Abstract
Resistance to thyroid hormone (RTH), a human syndrome, is characterized by high thyroid hormone (TH) and thyroid-stimulating hormone (TSH) levels. Mice with mutations in the thyroid hormone receptor beta (TRβ) gene that cannot bind steroid receptor coactivator 1 (SRC-1) and Src-1−/− mice both have phenotypes similar to that of RTH. Conversely, mice expressing a mutant nuclear corepressor 1 (Ncor1) allele that cannot interact with TRβ, termed NCoRΔID, have low TH levels and normal TSH. We hypothesized that Src-1−/− mice have RTH due to unopposed corepressor action. To test this, we crossed NCoRΔID and Src-1−/− mice to create mice deficient for coregulator action in all cell types. Remarkably, NCoRΔID/ΔID Src-1−/− mice have normal TH and TSH levels and are triiodothryonine (T3) sensitive at the level of the pituitary. Although absence of SRC-1 prevented T3 activation of key hepatic gene targets, NCoRΔID/ΔID Src-1−/− mice reacquired hepatic T3 sensitivity. Using in vivo chromatin immunoprecipitation assays (ChIP) for the related coactivator SRC-2, we found enhanced SRC-2 recruitment to TR-binding regions of genes in NCoRΔID/ΔID Src-1−/− mice, suggesting that SRC-2 is responsible for T3 sensitivity in the absence of NCoR1 and SRC-1. Thus, T3 targets require a critical balance between NCoR1 and SRC-1. Furthermore, replacement of NCoR1 with NCoRΔID corrects RTH in Src-1−/− mice through increased SRC-2 recruitment to T3 target genes.
INTRODUCTION
The maintenance of thyroid hormone (TH) levels is essential for development, metabolism, energy expenditure, and thermoregulation. TH levels are sustained by a negative-feedback loop within the hypothalamic-pituitary-thyroid (HPT) axis that operates to keep TH levels, both the predominant form, thyroxine (T4), and the active form, triiodothryonine (T3), within a precise range (1). Thyrotropin-releasing hormone (TRH) from the paraventricular nucleus of the hypothalamus (PVH) stimulates the release of thyroid-stimulating hormone (TSH) from the pituitary. TSH acts on the thyroid to synthesize and release TH, which then signals back to both the PVH and pituitary to suppress TRH and TSH levels, respectively (2). At the molecular level, T3 mediates its actions through its nuclear receptor isoforms, thyroid hormone receptor alpha (TRα) and TRβ (encoded by Thra and Thrb), which function as ligand-dependent transcription factors to either increase or decrease expression of target genes (3–5). Classically, on positively regulated TH targets, the presence of T3 allows the binding of coactivators, such as the steroid receptor coactivator family (SRC-1, -2, and -3) (6–8). These coactivators then recruit machinery to allow the activation of gene expression (9–12). In the absence of ligand, corepressors, such as nuclear corepressor 1 (NCoR1) or silencing mediator of retinoic acid (SMRT, or NCoR2), bind and recruit complexes to repress transcription (13–18). The processes by which negative TH targets are repressed or transcribed are not well understood, but active T3 repression does require SRC-1 (19–22).
In previous reports, our laboratory has demonstrated that NCoR1 also mediates T3 sensitivity and is critical in maintaining normal TH levels (23–25). NCoR1 binds to TR through two of its three nuclear-receptor-interacting domains, N2 and N3 (26–32). Mice globally expressing a conditional Ncor1 allele lacking N2 and N3, termed NCoRΔID, either during embryogenesis or postnatally, have decreased levels of T4 and T3 and normal TSH levels (24, 25). In addition to altering the set point of the HPT axis, NCoRΔID increases the sensitivity of peripheral tissues, such as the liver, to T3. This enhanced sensitivity leads to the increased activation of target genes in the presence of similar amounts of T3 (24). The role of NCoR1 in determining ligand sensitivity in the context of nuclear receptor signaling has been demonstrated by other groups using a mouse model with disrupted interaction between NCoR1 and histone deacetylase 3 (HDAC3) (33, 34).
Recently, it was hypothesized that the dominantly inherited human disorder resistance to thyroid hormone (RTH) that is caused by mutations in Thrb is due to an imbalance of corepressor recruitment in vivo (35, 36). Indeed, many of the Thrb mutations that cause RTH are characterized by impaired ability to bind T3 and therefore increased recruitment of corepressors (37). Therefore, we crossed the ThrbPV mouse, characterized by a knock-in frameshift mutation in the ligand-binding domain of the Thrb gene that prevents ThrbPV from binding T3, with NCoRΔID/ΔID mice (38). The ThrbPV model faithfully recapitulates RTH syndrome, including high TH and TSH levels (38, 39). Notably, NCoRΔID/ΔID ThrbPV mice had lower TH and TSH levels than their ThrbPV littermates, confirming that aberrant corepressor recruitment plays an important role in the development of RTH in vivo (40).
While RTH in humans is secondary to Thrb mutations in 85% of cases, two mouse models of RTH involve the coactivator SRC-1 (41). Mice null for SRC-1 (Src-1−/− mice) and mice with a mutated Thrb unable to interact with SRC-1 have high TH levels with inappropriately high TSH secretion (20, 21, 42). Thus, concurrent with the effects of NCoRΔID on ThrbPV mice, we hypothesized that both SRC-1 mouse models have RTH due to the unopposed recruitment of corepressors. We proposed that crossing NCoRΔID/ΔID mice to Src-1−/− mice would correct the high TH and TSH levels. Indeed, we show here that Src-1−/− mice expressing the NCoRΔID allele have normal TH and TSH levels. Furthermore, the pituitary, which is resistant to T3 in Src-1−/− mice, responds normally to T3 in NCoRΔID/ΔID Src-1−/− mice. Additionally, hepatic T3 targets, which cannot be upregulated by T3 in Src-1−/− mice, regain their sensitivity to T3 in NCoRΔID/ΔID Src-1−/− mice. Strikingly, NCoRΔID/ΔID Src-1−/− mice have increased levels of the coactivator SRC-2 and greater SRC-2 recruitment to the TR-binding sites of T3 target genes. Thus, in the absence of SRC-1 and NCoR1, SRC-2 is recruited to the TR-binding regions of T3 target genes, suggesting that the presence of NCoR1 in the absence of SRC-1 prevents additional coactivator recruitment. This implies that TR target genes respond to T3 based on the availability of specific corepressors and coactivators, explaining why individuals have different HPT axis set points and respond differently to similar amounts of T3.
MATERIALS AND METHODS
Animals.
All mouse breeding and housing were approved by the Beth Israel Deaconess Medical Center (BIDMC) Institutional Animal Care and Use Committee. Generation of mice containing the NCoRΔID allele (NCoRΔID) has been previously described (23, 24). Src-1−/− mice were rederived from previously described mice (a gift of B. W. O'Malley [42]). NCoRΔID/+ mice were crossed with Src-1+/− mice to create double-heterozygous (NCoRΔID/+ Src-1+/−) mice, which were bred to produce the mice used in this study: NCoR+/+ Src-1+/+ (wild type [WT]), NCoRΔID/ΔID Src-1+/+ (NCoRΔID/ΔID), NCoR+/+ Src-1−/− (Src-1−/−), and NCoRΔID/ΔID Src-1−/−. The mice were maintained on a mixed B6-129S strain background. To ensure validity and reproducibility, we studied both male and female cohorts for the euthyroid studies (n = 7 to 13 per genotype; 4 genotypes) and then developed three additional experimental cohorts (euthyroid, hypothyroid, and PTU + T3 [see below]) from the same parental lines. Importantly, NCoR1 and SRC-1 are not located on the same chromosome.
Animal experiments and sample collection.
All mouse experiments were approved by the BIDMC Institutional Animal Care and Use Committee. The animals were housed on a 12-h light, 12-h dark cycle and given standard rodent chow (F6 Rodent Diet 8664; Harlan Teklad, Indianapolis, IN) and water ad libitum unless otherwise specified. At the end of the experiments, the mice were euthanized by asphyxiation with CO2. Blood was drawn from the animals by cardiac puncture, and plasma was separated by centrifugation in EDTA-treated tubes and stored at −80°C until analysis. Tissues were rapidly collected, flash frozen in liquid nitrogen, and stored at −80°C until processed.
TSH suppression test.
Control male mice (chow mice) were maintained on the chow diet until 9 to 12 weeks of age. At 6 weeks of age, a subset of mice (PTU mice) was fed a low-iodine diet supplemented with 0.15% propylthiouracil (LoI/PTU) (Harlan Teklad TD.95125) for a minimum of 21 days (PTU mice) to induce hypothyroidism. The PTU mice were sacrificed on day 22 of the LoI/PTU diet. On day 21 of the LoI/PTU diet, a subset of mice (PTU + T3 mice) began to receive daily intraperitoneal (i.p.) injections of T3 (Sigma, St. Louis, MO) while they were maintained on a LoI/PTU diet: a low dose (0.2 μg/100 g body weight [BW]/day) for 7 days, followed by a medium dose (0.5 μg/100 g BW/day) for 7 days, followed by a high dose (1.0 μg/100 g BW/day) for 7 days (21, 43). The animals were sacrificed 14 to 15 h following the last injection of T3. The mice were euthanized as described above.
EchoMRI.
At 9 weeks of age, chow mice were subjected to magnetic resonance imaging (MRI) using EchoMRI (Echo Medical Systems, Houston, TX) to determine body composition.
Plasma analysis.
Total plasma T4 (TT4) and T3 (TT3) were measured by solid-phase radioimmunoassay (RIA) (Coat-a-Count; Diagnostic Products Corp., Los Angeles, CA) in 25 and 50 μl of plasma, respectively. Circulating TSH was measured via Milliplex MAP (rat thyroid hormone TSH panel; EMD Millipore, Billerica, MA) (25, 44).
Real-time quantitative PCR.
Total RNA was extracted from frozen tissues with Stat-60 reagent (Tel-Test, Friendswood, TX). Then, 0.25 μg of total RNA was reverse transcribed using a SuperScript Vilo cDNA synthesis kit from Invitrogen (Carlsbad, CA). TaqMan gene expression assays for all mRNAs were purchased from Applied Biosystems (Carlsbad, CA). Quantitative PCR (qPCR) was performed in duplicate or triplicate using the 800HT thermal cycler (Applied Biosystems). Relative mRNA levels were calculated using the standard curve method and normalized to peptidylprolyl isomerase A (cyclophilin; Ppia) in the heart or 18S rRNA (Rn18S) in the liver, pituitary, and PVH (24, 25, 45).
Western blot analysis.
Western blots were performed as previously described (24). Briefly, approximately 40 mg of liver was homogenized in 1 ml of cell lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerol phosphate, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail [Roche, Indianapolis, IN]). Samples were then sonicated four times for 5 s each time at power level 4 and centrifuged for 20 min at maximum speed. Twenty micrograms of total protein was resolved on 3 to 8% gradient Tris-acetate gels (Invitrogen) and blotted with the indicated primary antibodies, followed by appropriate horseradish peroxidase-conjugated secondary antibody, and developed using the ECL Plus Western blot detection system (Amersham, Piscataway, NJ). The primary antibodies used were an affinity-purified rabbit anti-NCoR1 antibody generated against the C-terminal portion of the NCoR1 molecule, anti-SRC-1 (128E7; Cell Signaling, Danvers, MA); anti-NCoR2 (PA1-843; Affinity Bioreagents/ThermoFisher); and anti-SRC-2 (A300-345A; Bethyl Laboratories, Montgomery, TX). The blots were then stripped (2% SDS, 62.5 mM Tris, pH 6.8, 0.8% 2-mercaptoethanol) and reblotted with anti-RNA polymerase II (05-952; EMD Millipore). The blots were scanned and quantified using ImageJ software (public domain; developed at the National Institute of Mental Health, Bethesda, MD), normalizing to anti-polymerase II.
ChIP.
Chromatin immunoprecipitation assay (ChIP) methods were adapted from previous studies (24, 46, 47). ChIP was performed on mouse livers from PTU + T3 mice of all genotypes. Thirty to 35 mg of minced mouse liver was double cross-linked by first cross-linking using 2 mM disuccinimidyl glutarate (DSG) in phosphate-buffered saline (PBS) at room temperature for 45 min. The livers were washed in PBS and then cross-linked using 1% formaldehyde for 5 min at room temperature. Cross-linking was stopped by adding 0.125 M glycine for 5 min, and the livers were washed again in PBS. We homogenized the livers in 10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, and 0.3% NP-40 with protease inhibitors. Following homogenization, crude nuclei were pelleted by centrifugation at 1,000 × g, washed with PBS, and then sonicated in 1-ml Adaptive Focused Acoustics tubes using a sonicator (Covaris, Woburn, MA) in a buffer of 50 mM Tris, pH 8.0, 10 mM EDTA, 0.25% SDS with protease inhibitors. This process achieves a DNA shear size of 200 to 500 bp. Following sonication, samples were spun to remove debris. Chromatin from mouse livers (n = 4 per genotype) was pooled into a single sample. The samples were diluted in 0.5× RIPA buffer with protease inhibitors and 20 μg glycogen incubated with the appropriate antibody overnight at 4°C with rocking. SRC-2 ChIP was performed with anti-SRC-2 A300-345A (Bethyl Laboratories, Montgomery, TX). SRC-1 ChIP was performed with anti-SRC-1 SRC-1 128E7 rabbit monoclonal antibody (MAb) 2191 (Cell Signaling Technology). We used IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) as a control for both ChIPs. Samples were precipitated with protein A-agarose beads (ThermoFisher). Cross-links were then reversed overnight at 65°C, and DNA was extracted using phenol-chloroform-isoamyl alcohol. DNA from 4 replicate precipitation reactions was used in quantitative PCR with SYBR green reaction chemistry (Applied Biosystems). The results are reported as fold enrichment (ChIP signal) normalized to the DNA input. Similar results were seen when we repeated the ChIPs with no antibody control (data not shown). Primer sequences were designed to regions of TR binding determined by in vivo chromatin affinity purification-DNA sequencing (ChAP-seq) using a biotinylated TRβ1 in mouse liver and are listed in Table 1 (47).
TABLE 1.
ChIP-qPCR primer sequences
| Set no. | Gene | Primer |
|
|---|---|---|---|
| Forward | Reverse | ||
| 1 | Abcb11 | TCTCACCAGGCTCTCTACCA | TTGCTTATAGGTCAATGGCCT |
| 2 | Thrsp A | CCCTCCACTTAGGCCAGGAC | GCCCTAGGTCACCTCCAGAG |
| 3 | Gpd2 Int1 A | GTCGTTGCTTTGGGCTGAGC | CCCATCGGCATACTCAGGTGG |
| 4 | Gpd2 Int1 B | CAGGATTCTTACGGAGAACGC | CCCAGGACCTACTGTGGATC |
| 5 | Gpd2 Int1 D | CCACTGACTTCCTGTCTCCC | GGCCACTGTGGCTGCATCAAG |
| 6 | β-Globin (Hbb1) | GTGTTGCCAAAAAGGATGC | GAAGCAAATGTGAGGAGCAACTG |
Statistical analyses.
Data are presented as sample means and standard errors of the mean (SEM). Sample numbers ranged from 4 to 13 and are stated specifically within the text or figure legends. The text or figure legends highlight how differences between groups were measured, which included unpaired t test, 1-way analysis of variance (ANOVA) with Tukey's multiple-comparison post hoc test, 2-way repeated-measures (RM) ANOVA with a Bonferroni post hoc test, and 2-way ANOVA with a Bonferroni post hoc test.
RESULTS
NCoRΔID/ΔID Src-1−/− mice develop normally.
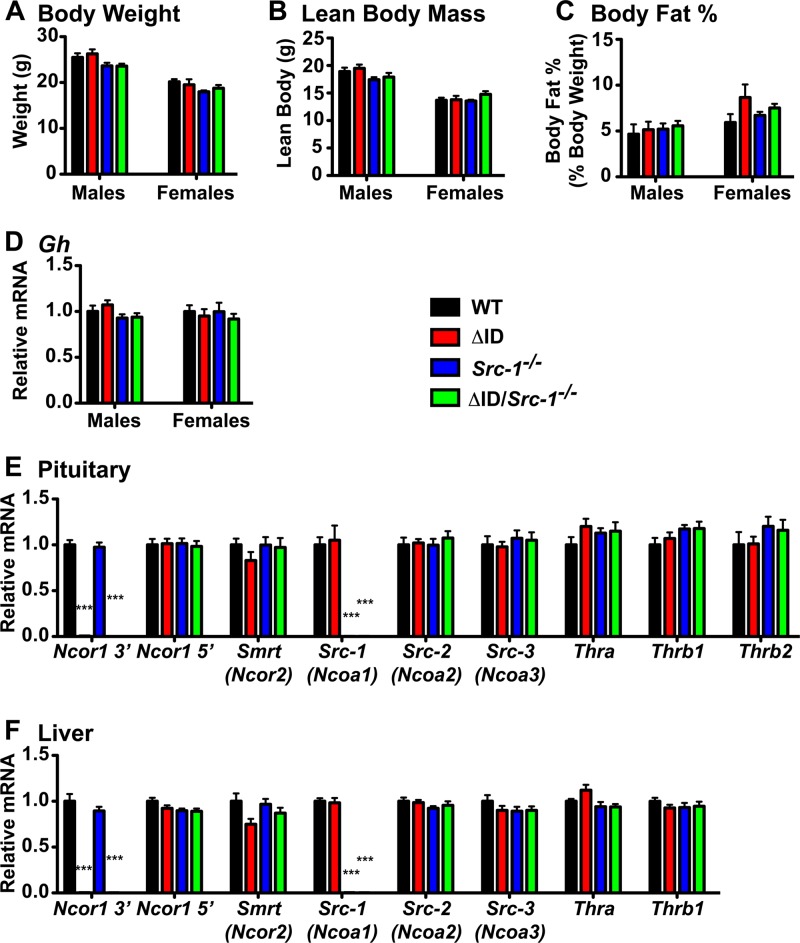
To determine the roles of corepressors and coactivators in establishing the set point of the thyroid axis, we crossed mice that globally express NCoRΔID (NCoRΔID/ΔID mice) with Src-1−/− mice (24, 42). Mice double homozygous for NCoRΔID and Src-1−/− (NCoRΔID/ΔID Src-1−/− mice) develop normally and are born at the expected Mendelian ratios. At 9 weeks of age, male and female NCoRΔID/ΔID Src-1−/− mice have body weights, lean masses, and body fat percentages similar to those of WT, NCoRΔID/ΔID, and Src-1−/− mice (Fig. 1A to C). NCoRΔID/ΔID Src-1−/− mice also have normal pituitary expression of growth hormone (Gh) mRNA levels compared to WT, NCoRΔID/ΔID, and Src-1−/− mice (Fig. 1D). Previously, we reported that NCoRΔID/ΔID mice weighed less than their WT littermates, but only at certain time points, which is consistent with the data reported here (24).
FIG 1.
Deletion of Src-1 and mutation of NCoR1 does not affect mRNA expression of other coregulators and thyroid hormone receptors. (A) Body weights of WT, NCoRΔID/ΔID (ΔID), Src-1−/−, and NCoRΔID/ΔID Src-1−/− (ΔID/Src-1−/−) mice at 9 weeks of age were measured in male and female mice. (B and C) Lean body mass (B) and body fat percentage (C) were measured in male and female mice at 9 weeks of age by EchoMRI. (D) Gh mRNA was measured by qPCR in the pituitaries of male and female mice, normalized to 18S rRNA, and expressed relative to the WT group. (A to D) Data are presented as means and SEM (n = 7 to 13 per genotype). Significance was tested with 1-way ANOVA. (E) In pituitaries from male WT, ΔID, Src-1−/−, and ΔID/Src-1−/− mice, qPCR was performed to quantify mRNA of the 3′ region of NCoR1 (Ncor1 3′), the 5′ region of NCoR1 (Ncor1 5′), SMRT (Ncor2), SRC-1 (Ncoa1), SRC-2 (Ncoa2), SRC-3 (Ncoa3), and thyroid hormone receptors (Thra, Thrb1, and Thrb2). (F) To quantify mRNA in the livers of the same male mice, qPCR was used to analyze the 3′ region of NCoR1 (Ncor1 3′), the 5′ region of NCoR1 (Ncor1 5′), SMRT (Ncor2), SRC-1 (Ncoa1), SRC-2 (Ncoa2), SRC-3 (Ncoa3), and thyroid hormone receptors (Thra and Thrb1). (E and F) mRNA was normalized to 18S rRNA and expressed relative to the WT group (n = 7 to 13 per genotype). Significance was tested by 1-way ANOVA with Tukey's multiple-comparison post hoc test. For Ncor1 3′, ***, P < 0.001 versus WT and Src-1−/− mice. For Src-1, ***, P < 0.001 versus WT and ΔID mice.
To assess the potential compensation by other nuclear cofactors for the loss of NCoR1 and SRC-1, expression of Ncor1 and Smrt (Ncor2) was tested in male pituitaries and livers (Fig. 1E and F). The expression levels of Smrt and the 5′ region of Ncor1 mRNA, which is common to both the WT allele and the NCoRΔID allele, were normal across all genotypes in both the pituitary and liver (23). Expression of the 3′ region of Ncor1 mRNA, which contains the N2 and N3 domains deleted from the NCoRΔID allele, was more than 99% lower in the pituitaries and livers of NCoRΔID/ΔID and NCoRΔID/ΔID Src-1−/− mice than in WT mice, consistent with our previous study (23). We also tested the expression of the three coactivators Src-1 (Ncoa1), Src-2 (Ncoa2; TIF-2), and Src-3 (Ncoa3) in the pituitaries and livers of all the mice (Fig. 1E and F). Expectedly, Src-1 expression was more than 99% lower in both the pituitaries and livers of Src-1−/− and NCoRΔID/ΔID Src-1−/− mice, respectively, than in those of WT mice. Src-2 and Src-3 expression remained unchanged compared to WT mice (Fig. 1E and F). Lastly, we measured mRNA expression of the different isoforms of Thr in both the pituitary (Thra, Thrb1, and Thrb2) and the liver (Thra and Thrb1) and found that the levels remained unchanged between genotypes (Fig. 1E and F).
Male and female NCoRΔID/ΔID Src-1−/− mice have normal TH levels.
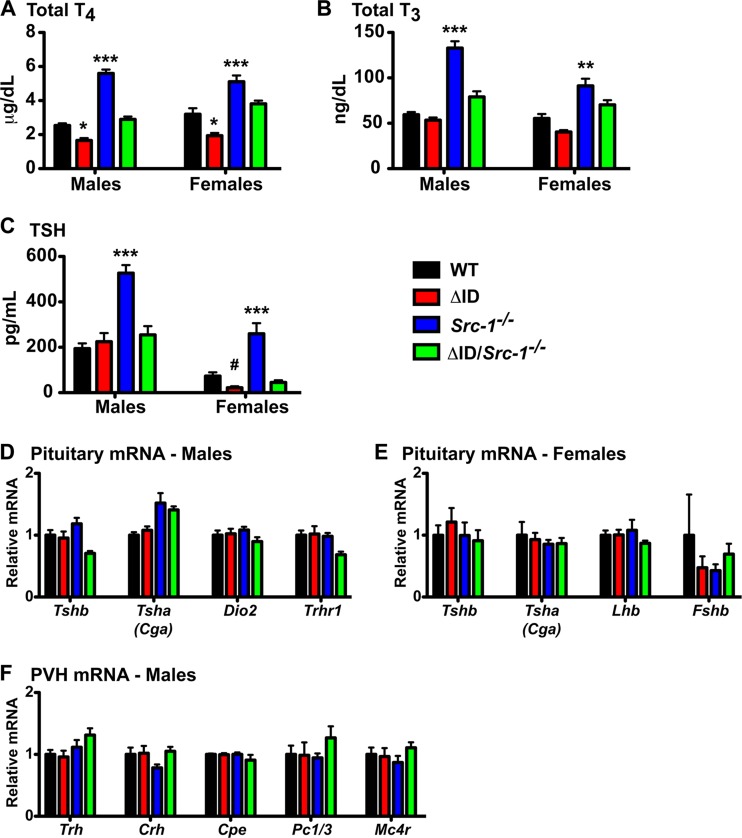
To measure the impact of replacing NCoR1 with NCoRΔID in Src-1−/− mice on thyroid hormone levels, we measured thyroid function in all genotypes in males and females (Fig. 2). As previously reported, TT4 levels were significantly reduced, by 34.6%, in male NCoRΔID/ΔID mice (1.66 ± 0.14 μm/dl) compared to WT mice (2.54 ± 0.13 μm/dl) (Fig. 2A) (24, 25, 40). We found that TT4 levels in male Src-1−/− mice (5.60 ± 0.23 μm/dl) were, as described previously, significantly increased, by 220% (20, 48–50). Strikingly, TT4 levels in male NCoRΔID/ΔID Src-1−/− mice (2.90 ± 0.17 μm/dl) were normal compared to WT mice. Similar to male mice, female NCoRΔID/ΔID mice had a significant (40%) decrease in TT4 levels (1.93 ± 0.16 μm/dl) compared to WT mice (3.20 ± 0.33 μm/dl) (Fig. 2A). We found that TT4 levels in female Src-1−/− mice (5.11 ± 0.36 μm/dl) were significantly increased, by 160% (20). Addition of NCoRΔID to Src-1−/− female mice normalized the TT4 level (3.81 ± 0.19 μm/dl) to that of WT mice (Fig. 2A).
FIG 2.
Disruption of the interaction between the TR and NCoR1 in Src-1−/− mice normalizes TH and TSH levels in male and female mice. Plasma total T4 (A), total T3 (B), and circulating TSH (C) were measured in male and female WT, NCoRΔID/ΔID (ΔID), Src-1−/−, and NCoRΔID/ΔID Src-1−/− (ΔID/Src-1−/−) mice. (A to C) n = 6 to 12. The data are presented as means and SEM. Significance was tested by 1-way ANOVA with Tukey's multiple-comparison post hoc test. *, P < 0.05 versus all; **, P < 0.01 versus all; ***, P < 0.001 versus all; #, P < 0.05 versus WT and Src-1−/−. (D) In pituitaries from male mice, mRNA expression was analyzed by qPCR for TSH subunits Tshb and Tsha (Cga), the type II deiodinase (Dio2), and the TRH receptor (Trhr1). (E) In pituitaries from female mice, mRNA expression was analyzed by qPCR for TSH subunits Tshb and Tsha (Cga), luteinizing hormone (Lhb), and follicle-stimulating hormone (Fshb). (F) In male mice, the PVH was microdissected, and mRNA was quantified by qPCR for thyrotropin-releasing hormone (Trh), corticotropin-releasing hormone (Crh), carboxypeptidase E (Cpe), prohormone convertase 1/3 (Pc1/3), and melanocortin 4 receptor (Mc4r). (D to F) mRNA was normalized to 18S rRNA and expressed relative to the WT group (n = 6 to 12). The data are presented as means and SEM. Significance was tested with 1-way ANOVA.
Although we did not see decreased TT3 levels in male NCoRΔID/ΔID mice (53.5 ± 2.8 ng/dl) on this background, TT3 levels in Src-1−/− mice (132.8 ± 7.6 ng/dl) were significantly increased, by 224%, compared to WT mice (59.4 ± 2.8 ng/dl), similar to previous reports (Fig. 2B) (20, 25, 48–50). Again, male NCoRΔID/ΔID Src-1−/− mice had TT3 levels (79.1 ± 6.1 ng/dl) similar to those of WT mice (Fig. 2B). In females, NCoRΔID/ΔID mice had TT3 levels (40.4 ± 2.1 ng/dl) similar to those of WT mice (55.3 ± 4.9 ng/dl) (Fig. 2B). TT3 levels in female Src-1−/− mice (91.1 ± 7.9 ng/dl) were significantly increased, by 165% (20). Female NCoRΔID/ΔID Src-1−/− mice had TT3 levels (70.3 ± 5.2 ng/ml) similar to those of WT mice (Fig. 2B). Whereas TT3 levels in male and female NCoRΔID/ΔID mice in this study were not significantly lower than those of WT mice, as we have previously reported, the TT3 assay used in this study is the same as we have used previously (24). We believe this difference is due to the background of the mice used in this study. Importantly, however, replacement of NCoR1 with the NCoRΔID allele in the Src-1−/− mouse normalizes TT3 levels.
Other groups have demonstrated a large increase in circulating TSH in male Src-1−/− mice, and here, we show a significant 271% increase (526.9 ± 35.2 pg/ml) in circulating TSH compared to WT mice (194.2 ± 22.9 pg/ml), while the TSH level was normal in NCoRΔID/ΔID mice (224.9 ± 38.0 pg/ml) (Fig. 2C). Male NCoRΔID/ΔID Src-1−/− mice have normal TSH levels (254.5 ± 38.8 pg/ml) compared to both WT and NCoRΔID/ΔID mice. In female mice, circulating TSH levels in NCoRΔID/ΔID mice (21.9 ± 6.3 pg/ml) were 70.1% lower than in WT mice (73.3 ± 16.5 pg/ml) as opposed to the 354% increase in TSH levels in Src-1−/− mice (259.8 ± 46.7 pg/ml) (Fig. 2C) (20). NCoRΔID/ΔID Src-1−/− female mice had normal plasma TSH levels (45.3 ± 9.6 pg/ml) similar to WT levels.
In order to analyze the complete HPT axis, we looked at pituitary function in male and female mice. Gene expression analysis in the pituitaries of male mice revealed normal expression of the TSH subunits Tsha (Cga) and Tshb, the type II deiodinase (Dio2), and the TRH receptor (Trhr1) across all genotypes (Fig. 2D). Gene expression in female mouse pituitaries of the TSH subunits Tsha (Cga) and Tshb remained unchanged between genotypes (Fig. 2E). Additionally, we tested the expression of luteinizing hormone (Lhb) and follicle-stimulating hormone (Fshb), and no differences were detected between genotypes (Fig. 2E). Additionally, we analyzed the mRNA expression from microdissected PVHs of male mice and found that the expression of Trh was also normal across genotypes, including Src-1−/− mice, despite their increased TH levels (Fig. 2F). Additionally, expression of corticotropin-releasing hormone (Crh), carboxypeptidase E (Cpe), prohormone convertase 1/3 (Pc1/3), and melanocortin 4 receptor (Mc4r) was the same in all genotypes (Fig. 2F).
Circulating TSH responds normally to changes in TH levels in NCoRΔID/ΔID Src-1−/− mice.
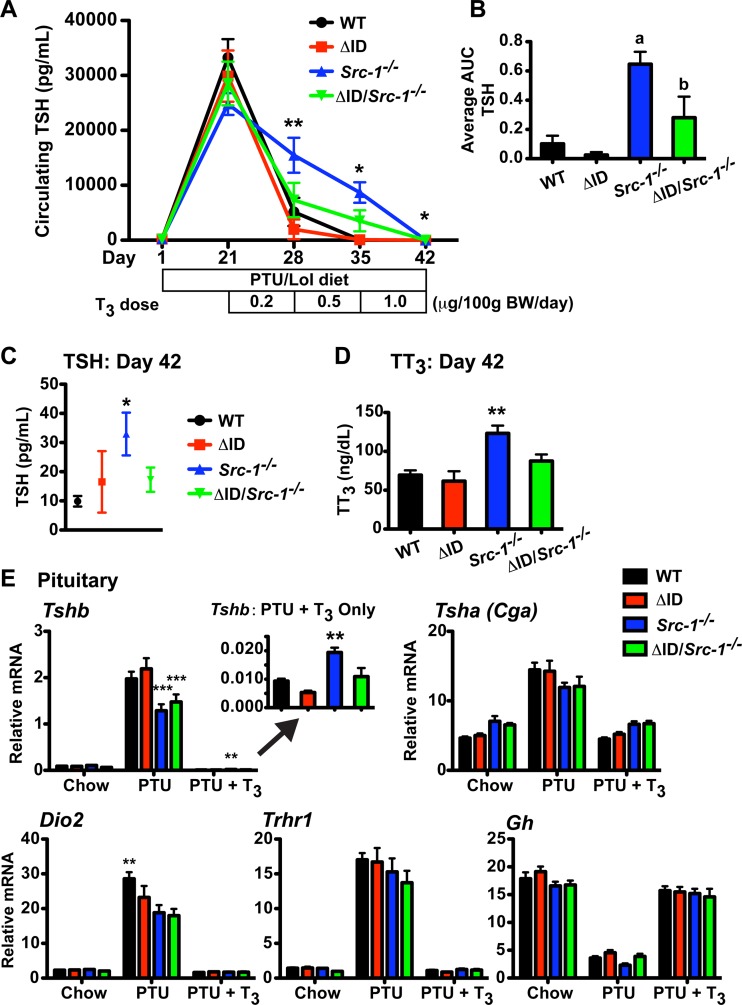
By expressing the NCoRΔID allele in male and female Src-1−/− mice, we were able to normalize TT4, TT3, and TSH levels, suggesting that the resistance present in Src-1−/− mice is due to unopposed NCoR1. To investigate the dynamic response of the HPT axis to changes in circulating TH levels, we performed a TSH suppression test on all groups of male mice (Fig. 3) (43). To accomplish this, mice were placed on a LoI/PTU diet to render the mice hypothyroid (PTU mice). After 21 days, the mice had undetectable TT4 levels (data not shown) and elevated plasma TSH concentrations greater than 24,000 pg/ml (Fig. 3A). The mice were then supplemented with daily injections of T3 for the next 3 weeks in increasing doses (0.2, 0.5, and 1.0 μg/100 g BW/day), and the fall in TSH levels was measured weekly. As previously demonstrated, the fall in plasma TSH levels in Src-1−/− mice is resistant to increasing doses of T3 compared to WT mice and also, as shown here, compared to NCoRΔID/ΔID mice (2-way RM ANOVA; P < 0.0001) (Fig. 3A and C) (20, 51). To further validate the sensitivity of NCoRΔID/ΔID Src-1−/− mice to T3, we averaged the area under the curve (AUC) for each mouse per genotype and found a significant difference between Src-1−/− mice (AUC, 0.65 ± 0.08) and the other genotypes of mice, WT (AUC, 0.10 ± 0.06), NCoRΔID/ΔID (AUC, 0.03 ± 0.01), and NCoRΔID/ΔID Src-1−/− (AUC, 0.28 ± 0.12) (P < 0.05) (Fig. 3B). Strikingly, NCoRΔID/ΔID Src-1−/− mice were as sensitive to T3 as WT mice, which carried through to the largest dose of 1.0 μg/100 g BW/day (Src-1−/− versus all, P < 0.05) (Fig. 3C). We also measured TT3 levels in all the mice following the largest dose of T3 (1.0 μg/100 g BW/day) and found that TT3 levels in Src-1−/− mice were significantly higher than in all other genotypes (Src-1−/− mice versus all, P < 0.01) (Fig. 3D).
FIG 3.
NCoRΔID/ΔID Src-1−/− mice suppress plasma TSH similarly to WT mice in response to T3. (A) The plasma TSH concentration was repeatedly measured in WT, NCoRΔID/ΔID (ΔID), Src-1−/−, and NCoRΔID/ΔID Src-1−/− (ΔID/Src-1−/−) male mice after 21 days of a LoI/PTU diet and consecutive 7-day treatments with increasing concentrations of T3 (0.2, 0.5, and 1.0 μg/100 g BW/day; n = 7 or 8). The data are presented as means ± SEM and were analyzed by repeated-measures 2-way ANOVA with the Bonferroni post hoc test. *, P < 0.05 for Src-1−/− versus all; **, P < 0.01 for Src-1−/− versus all. (B) AUC of each mouse averaged by genotype (n = 7 or 8). The data are presented as means and SEM and were analyzed by 1-way ANOVA with Tukey's multiple-comparison post hoc test. a, P < 0.01 for Src-1−/− versus WT and ΔID; b, P < 0.05 for Src-1−/− versus ΔID/Src-1−/−. (C and D) Circulating TSH levels and TT3 levels at day 42 following 7 days of treatment with the largest dose of T3. (C) n = 7 or 8. The data are presented as means ± SEM and were analyzed by repeated-measures 2-way ANOVA with a Bonferroni post hoc test. *, P < 0.05 for Src-1−/− versus all. (D) n = 7 or 8. The data are presented as means and SEM and were analyzed by 1-way ANOVA with Tukey's multiple-comparison post hoc test. **, P < 0.01 for Src-1−/− versus all. (E) Analysis of mRNA expression of TH-responsive targets in the pituitary by qPCR in the indicated groups. Expression of Tshb, Tsha, Dio2, Trhr1, and Gh was normalized to 18S rRNA (n = 7 to 12). The inset highlights Tshb mRNA expression at day 42 following 7 days of treatment with the largest dose of T3. The data are presented as means and SEM and were analyzed by 2-way ANOVA with a Bonferroni post hoc test. **, P < 0.01 versus all; ***, P < 0.001 versus WT and ΔID.
Next, we determined the response of pituitary gene expression to changes in circulating TH levels. Previous reports have found that expression of Tshb is resistant to T3 in Src-1−/− mice compared to WT mice, whereas NCoRΔID/ΔID mice are normal (24, 52). Mice of all genotypes were sacrificed after the final dose of T3 (1.0 μg/100 g BW/day; PTU + T3 mice) and compared to mice sacrificed after 21 days on a LoI/PTU diet (PTU mice) and control mice (chow mice). Expression of the TSH subunits Tshb and Tsha (Cga) increased following 21 days on a LoI/PTU diet and decreased with T3 in all genotypes (Fig. 3E). Tshb expression in PTU-treated mice versus chow mice increased as much in NCoRΔID/ΔID Src-1−/− mice as in WT and NCoRΔID/ΔID mice (23-fold versus 22- and 25-fold, respectively, compared with Src-1−/− mice at 12-fold). The repression of Tshb was also resistant to T3 in Src-1−/− mice, as expression decreased by only 66-fold between PTU and PTU + T3 mice compared with the rest of the genotypes, which decreased by at least 135-fold (P < 0.01) (Fig. 3E, inset). This resistance in Src-1−/− mice is even clearer when one considers the elevation in total T3 levels in the animals after T3 administration. Expression of Dio2, Trhr1, and Gh all responded as expected to PTU and T3, with no difference between genotypes, except for Dio2, where WT mice had a greater response to PTU than the other genotypes (P < 0.01) (Fig. 3E).
The hypothalamus and heart are normal in NCoRΔID/ΔID Src-1−/− mice.
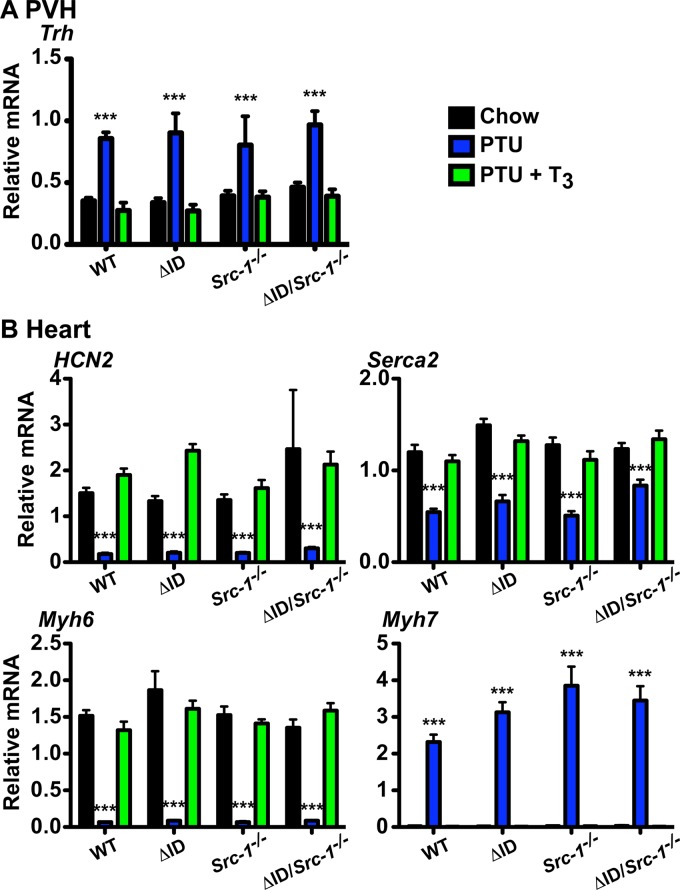
To explore the sensitivity of the hypothalamus and heart to TH, we examined gene expression in mice after the final dose of T3 (1.0 μg/100 g BW/day; PTU + T3 mice) and compared it to that in mice sacrificed after 21 days on a LoI/PTU diet (PTU mice) and control mice (chow mice). In the PVH, expression of Trh was significantly increased in all genotypes after 21 days on a LoI/PTU diet (Fig. 4A). Following treatment with T3, Trh levels were suppressed equally in all genotypes.
FIG 4.
Gene expression in the PVH and the heart. mRNA levels were measured by qPCR in WT, NCoRΔID/ΔID (ΔID), Src-1−/−, and NCoRΔID/ΔID Src-1−/− (ΔID/Src-1−/−) mice following 21 days of a LoI/PTU diet (PTU) and a LoI/PTU diet with T3 replacement for 21 days with increasing doses of T3 (PTU + T3) compared with the control (Chow). (A) Trh mRNA was measured in the PVH microdissected from mouse brain and was normalized to expression of 18S rRNA. (B) mRNA expression of known positive and negative TH targets in the heart. HCN2, Serca2, Myh6, and Myh7 were normalized to expression of cyclophilin. (A and B) n = 5 to 7. The data are presented as means and SEM and were analyzed by 2-way ANOVA with a Bonferroni post hoc test. ***, P < 0.001 versus chow and PTU + T3.
NCoRΔID/ΔID mice show normal sensitivity to T3 but slight derepression of Hcn2 and Serca-2 on a LoI/PTU diet, whereas Src-1−/− mice have normal expression of Serca-2 and the α and β myosin heavy-chain genes (Myh6 and Myh7, respectively) (24, 52). Here, we found no differences in the expression of Hcn2, Serca-2, Myh6, and Myh7 between genotypes in response to a chow diet, to a LoI/PTU diet, or to T3, suggesting that T3 action in the heart does not require NCoR1 or SRC-1 (Fig. 4B).
Expression of NCoRΔID in Src-1−/− mice reestablishes hepatic T3 sensitivity.
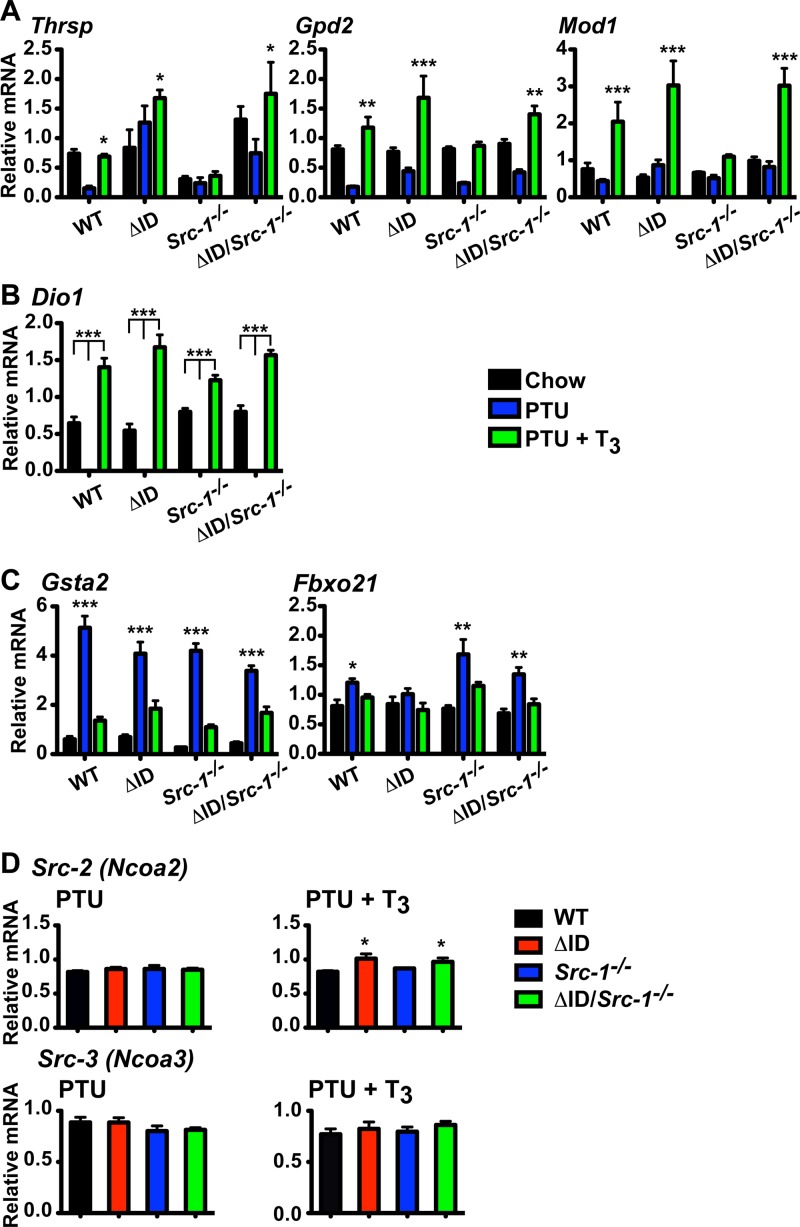
Unlike in the heart, mice expressing NCoRΔID have increased hepatic T3 sensitivity, which is characterized by increased expression of T3 target genes, such as the thyroid hormone-responsive SPOT 14 gene (Thrsp), the type I deiodinase gene (Dio1), the glycerol-3-phosphate dehydrogenase 2 gene (Gpd2), and the malic enzyme gene (Mod1), regardless of TH levels, and suggests that enhanced ligand-dependent activation occurs in the absence of NCoR1 (24). In contrast, Thrsp expression in Src-1−/− mice has previously been reported to be unresponsive to T3 (52). We therefore analyzed the expression of several hepatic genes across genotypes and found that Thrsp fails to respond to T3 administration in Src-1−/− mice (PTU+T3 versus PTU, 1.1-fold increase) compared to WT mice (PTU + T3 versus PTU, 4.6-fold increase) (Fig. 5A). Additionally, expression of Gpd2 and Mod1 is blunted in response to T3 in Src-1−/− mice (PTU + T3 versus PTU, 2.6-fold and 2.1-fold increases, respectively) compared to WT mice (PTU + T3 versus PTU, 6.7-fold and 4.7-fold increases, respectively). Replacing NCoR1 with NCoRΔID in Src-1−/− mice reestablishes the sensitivity of these genes' expression to T3 both in the context of the fold increase (PTU + T3 versus PTU, 2.6-fold increase [Thrsp], 3.5-fold increase [Gpd2], and 3.8-fold increase [Mod1]) (Fig. 5A) and overall expression. Interestingly, expression of Dio1, another positive TH target, is normal across genotypes (Fig. 5B). Furthermore, expression of negative TH targets, such as the glutathione S-transferase A2 gene (Gsta2) and the F-box protein 21 gene (Fbxo21), is unaffected by removal of normal NCoR1, SRC-1, or both (Fig. 5C). Thus, specific T3-positive targets in the liver are regulated by the amount of NCoR1 or SRC-1 present.
FIG 5.
Loss of TR-NCoR1 interaction in Src-1−/− mice reestablishes the T3 response in some positive TH targets in the liver. mRNA levels were measured by qPCR in livers from WT, NCoRΔID/ΔID (ΔID), Src-1−/−, and NCoRΔID/ΔID Src-1−/− (ΔID/Src-1−/−) mice following 21 days of a LoI/PTU diet (PTU) and a LoI/PTU diet with T3 replacement for 21 days with increasing doses of T3 (PTU + T3) compared with the control (Chow). (A to C) All genes were normalized to 18S rRNA (n = 6 to 13). The data are presented as means and SEM and were analyzed by 2-way ANOVA with a Bonferroni post hoc test. (A) T3 response in positive TH targets Thrsp, Gpd2, and Mod1 was reestablished with the addition of NCoRΔID. *, P < 0.05 versus PTU; **, P < 0.01 versus PTU; ***, P < 0.001 versus PTU. (B) Positive TH target type I deiodinase (Dio1) is not affected by changes in NCoR1 or SRC-1. ***, P < 0.001. (C) Expression of negative TH targets Gsta2 and Fbxo21. *, P < 0.05 versus chow and PTU + T3; **, P < 0.01 versus chow and PTU + T3; ***, P < 0.001 versus chow and PTU + T3. (D) Responses of SRC-2 (Ncoa2) and SRC-3 (Ncoa3) mRNAs to changes in TH levels under PTU and PTU + T3 conditions. The data were normalized to 18S rRNA (n = 6 to 13). The data are presented as means and SEM and were analyzed by 1-way ANOVA with Tukey's multiple-comparison post hoc test. *, P < 0.05 versus chow.
To understand how the loss of NCoR1 allows SRC-1-dependent positive TH targets to regain sensitivity in the absence of SRC-1, we looked at the expression of other coregulators. Expression levels were similar between genotypes for Ncor1, SMRT (Ncor2), and SRC-1 (Ncoa1) under chow, PTU, and PTU + T3 conditions (data not shown). However, we did find a small but significant increase in expression of SRC-2 (Ncoa2) in response to T3 in NCoRΔID/ΔID (1.23-fold increase; P < 0.05) and NCoRΔID/ΔID Src-1−/− (1.18-fold increase; P < 0.05) mice compared to WT mice, which suggests increased availability of SRC-2 in the presence of T3 and in the absence of NCoR1 and/or SRC-1 (Fig. 5D). In comparison, hepatic SRC-3 (Ncoa3) mRNA levels in mice do not change between genotypes under PTU or PTU + T3 conditions (Fig. 5D).
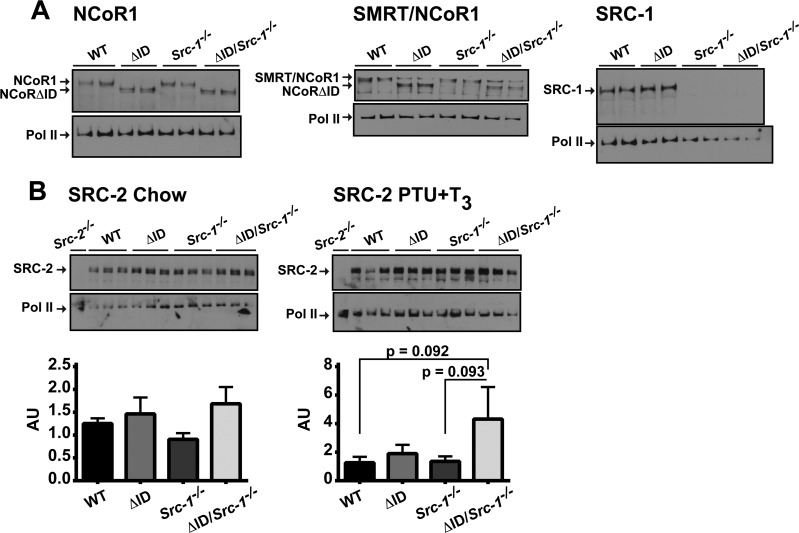
Given the small but elevated increase in Src-2 mRNA levels under PTU + T3 conditions, we next examined protein levels of coregulators across all genotypes. Western analysis of NCoR1 confirmed the presence of NCoRΔID in NCoRΔID/ΔID and NCoRΔID/ΔID Src-1−/− mice; however, the levels of NCoR1 and/or NCorΔID did not change among genotypes (Fig. 6A). We also confirmed the absence of SRC-1 in Src-1−/− and NCoRΔID/ΔID Src-1−/− mice, but the levels of SRC-1 between WT and NCoRΔID/ΔID mice were not different (Fig. 6A). Using an antibody that recognizes SMRT, NCoR1, and NCoRΔID, we assessed the levels of SMRT and found no difference between genotypes that lacked NCoR1, where the remaining 270-kDa band is SMRT (Fig. 6A). Upon examination of SRC-2 protein levels in all the genotypes under chow and PTU + T3 conditions, we found no significant difference in SRC-2 protein levels between genotypes under chow-fed conditions (Fig. 6B). However, under PTU + T3 conditions, SRC-2 protein levels trend toward increased levels in NCoRΔID/ΔID Src-1−/− mice compared to WT and Src-1−/− mice when quantified and normalized to RNA polymerase II levels (1-way ANOVA, P < 0.05; Tukey's multiple-comparison post hoc test, NCoRΔID/ΔID Src-1−/− versus WT, P = 0.092, and NCoRΔID/ΔID Src-1−/− versus Src-1−/−, P = 0.093) (Fig. 6B). We confirmed the specificity of the SRC-2 antibody by examining SRC-2 levels in WT and Src-2−/− mice (the livers were a gift from Bert O'Malley and Brian York).
FIG 6.
Under PTU + T3 conditions, SRC-2 protein levels trend toward increased levels in NCoRΔID/ΔID Src-1−/− mice compared to WT and Src-1−/− mice. (A) Whole-cell protein extracts were isolated from the livers of male WT, NCoRΔID/ΔID (ΔID), Src-1−/−, and NCoRΔID/ΔID Src-1−/− (ΔID/Src-1−/−) mice and subjected to Western analysis with anti-NCoR1, SMRT/NCoR1, and SRC-1. The blots were stripped and reprobed with anti-RNA polymerase II (Pol II). (B) Western analysis of SRC-2 protein levels in the livers of all genotypes under chow and PTU + T3 conditions compared to RNA polymerase II. The blots were scanned and quantified with ImageJ and normalized to RNA polymerase II levels (1-way ANOVA, P < 0.05; Tukey's multiple-comparison post hoc test, ΔID/Src-1−/− versus WT, P = 0.092, and ΔID/Src-1−/− versus Src-1−/−, P = 0.093). Extracts from Src-2−/− mice were used to confirm SRC-2 antibody specificity.
NCoRΔID/ΔID Src-1−/− mice have increased recruitment of SRC-2 to the promoters of T3-sensitive genes.
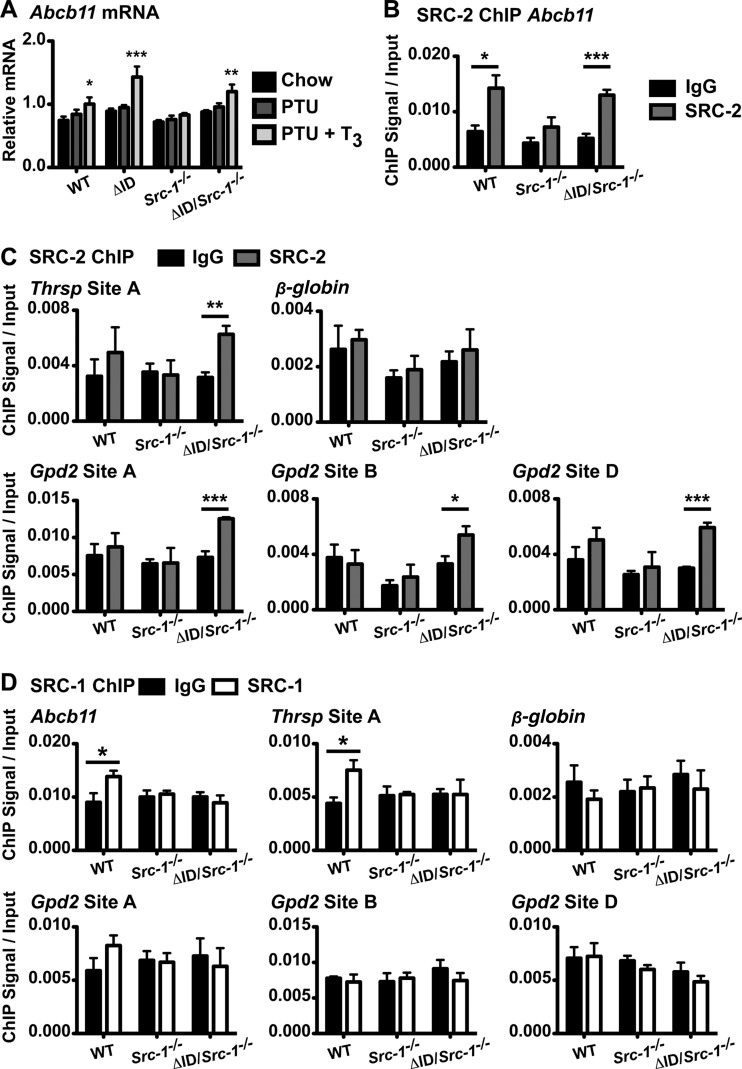
Mice null for SRC-2 have normal thyroid function (53). However, when mice are null for both SRC-1 and SRC-2, T4, T3, and TSH levels are even higher than in Src-1−/− mice, suggesting a partial compensatory role for SRC-2 in the regulation of the thyroid axis (53). Because of this and the trend of increased mRNA and protein levels of SRC-2 in NCoRΔID/ΔID Src-1−/− mice under PTU + T3 conditions, we asked if SRC-2 could serve as a substitute for SRC-1 in T3 gene activation. To confirm the performance of our SRC-2 antibody in ChIP, we analyzed the previously measured SRC-2 target Abcb11, which also binds TR (47, 54). Abcb11 is also a T3 target gene (Fig. 7A). Surprisingly, Abcb11 mRNA levels do not respond to T3 in Src-1−/− mice (Fig. 7A). Using ChIP, we show that SRC-2 is present at the Abcb11 promoter site compared to the IgG control in both WT and NCoRΔID/ΔID Src-1−/− mice given PTU + T3 but not in Src-1−/− mice, where Abcb11 expression is also blunted (Fig. 7B) (47).
FIG 7.
SRC-2 binds to T3 target gene promoters in the absence of SRC-1 and NCoR1. (A) In livers from male WT, NCoRΔID/ΔID (ΔID), Src-1−/−, and NCoRΔID/ΔID Src-1−/− (ΔID/Src-1−/−) mice, mRNA levels were measured by qPCR following 21 days of a LoI/PTU diet (PTU) and a LoI/PTU diet with T3 replacement for 21 days with increasing doses of T3 (PTU + T3) compared with control (Chow). Expression of the SRC-2 target gene Abcb11 was normalized to the expression of 18S rRNA (n = 6 to 13). The data are presented as means and SEM and were analyzed by 2-way ANOVA with a Bonferroni post hoc test. Chow versus PTU + T3, *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) SRC-2 ChIP was validated by ChIP-qPCR analysis using an anti-SRC-2 antibody (SRC-2) versus IgG for the proximal promoter of the SRC-2 target gene Abcb11 on the chromatin from livers of WT, Src-1−/−, and ΔID/Src-1−/− mice under PTU + T3 conditions. (C) Using the same chromatin, SRC-2 ChIP-qPCR analysis was performed for a TRβ binding site (site A) upstream of the Thrsp promoter, three intragenic sites (sites A, B, and D) within the first intron of the T3 target gene Gpd2, and a site on the β-globin gene (Hbb1) that does not bind TRβ. (D) ChIP-qPCR analysis was performed using an anti-SRC-1 antibody (SRC-1) or IgG on the same chromatin from panels B and C for the proximal promoter of the SRC-2 target gene Abcb11; Thrsp site A; Gpd2 sites A, B, and D; and a site in the β-globin gene (Hbb1) that does not bind TRβ. (B to D) n = 4. The results are reported as fold enrichment (ChIP signal) normalized to the DNA input. The data are presented as means and SEM and were analyzed by unpaired t test for antibody versus IgG. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To validate the role of SRC-2 in the reacquired T3 sensitivity of NCoRΔID/ΔID Src-1−/− mice, we performed ChIP for SRC-2 and examined its recruitment to a TRβ binding site upstream of the Thrsp promoter, to three intragenic TRβ binding sites within the first intron of Gpd2, and to a control site on the β-globin gene that does not bind TR, all previously described (47). All TRβ binding sites were identified using in vivo hepatic ChAP-seq in mice expressing a biotinylated-TRβ1 isoform in the liver only (47). Strikingly, there is increased recruitment of SRC-2 to the TRβ binding sites of these genes in NCoRΔID/ΔID Src-1−/− mice compared to the IgG control (Fig. 7C). In contrast, SRC-2 is not recruited in WT or Src-1−/− livers, suggesting that the presence of NCoR1 prevents its recruitment. Furthermore, SRC-2 is not recruited to the β-globin gene (Hbb1) (Fig. 7C). We also examined the recruitment of SRC-1 to these sites and found that NCoR1 allows recruitment of SRC-1 to the Abcb11 and Thrsp promoter in WT mice compared to IgG, but clearly, this is lost in mice that lack SRC-1 (Fig. 7D). SRC-1 is not recruited to TRβ binding sites on the Gpd2 and β-globin genes. Taken together, these data suggest that SRC-2 can promote T3 sensitivity in mice null for SRC-1 and expressing NCoRΔID.
DISCUSSION
The HPT axis is regulated through a feedback mechanism by which the hypothalamus releases TRH to increase TSH production in the pituitary, which stimulates TH release from the thyroid. In turn, TRH and TSH are suppressed by circulating TH (2). Recent studies by our group and others have shown that the circulating levels of TH and TSH are established by an interaction between TRβ isoforms and NCoR1 (24, 25, 40). Indeed, the correction of RTH by expressing NCoRΔID in ThrbPV mutant mice emphasizes the role of corepressors in establishing TH levels and the set point of the axis (40). Mice expressing a mutant form of Thrb that is unable to bind the coactivator SRC-1 are similar to this ThrbPV mutant, with high TH and TSH levels and increased pituitary Tshb mRNA levels (21). The proposed role of the coactivator SRC-1 and the corepressor NCoR1 in regulating TSH repression and establishing T3 sensitivity led us to hypothesize that the balance between coactivators and corepressors is important in determining the set point of the thyroid axis. Thus, we sought to express the mutant NCoRΔID allele in mice null for SRC-1 to create a mouse that does not have functional NCoR1 or SRC-1 in the context of TH signaling. Here, we show that these NCoRΔID/ΔID Src-1−/− mice have normal T4, T3, and TSH levels, demonstrating that the replacement of NCoR1 with NCoRΔID alleviated the functional loss of SRC-1 and reestablished a balance between corepressors and coactivators, which is necessary to set basal TH and TSH levels (40).
In the hypothyroid state, TSH levels are increased many hundredfold, and the ability of various doses of T3 to reduce TSH levels is a superb indicator of thyrotroph T3 sensitivity (43). Previous work has demonstrated that mice lacking a functional TRβ–SRC-1 interaction or null for SRC-1 are unable to suppress TSH levels to the same degree as WT mice (20, 21). In our studies, we saw the same resistance to T3 in Src-1−/− mice. Interestingly, the level of central resistance in Src-1−/− mice is more apparent when serum TSH rather than expression of the known T3 target genes Tshb and Trh is used as a readout. This raises the possibility that NCoR1 and SRC-1 are exerting their effects on other target genes that control TSH production translationally and posttranslationally. It is also important to stress that Src-1−/− mice have levels of Tshb and Trh mRNA expression similar to those of WT mice, which are inappropriately high considering the elevated T4 and T3 levels in the basal state. Thus, Src-1−/− mice are resistant to TH. The normalization of TH levels in NCoRΔID/ΔID Src-1−/− mice accompanied by now appropriate Tshb and Trh mRNA levels that are equivalent to those of the WT control animals demonstrates the need for balance between corepressors and coactivators in their regulation.
When NCoRΔID/ΔID Src-1−/− mice were given increasing doses of T3 after the induction of hypothyroidism, circulating TSH levels were suppressed to a degree similar to that in WT mice as opposed to the increased TSH levels in Src-1−/− mice. This correction was also seen in the context of pituitary gene expression, as Tshb mRNA levels fell with T3 administration equally in NCoRΔID/ΔID Src-1−/− and WT mice compared to the elevated Tshb mRNA levels in Src-1−/− mice. This is particularly striking considering that Src-1−/− mice had elevated TT3 levels following the 3 weeks of T3 therapy, demonstrating that these animals are indeed resistant to thyroid hormone at the level of gene expression. At the same time NCoRΔID/ΔID Src-1−/− mice have normal sensitivity consistent with SRC-1 and NCoR1 having a direct role in the regulation of the Tshb gene and demonstrating that the balance of coregulators is important for interpreting the dynamic changes in thyroid hormone levels.
At the level of the hypothalamus, Trh and other genes involved in the processing of the TRH peptide were relatively unaffected by the removal of functional NCoR1, SRC-1, or both. However, as discussed above, in the context of the elevated thyroid hormone levels, Src-1−/− mice have inappropriately elevated Trh mRNA levels whereas NCoRΔID/ΔID Src-1−/− mice have normal Trh mRNA and TT3 levels. We know that TRH is important to the set point of the HPT axis, as mice null for TRH have decreased T4 levels, slightly increased TSH levels, and reduced TSH bioactivity (55). The results from this study suggest that NCoR1 and SRC-1 are important to the regulation of Trh in the hypothalamus and also at the level of the pituitary (24, 25). This is indeed consistent with our recent findings, demonstrating that expression of NCoRΔID in the pituitary alone can alter TSH regulation by T3 (25).
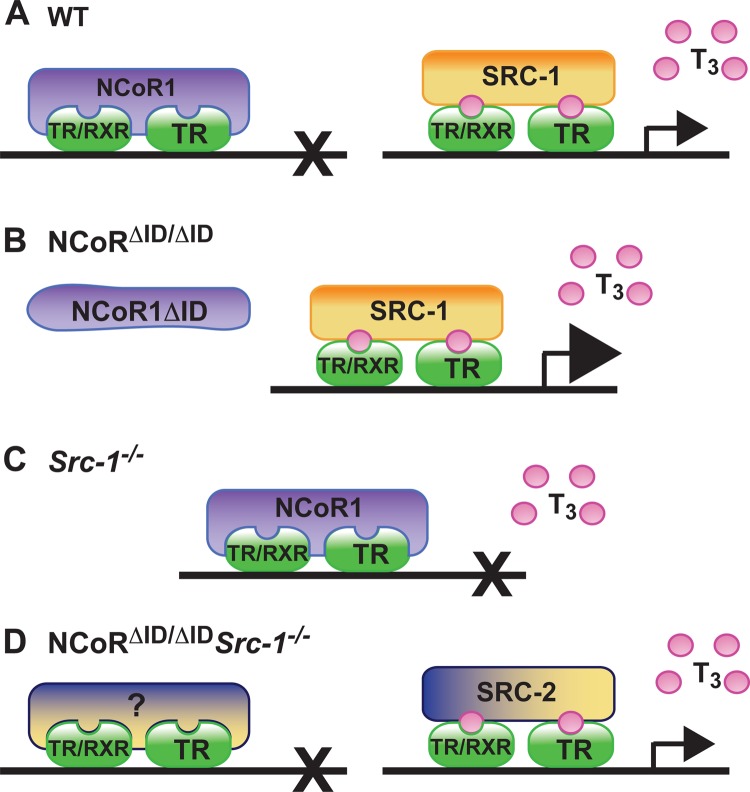
In addition to the pituitary, Src-1−/− mice are also unable to maintain normal levels of expression of TH targets in the liver despite the presence of increased circulating levels of TH. (Fig. 8A to C shows a model). In contrast, we have previously demonstrated that NCoR1 is critical for the T3 sensitivity of hepatic TR targets, such as Dio1, Gpd2, Mod1, and Thrsp, so that without available NCoR1, there is increased response to a set amount of T3 (23, 24). These data strongly suggest that the hepatic TRs may recruit both SRC-1 and NCoR1 in the presence of T3 and that the absence of NCoR1 prioritizes the recruitment of SRC-1, which then leads to increased gene expression (24). Here, we demonstrate that Src-1−/− mice fail to appropriately activate Thrsp, Gpd2, and Mod1 in the presence of increasing amounts of T3, consistent with decreased sensitivity. The replacement of NCoR1 with NCoRΔID in Src-1−/− mice reestablishes hepatic T3 sensitivity, which highlights the fact that the balance of NCoR1 and SRC-1 is critical. Moreover, once NCoR1 action is removed in NCoRΔID/ΔID Src-1−/− mice, target genes regain their T3 sensitivity, possibly by recruiting another coactivator, such as SRC-2, which has been shown to have a direct role in the regulation of hepatic genes, including Thrsp, Gpd2, and Mod1 (56). Therefore, we measured the expression of Src-2 mRNA in our mice and found that expression was higher in NCoRΔID/ΔID and NCoRΔID/ΔID Src-1−/− mice in response to T3. Furthermore, measurement of SRC-2 protein levels highlighted a trend toward increased SRC-2 in NCoRΔID/ΔID and NCoRΔID/ΔID Src-1−/− mice receiving T3. Finally, and most importantly, there was increased enrichment of SRC-2 in TR-binding regions of T3-sensitive genes in NCoRΔID/ΔID Src-1−/− mice in the presence of T3. Thus, in the absence of NCoR1 and SRC-1, positive regulation of key hepatic TH targets is mediated by SRC-2. Although we cannot rule out a compensatory role for SRC-3 or other coregulators when NCoR1 and SRC-1 are absent, we did not see changes in Src-3 mRNA expression between genotypes with T3. Moreover, previous studies have detected little overlap in the roles of SRC-1 and SRC-3 in hepatic metabolism (56). Interestingly, NCoR1 and SRC-1 do not appear crucial to the regulation of negative TH targets in the liver, as expression of Gsta2 and Fbxo21 responded similarly in all genotypes. Clearly, the roles of NCoR1 and SRC-1 in gene regulation appear to vary by tissue and gene target. Further work will be required to understand this specificity in vivo.
FIG 8.
In peripheral tissues, the balance of corepressors and coactivators is most important in determining T3 sensitivity in the liver. (A) On classic positive T3 targets in WT mice, NCoR1 is recruited to nuclear receptor heterodimers (TR/RXR) or homodimers (TR/TR) in the absence of ligand, and transcription is repressed. In the presence of T3, NCoR1 is released, and SRC-1 is recruited to mediate transcriptional activity. (B) When NCoR1 is not readily available, as exemplified in NCoRΔID/ΔID mice, there is increased response to T3, suggesting that recruitment of SRC-1 is also increased. (C) In Src-1−/− mice, when SRC-1 is absent, genes do not respond to T3, which is consistent with decreased sensitivity and suggests prioritized recruitment of NCoR1. (D) Replacement of NCoR1 with NCoRΔID in Src-1−/− mice reestablishes hepatic T3 sensitivity. Our work here describes the increased binding of SRC-2 in the liver in the absence of both SRC-1 and NCoR1 under increased T3 conditions, suggesting a compensatory role for SRC-2. NCoR1, nuclear corepressor 1; NCoRΔID/ΔID, mice expressing NCoRΔID; NCoRΔID/ΔID Src-1−/−, mice expressing NCoRΔID and also null for SRC-1; RXR, retinoid X receptor; SRC-1, steroid receptor coactivator 1 (Ncoa1); Src-1−/−, SRC-1 knockout mice; SRC-2, steroid receptor coactivator 2 (Ncoa2); T3, triiodothryonine (thyroid hormone); TR, thyroid hormone receptor.
While NCoR1 and SRC-1 have been shown to play critical roles in vivo in thyroid hormone and nuclear receptor action, their combinatorial role in ligand activation by TH has not been shown previously. Clearly, the set point of the HPT axis can be determined by a balance between corepressors and coactivators, suggesting that genetic differences in disease states could be modified depending upon the relative expression levels of these coregulators (57). The normalization of TSH secretion in NCoRΔID/ΔID Src-1−/− mice demonstrates that other coregulators, such SRC-2, can substitute for NCoR1 and SRC-1 in vivo but that NCoR1 and SRC-1 are likely the primary drivers of the set point. This balance of coregulators is also crucial to the regulation of T3 action in the liver and supports the notion that the primary role of corepressors and coactivators is to regulate the hormone sensitivity of nuclear receptors rather than to play a unique role in their ligand-independent or -dependent action (Fig. 8). In the presence of NCoRΔID, increased SRC-2 recruitment allows T3 sensitivity of genes in the absence of SRC-1. Thus, therapeutics targeting coregulators like NCoR1 and SRC-1 in a tissue-specific fashion could potentially be used to regulate T3 sensitivity to treat metabolic disease. Whether the effects on ligand sensitivity could be limited to T3 would need to be determined.
ACKNOWLEDGMENTS
We thank Bert O'Malley for the Src-1−/− mice and Bert O'Malley and Brian York for the Src-2−/− mouse liver samples. We thank Benjamin Rodwin, Nicholas F. O'Neill, and Paul D. Same for technical assistance.
This work was supported by National Institutes of Health grants DK078090 and DK056123 to A.N.H. and DK091403 and T3207516 to K.R.V.
Footnotes
Published ahead of print 18 February 2014
REFERENCES
- 1.Costa-e-Sousa RH, Hollenberg AN. 2012. Minireview: the neural regulation of the hypothalamic-pituitary-thyroid axis. Endocrinology 153:4128–4135. 10.1210/en.2012-1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiamolera MI, Wondisford FE. 2009. Minireview: thyrotropin-releasing hormone and the thyroid hormone feedback mechanism. Endocrinology 150:1091–1096. 10.1210/en.2008-1795 [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Lazar MA. 2000. The mechanism of action of thyroid hormones. Annu. Rev. Physiol. 62:439–466. 10.1146/annurev.physiol.62.1.439 [DOI] [PubMed] [Google Scholar]
- 4.Yen PM. 2001. Physiological and molecular basis of thyroid hormone action. Physiol. Rev. 81:1097–1142 [DOI] [PubMed] [Google Scholar]
- 5.Cheng SY, Leonard JL, Davis PJ. 2010. Molecular aspects of thyroid hormone actions. Endocr. Rev. 31:139–170. 10.1210/er.2009-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onate SA, Tsai SY, Tsai MJ, O'Malley BW. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354–1357. 10.1126/science.270.5240.1354 [DOI] [PubMed] [Google Scholar]
- 7.McKenna NJ, Lanz RB, O'Malley BW. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321–344. 10.1210/edrv.20.3.0366 [DOI] [PubMed] [Google Scholar]
- 8.McKenna NJ, O'Malley BW. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465–474. 10.1016/S0092-8674(02)00641-4 [DOI] [PubMed] [Google Scholar]
- 9.Leo C, Chen JD. 2000. The SRC family of nuclear receptor coactivators. Gene 245:1–11. 10.1016/S0378-1119(00)00024-X [DOI] [PubMed] [Google Scholar]
- 10.Hermanson O, Glass CK, Rosenfeld MG. 2002. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 13:55–60. 10.1016/S1043-2760(01)00527-6 [DOI] [PubMed] [Google Scholar]
- 11.McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, Inostroza J, Torchia J, Nolte RT, Assa-Munt N, Milburn MV, Glass CK, Rosenfeld MG. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357–3368. 10.1101/gad.12.21.3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Li Q. 2003. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol. Endocrinol. 17:1681–1692. 10.1210/me.2003-0116 [DOI] [PubMed] [Google Scholar]
- 13.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397–404. 10.1038/377397a0 [DOI] [PubMed] [Google Scholar]
- 14.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49–55. 10.1038/387049a0 [DOI] [PubMed] [Google Scholar]
- 15.Guenther MG, Barak O, Lazar MA. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091–6101. 10.1128/MCB.21.18.6091-6101.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43–48. 10.1038/387043a0 [DOI] [PubMed] [Google Scholar]
- 17.Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373–380. 10.1016/S0092-8674(00)80218-4 [DOI] [PubMed] [Google Scholar]
- 18.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. 2010. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 11:109–123. 10.1038/nrg2736 [DOI] [PubMed] [Google Scholar]
- 19.Fang S, Suh JM, Atkins AR, Hong SH, Leblanc M, Nofsinger RR, Yu RT, Downes M, Evans RM. 2011. Corepressor SMRT promotes oxidative phosphorylation in adipose tissue and protects against diet-induced obesity and insulin resistance. Proc. Natl. Acad. Sci. U. S. A. 108:3412–3417. 10.1073/pnas.1017707108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss RE, Xu J, Ning G, Pohlenz J, O'Malley BW, Refetoff S. 1999. Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. EMBO J. 18:1900–1904. 10.1093/emboj/18.7.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortiga-Carvalho TM, Shibusawa N, Nikrodhanond A, Oliveira KJ, Machado DS, Liao XH, Cohen RN, Refetoff S, Wondisford FE. 2005. Negative regulation by thyroid hormone receptor requires an intact coactivator-binding surface. J. Clin. Invest. 115:2517–2523. 10.1172/JCI24109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei L, Leblanc M, Barish G, Atkins A, Nofsinger R, Whyte J, Gold D, He M, Kawamura K, Li HR, Downes M, Yu RT, Powell HC, Lingrel JB, Evans RM. 2011. Thyroid hormone receptor repression is linked to type I pneumocyte-associated respiratory distress syndrome. Nat. Med. 17:1466–1472. 10.1038/nm.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astapova I, Lee LJ, Morales C, Tauber S, Bilban M, Hollenberg AN. 2008. The nuclear corepressor, NCoR, regulates thyroid hormone action in vivo. Proc. Natl. Acad. Sci. U. S. A. 105:19544–19549. 10.1073/pnas.0804604105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Astapova I, Vella KR, Ramadoss P, Holtz KA, Rodwin BA, Liao XH, Weiss RE, Rosenberg MA, Rosenzweig A, Hollenberg AN. 2011. The nuclear receptor corepressor (NCoR) controls thyroid hormone sensitivity and the set point of the hypothalamic-pituitary-thyroid axis. Mol. Endocrinol. 25:212–224. 10.1210/me.2010-0462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa-e-Sousa RH, Astapova I, Ye F, Wondisford FE, Hollenberg AN. 2012. The thyroid axis is regulated by NCoR1 via its actions in the pituitary. Endocrinology 153:5049–5057. 10.1210/en.2012-1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonas BA, Varlakhanova N, Hayakawa F, Goodson M, Privalsky ML. 2007. Response of SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) and N-CoR (nuclear receptor corepressor) corepressors to mitogen-activated protein kinase kinase kinase cascades is determined by alternative mRNA splicing. Mol. Endocrinol. 21:1924–1939. 10.1210/me.2007-0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makowski A, Brzostek S, Cohen RN, Hollenberg AN. 2003. Determination of nuclear receptor corepressor interactions with the thyroid hormone receptor. Mol. Endocrinol. 17:273–286. 10.1210/me.2002-0310 [DOI] [PubMed] [Google Scholar]
- 28.Astapova I, Dordek MF, Hollenberg AN. 2009. The thyroid hormone receptor recruits NCoR via widely spaced receptor-interacting domains. Mol. Cell. Endocrinol. 307:83–88. 10.1016/j.mce.2009.02.028 [DOI] [PubMed] [Google Scholar]
- 29.Zamir I, Zhang J, Lazar MA. 1997. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 11:835–846. 10.1101/gad.11.7.835 [DOI] [PubMed] [Google Scholar]
- 30.Webb P, Anderson CM, Valentine C, Nguyen P, Marimuthu A, West BL, Baxter JD, Kushner PJ. 2000. The nuclear receptor corepressor (N-CoR) contains three isoleucine motifs (I/LXXII) that serve as receptor interaction domains (IDs). Mol. Endocrinol. 14:1976–1985. 10.1210/me.14.12.1976 [DOI] [PubMed] [Google Scholar]
- 31.Cohen RN, Brzostek S, Kim B, Chorev M, Wondisford FE, Hollenberg AN. 2001. The specificity of interactions between nuclear hormone receptors and corepressors is mediated by distinct amino acid sequences within the interacting domains. Mol. Endocrinol. 15:1049–1061. 10.1210/me.15.7.1049 [DOI] [PubMed] [Google Scholar]
- 32.Cohen RN, Putney A, Wondisford FE, Hollenberg AN. 2000. The nuclear corepressors recognize distinct nuclear receptor complexes. Mol. Endocrinol. 14:900–914. 10.1210/me.14.6.900 [DOI] [PubMed] [Google Scholar]
- 33.Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bucan M, Ahima RS, Kaestner KH, Lazar MA. 2008. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456:997–1000. 10.1038/nature07541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.You SH, Liao X, Weiss RE, Lazar MA. 2010. The interaction between nuclear receptor corepressor and histone deacetylase 3 regulates both positive and negative thyroid hormone action in vivo. Mol. Endocrinol. 24:1359–1367. 10.1210/me.2009-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Refetoff S, Weiss RE, Usala SJ. 1993. The syndromes of resistance to thyroid hormone. Endocr. Rev. 14:348–399. 10.1210/edrv-14-3-348 [DOI] [PubMed] [Google Scholar]
- 36.Weiss RE, Tunca H, Knapple WL, Faas FH, Refetoff S. 1997. Phenotype differences of resistance to thyroid hormone in two unrelated families with an identical mutation in the thyroid hormone receptor beta gene (R320C). Thyroid 7:35–38. 10.1089/thy.1997.7.35 [DOI] [PubMed] [Google Scholar]
- 37.Parrilla R, Mixson AJ, McPherson JA, McClaskey JH, Weintraub BD. 1991. Characterization of seven novel mutations of the c-erbA beta gene in unrelated kindreds with generalized thyroid hormone resistance. Evidence for two “hot spot” regions of the ligand binding domain. J. Clin. Invest. 88:2123–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaneshige M, Kaneshige K, Zhu X, Dace A, Garrett L, Carter TA, Kazlauskaite R, Pankratz DG, Wynshaw-Boris A, Refetoff S, Weintraub B, Willingham MC, Barlow C, Cheng S. 2000. Mice with a targeted mutation in the thyroid hormone beta receptor gene exhibit impaired growth and resistance to thyroid hormone. Proc. Natl. Acad. Sci. U. S. A. 97:13209–13214. 10.1073/pnas.230285997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell CS, Savage DB, Dufour S, Schoenmakers N, Murgatroyd P, Befroy D, Halsall D, Northcott S, Raymond-Barker P, Curran S, Henning E, Keogh J, Owen P, Lazarus J, Rothman DL, Farooqi IS, Shulman GI, Chatterjee K, Petersen KF. 2010. Resistance to thyroid hormone is associated with raised energy expenditure, muscle mitochondrial uncoupling, and hyperphagia. J. Clin. Invest. 120:1345–1354. 10.1172/JCI38793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fozzatti L, Lu C, Kim DW, Park JW, Astapova I, Gavrilova O, Willingham MC, Hollenberg AN, Cheng SY. 2011. Resistance to thyroid hormone is modulated in vivo by the nuclear receptor corepressor (NCOR1). Proc. Natl. Acad. Sci. U. S. A. 108:17462–17467. 10.1073/pnas.1107474108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bottcher Y, Paufler T, Stehr T, Bertschat FL, Paschke R, Koch CA. 2007. Thyroid hormone resistance without mutations in thyroid hormone receptor beta. Med. Sci. Monit. 13:CS67–CS70 [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O'Malley BW. 1998. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science 279:1922–1925. 10.1126/science.279.5358.1922 [DOI] [PubMed] [Google Scholar]
- 43.Nikrodhanond AA, Ortiga-Carvalho TM, Shibusawa N, Hashimoto K, Liao XH, Refetoff S, Yamada M, Mori M, Wondisford FE. 2006. Dominant role of thyrotropin-releasing hormone in the hypothalamic-pituitary-thyroid axis. J. Biol. Chem. 281:5000–5007. 10.1074/jbc.M511530200 [DOI] [PubMed] [Google Scholar]
- 44.Castinetti F, Brinkmeier ML, Gordon DF, Vella KR, Kerr JM, Mortensen AH, Hollenberg A, Brue T, Ridgway EC, Camper SA. 2011. PITX2 and PITX1 regulate thyrotroph function and response to hypothyroidism. Mol. Endocrinol. 25:1950–1960. 10.1210/me.2010-0388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vella KR, Ramadoss P, Lam FS, Harris JC, Ye FD, Same PD, O'Neill NF, Maratos-Flier E, Hollenberg AN. 2011. NPY and MC4R signaling regulate thyroid hormone levels during fasting through both central and peripheral pathways. Cell Metab. 14:780–790. 10.1016/j.cmet.2011.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramadoss P, Unger-Smith NE, Lam FS, Hollenberg AN. 2009. STAT3 targets the regulatory regions of gluconeogenic genes in vivo. Mol. Endocrinol. 23:827–837. 10.1210/me.2008-0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramadoss P, Abraham BJ, Tsai L, Zhou Y, Costa-e-Sousa RH, Ye F, Bilban M, Zhao K, Hollenberg AN. 2014. Novel mechanism of positive versus negative regulation by thyroid hormone receptor beta 1 (TRbeta1) identified by genome-wide profiling of binding sites in mouse liver. J. Biol. Chem. 289:1313–1328. 10.1074/jbc.M113.521450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadow PM, Chassande O, Gauthier K, Samarut J, Xu J, O'Malley BW, Weiss RE. 2003. Specificity of thyroid hormone receptor subtype and steroid receptor coactivator-1 on thyroid hormone action. Am. J. Physiol. Endocrinol. Metab. 284:E36–E46 [DOI] [PubMed] [Google Scholar]
- 49.Alonso M, Goodwin C, Liao X, Ortiga-Carvalho T, Machado DS, Wondisford FE, Refetoff S, Weiss RE. 2009. In vivo interaction of steroid receptor coactivator (SRC)-1 and the activation function-2 domain of the thyroid hormone receptor (TR) beta in TRbeta E457A knock-in and SRC-1 knockout mice. Endocrinology 150:3927–3934. 10.1210/en.2009-0093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamiya Y, Zhang XY, Ying H, Kato Y, Willingham MC, Xu J, O'Malley BW, Cheng SY. 2003. Modulation by steroid receptor coactivator-1 of target-tissue responsiveness in resistance to thyroid hormone. Endocrinology 144:4144–4153. 10.1210/en.2003-0239 [DOI] [PubMed] [Google Scholar]
- 51.Sadow PM, Koo E, Chassande O, Gauthier K, Samarut J, Xu J, O'Malley BW, Seo H, Murata Y, Weiss RE. 2003. Thyroid hormone receptor-specific interactions with steroid receptor coactivator-1 in the pituitary. Mol. Endocrinol. 17:882–894. 10.1210/me.2002-0174 [DOI] [PubMed] [Google Scholar]
- 52.Takeuchi Y, Murata Y, Sadow P, Hayashi Y, Seo H, Xu J, O'Malley BW, Weiss RE, Refetoff S. 2002. Steroid receptor coactivator-1 deficiency causes variable alterations in the modulation of T(3)-regulated transcription of genes in vivo. Endocrinology 143:1346–1352. 10.1210/en.143.4.1346 [DOI] [PubMed] [Google Scholar]
- 53.Weiss RE, Gehin M, Xu J, Sadow PM, O'Malley BW, Chambon P, Refetoff S. 2002. Thyroid function in mice with compound heterozygous and homozygous disruptions of SRC-1 and TIF-2 coactivators: evidence for haploinsufficiency. Endocrinology 143:1554–1557. 10.1210/en.143.4.1554 [DOI] [PubMed] [Google Scholar]
- 54.Chopra AR, Kommagani R, Saha P, Louet JF, Salazar C, Song J, Jeong J, Finegold M, Viollet B, DeMayo F, Chan L, Moore DD, O'Malley BW. 2011. Cellular energy depletion resets whole-body energy by promoting coactivator-mediated dietary fuel absorption. Cell Metab. 13:35–43. 10.1016/j.cmet.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamada M, Saga Y, Shibusawa N, Hirato J, Murakami M, Iwasaki T, Hashimoto K, Satoh T, Wakabayashi K, Taketo MM, Mori M. 1997. Tertiary hypothyroidism and hyperglycemia in mice with targeted disruption of the thyrotropin-releasing hormone gene. Proc. Natl. Acad. Sci. U. S. A. 94:10862–10867. 10.1073/pnas.94.20.10862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeong JW, Kwak I, Lee KY, White LD, Wang XP, Brunicardi FC, O'Malley BW, DeMayo FJ. 2006. The genomic analysis of the impact of steroid receptor coactivators ablation on hepatic metabolism. Mol. Endocrinol. 20:1138–1152. 10.1210/me.2005-0407 [DOI] [PubMed] [Google Scholar]
- 57.Hollenberg AN. 2012. Metabolic health and nuclear-receptor sensitivity. N. Engl. J. Med. 366:1345–1347. 10.1056/NEJMcibr1114529 [DOI] [PubMed] [Google Scholar]