ABSTRACT
Using mass spectrometry, we identified p190RhoGAP (p190) as a binding partner of human papillomavirus 16 (HPV16) E7. p190 belongs to the GTPase activating protein (GAP) family and is one of the primary GAPs for RhoA. GAPs stimulate the intrinsic GTPase activity of the Rho proteins, leading to Rho inactivation and influencing numerous biological processes. RhoA is one of the best-characterized Rho proteins and is specifically involved in formation of focal adhesions and stress fibers, thereby regulating cell migration and cell spreading. Since this is the first report that E7 associates with p190, we carried out detailed interaction studies. We show that E7 proteins from other HPV types also bind p190. Furthermore, we found that conserved region 3 (CR3) of E7 and the middle domain of p190 are important for this interaction. More specifically, we identified two residues in CR3 of E7 that are necessary for p190 binding and used mutants of E7 with mutations of these residues to determine the biological consequences of the E7-p190 interaction. Our data suggest that the interaction of E7 with p190 dysregulates this GAP and alters the actin cytoskeleton. We also found that this interaction negatively regulates cell spreading on a fibronectin substrate and therefore likely contributes to important aspects of the HPV life cycle or HPV-induced tumorigenesis.
IMPORTANCE This study identifies p190RhoGAP as a novel cellular binding partner for the human papillomavirus (HPV) E7 protein. Our study shows that a large number of different HPV E7 proteins bind p190RhoGAP, and it identifies regions in both E7 and p190RhoGAP which are important for the interaction to occur. This study also highlights the likelihood that the E7-p190RhoGAP interaction may have important biological consequences related to actin organization in the infected cell. These changes could be an important contributor to the viral life cycle and during progression to cancer in HPV-infected cells. Importantly, this work also emphasizes the need for further study in a field which has largely been unexplored as it relates to the HPV life cycle and HPV-induced transformation.
INTRODUCTION
Human papillomaviruses (HPVs) are small, double-stranded DNA viruses which induce papillomas in cutaneous and mucosal epithelia and are also the etiological agents of many cervical and other anogenital cancers (1–5). More than 150 HPV types have been described, and many more are presumed to exist (6). Specific HPV types often preferentially infect distinct anatomical sites. HPVs associated with lesions that can progress to carcinogenesis are classified as “high-risk” types, the most common of which is HPV16. In contrast, HPVs associated with benign warts that regress with time are termed “low-risk” viruses (7).
The ability of HPVs to induce cellular immortalization and transformation is attributed primarily to the viral oncoproteins E6 and E7, which are consistently expressed in HPV-induced cancers (8–11). While E6 prevents apoptosis by inducing the degradation of the tumor suppressor p53 through the proteasome system, E7 disrupts cell cycle regulation by binding and inactivating the retinoblastoma tumor suppressor (pRb) (12, 13). In addition, both E6 and E7 alter other cellular signaling pathways by interacting with a plethora of cellular proteins and dysregulating their function, thereby enhancing the carcinogenic potential of the cell (14–19). To date, E7 has been reported to interact with over 50 cellular factors, although the biological significance of many of these interactions is unknown (20).
Here we demonstrate a novel interaction of HPV E7 with p190RhoGAP (p190), a Rho family GTPase activating protein (GAP). p190 belongs to a large family of proteins that stimulate the intrinsic GTPase activity of the Rho proteins, leading to Rho inactivation. Rho family GTPases serve as molecular switches, cycling between active, GTP-bound and inactive, GDP-bound states and transducing signals from the extracellular environment to elicit cellular responses such as changes in gene expression, morphology, and migration (21, 22). Of the known Rho proteins, Cdc42, Rac1, and RhoA are the most thoroughly characterized (23). The ability of Rho GTPases to associate with downstream effectors is held in balance by the opposing activities of guanine nucleotide exchange factors (GEFs), which encourage GTP loading, and GAPs, which catalyze the low-level GTPase activity of Rho (24). Precise spatial and temporal regulation of Rho family proteins is critical for numerous cellular processes; for instance, RhoA is crucial for efficient cell migration and cell spreading, and while some RhoA activity is required for migration, possibly to maintain sufficient adhesion to the substrate, high RhoA activity inhibits movement (25).
Through Rho-dependent and Rho-independent functions, p190 plays a critical role in regulating actin cytoskeleton dynamics and cell spreading; it also negatively controls tumor growth, transformation, metastasis, invasion, and angiogenesis, strongly suggesting that p190 may function as a tumor suppressor (26–31). Therefore, p190 appears to be an attractive target for a viral oncoprotein such as HPV E7. In this study, our aim was to characterize the interaction of E7 with p190 at the biochemical level and to determine the biological consequences of this association. We show that the C-terminal region of E7 is necessary and sufficient to associate with p190. Using a panel of surface-exposed mutants in the C terminus of E7, we identified two mutants unable to bind p190. Utilizing these two mutants of E7 and a dominant negative mutant of p190 as tools to probe the biological significance of the interaction, we first demonstrate that E7 binding to p190 contributes to an altered actin cytoskeleton. In addition, we show that E7-p190 interaction reduces cell spreading in E7-expressing cells on a fibronectin substrate.
MATERIALS AND METHODS
Plasmids.
The surface-exposed mutants within CR3 of HPV16 E7 were previously generated by site-directed mutagenesis and are described elsewhere (32). GFP-fused E7 constructs for HPV16, HPV6, HPV11, and HPV18 were described previously (33). Full-length E7 genes from HPV31, HPV33, HPV39, HPV45, HPV52, HPV55, HPV58, HPV59, HPV67, and HPV74 were PCR amplified from the viral genomes and cloned into the pCANmycEGFP vector by using EcoRI and XbaI restriction sites. The source for HPV genomes and the primer sequences used for HPV E7 cloning will be provided upon request. Hemagglutinin (HA)-tagged p190RhoGAP (p190) full-length and GTP-binding domain (GBD; amino acids 1 to 266) constructs were kind gifts from S. Parsons (University of Virginia, Charlottesville, VA) and were described previously (34). p190 MD (amino acids 379 to 1183) and GAP (amino acids 1180 to 1513) constructs were PCR amplified using the following primers: MD-F, ACTGTGGATCCTGGTTTGTTGTACTT; MD-R, ACTGTGAATTCTCACAGCTCATCATCACT; GAP-F, ACTGTGGATCCGATGATGAGCTGGGA; and GAP-R, ACTGTGAATTCTCAAGAAGACAACTGATT. These constructs were then cloned into the backbone of the pKH3 vector by using BamHI and EcoRI restriction sites. p190RA is the dominant negative p190(R1283A) mutant; it was a kind gift from Ian Macara (Vanderbilt University Medical Center, Nashville, TN).
Cell culture and transfection.
Human HaCat, CaSki, C33A, U2OS, and HT1080 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin (100 U/ml). For coimmunoprecipitation experiments, HT1080 cells were seeded into 10-cm plates at 2 × 106 cells per plate and transfected 24 h later with 8 μg of total DNA using X-tremeGENE HP (Roche, Laval, Quebec, Canada) according to the manufacturer's instructions. At 24 h posttransfection, cells were collected for coimmunoprecipitation experiments. U2OS cells stably expressing E7 or matched U2OS-neo control cells were obtained from Karl Munger (Brigham and Women's Hospital, Boston, MA) and maintained as previously described (35).
Antibodies.
For Western blots, rat anti-hemagglutinin (anti-HA) (Roche) was used at 1:2,000, and for coimmunoprecipitation experiments, 25 μl of mouse anti-HA hybridoma lysate (clone 12CA5) was utilized per sample. Green fluorescent protein (GFP) was detected using rabbit anti-GFP (Living Colors; Clontech, Mountain View, CA) at 1:2,000, and E7 was detected using mouse anti-E7 (8C9; Invitrogen, Burlington, Ontario, Canada) at 1:200. myc-tagged constructs were detected with a mouse anti-myc hybridoma lysate (clone 9E10), used at 1:200. Monoclonal anti-phosphotyrosine was used at 1:500 (Upstate). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit (Jackson Laboratories, Sacramento, CA), goat anti-rat (Pierce, Rockford, IL), and rabbit anti-mouse (Jackson Laboratories) were used as secondary antibodies.
Coimmunoprecipitation and Western blot analysis.
Cells were transfected at a 1:1 ratio with the myc-GFP fusion and an HA-tagged binding partner. Cells were harvested at 24 h posttransfection by scraping and were washed once with 1× phosphate-buffered saline (PBS). Cells were lysed in NP-40 (50 mM Tris, pH 7.8, 150 mM NaCl, 0.5% NP-40) lysis buffer supplemented with 1× mammalian protease inhibitor cocktail (Sigma, Oakville, Ontario, Canada). Typically, 1 mg of cell lysate was mixed with 100 μl of anti-myc hybridoma or 25 μl of anti-HA hybridoma and 100 μl of a 10% slurry of protein A-Sepharose resin (Sigma) and incubated at 4°C for 1 to 2 h on a nutator. Immunoprecipitates were washed three times with lysis buffer, resuspended in 2× lithium dodecyl sulfate (LDS) sample buffer, and then boiled for 5 min. Samples were analyzed by Western blotting. To assess the phosphorylation status of p190 in E7-expressing cells, U2OS cells were transfected with 2 μg of HA-p190 and 6 μg of GFP or GFP-E7. At 48 h posttransfection, the lysates were prepared in NP-40 lysis buffer and subjected to immunoprecipitation with anti-HA antibody, followed by immunoblotting as indicated.
Immunofluorescence and image analysis.
Cells were fixed with 3.7% formaldehyde for 20 min and then permeabilized with 0.2% Triton X-100 for 20 min at room temperature. Samples were washed extensively with 1× PBS, pH 7.4, and once with 0.1 M glycine (in PBS). The slides were then incubated with Alexa 568-phalloidin (Invitrogen) at 1:200 (in PBS) for 1 h at 37°C in a humidity chamber. The slides were washed extensively with PBS and then mounted on glass slides with ProLong antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) mounting medium (Invitrogen). Imaging was performed using a Nikon Eclipse Ti-S fluorescence microscope equipped with a QImaging Retiga 1300-coded monochrome 12-bit camera. Images were captured using Velocity software, version 7.0. Confocal images were acquired with a Fluoview 1000 laser scanning confocal microscope (Olympus Corp.), using a ×60 Plan Apochromat 1.42 oil objective. Multicolor images were acquired in the sequential acquisition mode to avoid cross-excitation. Quantification of phalloidin levels was carried out using images acquired on the fluorescence microscope and using ImageJ as previously described (36). Data are representative of a minimum of 75 cells for each sample.
Cell spreading.
Cells were trypsinized at 24 h posttransfection and washed two times with DMEM without FBS or antibiotics. The cells were resuspended in DMEM with 0.5% bovine serum albumin (BSA) (fatty acid free; Sigma) for 1 h at 37°C. Suspended cells were plated onto fibronectin-coated (10 μg/ml) coverslips for 20 min and then immediately fixed and stained with phalloidin as described above. Images were captured with a fluorescence microscope as described above, and cell area was determined using ImageJ.
RhoGTP assay.
Cellular RhoA-GTP was measured using a colorimetric G-Lisa RhoA activation assay kit as suggested by the manufacturer (Cytoskeleton).
RESULTS
HPV16 E7 associates with p190RhoGAP.
We initially observed that p190 associates with HPV16 E7 by mass spectrometry. HEK293 cells were transfected with epitope-tagged E7, and lysates were immunoprecipitated for the epitope tag and analyzed by liquid chromatography–matrix-assisted laser desorption ionization–tandem time of flight (LC-MALDI-TOF/TOF) mass spectrometry (data not shown). This identified p190 with a high confidence/coverage (84.3% sequence coverage). As an initial step in confirming this novel binding partner of E7, we utilized two human cervical cancer cell lines: CaSki cells are HPV16 positive and express the E6 and E7 oncoproteins, whereas C33A cells are HPV negative and served as a negative control. We carried out coimmunoprecipitation experiments by using anti-E7 antibody, followed by Western blotting with anti-p190 antibody. This experiment confirmed that p190 associates with E7 under endogenous conditions (Fig. 1).
FIG 1.

HPV16 E7 associates with p190RhoGAP (p190). Lysates from HPV16-positive (CaSki) and HPV-negative (C33A) cells were prepared and immunoprecipitated (IP) for E7 as described in Materials and Methods. Samples were immunoblotted as indicated.
C-terminal sequences of HPV16 E7 are necessary and sufficient to bind p190RhoGAP.
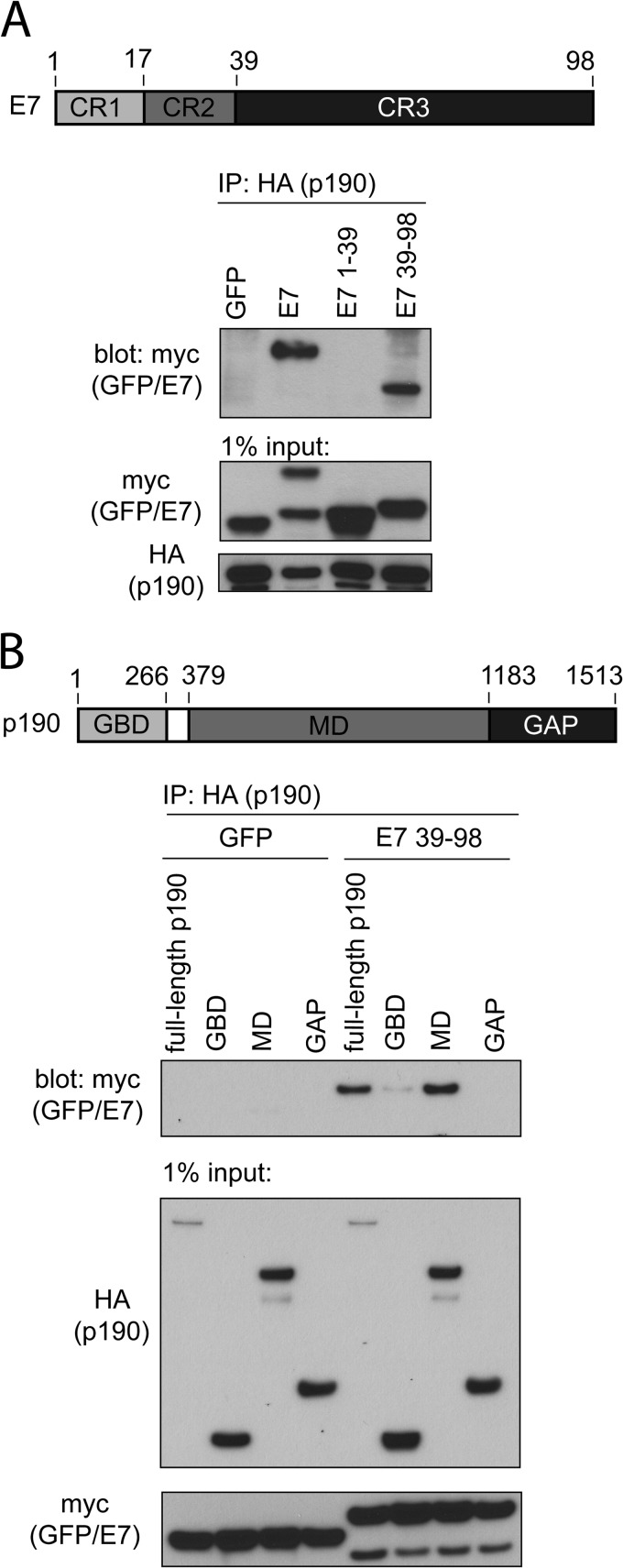
We next identified which regions within E7 and p190 are important for interaction. First, we utilized coimmunoprecipitation experiments and two fragments of HPV16 E7 fused to myc-GFP: residues 1 to 39 span the intrinsically disordered conserved region 1 (CR1) and CR2 of E7, whereas residues 39 to 98 contain CR3 of E7, which is the highly structured zinc-binding domain. We selected to carry out all transfection-based mapping studies in human fibrosarcoma HT1080 cells, since this cell line is transfected exceptionally well using commercially available transfection reagents. We cotransfected HT1080 cells with HA-tagged p190 and the individual myc-GFP-E7 constructs and carried out coimmunoprecipitation experiments using an anti-HA antibody. These experiments revealed that CR3 of E7 is both necessary and sufficient for interaction with p190 (Fig. 2A). In addition, we carried out these experiments in a reciprocal manner, immunoprecipitating for E7 and blotting for p190, with similar results (data not shown). Furthermore, we also mapped the region of p190 that is sufficient to associate with HPV16 E7. We utilized three fragments of p190: the GTP-binding domain (GBD) (residues 1 to 266), the middle domain (MD) (residues 379 to 1183), and the GAP domain (residues 1180 to 1513). Coimmunoprecipitation experiments with E7 residues 39 to 98 revealed that the middle domain, a region of p190 which is known to contain many protein-protein interaction motifs, is sufficient to associate with E7 (Fig. 2B) (37–39).
FIG 2.
Conserved region 3 (CR3) of HPV16 E7 and the middle domain (MD) of p190 are sufficient for interaction. (A) CR3 of HPV16 E7 (residues 39 to 98) is necessary and sufficient to associate with p190. HT1080 cells were transfected with full-length HA-tagged p190 and the indicated GFP or GFP-E7 fusion construct (E7, E7 1–39, and E7 39–98 are all GFP fusions). At 24 h posttransfection, cell lysates were prepared and subjected to coimmunoprecipitation as indicated. (B) The middle domain of p190 is sufficient to associate with E7 (residues 39 to 98). HT1080 cells were transfected with the indicated HA-tagged p190 constructs and GFP or GFP-E7 39–98. Samples were subjected to coimmunoprecipitation as indicated.
HPV16 E7 V55T and R66E mutants do not associate with p190RhoGAP.
We utilized a panel of E7 mutants targeting CR3 residues which are predicted to be solvent exposed (32) to map the surface of E7 required for p190 interaction. Initially, we cotransfected full-length p190 and each of the 21 individual point mutants of E7 in the context of residues 39 to 98, and we then carried out coimmunoprecipitation experiments. This approach revealed that two mutants, the V55T and R66E mutants, completely lost binding to p190 (Fig. 3A). Furthermore, we confirmed these findings using the V55T and R66E mutants in the context of full-length E7 (Fig. 3B). Interestingly, based on our previous structural modeling studies, the V55T and R66E mutants formed a small patch on the surface of E7 CR3 (32, 33). The identification of E7 mutants unable to bind p190 provided us with reagents to study the significance of the E7-p190 interaction, as described below.
FIG 3.
Residues V55 and R66 in HPV16 E7 are required for association with p190. (A) HT1080 cells were transfected with full-length HA-tagged p190 and GFP or GFP fused to residues 39 to 98 of HPV16 E7 containing the indicated mutations. Coimmunoprecipitation experiments were carried out at 24 h posttransfection, as indicated. (B) HT1080 cells were transfected with full-length HA-tagged p190 and full-length wild-type GFP-E7 or the V55T or R66E mutant, and samples were processed for coimmunoprecipitation as indicated for panel A.
E7 proteins from other HPV types interact with p190.
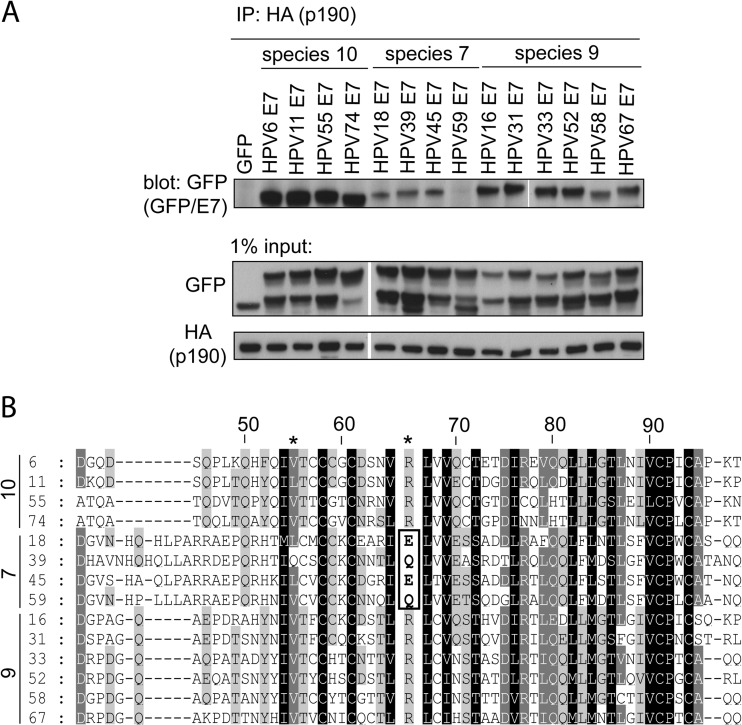
Since this is the first report illustrating that HPV16 E7 associates with p190, we next established whether E7 proteins of other HPVs similarly bind p190. For that purpose, we cloned 13 other E7 oncoproteins from their respective genomes. These included low-risk species 10 viruses HPV6, HPV11, HPV55, and HPV74, high-risk species 7 viruses HPV18, HPV39, HPV45, and HPV59, and high-risk species 9 viruses HPV16, HPV31, HPV33, HPV52, HPV58, and HPV67. We carried out coimmunoprecipitation experiments and found that all tested E7 proteins, except for HPV59, associated with p190, illustrating that this interaction is highly conserved (Fig. 4A). In this assay, low-risk species 10 E7 oncoproteins seemed to recover the most p190. On the other hand, high-risk species 7 E7 proteins, including that of HPV59, invariably recovered the least p190. Interestingly, this low binding is illustrated further by the observation that in reverse immunoprecipitation experiments, we could not detect the interaction between species 7 E7 proteins and p190 (data not shown). What is also intriguing is the observation that species 7 E7 oncoproteins contain glutamine or glutamate (Q or E) at the position corresponding to residue 66 in HPV16 (Fig. 4B). An arginine (R) at this position is perfectly conserved across all other tested E7 proteins and was also one of the two residues identified as important for p190 binding in HPV16 (Fig. 3A and B). Overall, these experiments illustrate that p190 is an evolutionarily conserved target of E7 proteins of multiple HPV species and likely represents an important target for an efficient HPV life cycle.
FIG 4.
The E7 proteins of multiple HPV species interact with p190. (A) E7 proteins from other HPV species and types associate with p190. HT1080 cells were transfected with HA-tagged p190 and GFP or GFP-E7 fusions, as indicated. At 24 h posttransfection, cell lysates were prepared and subjected to coimmunoprecipitation using anti-HA antibody. The samples were immunoblotted as indicated. (B) Multiple-sequence alignment of HPV E7 CR3 sequences. The residue numbering indicated on the top line corresponds to HPV16 E7 numbering. Indicated on the left of the alignment are the species and HPV type. Darker shading corresponds to more highly conserved residues. The positions of residues V55 and R66 are indicated at the top of the alignment (*). The E and Q residues of species 7 types at the position of R66 (HPV16) are highlighted with a box.
Expression of HPV16 E7 reduces stress fiber formation.
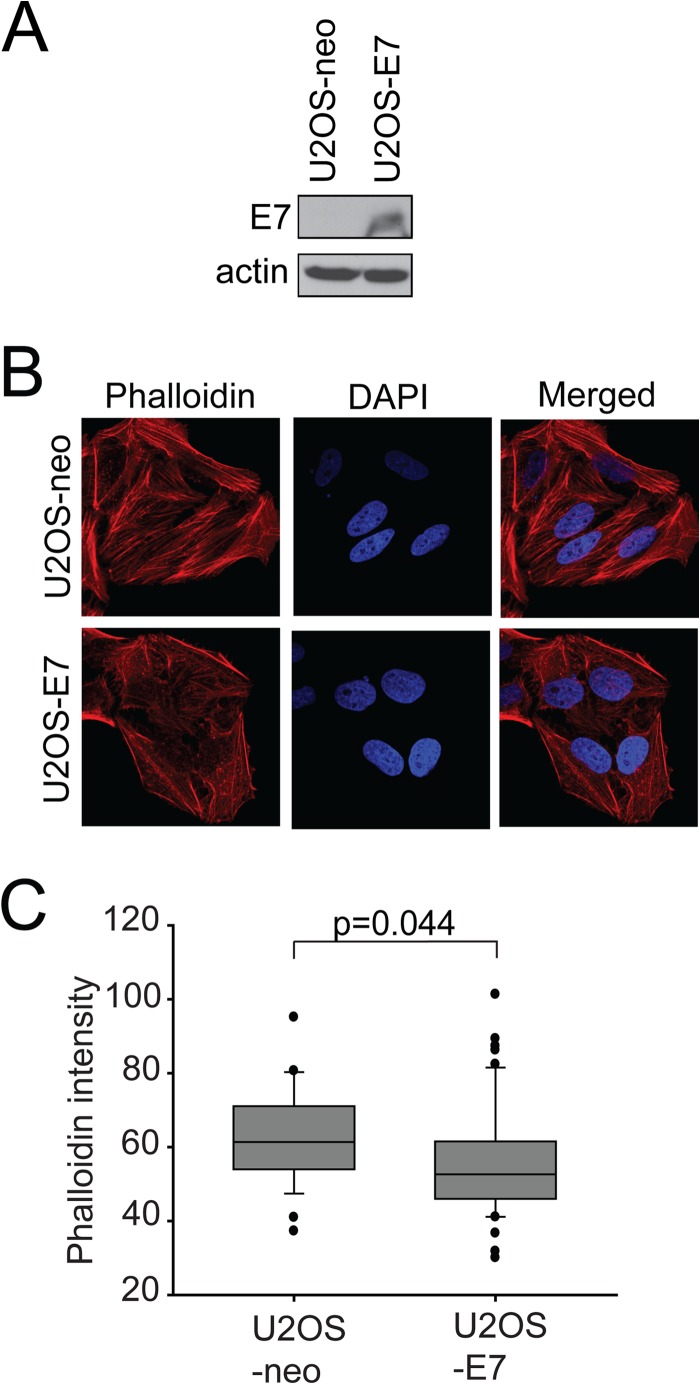
It has been well established that organization of actin filaments is controlled by the Rho family members (Rho, Rac, and Cdc42) (40, 41). In particular, RhoA regulates the assembly of actin stress fibers and focal contacts through activation of the downstream effectors mDia and ROCK kinase, while Cdc42 and Rac1 stimulate the formation of filopodia and lamellipodia, respectively, in migrating cells (42–45). It was previously shown in two independent publications that HPV16 E7- and HPV38 E7-expressing keratinocytes both contain reduced RhoA-GTP levels. The HPV16 E7 reduction in RhoA-GTP was shown to be due in part to E7 modulation of the cytoplasmic localization of p27, which can in turn bind and dysregulate RhoA (46). On the other hand, HPV38 E7 uses two distinct mechanisms to downregulate Rho activity: (i) mediating the activation of the CK2–MEK–extracellular signal-regulated kinase (ERK) pathway and (ii) directly binding to eukaryotic elongation factor 1A (eEF1A) and abolishing its effects on actin fiber formation (47). Due in part to these published observations and considering that p190 is a direct regulator of the Rho pathway, we wanted to assess the biological consequences of E7-p190 interaction. Attenuation of the RhoA pathway leads to a decrease in both actin stress fibers and focal adhesion formation (25). Therefore, we compared the status of stress fibers in cells expressing HPV16 E7 to those of control, GFP-expressing cells and cell expressing dominant negative p190 (p190RA). For the purposes of these functional assays, we utilized human osteosarcoma U2OS cells, since they were previously utilized for similar assays and were shown to behave similarly to human keratinocytes in these experiments (47). In addition, we also confirmed the results of these assays in human keratinocyte HaCat cells. U2OS or HaCat cells transfected with GFP or the indicated E7 construct were stained with Alexa 568-conjugated phalloidin to visualize F-actin. This staining revealed that control cells expressing GFP contained more actin stress fibers than did wild-type E7-expressing cells (Fig. 5C). In U2OS cells, the actin stress fibers were more frequently present throughout the cytoplasm of GFP cells, while such an organization was more frequently missing or altered in wild-type E7 cells. In those cells, F-actin was localized mainly at the periphery of the cells, in a cortical pattern (Fig. 5C). The phenotype observed in E7-expressing cells was similarly observed in cells expressing dominant negative p190 (Fig. 5A). Quantification of the actin stress fiber loss is presented in Fig. 5B and D, with statistically significant reductions observed in E7- and p190RA-expressing cells. In addition, a U2OS cell line stably expressing E7 (35) also displayed a significant disruption of actin stress fibers compared to the matched U2OS-neo control cell line (Fig. 6).
FIG 5.
Interaction with p190 is required for HPV16 E7 to disrupt actin stress fiber formation. (A) Dominant negative p190 disrupts actin stress fibers. U2OS cells were transfected with GFP or GFP-p190RA (dominant negative p190) and then stained with phalloidin-Alexa 568 to visualize F-actin. The nuclei were stained with DAPI, and images were acquired on a confocal microscope. (B) Quantification of phalloidin intensities in GFP- and p190RA-expressing U2OS or HaCat cells. Data are representative results from three independent experiments. (C) HPV16 E7 mutants unable to bind p190 do not efficiently disrupt actin stress fibers. U2OS cells were transfected with the indicated plasmids and stained for F-actin by use of phalloidin-Alexa 568. Nuclei were stained with DAPI. (D) Quantification of phalloidin staining in transfected U2OS or HaCat cells. Phalloidin intensities were quantified from images captured with a fluorescence microscope as described in Materials and Methods. Data are representative of three independent experiments.
FIG 6.
E7 affects F-actin levels in stable cell lines. (A) Expression of E7 in U2OS-neo and U2OS-E7 cells. Equal amounts of cell extracts from U2OS-neo and U2OS-E7 cells were analyzed by immunoblotting as indicated. (B) U2OS-neo and U2OS-E7 cells were stained with phalloidin-Alexa 568 to visualize F-actin. The nuclei were stained with DAPI, and images were acquired on a confocal microscope. (C) Quantification of phalloidin intensities in U2OS-neo and U2OS-E7 cells. Data are representative results from three independent experiments.
To assess the contribution of the E7-p190 interaction to the observed F-actin phenotype, we transfected U2OS cells with GFP-E7 V55T or R66E, the two mutants of E7 which did not bind p190, and assessed the levels of F-actin by staining the cells with phalloidin. We found that both V55T and R66E mutant-expressing cells had more stress fibers than wild-type E7 cells, with quantified phalloidin levels resembling those in the cells expressing GFP only (Fig. 5C and D). Overall, these data strongly suggest that expressed E7 phenotypically acts like dominant negative p190 and that association of E7 with p190 dysregulates this GAP and contributes to reduced F-actin levels.
The association of E7 with p190 influences cell spreading.
p190 was previously shown to play a crucial role in early events during cell spreading on fibronectin (28). Precise temporal regulation of RhoA by p190 upon cell adhesion to the extracellular matrix protein fibronectin is required for efficient cell spreading (28). As an additional confirmation of the biological significance of E7-p190 interaction, we assessed whether expression of E7 has any effects on cell spreading. We transfected U2OS or HaCat cells with GFP, wild-type E7 or the indicated E7 mutant, and dominant negative p190 and determined the areas of cell spreading on fibronectin-coated coverslips. Spreading was assessed 20 min after plating, as this was the time point at which control cells were well spread and at which p190 was previously shown to be essential in efficient cell spreading (28). We found that cells expressing wild-type E7, similarly to cells expressing the dominant negative p190 mutant, had a significantly decreased ability to spread. However, the two mutants of E7 which could not bind p190 (V55T and R66E) did not exhibit this effect (Fig. 7A and B). Interestingly, reduced cell spreading upon expression of dominant negative p190 was also observed in the HPV16-positive CaSki cells (Fig. 7B).
FIG 7.
E7 affects cell spreading. (A) U2OS cells were transiently transfected with GFP or GFP fusions of wild-type, V55T, or R66E E7 or dominant negative p190 (p190RA), plated on fibronectin for 20 min, and then stained for F-actin by use of phalloidin. Images were captured with a fluorescence microscope using a ×40 objective; representative images are shown. (B) The relative areas of individual U2OS, HaCat, and CaSki cells (transfected as indicated and processed as described for panel A) in digital images were measured with ImageJ. Data are representative of at least 75 cells and three independent experiments.
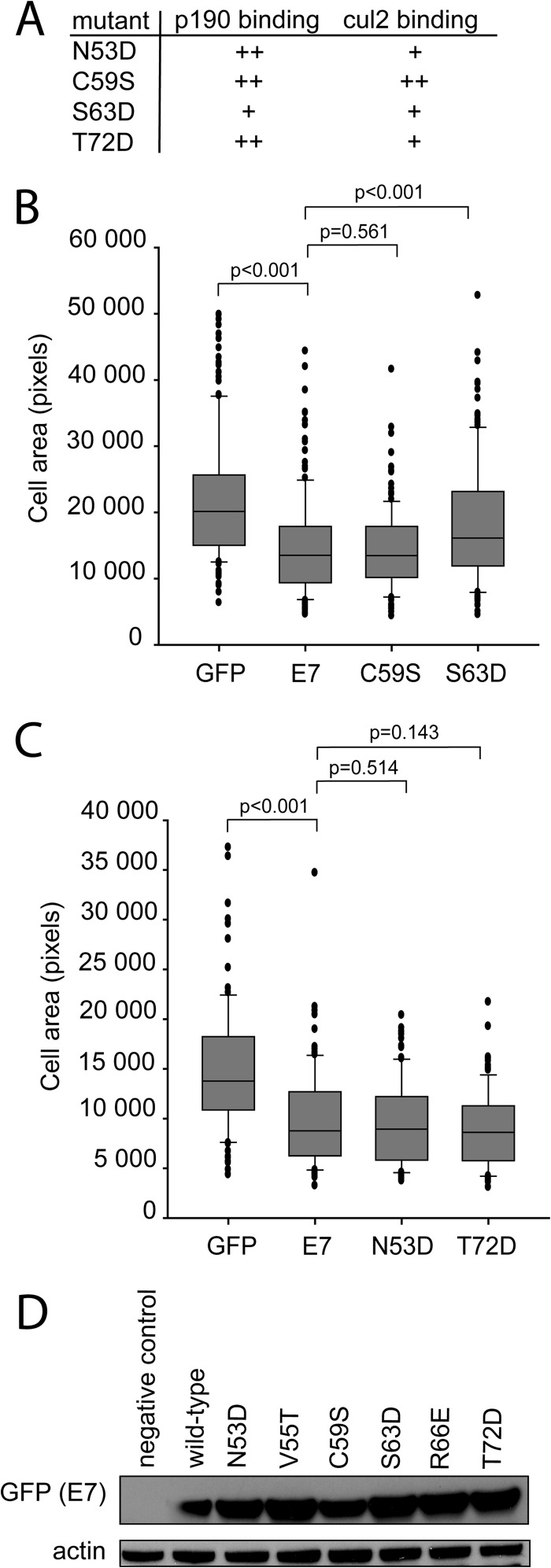
We also illustrated that the effects on cell spreading appear to be specific to the E7-p190 interaction by assessing the ability of additional mutants of E7 to spread. The C59S mutant is a mutant of E7 which binds p190 comparably to wild-type E7; when we tested it for effects on spreading, we found that it behaved like wild-type E7 (Fig. 8A and B). Since V55T and R66E mutants were previously shown to also have defects in the ability to bind the cullin 2 E3 ubiquitin ligase, we wanted to ascertain that the effects on cell spreading were due only to defects in p190 binding. In addition to the V55T and R66E mutants, N53D and T72D mutants were also previously shown to have reduced cullin 2 binding (33); however, the N53D and T72D mutants maintain wild-type p190 binding. When they were assessed in the cell spreading assay, the N53D and T72D mutants behaved like wild-type E7, demonstrating that the effects of E7 on cell spreading did not require interaction with cullin 2 and that only mutants with defects in p190 binding were impaired (Fig. 8A and C). These effects were not due to differences in expression levels, as all mutants of interest were expressed as well as wild-type E7 (Fig. 8D).
FIG 8.
The ability of E7 to associate with p190, but not cullin 2, appears to be necessary to reduce cell spreading. (A) p190 and cullin 2 (cul2) binding phenotypes of the mutants used in this study. The N53D, C59S, and T72D mutants bound p190 as well as wild-type E7 (++); the S63D mutant had reduced p190 binding (+). The C59S mutant bound cul2 comparably to the wild type (++), whereas the N53D, S63D, and T72D mutants all had reduced cul2 binding (+). (B) U2OS cells were transiently transfected with GFP or GFP fusions of wild-type, C59S, or S63D E7, plated on fibronectin for 20 min, and then stained for F-actin by use of phalloidin. Images were captured with a fluorescence microscope using a ×40 objective. The relative areas of individual cells in digital images were measured with ImageJ. Data are representative of at least 75 cells and two independent experiments. (C) U2OS cells were transiently transfected as indicated and processed as described for panel B. (D) U2OS cells were transfected with wild-type E7 or the indicated E7 mutant. At 24 h posttransfection, cell lysates were examined for E7 expression levels via Western blotting.
Expression of E7 or dominant negative p190 has the same effect on RhoA-GTP.
U2OS cells were transfected with GFP or wild-type E7, and cell lysates were analyzed for RhoA-GTP levels, similarly to previously described studies (28). Transfections with dominant negative p190 or wild-type p190 were used as controls. As previously reported (28), expression of wild-type p190 led to significantly decreased RhoA-GTP levels, whereas expression of dominant negative p190 had no effect on RhoA-GTP (Fig. 9A). Interestingly, in this assay the effects of wild-type E7 on RhoA-GTP were once again similar to those of dominant negative p190 (Fig. 9A). In addition, we also found that expression of E7 did not lead to changes in tyrosine phosphorylation of p190 (Fig. 9B). Together, these data further suggest that E7 has effects on p190 similar to those observed upon expression of dominant negative p190 and that E7 likely dysregulates p190.
FIG 9.
Effects on RhoA-GTP levels and p190 tyrosine phosphorylation. (A) Expression of wild-type p190, but not wild-type E7 or dominant negative p190, affects RhoA-GTP levels. U2OS cells were transfected with GFP, wild-type E7, p190, or dominant negative p190. At 24 h posttransfection, the cells were trypsinized, kept in suspension for 1 h, and then plated on fibronectin-coated tissue culture dishes for 20 min. The cells were collected and analyzed for RhoA-GTP levels as described in Materials and Methods. There was no statistical significance between wild-type E7 or p190RA and GFP. (B) E7 does not affect p190 tyrosine phosphorylation. U2OS cells were transfected with GFP or E7, and p190 tyrosine phosphorylation was assessed as described in Materials and Methods.
DISCUSSION
Many pathogens, including viruses, have evolved gene products to engage and subvert the actin cytoskeleton and, in particular, the Rho family GTPase signaling system (48–50). The actin cytoskeleton is highly dynamic and is manipulated primarily by members of the Rho family GTPases that control signal transduction pathways linking membrane receptors to the cytoskeleton. Rho family GTPases regulate numerous cellular processes, including F-actin polymerization, cell polarity, migration, spreading, and cell growth. Among the known Rho family GTPases, RhoA is responsible for the regulation of stress fiber formation. Rous sarcoma virus, simian virus 40 (SV40), polyomaviruses, adenoviruses, and HPVs have all been shown to encode viral gene products which manipulate the Rho family GTPase signaling pathway (46, 51–54). Of particular relevance for this report is the observation that the cutaneous betaherpesvirus HPV38 E7 protein affects actin stress fiber formation (47). Disruption of actin stress fibers was shown to be dependent on inhibition of RhoA activity (47). In addition, E7 of mucosal high-risk HPV16 has been implicated in dysregulating both RhoA and Rac1 GTPases (46, 54). The possible contribution and importance of the RhoA pathway in high-risk HPV-induced transformation are illustrated by recent findings that primary human keratinocytes can be immortalized effectively by treatment with a Rho kinase inhibitor (Y27632) and that this can substitute for E7 (55, 56). Interestingly, recent studies have implicated the cytoskeleton as being critical not only for processes such as migration and spreading but also for cell growth. Many cytoskeletal components and the elements of the Rho signaling pathway were discovered to play a central but largely complex role in control of cell proliferation (57).
In this report, we show that HPV16 E7 interacts with a potent regulator of RhoA, the p190RhoGAP protein. Interestingly, a recent study using mass spectrometry identified 1,079 cellular proteins targeted by various tumor virus oncoproteins; in that study, p190 was also identified as a potential target of HPV16 E7 (58). Interactions between another Rho GAP (p50RhoGAP) and SV40 small T antigen or Epstein-Barr virus (EBV) BFLF2 and BGLF3 were also identified in that study, suggesting that multiple tumor viruses target Rho during infection. We show that E7 binds p190 under endogenous conditions in HPV16-positive cervical cancer cells. We carried out extensive mapping studies, and we show that E7 proteins of diverse species and types, including high- and low-risk types, all bind p190, suggesting that p190 is an important target of E7. In another recent study which also utilized mass spectrometry to identify novel and unique interacting partners of a large number of HPV E7 proteins from different HPV types and which also included representatives of species 7, 9, and 10, only a few cellular proteins were found to be highly conserved targets of E7 (59). These included the pocket proteins pRb and p130, which are essential for control of the cell cycle and are also dysregulated by E7, as well as a 600-kDa E3 ubiquitin ligase, p600, whose role in E7 transformation and the HPV life cycle remains largely unknown (59). Our finding that E7-p190 interaction is highly conserved suggests that p190 is a crucial target of E7 in the HPV life cycle and perhaps could contribute to E7-induced transformation.
In addition, our mapping experiments highlight the importance of residue R66 (HPV16) for p190 interaction; we identified R66 as a necessary residue for interaction of p190 and HPV16 E7. When we examined the multiple-sequence alignment of the different HPV E7 proteins used in this report, we noticed that R66 is highly conserved (Fig. 4B), with the exception of the weakly interacting species 7 members. Members of species 7 do not contain a basic residue at the position corresponding to R66. A lack of arginine at this position could account for their reduced interaction with p190. This observation may serve as a potential tool to discriminate between strong and weak E7-p190 interactions in future studies. In addition to R66, residue V55 in HPV16 E7 was also necessary for p190 interaction. Our previous work has shown that R66E and V55T mutants both have a significant increase in the ability to transform primary baby rat kidney (BRK) cells (32). How the interaction with p190 influences BRK cell transformation remains an interesting topic to examine in future studies.
Coimmunoprecipitation experiments revealed that E7 CR3 is necessary and sufficient to interact with p190. Additionally, we showed that the middle domain of p190 is sufficient to associate with E7 residues 39 to 98. Interestingly, this region of p190 contains many protein-protein interaction motifs and contains the major site of tyrosine phosphorylation (Tyr 1105) (37, 60–62). Among the interactions made by this portion of p190, phosphotyrosine 1105 serves as the major binding site for p120RasGAP (60). Phosphorylation of Tyr 1105 in p190 by Src family kinases and other tyrosine kinases results in activation of p190's RhoGAP activity, with concomitant inhibition of p120RasGAP activity of bound p120. This phosphorylation event therefore coregulates signaling through the Ras and Rho pathways. Targeting of the middle domain by E7 could potentially have an impact on the ability of p190 to associate with binding partners, including tyrosine kinases and p120RasGAP. This observation serves as an intriguing possibility for future study, particularly for elucidating the mechanism by which E7 dysregulates p190.
As described in previous sections, a recent report has shown that human keratinocytes expressing HPV16 E7 have reduced RhoA-GTP levels (46). Additionally, a report examining cutaneous HPV38 E7 has clearly established that E7 reduces RhoA-GTP levels and downregulates levels of F-actin. Similarly to HPV38 E7 cells, we determined that HPV16 E7-expressing cells also showed fewer and more disorganized actin stress fibers and had reduced F-actin levels. However, E7 did not reduce RhoA-GTP levels under the conditions of our experiments (Fig. 9A). In addition, our results indicate that the E7-p190 interaction may be important for regulation of F-actin. The significance of E7-p190 interaction first became apparent when we observed that cells which expressed the two mutants of E7 which do not bind p190 also did not have defects in F-actin levels. Aside from possible F-actin regulation by the E7-p190 interaction, HPV16 E7 was also shown to directly associate with actin (63), suggesting that there may be multiple mechanisms for F-actin regulation in E7-expressing cells. Interestingly, as a control for our experiments, we also examined F-actin levels in cells expressing dominant negative p190 (Fig. 5A and B). We found that these cells also had decreased F-actin levels and appeared to be similar to E7-expressing cells, leading us to suggest that the interaction with E7 may inhibit p190's function.
To further understand the significance of E7-p190 interaction and to determine whether E7 negatively regulates p190's function, we assessed the ability of E7-expressing cells to spread on fibronectin-coated coverslips (Fig. 7). Cell spreading was previously shown to depend on functional p190, specifically during early events upon attachment to a substrate (28). Expression of dominant negative p190 leads to cells which are unable to spread efficiently (28). We utilized dominant negative p190 as a control in these experiments and assessed the ability of U2OS and HaCat cells transfected with GFP, p190RA, wild-type E7, or various E7 mutants to spread (Fig. 7 and 8). We determined that cells expressing wild-type E7 behaved similarly to cells expressing dominant negative p190, whereas the V55T and R66E mutants behaved similarly to GFP-expressing cells. In addition, mutants of E7 (C59S, N53D, and T72D) which maintained wild-type p190 binding ability behaved like wild-type E7 in this assay (Fig. 8). However, any reduction in p190 binding, as illustrated by the E7 S63D mutant (Fig. 3), could lead to changes in the ability of cells to spread (Fig. 8). In contrast, the ability of E7 to interact with cullin 2 was not required to affect cell spreading.
We also assessed the effects of E7 on RhoA-GTP levels in an assay in which the cells were kept in suspension and then plated on fibronectin-treated plates. Under such conditions, functional p190 is necessary to effectively decrease RhoA-GTP levels, preventing premature formation of actin stress fibers and allowing the cells to spread. In this assay, we found that expression of wild-type E7 and dominant negative p190 did not lead to a decrease in RhoA-GTP levels (Fig. 9A). Together, these experiments further suggest that E7 binding of p190 leads to dysregulation of this GAP and also highlight that E7 targeting of p190 has important biological consequences.
Our data strongly suggest that E7 negatively regulates the RhoGAP activity of p190; however, the mechanism by which this occurs remains unknown. Many reports in the past have focused on positive regulation of p190RhoGAP activity by phosphorylation of tyrosine 1105. However, we did not see any effects of E7 on this modification (Fig. 9B). In contrast to tyrosine phosphorylation, serine/threonine phosphorylation decreases p190 activity (64, 65). In certain circumstances, p190 activity can also be regulated such that there is an inhibition of RhoGAP and an increase in RacGAP activity (66, 67). In addition, negative regulation of p190 activity is also achieved by several different phosphatases which dephosphorylate p190, including protein tyrosine phosphatase 20 (PTP20) and protein tyrosine phosphatase receptor type z (Ptprz) (68, 69). Future work will focus on determining if any of the above-mentioned mechanisms are used by E7 to dysregulate p190 and what other effects this interaction may have on the Rho signaling pathways.
In conclusion, we have identified that p190, an upstream negative regulator of RhoA, is an interacting partner of the E7 proteins of multiple HPV types. Our data strongly suggest that p190 is dysregulated by E7, which leads to changes in the actin cytoskeleton. Our findings also suggest that E7's dysregulation of p190 influences cell properties which may be crucial for the viral life cycle, or perhaps for HPV-induced transformation. Most importantly, our work emphasizes the need for further studies in an area that has been largely unexplored in the HPV field.
ACKNOWLEDGMENTS
This work was supported by a grant from the Canadian Institute of Health Research to J.S.M. B.T. was supported by a Canadian Institute of Health Research Michael Smith Foreign Study Supplement and an Ontario Graduate Scholarship.
Footnotes
Published ahead of print 8 January 2014
REFERENCES
- 1.zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342–350. 10.1038/nrc798 [DOI] [PubMed] [Google Scholar]
- 2.Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ. 2006. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J. Pathol. 208:152–164. 10.1002/path.1866 [DOI] [PubMed] [Google Scholar]
- 3.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, Shah KV, Sidransky D. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92:709–720. 10.1093/jnci/92.9.709 [DOI] [PubMed] [Google Scholar]
- 4.Grussendorf-Conen EI. 1997. Anogenital premalignant and malignant tumors (including Buschke-Lowenstein tumors). Clin. Dermatol. 15:377–388. 10.1016/S0738-081X(96)00159-9 [DOI] [PubMed] [Google Scholar]
- 5.Masih AS, Stoler MH, Farrow GM, Wooldridge TN, Johansson SL. 1992. Penile verrucous carcinoma: a clinicopathologic, human papillomavirus typing and flow cytometric analysis. Mod. Pathol. 5:48–55 [PubMed] [Google Scholar]
- 6.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. 2004. Classification of papillomaviruses. Virology 324:17–27. 10.1016/j.virol.2004.03.033 [DOI] [PubMed] [Google Scholar]
- 7.zur Hausen H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55–F78 [DOI] [PubMed] [Google Scholar]
- 8.Halbert CL, Demers GW, Galloway DA. 1991. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65:473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedell MA, Jones KH, Grossman SR, Laimins LA. 1989. Identification of human papillomavirus type 18 transforming genes in immortalized and primary cells. J. Virol. 63:1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.zur Hausen H. 1999. Immortalization of human cells and their malignant conversion by high risk human papillomavirus genotypes. Semin. Cancer Biol. 9:405–411. 10.1006/scbi.1999.0144 [DOI] [PubMed] [Google Scholar]
- 11.Munger K, Phelps WC, Bubb V, Howley PM, Schlegel R. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63:4417–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werness BA, Levine AJ, Howley PM. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76–79. 10.1126/science.2157286 [DOI] [PubMed] [Google Scholar]
- 14.Huibregtse JM, Scheffner M, Howley PM. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gewin L, Myers H, Kiyono T, Galloway DA. 2004. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 18:2269–2282. 10.1101/gad.1214704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brehm A, Nielsen SJ, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. 1999. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 18:2449–2458. 10.1093/emboj/18.9.2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funk JO, Waga S, Harry JB, Espling E, Stillman B, Galloway DA. 1997. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 11:2090–2100. 10.1101/gad.11.16.2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen CL, Munger K. 2008. Direct association of the HPV16 E7 oncoprotein with cyclin A/CDK2 and cyclin E/CDK2 complexes. Virology 380:21–25. 10.1016/j.virol.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zerfass-Thome K, Zwerschke W, Mannhardt B, Tindle R, Botz JW, Jansen-Durr P. 1996. Inactivation of the cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein. Oncogene 13:2323–2330 [PubMed] [Google Scholar]
- 20.Chemes LB, Glavina J, Alonso LG, Marino-Buslje C, de Prat-Gay G, Sanchez IE. 2012. Sequence evolution of the intrinsically disordered and globular domains of a model viral oncoprotein. PLoS One 7:e47661. 10.1371/journal.pone.0047661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509–514. 10.1126/science.279.5350.509 [DOI] [PubMed] [Google Scholar]
- 22.Sastry SK, Burridge K. 2000. Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Exp. Cell Res. 261:25–36. 10.1006/excr.2000.5043 [DOI] [PubMed] [Google Scholar]
- 23.Takai Y, Sasaki T, Matozaki T. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153–208 [DOI] [PubMed] [Google Scholar]
- 24.Raftopoulou M, Hall A. 2004. Cell migration: Rho GTPases lead the way. Dev. Biol. 265:23–32. 10.1016/j.ydbio.2003.06.003 [DOI] [PubMed] [Google Scholar]
- 25.Nobes CD, Hall A. 1999. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144:1235–1244. 10.1083/jcb.144.6.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ridley AJ, Self AJ, Kasmi F, Paterson HF, Hall A, Marshall CJ, Ellis C. 1993. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J. 12:5151–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang JH, Gill S, Settleman J, Parsons SJ. 1995. c-Src regulates the simultaneous rearrangement of actin cytoskeleton, p190RhoGAP, and p120RasGAP following epidermal growth factor stimulation. J. Cell Biol. 130:355–368. 10.1083/jcb.130.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arthur WT, Burridge K. 2001. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol. Biol. Cell 12:2711–2720. 10.1091/mbc.12.9.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fincham VJ, Chudleigh A, Frame MC. 1999. Regulation of p190 Rho-GAP by v-Src is linked to cytoskeletal disruption during transformation. J. Cell Sci. 112:947–956 [DOI] [PubMed] [Google Scholar]
- 30.Wolf RM, Draghi N, Liang X, Dai C, Uhrbom L, Eklof C, Westermark B, Holland EC, Resh MD. 2003. p190RhoGAP can act to inhibit PDGF-induced gliomas in mice: a putative tumor suppressor encoded on human chromosome 19q13.3. Genes Dev. 17:476–487. 10.1101/gad.1040003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang DZ, Nur EKMS, Tikoo A, Montague W, Maruta H. 1997. The GTPase and Rho GAP domains of p190, a tumor suppressor protein that binds the M (r) 120,000 Ras GAP, independently function as anti-Ras tumor suppressors. Cancer Res. 57:2478–2484 [PubMed] [Google Scholar]
- 32.Todorovic B, Massimi P, Hung K, Shaw GS, Banks L, Mymryk JS. 2011. Systematic analysis of the amino acid residues of human papillomavirus type 16 E7 conserved region 3 involved in dimerization and transformation. J. Virol. 85:10048–10057. 10.1128/JVI.00643-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Todorovic B, Hung K, Massimi P, Avvakumov N, Dick FA, Shaw GS, Banks L, Mymryk JS. 2012. Conserved region 3 of human papillomavirus 16 E7 contributes to deregulation of the retinoblastoma tumor suppressor. J. Virol. 86:13313–13323. 10.1128/JVI.01637-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su L, Agati JM, Parsons SJ. 2003. p190RhoGAP is cell cycle regulated and affects cytokinesis. J. Cell Biol. 163:571–582. 10.1083/jcb.200308007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, Crum CP, Munger K. 2000. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. U. S. A. 97:10002–10007. 10.1073/pnas.170093297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallenius T, Vaahtomeri K, Kovac B, Osiceanu AM, Viljanen M, Makela TP. 2011. An association between NUAK2 and MRIP reveals a novel mechanism for regulation of actin stress fibers. J. Cell Sci. 124:384–393. 10.1242/jcs.072660 [DOI] [PubMed] [Google Scholar]
- 37.Hu KQ, Settleman J. 1997. Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J. 16:473–483. 10.1093/emboj/16.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang W, Sordella R, Chen GC, Hakre S, Roy AL, Settleman J. 2005. An FF domain-dependent protein interaction mediates a signaling pathway for growth factor-induced gene expression. Mol. Cell 17:23–35. 10.1016/j.molcel.2004.11.024 [DOI] [PubMed] [Google Scholar]
- 39.Wennerberg K, Forget MA, Ellerbroek SM, Arthur WT, Burridge K, Settleman J, Der CJ, Hansen SH. 2003. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr. Biol. 13:1106–1115. 10.1016/S0960-9822(03)00418-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bishop AL, Hall A. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241–255. 10.1042/0264-6021:3480241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall A. 2005. Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33:891–895. 10.1042/BST20050891 [DOI] [PubMed] [Google Scholar]
- 42.Amano M, Chihara K, Kimura K, Fukata Y, Nakamura N, Matsuura Y, Kaibuchi K. 1997. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science 275:1308–1311. 10.1126/science.275.5304.1308 [DOI] [PubMed] [Google Scholar]
- 43.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1:136–143. 10.1038/11056 [DOI] [PubMed] [Google Scholar]
- 44.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. 1992. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70:401–410. 10.1016/0092-8674(92)90164-8 [DOI] [PubMed] [Google Scholar]
- 45.Nobes CD, Hall A. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53–62. 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- 46.Charette ST, McCance DJ. 2007. The E7 protein from human papillomavirus type 16 enhances keratinocyte migration in an Akt-dependent manner. Oncogene 26:7386–7390. 10.1038/sj.onc.1210541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue J, Shukla R, Accardi R, Zanella-Cleon I, Siouda M, Cros MP, Krutovskikh V, Hussain I, Niu Y, Hu S, Becchi M, Jurdic P, Tommasino M, Sylla BS. 2011. Cutaneous human papillomavirus type 38 E7 regulates actin cytoskeleton structure for increasing cell proliferation through CK2 and the eukaryotic elongation factor 1A. J. Virol. 85:8477–8494. 10.1128/JVI.02561-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gouin E, Welch MD, Cossart P. 2005. Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 8:35–45. 10.1016/j.mib.2004.12.013 [DOI] [PubMed] [Google Scholar]
- 49.Munter S, Way M, Frischknecht F. 2006. Signaling during pathogen infection. Sci. STKE 2006:re5. 10.1126/stke.3352006re5 [DOI] [PubMed] [Google Scholar]
- 50.Favoreel HW, Enquist LW, Feierbach B. 2007. Actin and Rho GTPases in herpesvirus biology. Trends Microbiol. 15:426–433. 10.1016/j.tim.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 51.Fleissner E, Tress E. 1973. Chromatographic and electrophoretic analysis of viral proteins from hamster and chicken cells transformed by Rous sarcoma virus. J. Virol. 11:250–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McNutt NS, Culp LA, Black PH. 1973. Contact-inhibited revertant cell lines isolated from SV40-transformed cells. IV. Microfilament distribution and cell shape in untransformed, transformed, and revertant Balb-c 3T3 cells. J. Cell Biol. 56:412–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldman RD, Chang C, Williams JF. 1975. Properties and behavior of hamster embryo cells transformed by human adenovirus type 5. Cold Spring Harbor Symp. Quant. Biol. 39:601–614 [DOI] [PubMed] [Google Scholar]
- 54.Wu R, Coniglio SJ, Chan A, Symons MH, Steinberg BM. 2007. Up-regulation of Rac1 by epidermal growth factor mediates COX-2 expression in recurrent respiratory papillomas. Mol. Med. 13:143–150. 10.2119/2007-00005.Wu [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chapman S, Liu X, Meyers C, Schlegel R, McBride AA. 2010. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Invest. 120:2619–2626. 10.1172/JCI42297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, Haddad BR, Rhim JS, Dritschilo A, Riegel A, McBride A, Schlegel R. 2012. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 180:599–607. 10.1016/j.ajpath.2011.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mammoto A, Ingber DE. 2009. Cytoskeletal control of growth and cell fate switching. Curr. Opin. Cell Biol. 21:864–870. 10.1016/j.ceb.2009.08.001 [DOI] [PubMed] [Google Scholar]
- 58.Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M. 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495. 10.1038/nature11288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White EA, Sowa ME, Tan MJ, Jeudy S, Hayes SD, Santha S, Munger K, Harper JW, Howley PM. 2012. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc. Natl. Acad. Sci. U. S. A. 109:E260–E267. 10.1073/pnas.1116776109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roof RW, Haskell MD, Dukes BD, Sherman N, Kinter M, Parsons SJ. 1998. Phosphotyrosine (p-Tyr)-dependent and -independent mechanisms of p190 RhoGAP-p120 RasGAP interaction: Tyr 1105 of p190, a substrate for c-Src, is the sole p-Tyr mediator of complex formation. Mol. Cell. Biol. 18:7052–7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haskell MD, Nickles AL, Agati JM, Su L, Dukes BD, Parsons SJ. 2001. Phosphorylation of p190 on Tyr1105 by c-Src is necessary but not sufficient for EGF-induced actin disassembly in C3H10T1/2 fibroblasts. J. Cell Sci. 114:1699–1708 [DOI] [PubMed] [Google Scholar]
- 62.Koehler JA, Moran MF. 2001. Regulation of extracellular signal-regulated kinase activity by p120 RasGAP does not involve its pleckstrin homology or calcium-dependent lipid binding domains but does require these domains to regulate cell proliferation. Cell Growth Differ. 12:551–561 [PubMed] [Google Scholar]
- 63.Rey O, Lee S, Baluda MA, Swee J, Ackerson B, Chiu R, Park NH. 2000. The E7 oncoprotein of human papillomavirus type 16 interacts with F-actin in vitro and in vivo. Virology 268:372–381. 10.1006/viro.1999.0175 [DOI] [PubMed] [Google Scholar]
- 64.Maddox AS, Burridge K. 2003. RhoA is required for cortical retraction and rigidity during mitotic cell rounding. J. Cell Biol. 160:255–265. 10.1083/jcb.200207130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang W, Betson M, Mulloy R, Foster R, Levay M, Ligeti E, Settleman J. 2008. p190A RhoGAP is a glycogen synthase kinase-3-beta substrate required for polarized cell migration. J. Biol. Chem. 283:20978–20988. 10.1074/jbc.M802588200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ligeti E, Dagher MC, Hernandez SE, Koleske AJ, Settleman J. 2004. Phospholipids can switch the GTPase substrate preference of a GTPase-activating protein. J. Biol. Chem. 279:5055–5058. 10.1074/jbc.C300547200 [DOI] [PubMed] [Google Scholar]
- 67.Ligeti E, Settleman J. 2006. Regulation of RhoGAP specificity by phospholipids and prenylation. Methods Enzymol. 406:104–117. 10.1016/S0076-6879(06)06009-5 [DOI] [PubMed] [Google Scholar]
- 68.Shiota M, Tanihiro T, Nakagawa Y, Aoki N, Ishida N, Miyazaki K, Ullrich A, Miyazaki H. 2003. Protein tyrosine phosphatase PTP20 induces actin cytoskeleton reorganization by dephosphorylating p190 RhoGAP in rat ovarian granulosa cells stimulated with follicle-stimulating hormone. Mol. Endocrinol. 17:534–549. 10.1210/me.2002-0187 [DOI] [PubMed] [Google Scholar]
- 69.Tamura H, Fukada M, Fujikawa A, Noda M. 2006. Protein tyrosine phosphatase receptor type Z is involved in hippocampus-dependent memory formation through dephosphorylation at Y1105 on p190 RhoGAP. Neurosci. Lett. 399:33–38. 10.1016/j.neulet.2006.01.045 [DOI] [PubMed] [Google Scholar]