubiquitination assays were carried out by using 50 million BJAB (EBNA3C-negative), BJAB#7, and BJAB#10 (EBNA3C-positive) cells. H2AX antibody (2 μg) was used for pulldown experiments and subsequently detected by using ubiquitin- and H2AX-specific antibodies. The input was also analyzed as a loading control by using an anti-GAPDH antibody.
To support these results, we next performed in vivo ubiquitination experiments. HEK293 cells were cotransfected with plasmids expressing the HA-tagged ubiquitin moiety (HA-Ub), Flag-tagged EBNA3C, and Myc-tagged H2AX (Fig. 7C). The levels of H2AX ubiquitination were monitored by performing specific immunoprecipitation of H2AX molecules using an anti-Myc mouse monoclonal antibody (9E10) followed by WB analysis (Fig. 7C). The results demonstrated a significant difference in polyubiquitination levels of H2AX in the MG132-treated group compared to the DMSO control group (Fig. 7C, compare left and right panels). This is due to an impairment of proteasomal function, which resulted in an increase in the levels of ubiquitinated proteins. A dramatic enhancement in H2AX polyubiquitination was reproducibly observed in the presence of EBNA3C with MG132 treatment but not with the DMSO control (Fig. 7C, compare left and right panels, lanes 3). To support the above-described findings, ubiquitination experiments were also carried out with BJAB, BJAB#7, and BJAB#10 cells by performing H2AX immunoprecipitation using a specific anti-H2AX antibody (Fig. 7D). The results further demonstrated a relatively higher level of H2AX polyubiquitination in EBNA3C-expressing BJAB#7 and BJAB#10 cells than in BJAB control cells (Fig. 7D, compare lane 1 with lanes 2 and 3). This provided additional evidence to suggest that EBNA3C can facilitate H2AX destabilization through ubiquitin-mediated proteasomal degradation.
EBNA3C inhibits the growth-suppressive activity of H2AX.
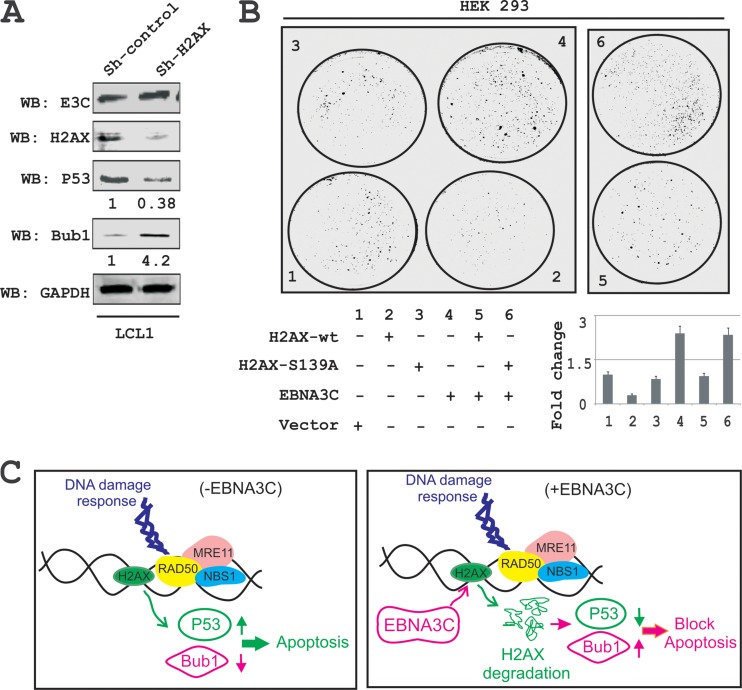
The spindle assembly checkpoint (SAC) and DDR are two critical mechanisms by which mammalian cells can maintain genomic stability (43). Moreover, H2AX was shown to induce cell cycle arrest via the p53/p21 pathway (44). In addition, knockdown of H2AX was shown to strongly suppress apoptosis in lung cancer cells (45). To elucidate the biological significance of H2AX degradation through EBNA3C in LCLs as well as to explore the underlying molecular mechanism of this DNA damage protein in EBV-induced B-cell lymphomagenesis, we used H2AX knockdown LCL1 cells compared to the control vector LCL1. Interestingly, H2AX knockdown resulted in a reduction in levels of the p53 tumor suppressor protein of about 2- to 3-fold, while the level of Bub1, a critical kinetochore protein essential for the SAC, was upregulated about 3- to 5-fold (Fig. 8A). These results now provide a potential molecular strategy for H2AX downregulation in LCLs and highlights a critical role for EBNA3C in regulating the DDR during B-cell lymphomagenesis.
FIG 8.
EBNA3C blocks H2AX-mediated growth-suppressive activity. (A) Knockdown in H2AX cells was evaluated to determine H2AX, EBNA3C, Bub1, and p53 expression levels using their respective antibodies. Experiments were independently performed three times, with similar results. (B) Colony formation assays was performed with HEK293 cells. Cells were transfected with the control vector, wild-type H2AX, the H2AX (S-A) mutant, EBNA3C, and EBNA3C combined with wild-type H2AX or H2AX (S-A). At 24 h posttransfection, the cells were selected with G418 for 2 weeks. On alternate days, the media and antibiotics were replaced. Cells were fixed with 3% PFA and stained with 0.1% crystal violet. The total number of colonies was measured by using Odyssey Image analysis software. (C) Schematic showing the role of EBNA3C in degradation of H2AX, which results in modulation of p53 and Bub1 levels. These changes can contribute to EBV-induced oncogenesis.
To specifically explore the inhibitory effects of EBNA3C on H2AX-mediated growth-suppressive properties, we carried out colony formation assays (CFAs) with HEK293 cells (Fig. 8B). Cells were transfected with either the vector control or vectors expressing wild-type H2AX, mutated H2AX (S139A), or EBNA3C alone or were cotransfected with combinations of EBNA3C and wild-type H2AX or EBNA3C and mutated H2AX (S139A) (Fig. 8B). At 24 h posttransfection, cells were further selected with G418 for 2 weeks. Both media and antibiotics were changed every 2 days. The total colony formation was scanned and quantitated by using Odyssey Image analysis software (Fig. 8B). The results showed that expression of wild-type H2AX caused an approximately 70% reduction in colony formation of HEK293 cells compared to the vector control. However, minimal effects were observed in the presence of the mutated version of H2AX (Fig. 8B, compare panels 1 to 3). This again signifies the importance of the Ser139 residue to the growth-suppressive properties of H2AX. Importantly, coexpression of EBNA3C with wild-type H2AX demonstrated an almost complete reversion of H2AX-mediated growth-suppressive activity, which was similar to that of the vector control (Fig. 8B, compare panels 1, 3, and 5). Interestingly, EBNA3C expression alone showed a dramatic enhancement of colony formation (Fig. 8B, panels 4 and 6), which further strengthens a role for EBNA3C as a dominant viral oncoprotein. Overall, these results describe a model where EBNA3C can play a critical role in regulating H2AX stability through ubiquitin-mediated degradation and can therefore contribute to modulating its activity during the DDR induced by viral infection. This ultimately drives establishment of the EBV latency program in infected primary B cells.
DISCUSSION
Viral infection and the cellular DDR have been widely debated and explored in recent years (24, 46). Viruses have evolved complex regulatory mechanisms to persist in infected cells by manipulating several DNA repair mechanisms (47). H2AX plays a key role in the DDR (48). H2AX is well known for its positive regulation of DNA repair and activation of the DNA damage checkpoints (49). H2AX also varies from H2A with a unique C-terminal region (23), which contains a highly conserved SQE motif (50). However, it has been suggested that H2AX is not utterly important for recognition of DNA damage but facilitates the damage response through interaction with proteins that recognize the phosphorylated form of H2AX (γ-H2AX) (49). However, previous studies by Bassing et al. reported that cells from H2AX-deficient mice are more prone to chromosomal instability and less efficient in DNA double-strand break repair (38).
Previously, it was reported that EBV latent proteins, EBNAs and LMPs, can contribute toward chromosomal instability and cellular DNA damage by inhibiting as well as facilitating degradation of DNA repair elements or through mislocalization (51, 52). Reports from our laboratory and others also revealed that EBNA3C can contribute to the oncogenic process through degradation of p53 or interaction with Chk2 (16, 20). However, limited information is available for demonstrating a role for EBNA3C in modulating H2AX.
Here we identified H2AX as an interacting partner of EBNA3C during EBV infection. We showed that in both EBV- as well as EBNA3C-positive B-cell lines, cellular H2AX levels were significantly downregulated. This initial observation was further corroborated by using an RNA interference strategy by knocking down EBNA3C expression in EBV-transformed LCLs. Using an in vitro model system for resting B-cell infection with an EBNA3C-deleted recombinant virus in comparison to wild-type EBV infection, we confirmed that EBNA3C was directly involved in regulating H2AX levels. The attenuation of H2AX levels was clearly more significant in wild-type virus infection, whereas EBNA3C-deleted EBV did not show any significant change in H2AX expression levels. These results supported our hypothesis that through modulation of the total H2AX levels, EBNA3C can contribute to EBV latency and cell proliferation.
These initial findings were further corroborated using HEK293 cells, further strengthening our initial hypothesis that EBNA3C is directly involved in deregulating H2AX expression. The reduction of H2AX levels was found to be inversely proportional to the increase in EBNA3C at the protein and transcript levels. While unable to bind DNA directly, EBNA3C was previously shown to be a strong transcriptional factor that can regulate the transcription of multiple cellular and viral genes. EBNA3C formed a stable complex with H2AX through interactions with its amino-terminal first 100 amino acid residues. Interestingly, we know that the N-terminal region of EBNA3C interacts with various cell cycle regulatory molecules, such as cyclin D1, cyclin A, p53, Mdm2, E2F1, IRF4, and Aurora kinase B (16, 30, 32, 34, 35).
Phosphorylation of Ser139 of H2AX (referred to as γ-H2AX) is indispensable for recruitment of DNA repair elements (53). Furthermore, H2AX is phosphorylated by ATM in response to double-strand breaks (53). More specifically, mutation at Ser139 completely abolished the interaction between H2AX and EBNA3C. This suggests that H2AX phosphorylation may play an important role in EBNA3C-regulated cell proliferation. This also suggests the involvement of EBNA3C in regulating the DDR and consequently also opens a new avenue for further exploration. Our reporter analysis of the H2AX promoter demonstrated that the N-terminal binding residues were sufficient for blocking H2AX transcription. Furthermore, H2AX localization is strictly confined to the nucleosomes (54). However, some reports demonstrated that H2AX also functions outside the nucleosomes (55, 56). Our results demonstrated that EBNA3C can localize with H2AX within similar nuclear compartments in EBV-transformed LCLs as well as EBNA3C-expressing stable cell lines. The staining intensity of H2AX in EBNA3C-negative and EBNA3C-positive cell lines showed a reduction in the levels of H2AX in the presence of EBNA3C, further supporting our hypothesis that EBNA3C can attenuate H2AX expression.
In addition to transcriptional regulation, our results also indicated that EBNA3C can regulate H2AX protein levels. Previous reports showed that H2AX is degraded by polyubiquitination through the proteasome degradation pathway and that inhibition of the proteasome can stabilize H2AX (41, 57, 58). We hypothesized that EBNA3C-mediated H2AX deregulation may occur through polyubiquitination and degradation, since our previous studies showed that EBNA3C recruits the ubiquitin-proteasome machinery for targeted proteolysis of multiple tumor suppressor proteins through its N-terminal residues (31, 59). Recently, Feng and Chen hypothesized that Ubc13 and RNF8 are involved in DDR-induced H2AX ubiquitination (60). How these proteins are involved in the functional regulation of H2AX levels in EBV-positive cells remains undetermined. In addition to this, monoubiquitination of H2AX was shown to be an essential epigenetic marker during the DNA damage response (61, 62). It would be interesting to determine whether EBNA3C could also initiate H2AX monoubiquitination to regulate nucleosomal activity and, thereby, gene transcription. For now, our results demonstrated that EBNA3C efficiently induces H2AX degradation, most likely through polyubiquitination.
To determine the biological implications of H2AX deregulation in EBV-driven oncogenesis, we generated LCLs knocked down for H2AX and evaluated the expression levels of the p53 tumor suppressor and the oncoprotein Bub1. Previously, Fragkos et al. suggested that H2AX was required for cell cycle arrest through p53 (44). There is a growing body of evidence for cross talk between the DDR elements and SAC components (43). Moreover, Bub1 was shown to be an essential mitotic protein in gammaherpesvirus-induced oncogenesis (63). Depletion of H2AX expression in LCLs led to a significant downregulation of p53 and upregulation of Bub1. This supports an antiproliferative role for H2AX in the context of EBV-induced transformation of B cells. Subsequent colony formation assays further supported the antiproliferative properties of H2AX. Our results also clearly demonstrated that EBNA3C can effectively neutralize the growth-suppressive effects of H2AX. Therefore, the essential latent antigen EBNA3C may have evolved to modulate the DDR and thus contribute to viral latency in addition to all its other functions. Overall, this study provides strong evidence to support a role for EBNA3C-mediated attenuation of total H2AX levels upon EBV infection (Fig. 8C) and may be important for targeted therapeutic strategies against EBV-associated B-cell lymphomas
ACKNOWLEDGMENTS
We are grateful to Alvaro N. A. Monteiro (H. Lee Moffitt Cancer Center, Tampa, FL), Toru Ouchi (Roswell Park Cancer Institute, Buffalo, NY), and Elliott Kieff (Harvard Medical School, Boston, MA) for kindly providing reagents.
This work was supported by NCI grant CA137894-05 and NIDDK grant DK050306-17.
E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
Footnotes
Published ahead of print 15 January 2014
REFERENCES
- 1.Epstein MA, Achong BG, Barr YM. 1964. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet i:702–703 [DOI] [PubMed] [Google Scholar]
- 2.Young LS, Murray PG. 2003. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene 22:5108–5121. 10.1038/sj.onc.1206556 [DOI] [PubMed] [Google Scholar]
- 3.Pegtel DM, Middeldorp J, Thorley-Lawson DA. 2004. Epstein-Barr virus infection in ex vivo tonsil epithelial cell cultures of asymptomatic carriers. J. Virol. 78:12613–12624. 10.1128/JVI.78.22.12613-12624.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan AS, To KF, Lo KW, Ding M, Li X, Johnson P, Huang DP. 2002. Frequent chromosome 9p losses in histologically normal nasopharyngeal epithelia from southern Chinese. Int. J. Cancer 102:300–303. 10.1002/ijc.10689 [DOI] [PubMed] [Google Scholar]
- 5.Cho WC. 2007. Nasopharyngeal carcinoma: molecular biomarker discovery and progress. Mol. Cancer 6:1. 10.1186/1476-4598-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson MP, Kurzrock R. 2004. Epstein-Barr virus and cancer. Clin. Cancer Res. 10:803–821. 10.1158/1078-0432.CCR-0670-3 [DOI] [PubMed] [Google Scholar]
- 7.Cohen JI. 2000. Epstein-Barr virus infection. N. Engl. J. Med. 343:481–492. 10.1056/NEJM200008173430707 [DOI] [PubMed] [Google Scholar]
- 8.Amon W, Farrell PJ. 2005. Reactivation of Epstein-Barr virus from latency. Rev. Med. Virol. 15:149–156. 10.1002/rmv.456 [DOI] [PubMed] [Google Scholar]
- 9.Rezk SA, Weiss LM. 2007. Epstein-Barr virus-associated lymphoproliferative disorders. Hum. Pathol. 38:1293–1304. 10.1016/j.humpath.2007.05.020 [DOI] [PubMed] [Google Scholar]
- 10.Gandhi MK, Tellam JT, Khanna R. 2004. Epstein-Barr virus-associated Hodgkin's lymphoma. Br. J. Haematol. 125:267–281. 10.1111/j.1365-2141.2004.04902.x [DOI] [PubMed] [Google Scholar]
- 11.Marchini A, Kieff E, Longnecker R. 1993. Marker rescue of a transformation-negative Epstein-Barr virus recombinant from an infected Burkitt lymphoma cell line: a method useful for analysis of genes essential for transformation. J. Virol. 67:606–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maruo S, Wu Y, Ishikawa S, Kanda T, Iwakiri D, Takada K. 2006. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc. Natl. Acad. Sci. U. S. A. 103:19500–19505. 10.1073/pnas.0604919104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halder S, Murakami M, Verma SC, Kumar P, Yi F, Robertson ES. 2009. Early events associated with infection of Epstein-Barr virus infection of primary B-cells. PLoS One 4:e7214. 10.1371/journal.pone.0007214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F, Tsang SF, Kurilla MG, Cohen JI, Kieff E. 1990. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J. Virol. 64:3407–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maruo S, Wu Y, Ito T, Kanda T, Kieff ED, Takada K. 2009. Epstein-Barr virus nuclear protein EBNA3C residues critical for maintaining lymphoblastoid cell growth. Proc. Natl. Acad. Sci. U. S. A. 106:4419–4424. 10.1073/pnas.0813134106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha A, Bamidele A, Murakami M, Robertson ES. 2011. EBNA3C attenuates the function of p53 through interaction with inhibitor of growth family proteins 4 and 5. J. Virol. 85:2079–2088. 10.1128/JVI.02279-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClellan MJ, Khasnis S, Wood CD, Palermo RD, Schlick SN, Kanhere AS, Jenner RG, West MJ. 2012. Downregulation of integrin receptor-signaling genes by Epstein-Barr virus EBNA 3C via promoter-proximal and -distal binding elements. J. Virol. 86:5165–5178. 10.1128/JVI.07161-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikitin PA, Luftig MA. 2011. At a crossroads: human DNA tumor viruses and the host DNA damage response. Future Virol. 6:813–830. 10.2217/fvl.11.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choudhuri T, Verma SC, Lan K, Murakami M, Robertson ES. 2007. The ATM/ATR signaling effector Chk2 is targeted by Epstein-Barr virus nuclear antigen 3C to release the G2/M cell cycle block. J. Virol. 81:6718–6730. 10.1128/JVI.00053-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, Allday MJ, Patel A, Dave SS, Kim W, Hu K, Guo J, Tainter D, Rusyn E, Luftig MA. 2010. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8:510–522. 10.1016/j.chom.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Summers KC, Shen F, Sierra Potchanant EA, Phipps EA, Hickey RJ, Malkas LH. 2011. Phosphorylation: the molecular switch of double-strand break repair. Int. J. Proteomics 2011:373816. 10.1155/2011/373816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jha HC, Upadhyay SK, AJ MP, Lu J, Cai Q, Saha A, Robertson ES. 2013. H2AX phosphorylation is important for LANA-mediated Kaposi's sarcoma-associated herpesvirus episome persistence. J. Virol. 87:5255–5269. 10.1128/JVI.03575-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. 2007. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 27:7028–7040. 10.1128/MCB.00579-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turnell AS, Grand RJ. 2012. DNA viruses and the cellular DNA-damage response. J. Gen. Virol. 93:2076–2097. 10.1099/vir.0.044412-0 [DOI] [PubMed] [Google Scholar]
- 25.Bajaj BG, Murakami M, Cai Q, Verma SC, Lan K, Robertson ES. 2008. Epstein-Barr virus nuclear antigen 3C interacts with and enhances the stability of the c-Myc oncoprotein. J. Virol. 82:4082–4090. 10.1128/JVI.02500-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight JS, Sharma N, Robertson ES. 2005. SCFSkp2 complex targeted by Epstein-Barr virus essential nuclear antigen. Mol. Cell. Biol. 25:1749–1763. 10.1128/MCB.25.5.1749-1763.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J, Verma SC, Murakami M, Cai Q, Kumar P, Xiao B, Robertson ES. 2009. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (KSHV) upregulates survivin expression in KSHV-associated B-lymphoma cells and contributes to their proliferation. J. Virol. 83:7129–7141. 10.1128/JVI.00397-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rios-Doria J, Fay A, Velkova A, Monteiro AN. 2006. DNA damage response: determining the fate of phosphorylated histone H2AX. Cancer Biol. Ther. 5:142–144. 10.4161/cbt.5.2.2530 [DOI] [PubMed] [Google Scholar]
- 29.Kang MA, So EY, Ouchi T. 2012. Deregulation of DNA damage response pathway by intercellular contact. J. Biol. Chem. 287:16246–16255. 10.1074/jbc.M111.337212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jha HC, Lu J, Saha A, Cai Q, Banerjee S, Prasad MA, Robertson ES. 2013. EBNA3C-mediated regulation of Aurora kinase B contributes to Epstein-Barr virus-induced B-cell proliferation through modulation of the activities of the retinoblastoma protein and apoptotic caspases. J. Virol. 87:12121–12138. 10.1128/JVI.02379-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saha A, Lu J, Morizur L, Upadhyay SK, AJ MP, Robertson ES. 2012. E2F1 mediated apoptosis induced by the DNA damage response is blocked by EBV nuclear antigen 3C in lymphoblastoid cells. PLoS Pathog. 8:e1002573. 10.1371/journal.ppat.1002573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi F, Saha A, Murakami M, Kumar P, Knight JS, Cai Q, Choudhuri T, Robertson ES. 2009. Epstein-Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology 388:236–247. 10.1016/j.virol.2009.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Cai Q, Lu J, Jha HC, Robertson ES. 2011. Upregulation of cellular Bcl-2 by the KSHV encoded RTA promotes virion production. PLoS One 6:e23892. 10.1371/journal.pone.0023892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee S, Lu J, Cai Q, Saha A, Jha HC, Dzeng RK, Robertson ES. 2013. The EBV latent antigen 3C inhibits apoptosis through targeted regulation of interferon regulatory factors 4 and 8. PLoS Pathog. 9:e1003314. 10.1371/journal.ppat.1003314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saha A, Halder S, Upadhyay SK, Lu J, Kumar P, Murakami M, Cai Q, Robertson ES. 2011. Epstein-Barr virus nuclear antigen 3C facilitates G1-S transition by stabilizing and enhancing the function of cyclin D1. PLoS Pathog. 7:e1001275. 10.1371/journal.ppat.1001275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886–895. 10.1016/S0960-9822(00)00610-2 [DOI] [PubMed] [Google Scholar]
- 37.Tarakanova VL, Stanitsa E, Leonardo SM, Bigley TM, Gauld SB. 2010. Conserved gammaherpesvirus kinase and histone variant H2AX facilitate gammaherpesvirus latency in vivo. Virology 405:50–61. 10.1016/j.virol.2010.05.027 [DOI] [PubMed] [Google Scholar]
- 38.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114:359–370. 10.1016/S0092-8674(03)00566-X [DOI] [PubMed] [Google Scholar]
- 39.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679. 10.1038/ncb1004 [DOI] [PubMed] [Google Scholar]
- 40.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. 2004. H2AX: the histone guardian of the genome. DNA Repair (Amst.) 3:959–967. 10.1016/j.dnarep.2004.03.024 [DOI] [PubMed] [Google Scholar]
- 41.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463–473. 10.1016/S0092-8674(00)00051-9 [DOI] [PubMed] [Google Scholar]
- 42.Nawaz Z, Lonard DM, Dennis AP, Smith CL, O'Malley BW. 1999. Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl. Acad. Sci. U. S. A. 96:1858–1862. 10.1073/pnas.96.5.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang C, Wang H, Xu Y, Brinkman KL, Ishiyama H, Wong ST, Xu B. 2012. The kinetochore protein Bub1 participates in the DNA damage response. DNA Repair (Amst.) 11:185–191. 10.1016/j.dnarep.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fragkos M, Jurvansuu J, Beard P. 2009. H2AX is required for cell cycle arrest via the p53/p21 pathway. Mol. Cell. Biol. 29:2828–2840. 10.1128/MCB.01830-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C, Xiong M, Luo Y, Li J, Zhang Y, Dong Y, Zhu Y, Niu T, Wang Z, Duan L. 2013. Genome-wide transcriptional analysis of apoptosis-related genes and pathways regulated by H2AX in lung cancer A549 cells. Apoptosis 18:1039–1047. 10.1007/s10495-013-0875-x [DOI] [PubMed] [Google Scholar]
- 46.Nikitin PA, Luftig MA. 2012. The DNA damage response in viral-induced cellular transformation. Br. J. Cancer 106:429–435. 10.1038/bjc.2011.612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaurushiya MS, Weitzman MD. 2009. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst.) 8:1166–1176. 10.1016/j.dnarep.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chadwick BP, Willard HF. 2001. Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Hum. Mol. Genet. 10:1101–1113. 10.1093/hmg/10.10.1101 [DOI] [PubMed] [Google Scholar]
- 49.Kinner A, Wu W, Staudt C, Iliakis G. 2008. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36:5678–5694. 10.1093/nar/gkn550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li A, Eirin-Lopez JM, Ausio J. 2005. H2AX: tailoring histone H2A for chromatin-dependent genomic integrity. Biochem. Cell Biol. 83:505–515. 10.1139/o05-114 [DOI] [PubMed] [Google Scholar]
- 51.Kamranvar SA, Masucci MG. 2011. The Epstein-Barr virus nuclear antigen-1 promotes telomere dysfunction via induction of oxidative stress. Leukemia 25:1017–1025. 10.1038/leu.2011.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39–85. 10.1146/annurev.biochem.73.011303.073723 [DOI] [PubMed] [Google Scholar]
- 53.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462–42467. 10.1074/jbc.C100466200 [DOI] [PubMed] [Google Scholar]
- 54.Downs JA, Nussenzweig MC, Nussenzweig A. 2007. Chromatin dynamics and the preservation of genetic information. Nature 447:951–958. 10.1038/nature05980 [DOI] [PubMed] [Google Scholar]
- 55.Bewersdorf J, Bennett BT, Knight KL. 2006. H2AX chromatin structures and their response to DNA damage revealed by 4Pi microscopy. Proc. Natl. Acad. Sci. U. S. A. 103:18137–18142. 10.1073/pnas.0608709103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Tseng M, Perdreau SA, Rossi F, Antonescu C, Besmer P, Fletcher JA, Duensing S, Duensing A. 2007. Histone H2AX is a mediator of gastrointestinal stromal tumor cell apoptosis following treatment with imatinib mesylate. Cancer Res. 67:2685–2692. 10.1158/0008-5472.CAN-06-3497 [DOI] [PubMed] [Google Scholar]
- 57.Bassing CH, Alt FW. 2004. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle 3:149–153. 10.4161/cc.3.2.689 [DOI] [PubMed] [Google Scholar]
- 58.Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW. 2002. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. U. S. A. 99:8173–8178. 10.1073/pnas.122228699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saha A, Murakami M, Kumar P, Bajaj B, Sims K, Robertson ES. 2009. Epstein-Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2. J. Virol. 83:4652–4669. 10.1128/JVI.02408-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng L, Chen J. 2012. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat. Struct. Mol. Biol. 19:201–206. 10.1038/nsmb.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bergink S, Jentsch S. 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458:461–467. 10.1038/nature07963 [DOI] [PubMed] [Google Scholar]
- 62.Lukas J, Bartek J. 2009. DNA repair: new tales of an old tail. Nature 458:581–583. 10.1038/458581a [DOI] [PubMed] [Google Scholar]
- 63.Xiao B, Verma SC, Cai Q, Kaul R, Lu J, Saha A, Robertson ES. 2010. Bub1 and CENP-F can contribute to Kaposi's sarcoma-associated herpesvirus genome persistence by targeting LANA to kinetochores. J. Virol. 84:9718–9732. 10.1128/JVI.00713-10 [DOI] [PMC free article] [PubMed] [Google Scholar]