Abstract
Apoptotic cells are swiftly engulfed by macrophages to prevent the release of noxious materials from dying cells. Apoptotic cells expose phosphatidylserine (PtdSer) on their surface, and macrophages engulf them by recognizing PtdSer using specific receptors and opsonins. Here, we found that mouse resident peritoneal macrophages expressing Tim4 and MerTK are highly efficient at engulfing apoptotic cells. Neutralizing antibodies against either Tim4 or MerTK inhibited the macrophage engulfment of apoptotic cells. Tim4-null macrophages exhibited reduced binding and engulfment of apoptotic cells, whereas MerTK-null macrophages retained the ability to bind apoptotic cells but failed to engulf them. The incubation of wild-type peritoneal macrophages with apoptotic cells induced the rapid tyrosine phosphorylation of MerTK, which was not observed with Tim4-null macrophages. When mouse Ba/F3 cells were transformed with Tim4, apoptotic cells bound to the transformants but were not engulfed. Transformation of Ba/F3 cells with MerTK had no effect on the binding or engulfment of apoptotic cells; however, Tim4/MerTK transformants exhibited strong engulfment activity. Taken together, these results indicate that the engulfment of apoptotic cells by resident peritoneal macrophages proceeds in two steps: binding to Tim4, a PtdSer receptor, followed by MerTK-mediated cell engulfment.
INTRODUCTION
Millions of harmful, useless, or senescent cells in our body undergo apoptosis every minute and are swiftly engulfed by macrophages for clearance (1, 2). Engulfment is required to prevent apoptotic cells from undergoing secondary necrosis and releasing noxious materials. Inefficient engulfment of apoptotic cells can activate the immune system and contribute to the development of systemic lupus erythematosus (SLE)-type autoimmune diseases (3). The engulfment of apoptotic cells is also important for digesting dead cell components into their building units for recycling and energy (4).
To facilitate recognition of apoptotic cells by phagocytes, they expose phosphatidylserine (PtdSer) on their surface (5, 6). We recently showed that a membrane protein (Xkr8) containing six transmembrane domains is activated by caspase and plays an indispensable role in apoptotic PtdSer exposure (7). Several molecules are reported to recognize PtdSer on apoptotic cells (1, 2). Milk-fat globule epidermal growth factor 8 (MFG-E8) is a soluble protein secreted from a subset of macrophages. It contains an epidermal growth factor domain that binds to the integrin-αvβ3 complex on macrophages and a discoidin domain that binds to PtdSer on apoptotic cells, thus serving as a bridging molecule between apoptotic cells and macrophages. Protein S and Gas6 (growth arrest-specific 6) also function as bridges by binding to receptors on macrophages and PtdSer on apoptotic cells (8). The receptors for protein S and Gas6 are type I membrane proteins of the TAM (Tyro 3, Axl, and MerTK) family, which have intracellular tyrosine kinase domains (9). Type I membrane proteins such as Tim (T-cell immunoglobulin and mucin domain-containing) family proteins, stabilins, and BAI1 also directly bind PtdSer and enhance the engulfment of apoptotic cells by phagocytes (10–12). In addition, many other molecules such as RAGE (receptor for advanced glycation end products) (13), complement C1q (14), and calreticulin (15) have been shown to be involved in the engulfment of apoptotic cells. Whether these molecules are redundant or have specific roles in the process remains to be determined.
Here we report that mouse resident peritoneal macrophages (rpMacs) that express both Tim4 and MerTK are highly capable of engulfing apoptotic cells. Tim4- or MerTK-null mutations prevented rpMac-mediated apoptotic cell engulfment. Tim4-null but not MerTK-null macrophages lost their ability to tether apoptotic cells. The apoptotic cell-induced tyrosine phosphorylation of MerTK, observed in wild-type macrophages, was abolished in Tim4-null macrophages. Tim4-transformed Ba/F3 cells were found to bind apoptotic cells, but their apoptotic cell engulfment activity was weak. In contrast, transformants expressing both Tim4 and MerTK efficiently engulfed apoptotic cells. These results indicate that mouse rpMacs require two PtdSer-dependent systems for efficient engulfment of apoptotic cells, the Tim4-mediated tethering of apoptotic cells and protein S/MerTK-mediated cell engulfment, and strongly confirm the “tether/tickle” hypothesis proposed by Henson's group (16, 17).
MATERIALS AND METHODS
Mice, cell lines, recombinant proteins, antibodies (Abs), and reagents.
C57BL/6J mice and MerTK−/− mice (18) were purchased from Japan SLC and the Jackson Laboratory, respectively. Tim4−/− mice were described previously (19). All mouse studies were approved by the ethics review committee for animal experimentation of the Graduate School of Medicine, Kyoto University.
Mouse interleukin-3 (IL-3)-dependent Ba/F3 cells were maintained in RPMI 1640 containing 10% fetal calf serum (FCS) and 45 U/ml mouse IL-3 as described previously (20). Human leucine zipper-tagged Fas ligand (FasL) was produced using COS7 cells as described previously (21) and concentrated by precipitation in 60%-saturated (NH4)2SO4, followed by dialysis against phosphate-buffered saline (PBS). Hamster anti-mouse Tim4 monoclonal antibody (MAb) (clone Kat5-18) was described previously (10). Biotin-conjugated goat anti-mouse MerTK was from R & D Systems. Allophycocyanin (APC)- and peridinin chlorophyll protein (PerCP)-Cy 5.5-labeled rat anti-mouse Mac1 MAb (CD11b, clone M1/70) were purchased from BioLegend and BD PharMingen, respectively. Mouse antiphosphotyrosine MAb (clone 4G10) and mouse anti-Flag MAb (clone M2) were from Merck Millipore and Sigma-Aldrich, respectively. APC-conjugated goat anti-hamster IgG was purchased from the Jackson Laboratory.
CellTracker Orange {CMRA; 9′-(4[and 5]-chloromethyl-2-carboxyphenyl)-7′-chloro-6′-oxo-1,2,2,4-tetramethyl-1,2-dihydropyrido[2′,3′-6]xanthene} and pHrodo Red succinimidyl ester (pHrodo) were purchased from Life Technology. Alexa Fluor 488-conjugated streptavidin was from Molecular Probes. Human protein S was purchased from Enzyme Research Laboratories.
Transformation of Ba/F3 cells.
Mouse retroviral vectors pMXspuro (22) and pENV-IRES-puro (23) were provided by T. Kitamura (Institute of Medical Science, University of Tokyo). Lentiviral expression vectors (CSII-EF, pCAG-HIVgp, pENV-IRES-puro, and pRSV-Rev) were from H. Miyoshi, Riken Resource Center. pMXs-Tim4 was previously described (10). Mouse MerTK cDNA (GenBank accession number NM_008587.1) was prepared by reverse transcription-PCR (RT-PCR) with mRNA from kidney, Flag tagged at the C terminus, and inserted into the CSII-EF vector.
Mouse MerTK was expressed in Ba/F3 cells by using a lentiviral vector system. In brief, human HEK293T cells were cotransfected with CSII-EF vector expressing Flag-tagged MerTK cDNA, pCAG-HIVgp, pRSV-Rev, and pENV-internal ribosome entry site (IRES)-puro. After culturing for 48 h, viruses in the cell supernatant were concentrated by centrifugation at 6,000 × g for 16 h at 4°C and used to spin-infect Ba/F3 cells as described previously (7). Transformants expressing MerTK were sorted with a FACSAria II instrument. To establish Ba/F3 transformants expressing Tim4, the Tim4 cDNA was placed downstream of the human EF-1α promoter of pNEF-BOS-EX, which carries a simian virus 40 (SV40) promoter-driven neomycin resistance gene in pEF-BOS-EX (24). The construct was then introduced into Ba/F3 cells by electroporation using a Super Electroporator NEPA21 type II system (Nepa Gene Co.), and the cells were cultured for 3 days. The Tim4-expressing cells were sorted with FACSAria II and were cultured in the presence of 800 μg/ml Geneticin (Gibco) at 0.3 cells/well in 96-well microtiter plates. Clones expressing high levels of Tim4 were expanded for further analysis.
Flow cytometry.
Cells were incubated on ice for 30 min with 1 μg/ml hamster anti-mouse Tim4 (clone Kat5-18) (10) and 1 μg/ml biotinylated anti-MerTK Ab in a mixture containing 50 μl of PBS and 2% FCS, followed by incubation with 1.0 μg/ml Alexa Fluor 488-conjugated streptavidin, 1 μg/ml PerCP-Cy 5.5-labeled rat anti-mouse Mac1, and 0.6 μg/ml APC-labeled anti-hamster IgG. The cells were then stained with 0.5 μM Sytox Blue (Life Technologies) to exclude dead cells and analyzed by flow cytometry with a FACSCanto II instrument (BD Biosciences).
Engulfment of apoptotic cells.
Engulfment of apoptotic cells was assayed with pHrodo-labeled prey (20, 25). In brief, thymocytes from 4- to 8-week-old C57BL/6J mice were treated with 100 units/ml FasL in Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS for 1.5 to 2 h at 37°C to induce apoptosis, washed with PBS, and incubated with 0.1 μg/ml pHrodo for 30 min at room temperature. After the reaction was stopped with 1 ml FCS, the cells were washed with PBS containing 10% FCS and were used as prey. At this stage, the annexin V+ propidium iodide-positive (PI+) cell population was usually less than 30%. In some cases, thymocytes were incubated with FasL in serum-free DMEM, labeled with pHrodo as described above, and washed with PBS containing 0.5% bovine serum albumin (BSA) and 0.25% globulin.
To prepare peritoneal macrophages, peritoneal cells (5 × 105) from wild-type and mutant mice at 8 to 14 weeks of age were incubated in 12-well plates at 37°C for 2 h in DMEM containing 10% FCS and were washed with PBS to remove nonadherent cells. The adherent cells were incubated at 37°C with 2 × 106 pHrodo-labeled apoptotic thymocytes in 1 ml of DMEM containing 10% FCS, washed with PBS, and then treated at 37°C with 0.25% trypsin–PBS containing 1 mM EDTA. Cells were collected by centrifugation at 500 × g for 5 min, suspended in 300 to 500 μl of CHES (N-cyclohexyl-2-aminoethanesulfonic acid)–fluorescence-activated cell sorter (FACS) buffer (20 mM CHES buffer [pH 9.0] containing 150 mM NaCl and 2% FCS) supplemented with 0.2 to 0.4 μg/ml APC-labeled rat anti-mouse Mac1, and analyzed by flow cytometry with a FACSCanto II instrument.
For the Ba/F3 engulfment of apoptotic cells (20), 1 × 105 Ba/F3 cells and 1 × 106 pHrodo-labeled apoptotic thymocytes were coincubated at 37°C in 0.6 ml of RPMI 1640 containing 10% FCS and 45 U/ml mouse IL-3. The cells were collected by centrifugation at 500 × g for 5 min, suspended in 500 μl of CHES-FACS buffer, and analyzed by flow cytometry as described above. For microscopic observation, Ba/F3 cells coincubated with pHrodo-labeled thymocytes were suspended in 500 μl of CHES-FACS buffer, transferred to Lab-Tek II chambered cover glasses (ThermoFisher Scientific), and examined by fluorescence microscopy (BioRevo BZ-9000; Keyence).
Binding of apoptotic cells to phagocytes.
The binding of apoptotic cells to phagocytes was assayed by using the CellTracker Orange-labeled cells as described previously (20). In brief, approximately 1 × 108 thymocytes were labeled by incubation in 5 ml of serum-free DMEM containing 10 μM CellTracker Orange at 37°C for 30 min and then incubated with FasL in DMEM containing 10% FCS at 37°C for 2 h to induce apoptosis. Peritoneal cells or Ba/F3 cells (1 × 105) were then coincubated in suspension with the CellTracker Orange-labeled apoptotic cells in PBS supplemented with 10% FCS, stained with 500 nM Sytox Blue, and analyzed by FACSCanto II. For peritoneal macrophages, the cells were stained with APC-conjugated anti-Mac1.
Immunoprecipitation and Western blotting.
Resident peritoneal cells (3 × 106 to 6 × 106 cells) on 3.5-cm-diameter plates were incubated at 37°C with 1.5 × 107 to 3 × 107 apoptotic thymocytes in 1 ml of DMEM containing 10% FCS, washed with cold PBS to remove apoptotic cells, and lysed by incubation at 4°C for 30 min in 1.5 ml of lysis buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EGTA, 1% Triton X-100, 5% glycerol, 2 mM Na2VO4, and a cocktail of protease inhibitors [cOmplete, Mini, EDTA free; Roche]). The lysates were centrifuged at 15,000 rpm for 10 min at 4°C, and the supernatants were used for immunoprecipitation.
Dynabeads protein G (10 μl) (Life Technologies) was conjugated with 2.5 μg of goat anti-mouse MerTK Ab according to the supplier's instruction. The macrophage cell lysates prepared from 6 × 106 cells were incubated overnight at 4°C with anti-MerTK-conjugated protein G beads in 1.5 ml lysis buffer. The beads were collected, washed with lysis buffer, and suspended in 30 μl of SDS-sample buffer (63 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 0.005% bromophenol blue [BPB], 2% β-mercaptoethanol). Proteins were eluted from the beads by heating the samples at 95°C for 5 min, and 15-μl aliquots were separated by electrophoresis on 7.5% polyacrylamide gels. After transferring the proteins to polyvinylidene difluoride (PVDF) membranes, the membranes were incubated at room temperature for 1 h in TBS-T (25 mM Tris-HCl [pH 7.5], 137 mM NaCl, 2.7 mM KCl, and 0.1% Tween 20) containing 5% BSA or skim milk to block nonspecific binding sites. The membranes were then incubated at 4°C overnight with 1,000-fold-diluted horseradish peroxidase (HRP)-conjugated antiphosphotyrosine MAb (4G10) or 0.5 μg/ml biotinylated anti-MerTK Ab in TBS-T containing 5% BSA or skim milk, followed by incubation with 0.71 μg/ml HRP-conjugated streptavidin. Proteins recognized by the antibody were visualized by a chemiluminescence reaction (Renaissance; NEN Life Science Products).
RESULTS
Tim4 and MerTK expression in resident peritoneal macrophages.
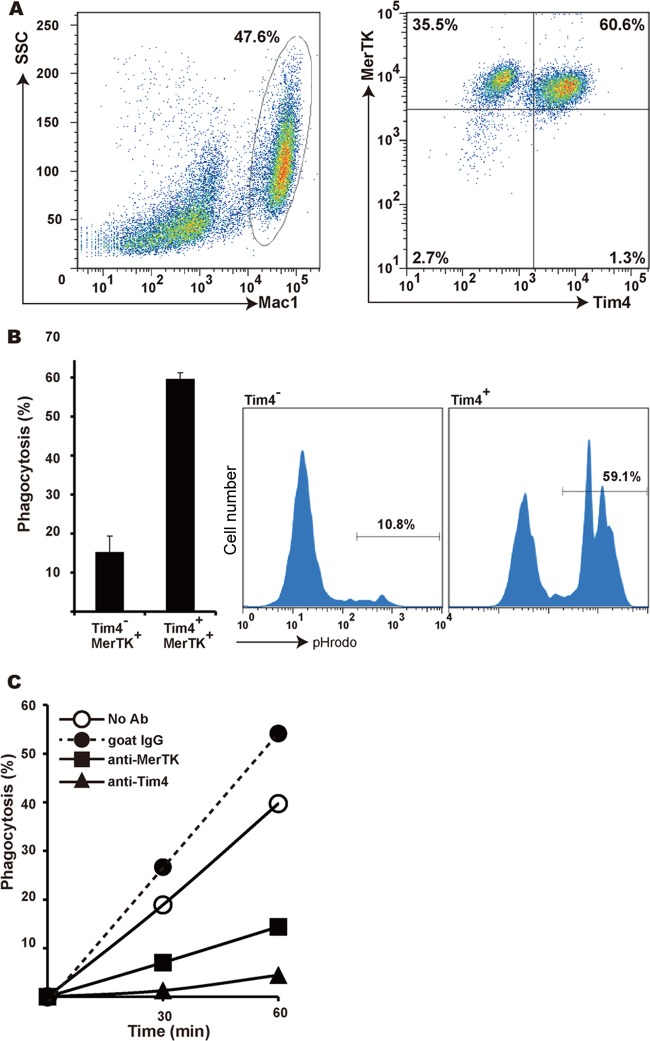
We previously reported that mouse rpMacs express Tim4 and that Tim4 is required for these macrophages to engulf apoptotic cells (19). In addition, Seitz et al. (26) showed that peritoneal macrophages require MerTK to engulf apoptotic cells. To examine whether the same rpMac population expresses Tim4 and MerTK, peritoneal cells were stained for Mac1, Tim4, and MerTK. Approximately 40% to 48% of the cells in the peritoneal cavity strongly expressed Mac1 and F4/80; among those cells, 36% were Tim4− MerTK+ and 61% were Tim4+ MerTK+, indicating that rpMacs consist of two populations, Tim4 and MerTK doubly positive cells and MerTK singly positive cells (Fig. 1A; see also Fig. S1 in the supplemental material). The ability of each macrophage population to engulf apoptotic cells was then examined. Mouse thymocytes were treated with FasL to induce approximately 70% of the cells to undergo apoptosis (as determined by annexin V staining) and were then labeled with pHrodo and used as prey for rpMacs. The majority of the pHrodo-positive macrophages, or the macrophages engulfing apoptotic cells, were Tim4+ MerTK+ cells, and very few Tim4− MerTK+ macrophages were seen (Fig. 1B). Neutralizing monoclonal antibodies against either Tim4 or MerTK strongly inhibited the uptake of apoptotic cells by the rpMacs (Fig. 1C). Taken together, these results suggest that of the two distinct rpMac populations, Tim4+ MerTK+ macrophages represent the population that engulfs apoptotic cells. These findings further implicate the involvement of both Tim4 and MerTK in the process.
FIG 1.
Tim4 and MerTK in mouse resident peritoneal macrophages. (A) Expression of Tim4 and MerTK in mouse resident peritoneal macrophages (rpMacs). Peritoneal cells from C57BL/6J mice were stained with PerCP-Cy5.5-conjugated anti-mouse Mac1 MAb, biotinylated goat anti-mouse MerTK Ab, and hamster anti-Tim4 MAb, followed by staining with Alexa Fluor 488-conjugated streptavidin and APC-conjugated anti-hamster IgG. The samples were then analyzed by flow cytometry. The staining profiles for Mac1 and for Tim4 and MerTK in Mac1-positive cells are shown. The Mac1+ cells represented in the left panel are analyzed in the right. Numbers indicate the percentages of Mac1+ cells in the left panel and of Tim4− MerTK+ and Tim4+ MerTK+ cells in the right panel. SSC, side scatter. (B) Engulfment of apoptotic cells by rpMacs. Resident peritoneal macrophages (5 × 105) were incubated with 2 × 106 pHrodo-labeled apoptotic thymocytes at 37°C for 60 min and stained with PerCP-Cy5.5-conjugated anti-mouse Mac1 MAb and with hamster anti-Tim4 MAb, followed by staining with APC-conjugated anti-hamster IgG, and analyzed by flow cytometry. Representative FACS profiles for the pHrodo-positive cells in Tim4+ MerTK+ and Tim4− MerTK+ populations in the Mac1+ cells are shown. The numbers indicate the percentage (phagocytosis) of the pHrodo-positive cells in each population. The experiments were performed in triplicate, and the average values with standard deviations (SD) (bars) are plotted. (C) Effect of Tim4 and MerTK neutralizing antibodies on rpMac engulfment of apoptotic cells. Resident peritoneal cells (5 × 105) were incubated with pHrodo-labeled apoptotic thymocytes (2 × 106) in the presence or absence of 10 μg/ml goat normal IgG, hamster anti-mouse Tim4 MAb, or goat anti-mouse MerTK Ab at 37°C for the indicated time. The percentage of pHrodo-positive macrophages was determined by flow cytometry and plotted.
Requirement of Tim4 and MerTK for the rpMac engulfment of apoptotic thymocytes.
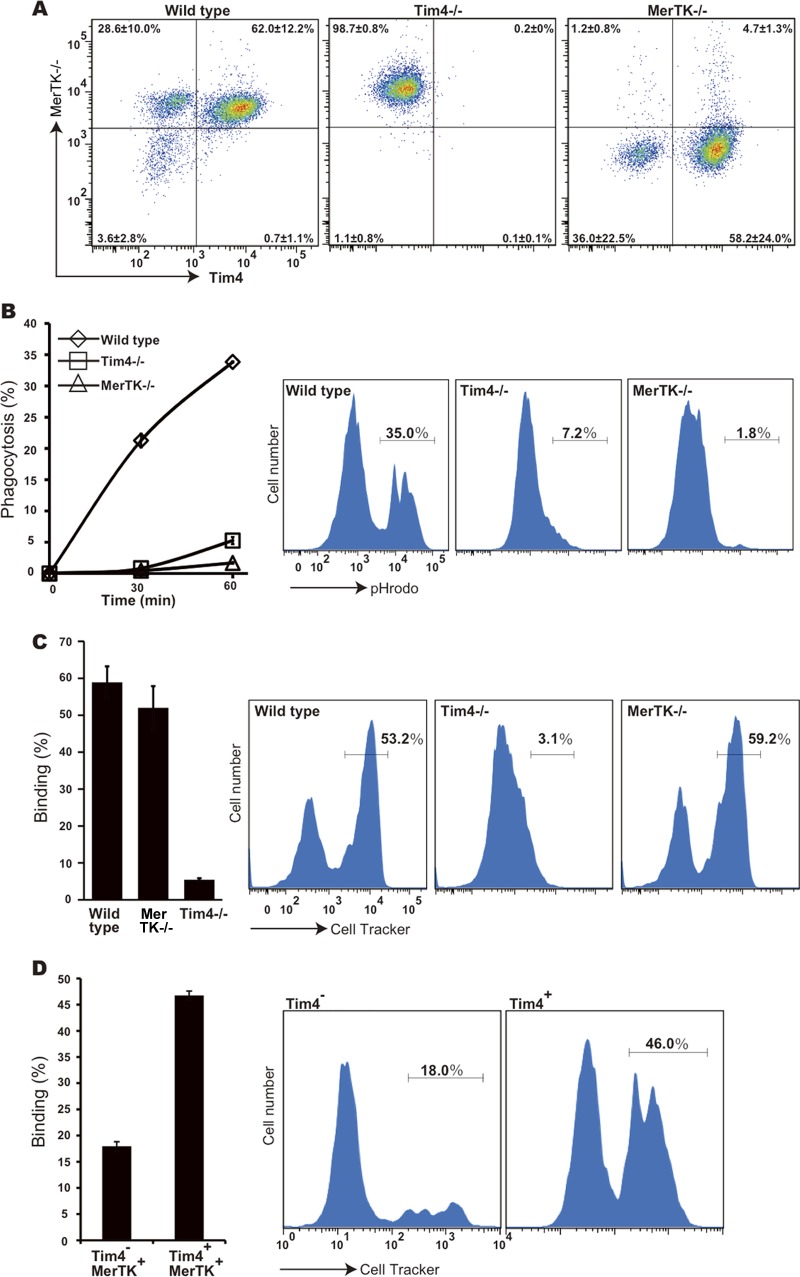
To confirm the requirement of Tim4 and MerTK for the rpMac engulfment of apoptotic cells, macrophages were prepared from Tim4−/− or MerTK−/− mice. In the Tim4−/− mice, the Neo gene replaces part of exons 1 and 2, encoding Tim4's extracellular region (19). In these mice, all the Mac1+ rpMacs were found to express MerTK but not Tim4 (Fig. 2A). MerTK−/− mice were previously established by deleting part of the MerTK cytoplasmic region (18), and yet its extracellular region was not detected in MerTK−/− macrophages as reported earlier (27), most likely due to the nonsense-mediated mRNA decay. As expected, the peritoneal cells of MerTK−/− mice were divided into two populations, Tim4+ Mac1+ cells and Tim4− Mac1+ cells. When rpMacs from either Tim4−/− or MerTK−/− mice were tested for their ability to engulf apoptotic cells, their engulfment activities were found to be severely reduced (Fig. 2B; see also Fig. S4 in the supplemental material), confirming that neither Tim4 nor MerTK alone is sufficient for apoptotic cell engulfment by these macrophages.
FIG 2.
Requirement of MerTK and Tim4 for apoptotic cell engulfment by rpMacs. (A) Expression of Tim4 and MerTK in wild-type and knockout rpMacs. Cells in the peritoneal cavity of wild-type, Tim4−/−, or MerTK−/− mice were incubated with PerCP-Cy5.5-conjugated anti-mouse Mac1 MAb, biotinylated goat anti-MerTK Ab, and hamster anti-Tim4 MAb, followed by staining with Alexa Fluor 488-conjugated streptavidin and APC-conjugated anti-hamster IgG. The stained cells were then analyzed by flow cytometry. The expression profiles of MerTK and Tim4 in the Mac1-positive population are shown. The experiments were carried out independently with 6 mice, and average percentages for MerTK− Tim4−, MerTK+ Tim4−, MerTK− Tim4+, and MerTK+ Tim4+ cell populations are indicated with SD. (B) Engulfment of apoptotic cells by rpMacs. Cells in the peritoneal cavity of wild-type, Tim4−/−, or MerTK−/− mice were incubated with pHrodo-labeled apoptotic thymocytes at 37°C for 30 or 60 min and analyzed by flow cytometry. The percentage of pHrodo-positive cells in the Mac1-positive population was determined by flow cytometry. The experiments were performed three times, and the average values are plotted with the SD (bars). Representative FACS profiles of pHrodo-positive cells in Mac1+ cells, obtained by incubation for 60 min, are shown at the right. (C) Binding of apoptotic thymocytes to rpMacs. CellTracker-labeled apoptotic thymocytes were incubated with peritoneal cells from wild-type, Tim4−/−, or MerTK−/− mice at 37°C for 30 min and analyzed by flow cytometry. The numbers indicate the percentages of CellTracker-positive cells. The experiments were done in triplicate, and the average values with the SD (bars) are plotted. At the right, representative CellTracker-staining profiles of the Mac1-positive cells are shown. (D) Binding of apoptotic cells by Tim4+ MerTK+ rpMacs. Resident peritoneal cells (1 × 105) were incubated at 37°C for 30 min with CellTracker-labeled apoptotic thymocytes (1 × 106) and stained with PerCP-Cy5.5-conjugated anti-mouse Mac1 MAb and hamster anti-Tim4 MAb, followed by staining with APC-conjugated anti-hamster IgG, and analyzed by flow cytometry. The numbers indicate the percentages of the CellTracker-positive cells in each population. The experiments were performed in triplicate, and the average values with SD (bars) are plotted. Representative FACS profiles for CellTracker-positive cells for each population are shown at the right.
Tim4 is a PtdSer receptor that tethers apoptotic cells (10), while MerTK is a receptor for the bridging proteins, Gas6 and protein S, that bind PtdSer (9, 28). Since protein S is abundant (about 300 nM or 25 μg/ml) in the serum (29) used for the engulfment assay, the macrophages expressing MerTK can bind apoptotic cells via protein S. To examine whether the MerTK−/− and Tim4−/− macrophages bind apoptotic cells, the macrophages were incubated with FasL-treated mouse thymocytes that were labeled with CellTracker Orange. As shown in Fig. 2C, approximately 50% to 60% of the Mac1+ rpMacs from wild-type and MerTK−/− mice were associated with CellTracker-labeled thymocytes, while there was minimal association of Tim4−/− macrophages with labeled apoptotic cells. Similar results were obtained under serum-free conditions (see Fig. S3 in the supplemental material), suggesting that Tim4 is required to tether apoptotic cells to rpMacs and that the MerTK-protein S interaction is not sufficient for recruiting apoptotic cells. In fact, of the two populations of wild-type rpMacs (Tim4+ MerTK+ and Tim4− MerTK+), the Tim4-positive population was found to strongly associate with the CellTracker-labeled cells (Fig. 2D).
Tim4-dependent tyrosine phosphorylation of MerTK.
MerTK is a type I membrane protein with a cytoplasmic tyrosine kinase domain (9). Todt (30) previously reported that when mouse rpMacs are exposed to apoptotic cells, MerTK is phosphorylated at tyrosine residues. We ascertained that if Tim4 were involved in the binding of apoptotic cells, then Tim4 expression might affect the tyrosine phosphorylation of MerTK. To examine this possibility, rpMacs were incubated with apoptotic thymocytes in the presence or absence of an anti-Tim4 neutralizing antibody. MerTK was immunoprecipitated with anti-MerTK antibodies and subjected to Western blot analysis with an antiphosphotyrosine MAb. As shown in Fig. 3A, rpMac incubation with apoptotic cells resulted in strong phosphorylation of MerTK, which was significantly inhibited by the presence of the anti-Tim4 Ab.
FIG 3.
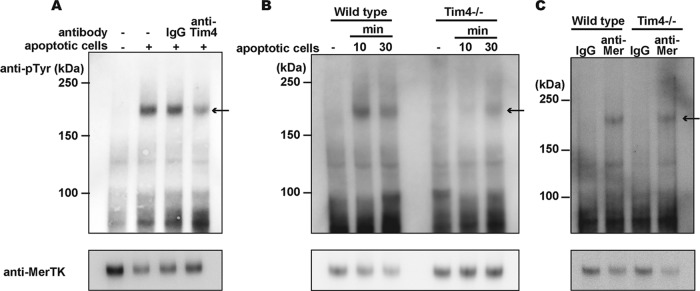
Involvement of Tim4 in apoptotic cell-induced tyrosine phosphorylation of MerTK. (A) Effect of the anti-Tim4 neutralizing antibody on the apoptotic cell-induced phosphorylation of MerTK. Resident peritoneal cells (6 × 106) were incubated for 30 min with 3 × 107 apoptotic thymocytes in the absence or presence of 10 μg/ml hamster control IgG or of an anti-Tim4 MAb. The rpMac cell lysates were subjected to immunoprecipitation with an anti-MerTK Ab, separated by SDS-PAGE, and analyzed by Western blotting using an antiphosphotyrosine MAb (4G10) (upper panel) or anti-MerTK Ab (lower panel). (B) Requirement of Tim4 expression for the apoptotic cell-induced phosphorylation of MerTK. Resident peritoneal cells (6 × 106) from wild-type or Tim4−/− mice were incubated with 3 × 107 apoptotic thymocytes at 37°C for 10 or 30 min. The cell lysates were immunoprecipitated with an anti-MerTK Ab and analyzed by Western blotting with an antiphosphotyrosine MAb (upper panel) or anti-MerTK Ab (lower panel). (C) Cross-linking-induced MerTK phosphorylation. Resident peritoneal cells (3 × 106) from wild-type or Tim4−/− mice were incubated with 30 μg/ml goat anti-MerTK Ab or goat control IgG at 37°C for 30 min, washed with PBS, and incubated with 30 μg/ml anti-goat IgG at 37°C for 10 min. The cell lysates were immunoprecipitated with anti-MerTK Ab and analyzed by Western blotting with antiphophotyrosine MAb (upper panel) or anti-MerTK Ab (lower panel).
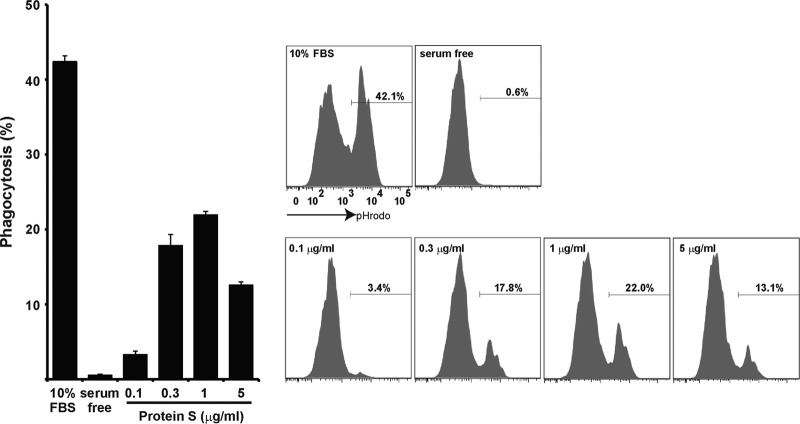
To further confirm the role of Tim4 in activating MerTK, the wild-type and Tim4−/− rpMacs were incubated with apoptotic cells. MerTK in the wild-type macrophages was strongly tyrosine phosphorylated after 10 min of incubation with apoptotic cells (Fig. 3B). This phosphorylation was transient and had decreased after 30 min of incubation. The Tim4−/− rpMacs did not exhibit clear tyrosine phosphorylation of MerTK after incubation with apoptotic cells for 10 min, and only weak phosphorylation was detected at 30 min. The tyrosine kinase activity of MerTK can also be activated by cross-linking MerTK with anti-MerTK Ab (30). Accordingly, when peritoneal macrophages were treated with goat anti-mouse MerTK, followed by rabbit anti-goat antibodies, MerTK was tyrosine phosphorylated. This cross-linking-induced tyrosine phosphorylation of MerTK was not affected by the Tim4-null mutation (Fig. 3C), suggesting that MerTK is functional without Tim4, and Tim4 indirectly affects the activation of MerTK during engulfment of apoptotic cells. Gas6 or protein S or both are known to bridge apoptotic cells to receptors on macrophages (28, 31). If Tim4 was sufficient for directly tethering apoptotic cells to rpMacs, bridging molecules might not be necessary for the MerTK-mediated engulfment of apoptotic cells. However, the engulfment of apoptotic cells by mouse rpMacs required FCS (Fig. 4). Furthermore, protein S promoted the engulfment of apoptotic cells in serum-free medium, resulting in a bell-shaped response curve, with the maximum response at 1.0 μg/ml. Since FCS contains approximately 25 μg/ml protein S (29), the serum requirement for the engulfment of apoptotic cells may be at least partially due to protein S present in the serum.
FIG 4.
Requirement of protein S for the engulfment of apoptotic cells by rpMacs. Resident peritoneal cells (5 × 105) were incubated with 2 × 106 pHrodo-labeled apoptotic thymocytes in 10% FCS or in serum-free DMEM containing the indicated concentration of protein S at 37°C for 60 min and were then subjected to flow cytometry. The pHrodo-staining profiles of the Mac1-positive population are shown at the right with numbers indicating the percentages of pHrodo-positive macrophages. The experiments were performed three times, and the average percentages of the pHrodo-positive macrophages are plotted with SD (bars).
Reconstitution of apoptotic cell engulfment with the mouse Ba/F3 pro-B cell line.
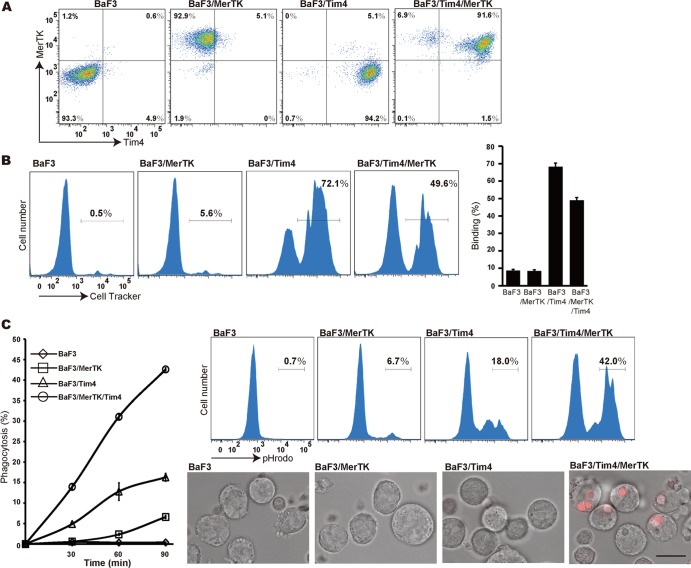
The above results indicated that both Tim4 and MerTK are indispensable for the engulfment of apoptotic cells by mouse rpMacs. We recently developed an engulfment system, using the mouse B cell line, Ba/F3, that grew in suspension, and showed that engulfment proceeds via two distinct steps, tethering and engulfment (20). To study the roles of Tim4 and MerTK, Ba/F3 cells, which endogenously expressed neither Tim4 nor MerTK, were transformed with mouse Tim4, mouse MerTK, or both (Fig. 5A). As shown in Fig. 5B, the CellTracker-labeled apoptotic cells bound strongly to Tim4-expressing Ba/F3 (BaF3-Tim4 and BaF3-Tim4/MerTK) but not to the parental Ba/F3 or the MerTK transformants, in agreement with the results obtained with rpMacs. When the engulfment of apoptotic cells was assayed with pHrodo-labeled thymocytes, both the MerTK and Tim4 transformants exhibited limited engulfment activity. In contrast, Ba/F3 transformants that coexpressed Tim4 and MerTK exhibited strongly enhanced engulfment activity (Fig. 5C). Microscopic observation of the Ba/F3 cells incubated with apoptotic cells indicated that Tim4-expressing cells rarely contained a pHrodo-positive cell after a 90-min incubation with apoptotic cells whereas approximately half of the Ba/F3 transformants expressing both Tim4 and MerTK contained at least one pHrodo-positive cell, confirming that Tim4 and MerTK cooperate to engulf apoptotic cells.
FIG 5.
Reconstitution of Tim4- and MerTK-mediated phagocytosis with mouse Ba/F3 cells. (A) Expression of Tim4 and MerTK in Ba/F3 transformants. Ba/F3 stable transformants expressing MerTK, Tim4, or both were stained with a biotinylated anti-MerTK Ab and hamster anti-Tim4 MAb, followed by Alexa Fluor 488-conjugated streptavidin and APC-conjugated anti-hamster IgG. The cells were analyzed by flow cytometry. (B) Binding of apoptotic thymocytes to Ba/F3 transformants. Ba/F3 transformants (1 × 105 cells) expressing Tim4, MerTK, or Tim4 and MerTK were incubated with CellTracker Orange-labeled apoptotic thymocytes (1 × 106 cells) at 37°C for 30 min and were subjected to flow cytometry. Numbers indicate the percentages of the CellTracker-positive cells. Experiments were performed in triplicate, and the average values are plotted with SD (bars) in the right panel. (C) Engulfment of apoptotic cells by Ba/F3 transformants. The Ba/F3 transformants expressing Tim4, MerTK, or both were incubated at 37°C for the indicated times with pHrodo-labeled apoptotic thymocytes and were analyzed by flow cytometry. The numbers indicate the percentages of pHrodo-positive cells. The experiments were performed in triplicate, and the average values are plotted with SD (bars). A representative FACS profile obtained with Ba/F3 cells that were incubated for 90 min with apoptotic cells is shown in the right panel. Ba/F3 cells were incubated with pHrodo-labeled apoptotic thymocytes at 37°C for 90 min, transferred to Lab-Tek chambered cover glasses, and examined by fluorescence microscopy. Bright-field and pHrodo (red) merged images are shown. Scale bar, 15 μm.
DISCUSSION
Macrophages and immature dendritic cells that efficiently engulf apoptotic cells by recognizing PtdSer exposed on the apoptotic cells are highly heterogeneous (32, 33). Many soluble and transmembrane proteins have been shown to mediate the PtdSer-dependent engulfment of apoptotic cells (34); however, the mechanism(s) by which these molecules function in different macrophage populations has not been thoroughly investigated. Here we found that the efficient engulfment of apoptotic cells by mouse rpMacs requires the expression of Tim4 and MerTK. Tim4 is a type I membrane protein that functions as a receptor for apoptotic cells by directly binding to the PtdSer on apoptotic cells (10). Accordingly, apoptotic cells strongly bound to Tim4-expressing Ba/F3 cells and wild-type rpMacs. Tim4 deficiency completely blocked the binding of apoptotic cells to the rpMacs, but the null mutation of MerTK had no effect on this process. These results agree with previous observations that peritoneal macrophages from MerTK−/− mice still bind apoptotic cells (35) and suggest that Tim4 functions to tether apoptotic cells to the phagocytic rpMac.
MerTK does not directly recognize apoptotic cells, and protein S and Gas6, ligands for MerTK that bind PtdSer, are proposed to serve as bridges between apoptotic cells and macrophages expressing MerTK (9). Yet MerTK-expressing Ba/F3 cells or Tim4−/− peritoneal macrophages that express MerTK did not bind apoptotic cells in the presence of FCS, which contains protein S. The affinity of protein S for PtdSer (Kd [dissociation constant] of approximately 28 nM) (36) is 1/10 that of Tim4 for PtdSer (Kd of 2 nM), and its affinity for MerTK is also quite low (37), which may explain the limited binding of apoptotic cells to MerTK-expressing cells in the absence of Tim4. An example of a well-characterized two-receptor system is the tumor necrosis factor (TNF) receptor system, in which signal-transducing TNF-R1 exhibits a low affinity for TNF-α, while non-signal-transducing TNF-R2 exhibits a high affinity for TNF-α. A ligand-passing model was proposed (38) in which TNF-α first binds to TNF-R2 and is then delivered to TNF-R1 to initiate signal transduction. We propose that, in similarity to the TNF receptor system, Tim4 in rpMacs recruits apoptotic cells to increase the local concentration of apoptotic cells at the cell surface. The recruited apoptotic cells are then passed to MerTK via protein S for engulfment. In migration of T cells in the endothelium system, T cells first bind to endothelial cells at their focal region where high-affinity lymphocyte function-associated antigen 1 (LFA-1) is localized. This causes clustering of intermediate-affinity LFA-1 at the leading edge, allowing this region to bind to endothelium cells for cell's rolling (39). Similarly, binding of apoptotic cells to macrophages via Tim4 may cause clustering of PtdSer, which allows the dead cells to bind to MerTK via protein S.
The TAM family kinases have been shown to physically interact with other receptors; Axl interacts with interferon receptor type I (IFN-RI) in dendritic cells to regulate the IFN response (40), and integrin-α5 interacts with MerTK to promote the engulfment of apoptotic cells in HEK293T cells (41). On the other hand, the immunoprecipitation of MerTK followed by Western blotting with an anti-Tim4 MAb showed no clear evidence of an association between MerTK and Tim4 (C. Nishi, K. Segawa, and S. Nagata, unpublished observation). However, the coincubation of peritoneal macrophages with apoptotic cells consistently reduced the MerTK protein level in a Tim4-dependent manner, indicating that apoptotic cells are internalized together with Tim4 and MerTK (Fig. 3). Additional studies will be required to understand the molecular mechanism underlying Tim4's enhancement of the MerTK-mediated cell engulfment.
Thioglycolate-elicited peritoneal macrophages (thio-pMacs) also express MerTK and require MerTK for the efficient engulfment of apoptotic cells (26, 42) (see Fig. S2 in the supplemental material). Interestingly, thio-pMacs do not express Tim4 or any other Tim family members (Tim-1 and Tim-3) that bind PtdSer (10). In contrast to MerTK−/− rpMacs, MerTK−/− thio-pMacs did not bind apoptotic cells (data not shown), suggesting that these macrophages may not require a MerTK-independent tethering step to engulf apoptotic cells. thio-pMacs express MFG-E8 (43, 44) and integrin-αvβ3 or integrin-αvβ5, both of which bind MFG-E8 and synergize with MerTK to enhance apoptotic cell engulfment (41). On the other hand, MFG-E8 did not enhance the apoptotic cell uptake by rpMacs, and the expression of integrin-αvβ3 in the Tim4- and MerTK-expressing Ba/F3 cells had no effect on their ability to engulf apoptotic cells (data not shown). These results suggest that the Tim4-MerTK and integrin-MerTK systems are independent and function differently in different cell types. It is possible that the synergistic effects of integrin-αvβ3 (or integrin-αvβ5) and MerTK mitigate the requirement of a separate tethering step in thio-pMacs.
Tim4 and MerTK are coexpressed not only in rpMacs but also in splenic tingible-body macrophages and thymic macrophages. Requirement of MerTK for the engulfment of apoptotic cells by tingible-body macrophages and thymic macrophages has been previously reported (35, 45). Whether Tim4 or other Tim family members are required for the engulfment of apoptotic cells in these and other macrophage populations remains to be determined. In this regard, the engulfment system using Ba/F3 cells may well be suitable to examine the contribution of the proposed molecules in engulfment of apoptotic cells. Finally, MerTK−/− mice develop an SLE-type autoimmune disease (46), and it was recently shown that the Tim4 and MFG-E8-null mutations synergistically affect the development of autoimmunity in mice (19). It will be interesting to determine whether the Tim4-null mutation enhances the autoimmunity that develops in MerTK−/− mice.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Fujii for secretarial assistance.
This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture in Japan. S.T. is a research fellow of the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print 10 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01394-13.
REFERENCES
- 1.Nagata S, Hanayama R, Kawane K. 2010. Autoimmunity and the clearance of dead cells. Cell 140:619–630. 10.1016/j.cell.2010.02.014 [DOI] [PubMed] [Google Scholar]
- 2.Ravichandran KS. 2011. Beginnings of a good apoptotic meal: the find-me and eat-me signaling pathways. Immunity 35:445–455. 10.1016/j.immuni.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muñoz LE, Lauber K, Schiller M, Manfredi AA, Herrmann M. 2010. The role of defective clearance of apoptotic cells in systemic autoimmunity. Nat. Rev. Rheumatol. 6:280–289. 10.1038/nrrheum.2010.46 [DOI] [PubMed] [Google Scholar]
- 4.von Figura K, Hasilik A. 1986. Lysosomal enzymes and their receptors. Annu. Rev. Biochem. 55:167–193 [DOI] [PubMed] [Google Scholar]
- 5.Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. 1992. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148:2207–2216 [PubMed] [Google Scholar]
- 6.Martin SJ, Finucane DM, Amarante-Mendes GP, O'Brien GA, Green DR. 1996. Phosphatidylserine externalization during CD95-induced apoptosis of cells and cytoplasts requires ICE/CED-3 protease activity. J. Biol. Chem. 271:28753–28756. 10.1074/jbc.271.46.28753 [DOI] [PubMed] [Google Scholar]
- 7.Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. 2013. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341:403–406. 10.1126/science.1236758 [DOI] [PubMed] [Google Scholar]
- 8.Qingxian L, Qiutang L, Qingjun L. 2010. Regulation of phagocytosis by TAM receptors and their ligands. Front Biol (Beijing) 5:227–237. 10.1007/s11515-010-0034-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemke G, Rothlin CV. 2008. Immunobiology of the TAM receptors. Nat. Rev. Immunol. 8:327–336. 10.1038/nri2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. 2007. Identification of Tim4 as a phosphatidylserine receptor. Nature 450:435–439. 10.1038/nature06307 [DOI] [PubMed] [Google Scholar]
- 11.Kim S, Park S-Y, Kim S-Y, Bae D-J, Pyo J-H, Hong M, Kim I-S. 2012. Cross talk between engulfment receptors stabilin-2 and integrin αvβ5 orchestrates engulfment of phosphatidylserine-exposed erythrocytes. Mol. Cell. Biol. 32:2698–2708. 10.1128/MCB.06743-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS. 2007. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450:430–434. 10.1038/nature06329 [DOI] [PubMed] [Google Scholar]
- 13.He M, Kubo H, Morimoto K, Fujino N, Suzuki T, Takahasi T, Yamada M, Yamaya M, Maekawa T, Yamamoto Y, Yamamoto H. 2011. Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells. EMBO Rep. 12:358–364. 10.1038/embor.2011.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Païdassi H, Tacnet-Delorme P, Garlatti V, Darnault C, Ghebrehiwet B, Gaboriaud C, Arlaud GJ, Frachet P. 2008. C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J. Immunol. 180:2329–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, Bratton DL, Oldenborg PA, Michalak M, Henson PM. 2005. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 123:321–334. 10.1016/j.cell.2005.08.032 [DOI] [PubMed] [Google Scholar]
- 16.Henson PM, Bratton DL, Fadok VA. 2001. Apoptotic cell removal. Curr. Biol. 11:R795–R805. 10.1016/S0960-9822(01)00474-2 [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann PR, deCathelineau AM, Ogden CA, Leverrier Y, Bratton DL, Daleke DL, Ridley AJ, Fadok VA, Henson PM. 2001. Phosphatidylserine (PS) induces PS receptor-mediated macropinocytosis and promotes clearance of apoptotic cells. J. Cell Biol. 155:649–659. 10.1083/jcb.200108080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Q, Gore M, Zhang Q, Camenisch T, Boast S, Casagranda F, Lai C, Skinner MK, Klein R, Matsushima GK, Earp HS, Goff SP, Lemke G. 1999. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398:723–728. 10.1038/19554 [DOI] [PubMed] [Google Scholar]
- 19.Miyanishi M, Segawa K, Nagata S. 2012. Synergistic effect of Tim4 and MFG-E8 null mutations on the development of autoimmunity. Int. Immunol. 24:551–559. 10.1093/intimm/dxs064 [DOI] [PubMed] [Google Scholar]
- 20.Toda S, Hanayama R, Nagata S. 2012. Two-step engulfment of apoptotic cells. Mol. Cell. Biol. 32:118–125. 10.1128/MCB.05993-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiraishi T, Suzuyama K, Okamoto H, Mineta T, Tabuchi K, Nakayama K, Shimizu Y, Tohma J, Ogihara T, Naba H, Mochizuki H, Nagata S. 2004. Increased cytotoxicity of soluble Fas ligand by fusing isoleucine zipper motif. Biochem. Biophys. Res. Commun. 322:197–202. 10.1016/j.bbrc.2004.07.098 [DOI] [PubMed] [Google Scholar]
- 22.Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. 2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31:1007–1014. 10.1016/S0301-472X(03)00260-1, [DOI] [PubMed] [Google Scholar]
- 23.Morita S, Kojima T, Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066. 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- 24.Murai K, Murakami H, Nagata S. 1998. Myeloid-specific transcriptional activation by murine myeloid zinc finger protein-2. Proc. Natl. Acad. Sci. U. S. A. 95:3461–3466. 10.1073/pnas.95.7.3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miksa M, Komura H, Wu R, Shah KG, Wang P. 2009. A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J. Immunol. Methods 342:71–77. 10.1016/j.jim.2008.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seitz HM, Camenisch TD, Lemke G, Earp HS, Matsushima GK. 2007. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. J. Immunol. 178:5635–5642 [DOI] [PubMed] [Google Scholar]
- 27.Behrens EM, Gadue P, Gong SY, Garrett S, Stein PL, Cohen PL. 2003. The mer receptor tyrosine kinase: expression and function suggest a role in innate immunity. Eur. J. Immunol. 33:2160–2167. 10.1002/eji.200324076 [DOI] [PubMed] [Google Scholar]
- 28.Rothlin CV, Lemke G. 2010. TAM receptor signaling and autoimmune disease. Curr. Opin. Immunol. 22:740–746. 10.1016/j.coi.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rezende SM, Simmonds RE, Lane DA. 2004. Coagulation, inflammation, and apoptosis: different roles for protein S and the protein S-C4b binding protein complex. Blood 103:1192–1201. 10.1182/blood-2003-05-1551 [DOI] [PubMed] [Google Scholar]
- 30.Todt JC. 2004. The receptor tyrosine kinase MerTK activates phospholipase C 2 during recognition of apoptotic thymocytes by murine macrophages. J. Leukoc. Biol. 75:705–713. 10.1189/jlb.0903439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson HA, Maylock CA, Williams JA, Paweletz CP, Shu H, Shacter E. 2003. Serum-derived protein S binds to phosphatidylserine and stimulates the phagocytosis of apoptotic cells. Nat. Immunol. 4:87–91. 10.1038/ni871 [DOI] [PubMed] [Google Scholar]
- 32.den Haan JMM, Martinez-Pomares L. 2013. Macrophage heterogeneity in lymphoid tissues. Semin. Immunopathol. 35:541–552. 10.1007/s00281-013-0378-4 [DOI] [PubMed] [Google Scholar]
- 33.Gordon S, Plűddemann A. 2013. Tissue macrophage heterogeneity: issues and prospects. Semin. Immunopathol. 35:533–540. 10.1007/s00281-013-0386-4 [DOI] [PubMed] [Google Scholar]
- 34.Ravichandran KS. 2010. Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. J. Exp. Med. 207:1807. 10.1084/jem.20101157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott RS, McMahon EJ, Pop SM, Reap EA, Caricchio R, Cohen PL, Earp HS, Matsushima GK. 2001. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 411:207–211. 10.1038/35075603 [DOI] [PubMed] [Google Scholar]
- 36.Baroni M, Pavani G, Marescotti D, Kaabache T, Borgel D, Gandrille S, Marchetti G, Legnani C, D'Angelo A, Pinotti M, Bernardi F. 2010. Membrane binding and anticoagulant properties of protein S natural variants. Thromb. Res. 125:e33–e39. 10.1016/j.thromres.2009.09.015 [DOI] [PubMed] [Google Scholar]
- 37.Nagata K, Ohashi K, Nakano T, Arita H, Zong C, Hanafusa H, Mizuno K. 1996. Identification of the product of growth arrest-specific gene 6 as a common ligand for Axl, Sky, and Mer receptor tyrosine kinases. J. Biol. Chem. 271:30022–30027. 10.1074/jbc.271.47.30022 [DOI] [PubMed] [Google Scholar]
- 38.Tartaglia LA, Pennica D, Goeddel DV. 1993. Ligand passing: the 75-kDa tumor necrosis factor (TNF) receptor recruits TNF for signaling by the 55-kDa TNF receptor. J. Biol. Chem. 268:18542–18548 [PubMed] [Google Scholar]
- 39.Smith A, Stanley P, Jones K, Svensson L, McDowall A, Hogg N. 2007. The role of the integrin LFA-1 in T-lymphocyte migration. Immunol. Rev. 218:135–146. 10.1111/j.1600-065X.2007.00537.x [DOI] [PubMed] [Google Scholar]
- 40.Rothlin C, Ghosh S, Zuniga E, Oldstone M, Lemke G. 2007. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 131:1124–1136. 10.1016/j.cell.2007.10.034 [DOI] [PubMed] [Google Scholar]
- 41.Wu Y, Singh S, Georgescu M-M, Birge RB. 2005. A role for Mer tyrosine kinase in alphavbeta5 integrin-mediated phagocytosis of apoptotic cells. J. Cell Sci. 118:539–553. 10.1242/jcs.01632 [DOI] [PubMed] [Google Scholar]
- 42.Gal A, Li Y, Thompson DA, Weir J, Orth U, Jacobson SG, Apfelstedt-Sylla E, Vollrath D. 2000. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat. Genet. 26:270–271. 10.1038/81555 [DOI] [PubMed] [Google Scholar]
- 43.Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. 2002. Identification of a factor that links apoptotic cells to phagocytes. Nature 417:182–187. 10.1038/417182a [DOI] [PubMed] [Google Scholar]
- 44.Nandrot EF, Anand M, Almeida D, Atabai K, Sheppard D, Finnemann SC. 2007. Essential role for MFG-E8 as ligand for alphavbeta5 integrin in diurnal retinal phagocytosis. Proc. Natl. Acad. Sci. U. S. A. 104:12005–12010. 10.1073/pnas.0704756104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahman ZSM, Shao W-H, Khan TN, Zhen Y, Cohen PL. 2010. Impaired apoptotic cell clearance in the germinal center by Mer-deficient tingible body macrophages leads to enhanced antibody-forming cell and germinal center responses. J. Immunol. 185:5859–5868. 10.4049/jimmunol.1001187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen PL, Caricchio R, Abraham V, Camenisch TD, Jennette JC, Roubey RAS, Earp HS, Matsushima G, Reap EA. 2002. Delayed apoptotic cell clearance and lupus-like autoimmunity in mice lacking the c-mer membrane tyrosine kinase. J. Exp. Med. 196:135–140. 10.1084/jem.20012094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.