Abstract
When DNA double-strand breaks occur, the cell cycle stage has a major influence on the choice of the repair pathway employed. Specifically, nonhomologous end joining is the predominant mechanism used in the G1 phase of the cell cycle, while homologous recombination becomes fully activated in S phase. Studies over the past 2 decades have revealed that the aberrant joining of replication-associated breaks leads to catastrophic genome rearrangements, revealing an important role of DNA break repair pathway choice in the preservation of genome integrity. 53BP1, first identified as a DNA damage checkpoint protein, and BRCA1, a well-known breast cancer tumor suppressor, are at the center of this choice. Research on how these proteins function at the DNA break site has advanced rapidly in the recent past. Here, we review what is known regarding how the repair pathway choice is made, including the mechanisms that govern the recruitment of each critical factor, and how the cell transitions from end joining in G1 to homologous recombination in S/G2.
INTRODUCTION
DNA double-strand breaks (DSBs) are exceedingly dangerous chromosomal lesions. Failure to accurately repair DSBs can lead to gross chromosome rearrangements or mutations at the break site, which can cause cell death, cell transformation, and tumorigenesis. Two mechanistically distinct pathways have evolved to eliminate DSBs from the genome: nonhomologous DNA end joining (NHEJ) and homologous recombination (HR), both of which are conserved in all kingdoms of life. NHEJ entails the tethering of the broken DNA ends and their ligation (1). NHEJ is active throughout the cell cycle. While NHEJ accurately repairs “clean” DSBs whose ends are compatible and harbor undamaged terminal nucleotides, it is also capable of joining mismatched termini or termini that harbor damaged, otherwise-unligatable terminal nucleotides. In the latter case, joining is associated with DNA sequence loss. Moreover, when ends from two different chromosomes are joined, a chromosomal translocation ensues. In HR, the intact sister chromatid is most often engaged as the information donor. This process is normally accurate but requires that cells be in the S or G2 phase of the cell cycle, when DNA replication generates the sister chromatid to direct the repair process.
How DSB repair pathway choice is determined at the molecular level has been the subject of intense study for quite some time. It has become clear that whether or not the DNA ends have undergone extensive 5′-to-3′ nucleolytic resection exerts a major impact on this choice, such that ends that bear long 3′ DNA tails become destined for HR repair (2, 3). As first revealed in genetic studies performed with the budding yeast Saccharomyces cerevisiae, three distinct nucleases function in the DSB end resection process: the 5′-to-3′ exonuclease I (Exo1), the flap endonuclease Dna2 in conjunction with the RecQ family helicase Sgs1 (BLM in humans), and Mre11, part of the Mre11-Rad50-Xrs2 (MRX) complex (MRE11-RAD50-NBS1, or MRN, in humans) (4, 5). MRX is required for the initiation of resection near the DSB, whereas Exo1 and Dna2 carry out long-range resection. There is evidence of cross talk between these pathways, as MRN and BLM both stimulate Exo1 in vitro (6, 7). The single-stranded DNA (ssDNA) binding protein RPA (replication protein A) coats the 3′ tails generated during resection, preventing the formation of secondary structures (8). RPA also stimulates Exo1 and directs DNA2 cleavage to the 5′ flap after unwinding by BLM (6, 9).
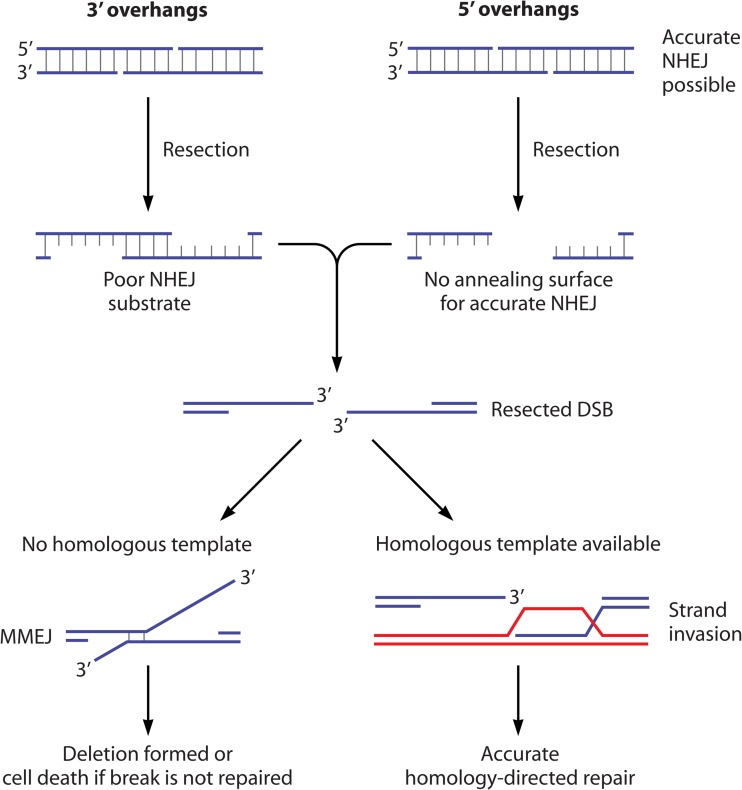
At DSBs with 5′ overhangs, resection eliminates the ability to align the ends for precise NHEJ (Fig. 1). DSBs with 3′ overhangs retain base-pairing potential after resection begins, but resection introduces extended gaps adjacent to the terminal nucleotides (Fig. 1). These DNA gaps have been shown to strongly inhibit NHEJ (10). A single-base gap reduces the NHEJ efficiency of otherwise-compatible overhangs severalfold, and accurate joining is prevented when the gap reaches 3 or 4 nucleotides (10). Thus, the onset of resection blocks restorative repair by NHEJ and instead favors DSB repair by HR. While accurate repair of the break by recombination is the preferred outcome, an error-prone backup pathway, referred to as the microhomology-mediated end joining (MMEJ) pathway, can also restore the DNA duplex (11). This pathway is mainly relevant when the ssDNA tails formed by resection fail to base pair fully, and microhomologies in the ssDNA tails instead form a short DNA hybrid to initiate the joining process (Fig. 1). MMEJ leads to deletion of the intervening sequence and is thus a highly mutagenic outcome.
FIG 1.
The effects of nucleolytic resection on NHEJ of DNA ends that bear 5′ or 3′ overhangs. With ends that harbor 3′ overhangs (top left), NHEJ becomes inefficient once the gap size stemming from resection becomes 2 nucleotides or greater. Accurate NHEJ of ends with 5′ overhangs (top right) is eliminated by resection. After resection, repair normally proceeds by HR (bottom right). Occasionally, and especially when a homologous template is unavailable, deletion-prone MMEJ occurs (bottom left).
The finding that resection can be inhibited by holding yeast cells in G1 or inhibiting cyclin-dependent kinase (CDK) activity provided the first major clue into the mechanism by which pathway choice is regulated (12, 13). Later work identified Sae2 (CtIP in humans), an MRX/MRN interaction partner, as the critical CDK target responsible for this phenomenon (14, 15). A model has emerged in which CDK phosphorylates CtIP at the G1-S transition to activate resection. In Schizosaccharomyces pombe, periodic expression of the CtIP homolog Ctp1 is also relevant for the cell cycle-dependent regulation of resection (16), and this mechanism seems to be conserved in mammalian cells (17). A second CDK target is the Dna2 nuclease, whose phosphorylation stimulates its recruitment to DSBs (18). The above findings suggested a straightforward model in which a phospho-dependent switch at the G1-S transition turns on resection. However, further investigations have revealed much greater complexity in the determination of DSB repair pathway choice, which involves DNA damage checkpoint proteins, the breast cancer susceptibility gene BRCA1, telomeric factors, ubiquitylation and SUMOylation cascades mediated by RING finger proteins, and a variety of histone modifications. While the basic mechanisms of HR and NHEJ are conserved, many of the mammalian factors that have been characterized in pathway choice do not have a clear ortholog in S. cerevisiae, suggesting that this aspect of the DNA damage response has diverged considerably in higher organisms. Indeed, blocking of resection in the G1 cell cycle phase seems to be more robust in mammalian cells than in yeast, as residual resection can occur during G1 arrest in yeast (19, 20). Here, we review these findings and strive to integrate them in a mechanistic model.
REGULATION OF THE ONSET OF RESECTION BY 53BP1 AND BRCA1
A series of studies published in 2010 implicated 53BP1, a target of the ATM kinase that forms nuclear foci upon DSB induction, and the tumor suppressor BRCA1 in DNA end resection control. 53BP1 was shown to negatively regulate resection in G1 (21). Importantly, BRCA1 promotes the removal of 53BP1 in S phase to allow resection (22). Consequently, in cells lacking BRCA1, resection is not upregulated in S phase and inappropriate NHEJ occurs at replication-associated DSBs, leading to gross chromosomal rearrangements. In mice, deletion of 53BP1 suppresses the embryonic lethality and prevents the chromosomal rearrangements seen in BRCA1−/− animals, emphasizing the importance of BRCA1-dependent removal of 53BP1 to facilitate the transition from NHEJ to HR (22–25).
The above findings raise new questions. Numerous proteins have been found that colocalize with γ-H2AX foci, which mark DNA damage sites. Some of these factors function upstream of both 53BP1 and BRCA1 recruitment, and how they promote or suppress either of the two repair pathways is the subject of ongoing investigations. What role do chromatin modifications play in recruitment of key factors? What is the molecular switch that activates BRCA1's ability to remove 53BP1 exclusively in S/G2? Moreover, what is the mechanism by which 53BP1 blocks the resection nucleases? Below, we analyze recent findings that help address these questions.
UPSTREAM ACTION OF RNF8 AND RNF168 UBIQUITIN LIGASES
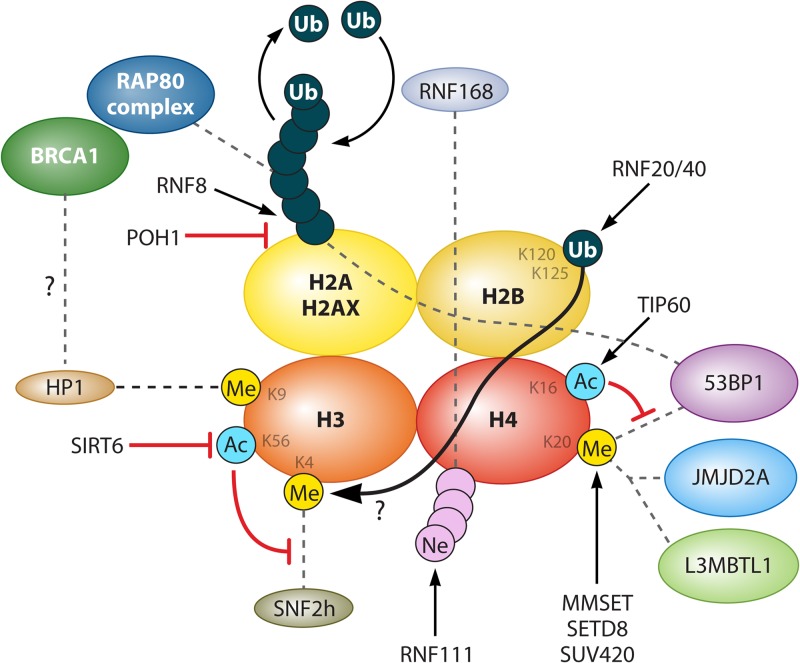
ATM, MDC1, the MRN complex, and the RING finger E3 ubiquitin ligases RNF8 and RNF168 are among the earliest factors found in DNA damage foci (26). 53BP1 and BRCA1 appear later, and their recruitment is dependent on the aforementioned upstream factors (27). RNF8 is recruited via an interaction between its FHA domain and phosphosites on MDC1 (27, 28). Acting with its partner E2 enzyme UBC13 (29), RNF8 ubiquitinates γ-H2AX (28) and H2A (27). This, along with other chromatin modifications that occur near the DSBs, is summarized in Fig. 2. RNF168 is then recruited, which also partners with UBC13 and further propagates the ubiquitination of histone γ-H2AX and of lysines 13 and 15 of histone H2A (30, 31). It should be noted that RNF8 and RNF168 form K63-linked ubiquitin chains which, unlike K48-linked chains, do not lead to protein degradation by the proteasome. Instead, the ubiquitinated targets act as recruitment platforms for downstream factors, which will be discussed in detail below. RNF168 recruitment is partially dependent on RNF111 and UBE2M, which are E3 and E2 enzymes, respectively, that conjugate the ubiquitin-like protein NEDD8 to a cluster of lysine residues on the N terminus of histone H4 (32). RNF168 physically interacts with “polyneddylated” H4, which is thought to be crucial for its DSB recruitment (32).
FIG 2.
Histone modifications involved in 53BP1 or BRCA1 recruitment to DSBs. Proteins that add or remove each modification are indicated by arrows. Dotted lines indicate protein-protein interactions. A question mark indicates a relationship that has been documented in the literature but whose mechanism is unknown.
Two deubiquitinating enzymes (DUBs), USP34 and OTUB1, regulate the level of RNF168 and the activity of the UBC13-RNF168 complex, respectively. The level of RNF168 in cells is controlled by its ubiquitin-dependent proteolysis. USP34 enhances the abundance of RNF168 by deubiquitinating it (33). Accordingly, cells deficient in USP34 are unable to mount DNA damage-induced 53BP1 and BRCA1 foci (33). In this regard, USP34 acts as a positive regulator of RNF168. OTUB1, on the other hand, negatively regulates RNF168 activity, although the mechanism is, surprisingly, independent of its DUB activity (34, 35). OTUB1 physically interacts with and attenuates the ubiquitin-conjugating activity of UBC13, the E2 partner for both RNF8 and RNF168. While OTUB1 restricts the activity of both UBC13-RNF8 and UBC13-RNF168 in vitro, only RNF168-dependent events seem to be affected in vivo. The end result is that OTUB1 negatively regulates HR. Its depletion alleviates the HR defect seen in cells treated with an ATM inhibitor, and OTUB1 overexpression inhibits HR (35). Finally, RNF168 activity is kept in check by the E3 ubiquitin ligases TRIP12 and UBR5, which prevent histone ubiquitination from spreading beyond the region surrounding the DSB (36).
Recently, RNF168 has also been shown to interact with the A and B isoforms of the E2 ubiquitin-conjugating enzyme RAD6 (37). These are orthologs of yeast Rad6, which helps mediate postreplication DNA repair, proteolysis via the N-end rule pathway, and other processes (38). Both RAD6 isoforms are required for 53BP1 and BRCA1 recruitment to DSBs (37). Liu et al. (37) implicated the RAD6A/B-RNF168 complexes in ubiquitination of the linker histone H1.2. However, the biological relevance of H1.2 ubiquitination remains to be established. It seems likely that other proteins are targeted by ubiquitination during the DNA damage response, and further work will be needed to identify and characterize the contribution of novel targets.
Besides ubiquitination, SUMOylation also plays a role in the RNF8-RNF168 pathway. SUMO, UBC9 (the SUMO E2-conjugating enzyme), and the SUMO ligases PIAS1 and PIAS4 all localize to DNA damage, and the recruitment of both 53BP1 and BRCA1 requires PIAS1 and PIAS4 (39, 40). While RNF8 localization occurs efficiently in the absence of either PIAS protein, PIAS4 is required for RNF168 localization and establishment of H2A ubiquitin adducts (39). While these results identify the step in the cascade where PIAS4 acts, it is unclear where PIAS1 functions. How PIAS1 and PIAS4 are recruited to DNA damage is also unknown, beyond the requirement of MDC1 for their localization (39). BRCA1 and 53BP1 have both been identified as likely PIAS1/PIAS4 targets, but how SUMOylation affects the properties of these factors is not yet known (39, 40). Given that SUMOylation affects H2A ubiquitination, an event upstream of 53BP1 and BRCA1 recruitment, there are likely other SUMOylation targets that remain to be identified.
THE RAP80 COMPLEX AND RNF20/RNF40-DEPENDENT RECRUITMENT OF BRCA1
Ubiquitination of H2A by RNF8 and RNF168 is thought to provide a recruitment platform for RAP80 and its associated proteins, BRCC36, Abraxas, MERIT40, and BRCC45 (27, 41–46). RAP80 directly interacts with ubiquitin and presumably with ubiquitinated H2A, thereby nucleating the assembly of a higher-order complex that harbors the aforementioned partner proteins (Fig. 2) (42, 44, 45). All members of the RAP80 complex are required for each other's stability and for the optimal recruitment of BRCA1 to DSBs, which is contingent upon its direct interaction with Abraxas (41, 43, 45). BRCC36 is a DUB that specifically targets K63-linked ubiquitin chains (47). The ubiquitin content of γ-H2AX foci peaks early and then it decreases in a manner that is dependent on the RAP80 protein complex, suggesting that the latter counteracts the actions of RNF8 and RNF168 (47). While loss of RAP80 reduces BRCA1 foci, it, paradoxically, increases long-range resection as well (48). A possible explanation is that deubiquitination of H2A affects the resection nucleases differently, e.g., stimulating MRN-dependent resection near the DSB but negatively regulating long-range resection by BLM-DNA2 and/or EXO1. This is an important topic that should be addressed.
It has been suggested that removal of K63-linked Ub chains from H2A by the RAP80 complex is part of an intricate process that helps convert them to K6-linked chains catalyzed by the E3-ubiquitin ligase activity of the BRCA1-BARD1 complex (41). Consistent with this idea, RAP80 has affinity for K63- and K6-linked but not K48-linked ubiquitin chains (44). However, a recent report showed that a BRCA1 RING domain mutant that was defective in E3 ligase activity supported resection at the wild-type level (49). This study revealed that, if BRCA1 does indeed form K6-linked chains at DSB sites, they are dispensable for efficient resection.
The RING finger E3 ubiquitin ligases RNF20 and RNF40 also function upstream of BRCA1 recruitment, but the mechanism is less clear. These ligases modify histone H2B on lysines 120 and 125 (Fig. 2) (50). RNF20 knockdown causes a delay in the recruitment of the general recombinase RAD51 and its partner proteins to DNA damage, and an H2B point mutant that cannot be ubiquitinated exerts a stronger effect in this regard (50, 51). NBS1, an integral component of the MRN complex, interacts directly with RNF20, and this interaction is required for resection (51). However, H2B ubiquitination occurs efficiently in the absence of MRN, arguing against an involvement of MRN in RNF20-mediated H2B modification (51). RNF20 is also required for methylation of lysine 4 in histone H3 (51). This methylation mark, alongside the deacetylation of lysine 56 in H3 by SIRT6, leads to recruitment of the SNF2h nucleosome remodeler, which is required for optimal resection efficiency (51, 52). Intriguingly, the RNF20 resection defect can be overcome by treating cells with chloroquine, which leads to chromatin relaxation (51). Thus, RNF20 and RNF40 may promote histone eviction, an important step, considering that nucleosomes pose an obstacle to resection nucleases, as revealed in biochemical reconstitution studies (53). Indeed, chromatin immunoprecipitation experiments have yielded results suggesting that histones are removed near the DSB site within a time frame consistent with resection (54, 55). BRCA1 recruitment is also reduced in the absence of HP1, which interacts with H3 methylated on K9 (56). Further work is needed to determine the mechanism by which HP1 influences BRCA1 activity.
RECRUITMENT OF 53BP1
How 53BP1 is recruited to DSB sites has been debated. While it has been reported that 53BP1 recruitment requires its interaction with the tandem BRCT domains of MDC1 (57, 58), other studies have found the proposed MDC1 interaction domain to be dispensable in this regard (59, 60). 53BP1 also interacts with histone H4 dimethylated on lysine 20 (H4K20me2) via its Tudor domain (Fig. 2), but early studies did not detect an increase in H4K20me2 upon DNA damage (61). This suggested a model in which 53BP1 is initially recruited by MDC1 and is then retained in the chromatin region surrounding the break through an interaction with H4K20me2, which is presumed to be exposed following damage (61). This model is consistent with recent reports implicating PR-SET7/SETD8 and SUV420 in 53BP1 recruitment (62, 63). An alternate model has been proposed in which the methyltransferase MMSET promotes a localized increase in H4K20me2 near the DSB site (64). Like 53BP1, MMSET is phosphorylated by ATM upon the occurrence of DNA damage and appears to be recruited via an interaction with the BRCT domains of MDC1 (64). Further work will be needed to resolve conflicting data in this area. 53BP1 also harbors a C-terminal ubiquitin-interacting motif that recognizes histone H2A ubiquitinated on K15, a modification that is catalyzed by RNF168 (Fig. 2) (59). This finding helped explain why RNF8 and RNF168 are required for 53BP1 recruitment, but the RAP80 complex, which functions immediately downstream, is not (47). Furthermore, acetylation of histone H4 on lysine 16 by TIP60 has been shown to reduce the affinity of 53BP1 for H4K20me2 (Fig. 2) (65). Thus, three separate histone modifications—methylation, ubiquitination, and acetylation—are involved in regulating the recruitment and retention of 53BP1 (59).
Additional layers of regulation are afforded by two proteins, JMJD2A and L3MBTL1, which compete with 53BP1 for binding to H4K20me2 (Fig. 2) (66–68). JMJD2A interacts with H4K20me2 via its tandem Tudor domains, whereas L3MBTL1 utilizes an MBT domain to accomplish this feat. The RNF8-RNF168 pathway also has a role in the removal of JMJD2A and L3MBTL1 from DSB regions. Specifically, RNF8 and RNF168 directly ubiquitinate JMJD2A to trigger its degradation (68), and RNF168 also ubiquitinates VCP, which has ATPase activity, causing it to relocalize to chromatin, where it removes L3MBTL1 (66). The proteasome-associated DUB POH1 antagonizes RNF8-RNF168 activity on JMJD2A, promoting the stability of the latter and attenuating 53BP1 recruitment (67). POH1 likely targets other proteins too, as it has been implicated in RAD51 accumulation, a process that is independent of (and is, in fact, antagonized by) 53BP1 (67). Advanced microscopy has shown that, as cells transition into S/G2, POH1 promotes the removal of RAP80 from DSB sites (49). As discussed above, the recruitment of RAP80 is dependent on RNF8/RNF168-mediated ubiquitination of histone H2A. Interestingly, the DUB activity of BRCC36, an integral component of the RAP80 complex, counteracts RNF168 to help achieve an equilibrium state of ubiquitination. An attractive model is that POH1 removes the initial ubiquitin conjugated to H2A, terminating the chain extension/reduction cycle maintained by RNF168 and RAP80 (Fig. 2).
The BLM helicase has recently also been implicated in 53BP1 recruitment. Cells from Bloom syndrome patients fail to recruit 53BP1 to DNA damage sites (69). BLM-deficient cells also show evidence of increased resection during microhomology-mediated end joining, which is surprising given that BLM helps catalyze one branch of long-range resection (69). These results suggest a dual role for BLM: an early antiresection function with 53BP1 and a late proresection role with DNA2 after 53BP1 has been removed. Separation-of-function BLM mutants will be critical in further characterizing this dichotomy.
RIF1 AND PTIP, KEY EFFECTORS OF 53BP1
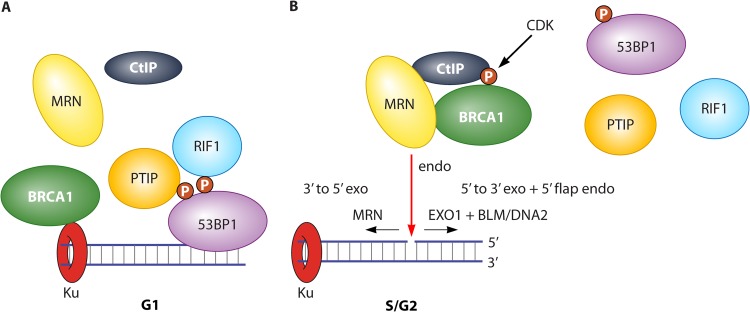
The N terminus of 53BP1 is phosphorylated by ATM after DNA damage, but the phosphorylation sites are not required for 53BP1 localization to nuclear foci. Instead, these sites are necessary for resection attenuation, suggesting that phosphorylation of 53BP1 is important for the recruitment of effector molecules (70). A series of papers published in 2013 identified RIF1 as a 53BP1 effector (Fig. 3A) (71–74). Like 53BP1, RIF1 is required for NHEJ and is removed from foci in S/G2 in a BRCA1- and CtIP-dependent manner. 53BP1 and RIF1 coimmunoprecipitate from cell extracts (73), but a direct interaction in vitro with purified proteins has not yet been reported, so it remains possible that an adapter links these two proteins together.
FIG 3.
Model for DSB occupancy in G1 and S/G2. (A) In G1 cells, 53BP1 is phosphorylated by ATM and becomes localized at the break. RIF1 and PTIP are recruited in a phospho-dependent manner and block resection via an unknown mechanism. BRCA1 is bound to Ku but in a quantity so low that they do not appear as visual foci. (B) In S/G2, CtIP is phosphorylated by CDK, inducing the formation of a complex with BRCA1 and MRN. This complex displaces 53BP1 and initiates resection. Ku may be removed by coordinated endo- and exonuclease activities that are initiated from a nick in a manner analogous to the postulated mechanism of Spo11 removal.
The observation that loss of RIF1 only partially restores HR in cells lacking BRCA1 (75) suggested that another protein may be involved in the attenuation of HR to favor NHEJ. Indeed, PTIP has recently been identified as an additional 53BP1 effector (76). Using its tandem BRCT domains, PTIP interacts directly with 53BP1 in a manner that is dependent on the ATM-mediated phosphorylation of serine 25 of the latter (76). Like RIF1, loss of PTIP restores RPA foci in BRCA1-deficient cells, indicating that PTIP also plays a role in blocking resection (76). PTIP has also been reported to bind γ-H2AX, but the functional significance of this is not yet clear (77).
The mechanisms by which RIF1 and PTIP prevent resection and promote NHEJ have not yet been elucidated. However, it is known that RIF1 interacts with the BLM helicase and is required for recruitment of BLM to foci (75, 78). Conceivably, RIF1 may inhibit the ability of BLM to unwind DNA for resection by DNA2. Or, RIF1 and/or PTIP may stabilize NHEJ proteins, such as the Ku heterodimer on DNA ends, which would prevent access of the ends for the resection nucleases (see below). The DNA end structure may help determine the activities of RIF1 and PTIP. A key difference between the two effectors is that PTIP seems specific to the inappropriate S-phase NHEJ events that characterize cells lacking BRCA1, while RIF1 is additionally required for class switch recombination, a specialized form of NHEJ that occurs at DSBs generated by cytosine deamination in developing B cells. Interestingly, V(D)J recombination, a form of NHEJ that joins hairpin-capped ends during lymphocyte development, is unaffected by the loss of RIF1 (71) but is defective in cells lacking PTIP (79). Also, NHEJ of blunt-ended leading-strand telomeres is more dependent on 53BP1 than lagging-strand telomeres, which have 3′ overhangs (80). Uncovering the mechanisms of resection inhibition by RIF1 and PTIP will be a top priority in the near future.
THE G1-S TRANSITION: CtIP PHOSPHORYLATION AND REMOVAL OF 53BP1 BY BRCA1
The mechanism by which BRCA1 helps mediate 53BP1 removal as cells transition from G1 into S phase is unclear. However, it is important to note that the loss of BRCA1 leads to the DSB recruitment of 53BP1 in G2 (73), suggesting that the 53BP1 recruitment platform remains intact in S/G2 but is somehow masked by BRCA1. Similarly, 53BP1 knockdown leads to ectopic BRCA1 nuclear foci in G1 (73), indicating that the potential for BRCA1 recruitment exists in G1 but is blocked in a 53BP1-dependent manner.
How, then, do cells switch from pro-NHEJ 53BP1 in G1 to proresection BRCA1 in S/G2? Existing evidence hints at CtIP as the key mediator of this switch. CtIP phosphorylation by CDK is required for the initiation of resection in S phase (81). A G2-specific complex containing CtIP, MRN, and BRCA1 has been reported (82, 83). CDK activity is required for the assembly of this higher-order complex (82). In a unified model, formation of the CtIP-MRN-BRCA1 complex triggers 53BP1-RIF1 removal and resection initiation (Fig. 3B). The finding that RIF1 could not be removed from foci in a CtIP phosphorylation site mutant is certainly consistent with this idea (73). Depletion of 53BP1 or RIF1 restores resection in BRCA1-deficient cells but not in cells lacking CtIP (34). These results reveal that CtIP has other functions beyond 53BP1-RIF1 removal (81) and that removal of 53BP1-RIF1 is the critical function of BRCA1 in resection. The observation that phosphorylated CtIP is ubiquitinated by BRCA1 led to the suggestion that this CtIP modification promotes resection (84). However, the recent finding that a BRCA1 RING domain mutant is still competent for resection casts doubt on models in which ubiquitination of downstream targets by BRCA1 is important for 53BP1 removal (49). Instead, 53BP1 displacement seems to be a separate activity of BRCA1, with the E3 ubiquitin ligase activity provided by RNF8 and RNF168 being most relevant to resection.
The observation that BRCA1 foci are impaired but not abolished in the absence of the RAP80 complex indicates that there is another means of BRCA1 recruitment independent of the BRCA1-Abraxas interaction (41, 43, 48, 85). Specifically, the FHA and BRCT domains of NBS1 interact with MDC1 that has been phosphorylated by casein kinase 2, and this interaction could be sufficient to tether the BRCA1-CtIP-MRN ensemble to the damage site when the RAP80 complex is missing (86).
Evidence from yeast has identified the chromatin remodeler Fun30 (SMARCAD1 in humans) as another possible player in 53BP1 removal (87–89). Fun30 is especially important for resection when Rad9, the yeast ortholog of 53BP1, is present (87). It will be important to investigate genetic interactions between SMARCAD1 and members of the 53BP1-BRCA1 pathway.
REMOVAL OF Ku FROM DSB ENDS
Ku is a conserved, toroidal heterodimeric molecule consisting of two subunits, Ku70 and Ku80. It is abundant in the nucleus and has high affinity for DNA ends. The engagement of DNA ends by Ku represents the key first step in NHEJ (90). Genetic experiments in yeast have shown that Ku occludes DNA ends from Exo1. Ku and MRX/MRN are among the first proteins recruited to DSBs (91, 92). Importantly, the MRX nuclease activity and Sae2 (orthologous to CtIP) function in conjunction to remove Ku (92–95). Insights into how the MRX nuclease might release Ku from DNA ends have emanated from studies on meiotic recombination initiation. Early on in the meiotic program, the topoisomerase-like protein Spo11 acts in a genome-wide fashion to introduce DSBs into chromosomes to trigger HR. This serves to tie homologs together to prepare them for segregation in the first meiotic division. After mediating DNA strand breakage, Spo11 remains covalently bound to the DSB ends. A plausible model for Spo11 removal involves endonucleolytic incision of the DNA adjacent to the end by the MRX complex, followed by bidirectional exonucleolytic digestion from the nick, with MRX acting 3′ to 5′ (toward the break end) and Exo1 or Dna2/BLM performing long-range 5′-to-3′ resection (96). The use of small-molecule inhibitors of MRN endo- and exonuclease activities has provided support for a similar model at mitotic DSBs, where Spo11 is absent but Ku is likely to be bound (97). Whether or not the human MRN complex can overcome resection inhibition by Ku in a manner analogous to Spo11 is being addressed by different groups (98, 99). It is also unclear whether Ku exerts as much of an inhibitory effect on resection in mammalian cells as it does in yeast (24).
The identification of an interaction between BRCA1 and Ku provided support for a mechanism by which MRN, in complex with BRCA1 and phospho-CtIP, mediates Ku removal (Fig. 3B) (100). More recent results have shown that the BRCA1-Ku interaction is specific to the G1 phase, however, and is important for NHEJ fidelity (101). These results reveal that BRCA1 is in fact present at the DSB site during G1, even though it does not form microscopically visible foci until S/G2. It will be critically important to determine whether the Ku-interacting form of BRCA1 is part of the aforementioned complex with MRN and CtIP or whether separate complexes that fulfill different functions exist. It is possible that BRCA1 associates only with Ku in G1 but forms a distinct complex with MRN and CtIP upon entry into S phase in order to activate the MRN nuclease activity to initiate resection. The N terminus of BRCA1 interacts with both Ku and Nbs1, but mutants differentially affected for either interaction are not yet available (82, 100, 101). Whether the MRN nuclease activity is required for Ku removal in mammalian cells remains an open question, but recent advances in assessing Ku localization should allow this important experiment to be done (102).
Another possibility is that BRCA1, in a manner analogous to 53BP1, functions through effector molecules to promote resection. One candidate for such a factor is the telomere protein TRF1, which has recently been shown to localize to foci downstream of BRCA1 to promote resection (103). Alleles that separate the telomeric function of TRF1 from this resection role will be beneficial for the further characterization of this factor in resection.
CONCLUDING REMARKS
In Escherichia coli, the RecBCD complex, which harbors both helicase and nuclease activities, is the major protein machine that mediates resection. An early assumption was that eukaryotes employ a similar compact, efficient resection enzyme ensemble. The failure of early radiation sensitivity screens in yeast to identify “the” resection nuclease was a clue that resection is a great deal more complicated in eukaryotes. It is fair to say that no one anticipated the level of complexity that has been uncovered to date, though. Besides the involvement of three distinct nucleases, layers upon layers of intricate regulation have been identified in regard to making the correct choice between NHEJ and HR.
ACKNOWLEDGMENTS
The research in our laboratory has been supported by research grants and a program project grant (SBDR) from the National Institutes of Health.
Footnotes
Published ahead of print 27 January 2014
REFERENCES
- 1.Chiruvella KK, Liang Z, Wilson TE. 2013. Repair of double-strand breaks by end joining. Cold Spring Harbor Perspect. Biol. 5:a012757. 10.1101/cshperspect.a012757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47:497–510. 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- 3.Symington LS, Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45:247–271. 10.1146/annurev-genet-110410-132435 [DOI] [PubMed] [Google Scholar]
- 4.Mimitou EP, Symington LS. 2011. DNA end resection: unraveling the tail. DNA Repair (Amst.) 10:344–348. 10.1016/j.dnarep.2010.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niu H, Raynard S, Sung P. 2009. Multiplicity of DNA end resection machineries in chromosome break repair. Genes Dev. 23:1481–1486. 10.1101/gad.1824209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25:350–362. 10.1101/gad.2003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. U. S. A. 105:16906–16911. 10.1073/pnas.0809380105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Lisby M, Symington LS. 2013. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 50:589–600. 10.1016/j.molcel.2013.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467:108–111. 10.1038/nature09318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daley JM, Laan RL, Suresh A, Wilson TE. 2005. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 280:29030–29037. 10.1074/jbc.M505377200 [DOI] [PubMed] [Google Scholar]
- 11.McVey M, Lee SE. 2008. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 24:529–538. 10.1016/j.tig.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aylon Y, Liefshitz B, Kupiec M. 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23:4868–4875. 10.1038/sj.emboj.7600469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431:1011–1017. 10.1038/nature02964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455:689–692. 10.1038/nature07215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huertas P, Jackson SP. 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284:9558–9565. 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28:134–146. 10.1016/j.molcel.2007.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, Lee WH. 2006. CtIP activates its own and cyclin D1 promoters via the E2F/RB pathway during G1/S progression. Mol. Cell. Biol. 26:3124–3134. 10.1128/MCB.26.8.3124-3134.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Niu H, Chung WH, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G. 2011. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat. Struct. Mol. Biol. 18:1015–1019. 10.1038/nsmb.2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barlow JH, Lisby M, Rothstein R. 2008. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell 30:73–85. 10.1016/j.molcel.2008.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zierhut C, Diffley JF. 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 27:1875–1885. 10.1038/emboj.2008.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 2010. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med. 207:855–865. 10.1084/jem.20100244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141:243–254. 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17:688–695. 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, Chen HT, Fernandez-Capetillo O, D'Andrea A, Nussenzweig A. 2012. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell 46:125–135. 10.1016/j.molcel.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, Deng CX, Finkel T. 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell 35:534–541. 10.1016/j.molcel.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bekker-Jensen S, Mailand N. 2010. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst.) 9:1219–1228. 10.1016/j.dnarep.2010.09.010 [DOI] [PubMed] [Google Scholar]
- 27.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887–900. 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- 28.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131:901–914. 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang B, Elledge SJ. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. U. S. A. 104:20759–20763. 10.1073/pnas.0710061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136:435-446. 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- 31.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136:420–434. 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- 32.Ma T, Chen Y, Zhang F, Yang CY, Wang S, Yu X. 2013. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol. Cell 49:897–907. 10.1016/j.molcel.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sy SM, Jiang JWSO, Deng Y, Huen MS. 2013. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res. 41:8572–8580. 10.1093/nar/gkt622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juang YC, Landry MC, Sanches M, Vittal V, Leung CC, Ceccarelli DF, Mateo AR, Pruneda JN, Mao DY, Szilard RK, Orlicky S, Munro M, Brzovic PS, Klevit RE, Sicheri F, Durocher D. 2012. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 45:384–397. 10.1016/j.molcel.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, O'Donnell L, Kumakubo A, Munro M, Sicheri F, Gingras AC, Natsume T, Suda T, Durocher D. 2010. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466:941–946. 10.1038/nature09297 [DOI] [PubMed] [Google Scholar]
- 36.Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grofte M, Bartkova J, Poulsen M, Oka Y, Bekker-Jensen S, Mailand N, Neumann B, Heriche JK, Shearer R, Saunders D, Bartek J, Lukas J, Lukas C. 2012. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150:697–709. 10.1016/j.cell.2012.06.039 [DOI] [PubMed] [Google Scholar]
- 37.Liu C, Wang D, Wu J, Keller J, Ma T, Yu X. 2013. RNF168 forms a functional complex with RAD6 during the DNA damage response. J. Cell Sci. 126:2042–2051. 10.1242/jcs.122945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Game JC, Chernikova SB. 2009. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair (Amst.) 8:470–482. 10.1016/j.dnarep.2009.01.007 [DOI] [PubMed] [Google Scholar]
- 39.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462:935–939. 10.1036/nature08657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, Ng T, Solomon E. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462:886–890. 10.1038/nature08593 [DOI] [PubMed] [Google Scholar]
- 41.Feng L, Huang J, Chen J. 2009. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 23:719–728. 10.1101/gad.1770609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H, Chen J, Yu X. 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316:1202–1205. 10.1126/science.1139621 [DOI] [PubMed] [Google Scholar]
- 43.Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, Greenberg RA. 2009. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 23:740–754. 10.1101/gad.1739609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316:1198–1202. 10.1126/science.1139516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316:1194–1198. 10.1126/science.1139476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M, Chen J, Yu X. 2009. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol. Cell. Biol. 29:849–860. 10.1128/MCB.01302-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. 2009. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl. Acad. Sci. U. S. A. 106:3166–3171. 10.1073/pnas.08007485106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. 2011. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 25:685–700. 10.1101/gad.2011011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kakarougkas A, Ismail A, Katsuki Y, Freire R, Shibata A, Jeggo PA. 2013. Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res. 41:10298–10311. 10.1093/nar/gkt802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, Shkedy D, Smorodinsky NI, van Vliet N, Kuster B, Mann M, Ciechanover A, Dahm-Daphi J, Kanaar R, Hu MC, Chen DJ, Oren M, Shiloh Y. 2011. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell 41:529–542. 10.1016/j.molcel.2011.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K. 2011. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell 41:515–528. 10.1016/j.molcel.2011.02.002 [DOI] [PubMed] [Google Scholar]
- 52.Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, D'Urso A, Naar AM, Kingston R, Rippe K, Mostoslavsky R. 2013. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 51:454–468. 10.1016/j.molcel.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adkins NL, Niu H, Sung P, Peterson CL. 2013. Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 20:836–842. 10.1038/nsmb.2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonetti D, Anbalagan S, Lucchini G, Clerici M, Longhese MP. 2013. Tbf1 and Vid22 promote resection and non-homologous end joining of DNA double-strand break ends. EMBO J. 32:275–289. 10.1038/emboj.2012.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB. 2013. Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proc. Natl. Acad. Sci. U. S. A. 110:16874–16879. 10.1073/pnas.1306160110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee YH, Kuo CY, Stark JM, Shih HM, Ann DK. 2013. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res. 41:5784–5798. 10.1093/nar/gkt231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eliezer Y, Argaman L, Rhie A, Doherty AJ, Goldberg M. 2009. The direct interaction between 53BP1 and MDC1 is required for the recruitment of 53BP1 to sites of damage. J. Biol. Chem. 284:426–435. 10.1074/jbc.M807375200 [DOI] [PubMed] [Google Scholar]
- 58.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961–966. 10.1038/nature01446 [DOI] [PubMed] [Google Scholar]
- 59.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, Durocher D. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499:50–54. 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zgheib O, Pataky K, Brugger J, Halazonetis TD. 2009. An oligomerized 53BP1 tudor domain suffices for recognition of DNA double-strand breaks. Mol. Cell. Biol. 29:1050–1058. 10.1128/MCB.01011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127:1361–1373. 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hartlerode AJ, Guan Y, Rajendran A, Ura K, Schotta G, Xie A, Shah JV, Scully R. 2012. Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks. PLoS One 7(11):e49211. 10.1371/journal.pone.0049211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsiao KY, Mizzen CA. 2013. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J. Mol. Cell Biol. 5:157–165. 10.1093/jmcb/mjs066 [DOI] [PubMed] [Google Scholar]
- 64.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. 2011. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470:124–128. 10.1038/nature09658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20:317–325. 10.1038/nsmb.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP. 2011. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 18:1345–1350. 10.1038/nsmb.2188 [DOI] [PubMed] [Google Scholar]
- 67.Butler LR, Densham RM, Jia J, Garvin AJ, Stone HR, Shah V, Weekes D, Festy F, Beesley J, Morris JR. 2012. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 31:3918–3934. 10.1038/emboj.2012.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, Sixma TK, Richard S. 2012. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31:1865–1878. 10.1038/emboj.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grabarz A, Guirouilh-Barbat J, Barascu A, Pennarun G, Genet D, Rass E, Germann SM, Bertrand P, Hickson ID, Lopez BS. 2013. A role for BLM in double-strand break repair pathway choice: prevention of CtIP/Mre11-mediated alternative nonhomologous end-joining. Cell Rep. 5:21–28. 10.1016/j.celrep.2013.08.034 [DOI] [PubMed] [Google Scholar]
- 70.Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, Nussenzweig A, Nussenzweig MC. 2011. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell 42:319–329. 10.1016/j.molcel.2011.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49:858–871. 10.1016/j.molcel.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, Nussenzweig A, Casellas R, Robbiani DF, Nussenzweig MC. 2013. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339:711–715. 10.1126/science.1230624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49:872–883. 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 74.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339:700–704. 10.1126/science.1231573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng L, Fong KW, Wang J, Wang W, Chen J. 2013. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J. Biol. Chem. 288:11135–11143. 10.1074/jbc.M113.457440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, Wesemann DR, Lee JE, Tubbs A, Sleckman BP, Daniel JA, Ge K, Alt FW, Fernandez-Capetillo O, Nussenzweig MC, Nussenzweig A. 2013. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153:1266–1280. 10.1016/j.cell.2013.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan W, Shao Z, Li F, Niu L, Shi Y, Teng M, Li X. 2011. Structural basis of gammaH2AX recognition by human PTIP BRCT5-BRCT6 domains in the DNA damage response pathway. FEBS Lett. 585:3874–3879. 10.1016/j.febslet.2011.10.045 [DOI] [PubMed] [Google Scholar]
- 78.Xu D, Muniandy P, Leo E, Yin J, Thangavel S, Shen X, Ii M, Agama K, Guo R, Fox D, III, Meetei AR, Wilson L, Nguyen H, Weng NP, Brill SJ, Li L, Vindigni A, Pommier Y, Seidman M, Wang W. 2010. Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J. 29:3140–3155. 10.1038/emboj.2010.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Callen E, Faryabi RB, Luckey M, Hao B, Daniel JA, Yang W, Sun HW, Dressler G, Peng W, Chi H, Ge K, Krangel MS, Park JH, Nussenzweig A. 2012. The DNA damage- and transcription-associated protein paxip1 controls thymocyte development and emigration. Immunity 37:971–985. 10.1016/j.immuni.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lottersberger F, Bothmer A, Robbiani DF, Nussenzweig MC, de Lange T. 2013. Role of 53BP1 oligomerization in regulating double-strand break repair. Proc. Natl. Acad. Sci. U. S. A. 110:2146–2151. 10.1073/pnas.1222617110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. 2007. Human CtIP promotes DNA end resection. Nature 450:509–514. 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen L, Nievera CJ, Lee AY, Wu X. 2008. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 283:7713–7720. 10.1074/jbc.M710245200 [DOI] [PubMed] [Google Scholar]
- 83.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. 2006. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 20:34–46. 10.1101/gad.1381306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu X, Fu S, Lai M, Baer R, Chen J. 2006. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 20:1721–1726. 10.1101/gad.1431006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yin Z, Menendez D, Resnick MA, French JE, Janardhan KS, Jetten AM. 2012. RAP80 is critical in maintaining genomic stability and suppressing tumor development. Cancer Res. 72:5080–5090. 10.1158/0008-5472.CAN-12-1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chapman JR, Jackson SP. 2008. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 9:795–801. 10.1038/embor.2008.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G. 2012. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489:576–580. 10.1038/nature11355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, van Attikum H, Llorente B. 2012. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489:581–584. 10.1038/nature11353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE. 2012. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol. Cell. Biol. 32:4727–4740. 10.1128/MCB.00566-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Deriano L, Roth DB. 2013. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 47:433–455. 10.1146/annurev-genet-110711-155540 [DOI] [PubMed] [Google Scholar]
- 91.Mirzoeva OK, Petrini JH. 2001. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21:281–288. 10.1128/MCB.21.1.281-288.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu D, Topper LM, Wilson TE. 2008. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178:1237–1249. 10.1534/genetics.107.083535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Langerak P, Mejia-Ramirez E, Limbo O, Russell P. 2011. Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7(9):e1002271. 10.1371/journal.pgen.1002271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mimitou EP, Symington LS. 2010. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 29:3358–3369. 10.1038/emboj.2010.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29:3370–3380. 10.1038/emboj.2010.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neale MJ, Pan J, Keeney S. 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436:1053–1057. 10.1038/nature03872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, Ismail A, Ismalaj E, Petricci E, Neale MJ, Bristow RG, Masson JY, Wyman C, Jeggo PA, Tainer JA. 2013. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53:7–18. 10.1016/j.molcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sun J, Lee KJ, Davis AJ, Chen DJ. 2012. Human Ku70/80 protein blocks exonuclease 1-mediated DNA resection in the presence of human Mre11 or Mre11/Rad50 protein complex. J. Biol. Chem. 287:4936–4945. 10.1074/jbc.M111.306167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang SH, Zhou R, Campbell J, Chen J, Ha T, Paull TT. 2013. The SOSS1 single-stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J. 32:126–139. 10.1038/emboj.2012.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wei L, Lan L, Hong Z, Yasui A, Ishioka C, Chiba N. 2008. Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Mol. Cell Biol. 28:7380–7393. 10.1128/MCB.01075-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang G, Plo I, Wang T, Rahman M, Cho JH, Yang E, Lopez BS, Xia F. 2013. BRCA1-Ku80 protein interaction enhances end-joining fidelity of chromosomal double-strand breaks in the G1 phase of the cell cycle. J. Biol. Chem. 288:8966–8976. 10.1074/jbc.M112.412650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Britton S, Coates J, Jackson SP. 2013. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 202:579–595. 10.1083/jcb.201303073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McKerlie M, Walker JR, Mitchell TR, Wilson FR, Zhu XD. 2013. Phosphorylated (pT371)TRF1 is recruited to sites of DNA damage to facilitate homologous recombination and checkpoint activation. Nucleic Acids Res. 41:10268–10282. 10.1093/nar/gkt775 [DOI] [PMC free article] [PubMed] [Google Scholar]