Abstract
In the fission yeast Schizosaccharomyces pombe, centromeric heterochromatin is maintained by an RNA-directed RNA polymerase complex (RDRC) and the RNA-induced transcriptional silencing (RITS) complex in a manner that depends on the generation of short interfering RNA. In association with the telomerase RNA component (TERC), the telomerase reverse transcriptase (TERT) forms telomerase and counteracts telomere attrition, and without TERC, TERT has been implicated in the regulation of heterochromatin at locations distinct from telomeres. Here, we describe a complex composed of human TERT (hTERT), Brahma-related gene 1 (BRG1), and nucleostemin (NS) that contributes to heterochromatin maintenance at centromeres and transposons. This complex produced double-stranded RNAs homologous to centromeric alpha-satellite (alphoid) repeat elements and transposons that were processed into small interfering RNAs targeted to these heterochromatic regions. These small interfering RNAs promoted heterochromatin assembly and mitotic progression in a manner dependent on the RNA interference machinery. These observations implicate the hTERT/BRG1/NS (TBN) complex in heterochromatin assembly at particular sites in the mammalian genome.
INTRODUCTION
Telomeres and centromeres are both tightly condensed heterochromatic areas within the genome, and the maintenance of heterochromatin is important for overall genome stability. In Schizosaccharomyces pombe, heterochromatin near centromeres is maintained by the RNA-directed RNA polymerase complex (RDRC) and the RNA-induced transcriptional silencing (RITS) complex (1, 2). Specifically, inhibition of RNA-dependent RNA polymerase (RdRP) activity leads to loss of small interfering RNAs (siRNAs) that are associated with the RITS complex and correlates with loss of transcriptional silencing and heterochromatin at centromeres (3). In addition, when RdRP activity is inhibited, siRNAs that are usually associated with the RITS complex are lost (4). These observations implicate RdRPs as a component of a loop coupling heterochromatin assembly to siRNA production.
In Caenorhabditis elegans, the Argonaute CSR-1, the RdRP EGO-1, and the Dicer-related helicase DRH-3 localize to chromosomes and are required for proper chromosome segregation, and in the absence of these factors, chromosomes fail to properly align in mitotic phase (5, 6). Moreover, a conserved germ line-specific nucleotidyltransferase, CDE-1, localizes specifically to mitotic chromosomes in embryos in a manner that requires the RdRP EGO-1, which physically interacts with CDE-1, and the Argonaute protein CSR-1 (5, 6). Although it is clear that RdRP and components of the RNA interference (RNAi) machinery are necessary to regulate heterochromatin in S. pombe and C. elegans, it is believed that heterochromatin is regulated in mammals through different mechanisms (7).
Telomerase is a ribonucleoprotein complex that elongates telomeres. Human telomerase reverse transcriptase (hTERT) acts as an RNA-dependent DNA polymerase (RdDP) and synthesizes telomere DNA from a noncoding RNA (ncRNA) template, hTERC. Telomere homeostasis mediated by TERT maintains genomic stability and regulates cell life span by maintaining telomeres. However, several lines of evidence suggest that hTERT has functions independent of its known role in telomere maintenance (8–12). Specifically, hTERT expression reduces the number of dicentric chromosomes and suppresses aneuploidy in a manner independent of telomere length (13). In consonance with these observations, hTERT forms several intracellular complexes, only some of which contribute to telomere maintenance (14, 15). For example, hTERT forms a complex with Brahma-related gene 1 (BRG1, or SMARC4) that modulates the transcription of Wnt-dependent genes, such as Myc and cyclin D1 (11). hTERT also interacts with the RNA component of mitochondrial RNA processing endoribonuclease (RMRP) to exert RdRP activity, which produces double-stranded RNAs (dsRNAs) that are processed into siRNAs by the RNase III-like enzyme Dicer (10). In addition, we showed that the hTERT/BRG1 complex includes the nucleolar GTP-binding protein, nucleostemin (NS) (this complex will be referred to as TBN), which is essential for maintaining the function of tumor initiating cells (TICs) through telomere-independent mechanisms (15). Together, these studies suggest that hTERT forms multiple complexes with distinct functions in chromosome stability and the maintenance of heterochromatin. Here, we investigated whether hTERT regulates the assembly of heterochromatin in mammalian cells, specifically in complex with BRG1 and NS.
MATERIALS AND METHODS
Generation of an anti-hTERT MAb.
Sense and antisense oligonucleotides corresponding to 304 amino acids (aa) to 460 aa of hTERT were purchased from Integrated DNA Technologies Co., Ltd. (IDT) and cloned into plasmid pET-30a(+) (Novagen). A recombinant, carboxyl-terminal His-tagged hTERT protein of 157 amino acids of hTERT (position 304 to 460) was overexpressed in Escherichia coli and purified by passage through a nickel-agarose column. The recombinant purified hTERT was used as an immunogen to stimulate the production of anti-hTERT monoclonal antibody (MAb) in mice using standard methodologies (16). A sequential screening strategy was used to identify hybridomas producing anti-hTERT MAb. In the initial screen, an enzyme-linked immunosorbent assay (ELISA) was employed to detect hybridomas that produced MAb that bound plate-bound purified His-hTERT. The reactive supernatants were then used in validation experiments described in the legend of Fig. 1.
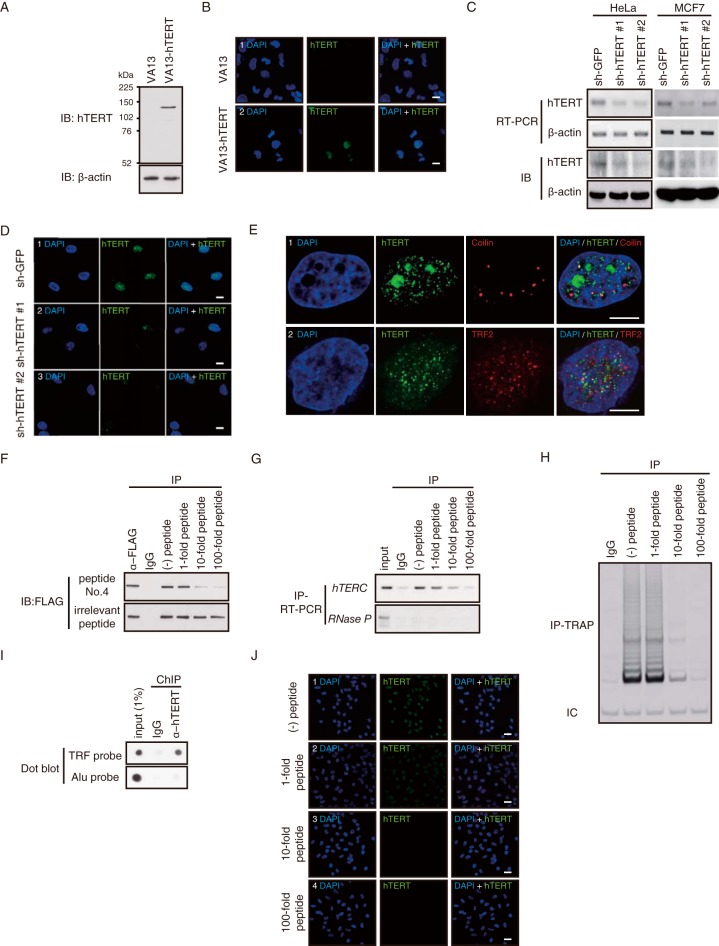
FIG 1.
Validation of the anti-hTERT MAb. (A) Immunoblot (IB) of ectopically expressed hTERT using an anti-hTERT MAb. hTERT was transiently expressed in VA13 cells and immunoblotted with the 10E9-2 anti-hTERT MAb. (B) Immunofluorescence (IF) of hTERT and DAPI in VA13 cells. VA13 and VA13-hTERT cells were immunostained with the 10E9-2 MAb followed by DAPI staining. Scale bar, 10 μm. (C) RT-PCR or IB of hTERT expression in HeLa or MCF7 cells expressing control (shRNA to GFP) or hTERT-specific shRNAs. β-Actin, internal control. (D) IF of hTERT in HeLa cells expressing sh-GFP as a control, sh-hTERT 1, or sh-hTERT 2. Representative images are shown. Scale bar, 10 μm. (E) Colocalization of hTERT with coilin or TRF2. HeLa cells were immunostained with anti-hTERT MAb and anticoilin antibodies (row 1) or anti-TRF2 antibodies (row 2) followed by DAPI staining. Scale bar, 10 μm. (F) Immunoprecipitation (IP) of overexpressed hTERT using anti-hTERT MAb. FLAG-tagged hTERT was transiently expressed in 293T cells, and immune complexes were isolated using anti-hTERT MAb incubated with or without peptide 4 or irrelevant peptide (peptide 5) and immunoblotted with the FLAG-M2 antibody. (G) IP of endogenous hTERT using anti-hTERT MAb. Immune complexes were isolated from HeLa cells using anti-hTERT MAb incubated with or without antigen peptides, and hTERC and RNase P RNA were detected by RT-PCR. (H) IP-TRAP of endogenous hTERT from HeLa cells. IC, internal control. (I) ChIP performed in HeLa cells using the anti-hTERT MAb. Dot blot signals were detected with the indicated γ-32P-labeled probes. (J) IF of hTERT and DAPI in HeLa cells. Cells were immunostained with anti-hTERT MAb incubated with no peptide [(−) peptide] or a 1-, 10-, or 100-fold molar excess of peptide 4. Representative images are shown. Scale bar, 10 μm.
Absorption of anti-hTERT MAb.
An anti-hTERT MAb was first incubated with no peptide or a 1-fold, 10-fold, or 100-fold molar excess of peptide 4 (see Fig. S1 in the supplemental material). After 1 h of incubation at 4°C, the MAb was used for immunofluorescence (IF) or immunoprecipitation (IP) experiments.
Peptide array.
Seventy-five peptides derived from a truncated version of hTERT (aa 304 to 460) covalently bound to a continuous cellulose membrane. The panel of peptides was then probed with the anti-hTERT MAb, and bound antibody was detected using Pep Spot (Funakoshi) according to the manufacturer's protocol.
Cell culture and stable expression of hTERT and GFP-hTERT.
The human cell lines 293T, HeLa, MCF7, and VA13 and mouse embryonic fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (IFS). The animal experiment protocols were approved by the Committee for Ethics in Animal Experimentation, and the experiments were conducted in accordance with the Guideline for Animal Experiments of the National Cancer Center. HeLa cells and VA13 cells transiently transfected with pNKFLAG-Z-hTERT (10) were used for sucrose density gradient centrifugation and immunoblotting (IB). Amphotropic retroviruses were created as previously described (17) using the vector pBH-hTERT or pMIG-hTERT-GFP (where GFP is green fluorescent protein) (a generous gift from Akira Orimo). Plasmids were transfected using Fugene HD (Roche Diagnostics). After infection, VA13-hTERT cells were selected with hygromycin (100 μg/ml) for 7 days.
Mitotic cell synchronization.
The mitotic cell synchronization protocol described by Summers et al. (18) was used. Briefly, cells were switched to medium containing 2.5 mM thymidine (Nacalai Tesque) and incubated for 24 h. Six hours after release, the cells were incubated in medium containing 0.1 μg/ml nocodazole (Invitrogen) for 14 h. After gentle shaking of the dishes, mitotic cells were retrieved.
RT-PCR and quantitative RT-PCR (qRT-PCR).
Total cellular RNA was isolated using TRIzol (Invitrogen), treated with DNase (Promega), and subjected to reverse transcription-PCR (RT-PCR). The RT reaction was performed for 60 min at 42°C, followed immediately by PCR (94°C for 30 s, 60°C for 30 s, and 72°C for 30 s). Cycle numbers for PCR are shown in Table S1 in the supplemental material. The alphoid reverse primer was labeled with [γ-32P]ATP. satellite I primers were used instead of alphoid primers to obtain unequivocal PCR products for VA13 cells. Primers used for RT-PCR are listed in Table S1.
Quantitative RT-PCR was performed with a LightCycler 480II (Roche) using LightCycler 480 SYBR green I Master (Roche) according to the manufacture's protocols. Quantitative RT-PCR of Satellite 2 (Sat2) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed using an Epitect ChIP Antibody Kit for human histone H3 trimethylated at lysine 9 (H3K9me3) (Qiagen) according to the manufacturer's protocols. Human GAPDH, human β-actin, and mouse β-actin genes were used as reference genes. Primers used for qRT-PCR are listed in Table S2 in the supplemental material.
Stable expression of shRNA.
We used the pLKO.1-puro vector and the sequences listed in Table S3 in the supplemental material to create short hairpin RNA (shRNA) vectors specific for hTERT, BRG1 (15), NS (15), GFP, AGO2, and DICER1 (10). These vectors were used to make amphotropic lentiviruses, and polyclonal cell populations were purified by selection with puromycin (2 μg/ml for 3 days).
Antibodies.
The following antibodies were used for immunoblotting: anti-FLAG M2 (A2220; Sigma-Aldrich), anti-β-actin AC-15 (A5441; Sigma-Aldrich), anti-NS (A300-600A; Bethyl Laboratories), anti-BRG1 (a gift from Tsutomu Ohta, National Cancer Center), anti-phospho-histone H3 (Ser10) (pHH3Ser10, (06-570; Millipore), anti-H3K9me3 (07-442; Millipore), anti-histone H3 (06-755; Millipore), anti-DICER 13D6 (ab14601; Abcam), anti-AGO2 4G8 (015-22031; Wako), and anti-hTERT MAb (clone 10E9-2).
The following antibodies were used for immunoprecipitation (IP), IP–RT-PCR, IP-telomere repeat amplification protocol (TRAP), and chromatin immunoprecipitation (ChIP): anti-hTERT MAb (clone 10E9-2), anti-AGO2 4G8 (015-22031; Wako), anti-H3K9me3 (ab8898; Abcam), ChromPure mouse IgG (015-000-003; Jackson ImmunoResearch), and ChromPure rabbit IgG (011-000-003; Jackson ImmunoResearch).
The mouse MAbs used for immunofluorescence analysis were anti-hTERT MAb (clone 10E9-2), anticoilin IH10 (ab87913; Abcam), anti-centromere protein A (CENPA) 3-19 (D115-3; Medical and Biological Laboratories Co., Ltd.), and anti-AGO2 4G8 (015-22031; Wako). The rabbit MAb used was anti-CENPA EP800Y (04-205; Millipore). The rabbit polyclonal antibodies used were anti-NS (A300-600A; Bethyl laboratories), anti-BRG1 (a gift from Tsutomu Ohta, National Cancer Center), anti-pHH3Ser10 (06-570; Millipore), anti-α-tubulin DM1A (T6199; Sigma-Aldrich), anti-H3K9me3 (07-442; Millipore), and anti-HP1β (07-333; Millipore). The rat MAb used was anti-α-tubulin YL1/2 (ab6160; Abcam). The goat polyclonal antibody used was anti-TRF2 (IMG-148A; Imgenex). The following secondary antibodies were used: Alexa Fluor 488-, 568-, or 647-conjugated goat anti-mouse, -rabbit, or -rat IgG (Invitrogen) and Alexa Fluor 488- or 568-conjugated donkey anti-mouse or -goat IgG (Invitrogen).
Immunoblotting.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing 1% NP-40, 1 mM EDTA, 50 mM Tris-HCl (pH 7.4), and 150 mM NaCl. After sonication, lysates were cleared of insoluble material by centrifugation at 21,000 × g at 4°C for 15 min. Proteins (100 μg) were subjected to SDS-PAGE in 6%, 8%, or 12% polyacrylamide gels, followed by immunoblot analysis.
Immunoprecipitation.
For IP followed by immunoblotting, cells were lysed in lysis buffer A (LBA) containing 0.5% NP-40, 20 mM Tris-HCl (pH 7.4), and 150 mM NaCl. After sonication, lysates were cleared of insoluble material by centrifugation at 21,000 × g at 4°C for 15 min, followed by the addition of 20 μl of prewashed 1:1 slurry of anti-FLAG M2 Affinity gel (Sigma-Aldrich) or 4 μg of antibody and 30 μl of Pierce protein A plus agarose (Thermo Scientific) and incubation overnight at 4°C. The bead complexes were washed three times with LBA and eluted in 2× SDS loading buffer containing 20% β-mercaptoethanol, 20% glycerol, 4% SDS, and 100 mM Tris-HCl (pH 7.4), followed by SDS-PAGE and immunoblot analysis.
For IP followed by RT-PCR, cells were lysed in LBA. After a 30-min incubation on ice, lysates were cleared of insoluble material by centrifugation at 21,000 × g at 4°C for 15 min. The whole-cell lysates were incubated overnight with 4 μg of antibody and 30 μl of Pierce protein A plus agarose at 4°C. The protein A bead complexes were then washed three times with LBA, eluted in TRIzol, and analyzed by RT-PCR.
Sucrose density gradient centrifugation.
HeLa cells overexpressing FLAG-hTERT were lysed in LBA and incubated on ice for 30 min. Lysates were cleared of insoluble material by centrifugation at 21,000 × g at 4°C for 30 min. Five hundred microliters of lysate (5 μg/μl protein concentration) was loaded onto 5 ml of a preformed 15 to 37.5% sucrose-LBA gradient and centrifuged for 17 h at 130,000 × g using a P40ST rotor in an ultracentrifuge (Hitachi). After centrifugation, the contents were aliquoted into 10 consecutive fractions (F1 to F10) with a volume of 500 μl each, and the fractions were sequentially numbered from high density (F1) to low density (F10). Seventy-five microliters of each fraction was used for immunoblotting.
TRAP assay.
A telomeric repeat amplification protocol (TRAP) was used to detect telomere-specific reverse transcriptase activity as described previously (19).
Immunofluorescence.
HeLa, VA13, or MCF7 cells (2.8 × 104 cells) were seeded onto eight-well culture slides (BD). The cells were fixed using a protocol based on that described by Thibault and Buschmann (20). Briefly, 1 day after seeding, cells were washed twice with phosphate-buffered saline not containing either calcium ions or magnesium ions [PBS(−)] and fixed in PBS(−) containing 1% paraformaldehyde and 0.5% Triton X-100 at 37°C for 20 min. Then, the cells were washed three times in PBS(−), followed by a 30-min incubation in PBS(−) containing 100 mM glycine at room temperature. All immunocytochemical reactions were performed at room temperature, unless otherwise specified. The fixed cells were incubated for 2 h or at 4°C overnight with PBS(−) containing 5% bovine serum albumin Cohn fraction V (WAKO) to block nonspecific reactions. The fixed cells were incubated first with primary antibody at room temperature for 2 h or at 4°C overnight. After cells were washed four times with PBS(−), they were incubated with secondary antibodies for 1.5 h. After another four washes in PBS(−), the cells were mounted in Vectashield containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories) and imaged under a laser scanning microscope (TCS SP2; Leica Microsystems). To analyze the mitotic index in the cells expressing shRNAs, HeLa cells were infected with lentiviruses to express shRNAs, and polyclonal cell populations were purified by selection with puromycin. Four days after infection, the cells were fixed. For analyzing DNA fluorescence intensity, the cells were treated with PBS(−) containing 1 mM Hoechst 33342 (Invitrogen) for 20 min at room temperature, followed by two washes in PBS(−). The cells were then observed under a spinning-disk confocal microscope (IX-81 with DSU; Olympus Corporation).
DNA content analysis.
DNA content analysis was performed with a JSAN cell sorter (Bay Bioscience) using a Cycletest Plus DNA reagent kit (BD Bioscience) according to the manufacturer's protocols.
IP-RdRP assay.
IP followed by an RdRP (IP-RdRP) assay was modified from the original protocol (21). For the assay, 1 × 107 cells were lysed in 1 ml of lysis buffer A (0.5% NP-40, 20 mM Tris-HCl [pH 7.4], and 150 mM NaCl). After sonication, lysates were cleared of insoluble material by centrifugation at 21,000 × g at 4°C for 15 min. The lysate was preabsorbed with 40 μl of Pierce protein A plus agarose (Thermo Scientific) for 30 min. Preabsorbed lysate was mixed with 10 μg of anti-hTERT MAb (clone 10E9-2), anti-BRG1 antibody (ab4081; Abcam), or anti-NS antibody (A300-599A; Bethyl Laboratories) and 40 μl of Pierce protein A plus agarose and incubated overnight at 4°C. Immune complexes were washed four times with 1× acetate buffer (10 mM HEPES-KOH [pH 7.8], 100 mM potassium acetate, and 4 mM MgCl2) containing 10% glycerol, 0.1% Triton X, and 0.06× protease inhibitor and once with AGC solution [1× acetate buffer, 10% glycerol, and 0.02% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) containing 2 mM CaCl2. The bead suspension was treated with 0.25 unit/μl MNase at 25°C for 15 min. The immunoprecipitates were subsequently washed twice with AGC solution containing 3 mM EGTA and once with 1× acetate buffer containing 0.02% CHAPS. Finally, 40 μl of a reaction mixture was prepared from 20 μl of the bead suspension, 6 μl of [α-32P]UTP (3,000 Ci/mmol), a final 25 ng/μl of RNA template, and supplements and incubated at 32°C for 2 h. The final concentrations of ribonucleotides were 1 mM ATP, 0.2 mM GTP, 10.5 μM UTP, and 0.2 mM CTP. The resulting products were treated with proteinase K to stop the reaction, purified several times with phenol-chloroform until the white interphase was gone, and precipitated using ethanol. For a UTP incorporation assay, RdRP products were treated with RNase I (2 U; Promega) at 37°C for 2 h to digest single-stranded RNAs (ssRNAs) and RNase V1 (0.2 U; Ambion) at 37°C for 1 h to digest dsRNAs, followed by proteinase K treatment, phenol-chloroform purification, and ethanol precipitation. In the Northern blotting experiment that examined antisense RNA synthesized by hTERT, nonlabeled UTP was used instead of [α-32P]UTP. Synthetic RNAs used as templates for IP-RdRP assays were the following: synthetic RNA 1, 5′-GGGUUUAAAAUGUAAUAGGACCCACAUGAUCCCA-3′; synthetic RNA 2, 5′-GGGAUCAUGUGGGUCCUAUUACAUUUUAAACCCA-3′.
Northern blotting.
Small RNAs (<200 nucleotides in length) were isolated using a mirVana miRNA Isolation Kit (Ambion) according to the manufacturer's protocol. Ten micrograms of small RNA or the product of the IP-RdRP assay was separated on denaturing 20% or 10% polyacrylamide gels and then blotted onto Hybond-N+ membranes (GE Healthcare) using a Trans-Blot SD semidry transfer cell (Bio-Rad). Hybridization was performed in Church buffer (0.5 M NaHPO4 [pH 7.2], 1 mM EDTA, and 7% SDS) containing 1 × 106 cpm/ml of 32P-labeled sense probe derived from nucleotides 229 to 267 of RMRP, antisense probe derived from nucleotides 1 to 70 of hLINE1, or antisense probe derived from nucleotides 120 to 171 of alphoid for 14 h. The membranes were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and the signals were detected by autoradiography.
Chromatin immunoprecipitation (ChIP).
HeLa cells (1 × 107 cells) were cross-linked with 1% paraformaldehyde (Nacalai Tesque) in culture medium for 10 min at room temperature. After aldehydes were quenched with PBS(−) containing 200 mM glycine (Sigma-Aldrich) for 5 min followed by a PBS(−) wash, the fixed cells were harvested with 1 ml of NP-40 buffer (10 mM Tris-HCl [pH 8.0], 10 mM NaCl, and 0.5% NP-40) using a cell scraper (Costar) and centrifuged at 1,000 × g at 4°C for 3 min. The cell pellets were resuspended in 100 μl of SDS lysis buffer B (50 mM Tris-HCl [pH 8.0], 1% SDS, and 10 mM EDTA) and 400 μl of ChIP dilution buffer (50 mM Tris-HCl [pH 8.0], 167 mM NaCl, 1.1% Triton X-100, and 0.11% sodium deoxycholate) containing 1% protease inhibitor cocktail [4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), aprotinin, E-64, leupeptin hemisulfate monohydrate; Nacalai Tesque]. The lysate was sonicated using a sonication system (Bioruptor, CosmoBio). The sonicated lysate was centrifuged at 20,000 × g at 4°C for 10 min, and the supernatant was transferred to new tubes. ChIP dilution buffer (500 μl) was added to the supernatant, and 100 μl of the supernatant was set aside for input. One hundred microliters of 1× RIPA buffer A (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, and 0.1% sodium deoxycholate) was added to the supernatant. The supernatant was precleared using protein G-Sepharose beads (GE Healthcare) at 4°C for 2 h, followed by centrifugation at 9,000 × g at 4°C for 1 min. The supernatant was then transferred to new tubes. Antibodies were added to the supernatant, and the tubes were rotated overnight at 4°C. Protein G-Sepharose beads blocked by DMEM supplemented with 10% IFS were then added, and the samples were rotated for another 2 h at 4°C. The antibody-protein G-Sepharose complex was washed with 1× RIPA buffer A at 4°C for 5 min followed by a wash with 1× RIPA buffer B (50 mM Tris-HCl [pH 8.0], 500 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, and 0.1% sodium deoxycholate) and two washes with TE buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA) at 4°C for 5 min. ChIP elution buffer (200 μl; 10 mM Tris-HCl [pH 8.0], 300 mM NaCl, 5 mM EDTA, and 0.5% SDS) was added and incubated at 65°C overnight. After the incubation, 4 μg of RNase A (Invitrogen) was added and incubated at 37°C for 30 min. Proteinase K (20 μg) was then added, and the sample was incubated at 55°C for 1 h. DNA was then purified using a standard phenol-chloroform extraction followed by ethanol precipitation and was resuspended in TE buffer.
Dot blot analysis.
DNA retrieved by ChIP was denatured at 100°C for 10 min in 400 mM NaOH and 10 mM EDTA. Denatured samples were dotted onto a Hybond-XL membrane (Amersham) using a Bio-Dot Apparatus (Bio-Rad). The membrane was baked at 80°C and prehybridized with Church buffer (0.5 M NaHPO4 [pH 7.2], 1 mM EDTA, 3% SDS) at 37°C for 2 h. After prehybridization, γ-32P-labeled probes were added and hybridized at 37°C overnight. After hybridization, the membrane was rinsed twice with 10 ml of 2× SSC containing 0.1% SDS followed by two washes with 50 ml of 2× SSC containing 0.1% SDS for 10 min, two washes with 50 ml of 1× SSC containing 0.1% SDS for 10 min, and four washes with 50 ml of 0.1× SSC containing 0.1% SDS for 5 min. The washing steps were done at 37°C. Probes labeled with [γ-32P]ATP using T4 polynucleotide kinase (TaKaRa Bio, Inc.) were the following: TRF probe, (5′-CCCTAACCCTAACCCTAA-3′; Alu probe, 5′-CGGAGTCTCGCTCTGTCGCCCAGGCTGGAGTGCAGTGGCGCGA-3′; alphoid probe, 5′-AGTTTCTGAGAATCATTCTGTCTAG-3′; hLINE1 probe, 5′-CGGAAGAGTGTCTGGAGCAA-3′.
AZA, 5-aza-dC, and TSA treatment.
For treatment of 5-azacytidine (AZA) and trichostatin A (TSA), HeLa cells were switched to medium containing 10 μM AZA (Sigma-Aldrich) for 24 h followed by treatment with 0.5 μM TSA (Sigma-Aldrich) for 12 h. For treatment of 5-aza-deoxycytidine (5-aza-dC) and TSA, HeLa cells were switched to medium containing 0.3 μM 5-aza-dC (Sigma-Aldrich) twice for 48 h and 24 h followed by treatment of 0.5 μM TSA for 24 h.
RESULTS
hTERT localizes to both mitotic spindles and centromeres.
TERT protein is expressed at low levels even in human cancer cell lines (22). To facilitate the study of hTERT, we generated a new anti-hTERT monoclonal antibody (MAb) (clone 10E9-2) by immunizing mice with a recombinant truncated version of hTERT (amino acids 304 to 460). Using this MAb, we detected ectopically expressed hTERT in VA13 cells, known to lack hTERT, by immunoblotting (IB) (Fig. 1A) and immunofluorescence (IF) (Fig. 1B). We also demonstrated that the 10E9-2 MAb detected endogenous TERT in the HeLa and MCF7 human cancer cell lines (Fig. 1C and D). Expression of two different hTERT-specific short hairpin RNAs (shRNAs) in these cells suppressed hTERT mRNA expression and endogenous protein expression in both cell lines (Fig. 1C and D). We found that hTERT colocalized with coilin, a marker for Cajal bodies (14), or TRF2, a component of shelterin complex which locates at telomeres during S phase (23, 24), in HeLa cells (Fig. 1E). Because we used asynchronously dividing cells, not all of TRF2 colocalized with hTERT as reported previously (25). To confirm the specificity of 10E9-2, we mapped the epitope recognized by this MAb by testing reactivity to 75 peptides spanning amino acids 304 to 460 of hTERT (see Fig. S1A in the supplemental material) and found that peptide 4 (sequence, TSRPPRPWDT) was recognized by this MAb (see Fig. S1B). We then showed that the addition of peptide 4 but not an irrelevant peptide (peptide 5) (see Fig. S1B) inhibited the ability to isolate exogenous (Fig. 1F) or endogenous hTERT immune complexes (Fig. 1F to H) as assessed by IB, measuring hTERC levels, or TRAP assay. Furthermore, we performed a ChIP assay using this MAb and detected a strong signal by a telomere restriction fragment (TRF) probe (Fig. 1I) (26). When we performed IF, peptide 4 inhibited the signal from this antibody binding to endogenous hTERT (Fig. 1J). Taken together, these observations confirm that the 10E9-2 MAb recognizes endogenous hTERT.
To determine the subcellular localization of hTERT, we performed IF experiments in VA13, VA13-hTERT, and human cancer cell lines. Although we failed to detect hTERT in VA13 cells in any phase of the cell cycle (Fig. 1B and 2A), we found that hTERT localized to mitotic spindles, as assessed by colocalization with α-tubulin, in VA13-hTERT cells (Fig. 2A) and HeLa cells (Fig. 2B). The expression of hTERT-specific shRNAs (Fig. 2C, upper panels) or introduction of peptide 4 (Fig. 2C, lower panels) eliminated this staining pattern. We also confirmed that hTERT colocalized with CENPA, a centromeric marker (Fig. 2D and E), during mitosis, which indicates that hTERT localized not only on mitotic spindles but also at centromeres. Although hTERT associates with telomeres during S phase (26), TRF1 and TRF2 create a closed configuration of telomeres during mitosis that prevents telomerase from elongating telomeres (27). Based on these observations, we concluded that TERT is localized to mitotic spindles and centromeres in mitosis.
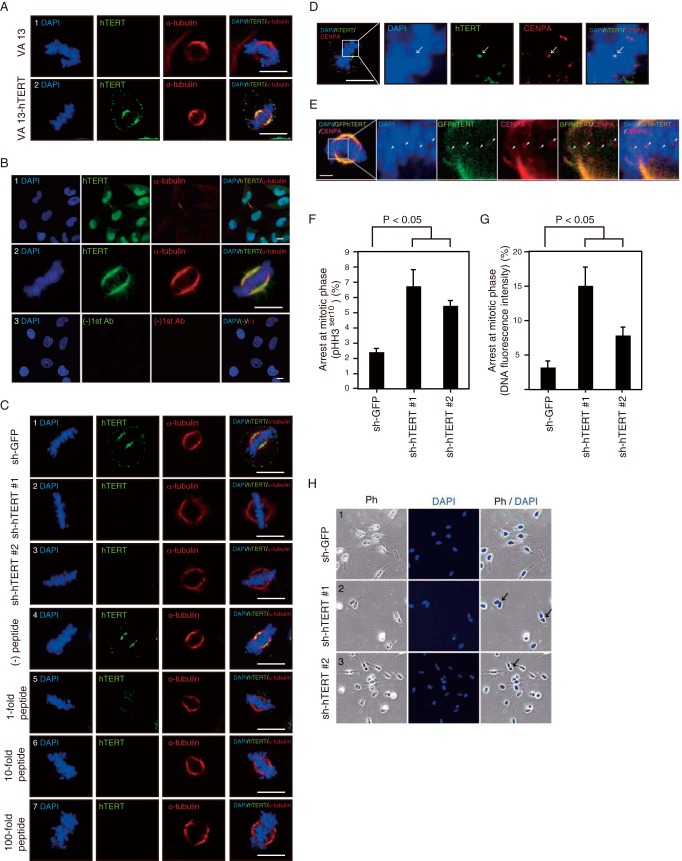
FIG 2.
hTERT localizes to both mitotic spindles and centromeres. (A) Immunofluorescence (IF) of hTERT, α-tubulin, and DAPI in VA13 cells. VA13 and VA13-hTERT cells were immunostained with the 10E9-2 anti-hTERT MAb and anti-α-tubulin antibodies followed by DAPI staining. Scale bar, 10 μm. (B) IF of hTERT, α-tubulin, and DAPI in HeLa cells in interphase (rows 1 and 3) or in mitosis (row 2). HeLa cells were not immunostained [(−)1st Ab] or were immunostained with anti-hTERT MAb and anti-α-tubulin antibodies followed by DAPI staining. Scale bar, 10 μm. (C) IF of hTERT, α-tubulin, and DAPI in HeLa cells. Suppression of hTERT (sh-GFP as a control) was observed by immunofluorescence. MAb was incubated with no peptide [(−) peptide] and a 1-, 10-, or 100-fold molar excess of antigen peptide. HeLa cells were immunostained with anti-hTERT MAb and anti-α-tubulin antibodies followed by DAPI staining. Representative images are shown. Scale bar, 10 μm. (D) Colocalization of hTERT and CENPA. HeLa cells were immunostained with anti-hTERT MAb and anti-CENPA antibodies followed by DAPI staining. The image in the rectangle shown in the left panel is magnified and shown on the right. Arrow denote colocalization of hTERT with CENPA. Scale bar, 10 μm. (E) Colocalization of GFP-hTERT and CENPA. HeLa-GFP-hTERT cells were immunostained with anti-CENPA followed by DAPI staining. The image in the rectangle shown in the left panels is magnified and shown on the right. Arrowheads denote colocalization of GFP-hTERT with CENPA. Scale bar, 5 μm. (F) A graph of the mitotic arrest according to pHH3Ser10 immunofluorescence in HeLa cells expressing shRNAs. The P value for sh-hTERT versus sh-GFP was <0.05. (G) A graph of the mitotic arrest according to DNA fluorescence intensity and the morphology of HeLa cells expressing shRNAs. The P value for sh-hTERT versus sh-GFP was <0.05. (H) Binucleate formation of HeLa cells with sh-hTERT. HeLa cells expressing sh-GFP, sh-hTERT 1, or sh-hTERT 2 were stained with DAPI. Arrow denote binucleate cells. Ph, phase-contrast image.
To determine whether the association of hTERT with mitotic spindles and centromeres exerts an influence on their function, we assessed mitotic progression in cells expressing control or hTERT-specific shRNAs by measuring phospho-histone H3Ser10 (pHH3Ser10) (Fig. 2F) or DNA fluorescence intensity (Fig. 2G). We found that the percentage of cells arrested in mitosis was increased 2- to 5-fold in cells in which hTERT was suppressed compared to cells expressing shRNA targeting GFP (sh-GFP) (P values of <0.05) (Fig. 2F and G). At the same time, we found an increased ratio of binucleate cells in HeLa cells expressing sh-hTERT (sh-hTERT 1, 3.6%, or 16/443 cells; sh-hTERT 2, 2.6%, or 9/350 cells) compared to those expressing sh-GFP (0.34%, 2/591 cells) (Fig. 2H). We confirmed that under these conditions, expression of these shRNAs did not alter cell viability (10). These observations demonstrate that the suppression of hTERT inhibits mitotic progression.
hTERT regulates gene expression from centromeres and retrotransposons and is essential for heterochromatin assembly.
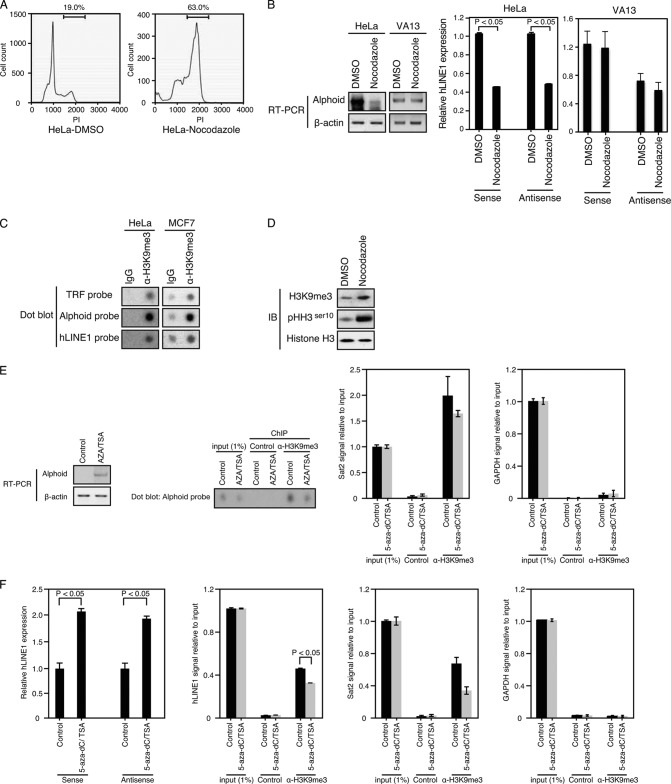
The maintenance of heterochromatin at centromeres and other regions is essential for proper chromosomal segregation during mitosis in S. pombe (28), C. elegans (5, 6), and human cells (29). Using nocodazole treatment to arrest cells in mitosis as assessed by staining for pHH3Ser10 (Fig. 3D) and examining the cell cycle distribution of treated cells (Fig. 3A), we monitored the expression of transcripts from centromeric alpha-satellite (alphoid) repeat elements (30) and the noncentromeric long interspersed repeat element 1 (LINE1) (31) in HeLa cells or VA13 cells. We confirmed that the expression of alphoid and hLINE1 transcripts was tightly suppressed during mitosis in HeLa cells (Fig. 3B). In consonance with these findings, we found baseline levels of histone H3 trimethylated at Lys9 (H3K9me3), a marker for silent chromatin (32), at telomeres and alphoid and hLINE1 elements in cycling cells (Fig. 3C) and increased levels of H3K9me3 in HeLa cells treated with nocodazole (Fig. 3D). To confirm that the expression of alphoid or hLINE1 RNA levels represented the heterochromatic state of these regions, we exposed these cells to trichostatin A (TSA) and two inhibitors of DNA methyltransferase, 5-azacytidine (AZA) and 5-aza-deoxycytidine (5-aza-dC) (33), and found that cells treated with these agents overexpressed alphoid or hLINE1 RNA and showed downregulation of H3K9me3 marks at alphoid and hLINE1 regions (Fig. 3E and F). These observations confirmed that alphoid and hLINE1 are regulated by the state of heterochromatin.
FIG 3.
Expression of alphoid and hLINE1 is regulated by the state of heterochromatin. (A) DNA content analysis of cells treated with DMSO or nocodazole. The cell fractions in G2/M phase were 19% (DMSO) and 63% (nocodazole). (B) RT-PCR of alphoid and qRT-PCR of hLINE1 RNA expression levels in HeLa or VA13 cells treated with nocodazole (manipulated) or DMSO (unmanipulated). The alphoid DNA PCR products are 171 bp, or multiples of 171 bp (56). A representative band is shown. β-Actin, internal control. P values for nocodazole versus DMSO were <0.05. (C) ChIP performed in HeLa or MCF7 cells using anti-H3K9me3 antibody. Dot blot signals were detected with the indicated γ-32P-labeled probes. (D) Immunoblot (IB) analysis using the indicated antibodies in HeLa cells treated with nocodazole or DMSO. Histone H3, internal control. (E) RT-PCR of alphoid and ChIP using anti-H3K9me3 Ab. Dot blot signals were detected with the γ-32P-labeled alphoid probes. HeLa cells were treated with AZA (10 μM) followed by TSA (0.5 μM). Satellite 2 (Sat2) (positive control) and GAPDH (negative control) were used as controls for ChIP using anti-H3K9me3 Ab. (F) qRT-PCR of hLINE1 and ChIP using anti-H3K9me3 Ab. HeLa cells were treated with 5-aza-dC (0.3 μM) followed by TSA (0.5 μM). α, anti.
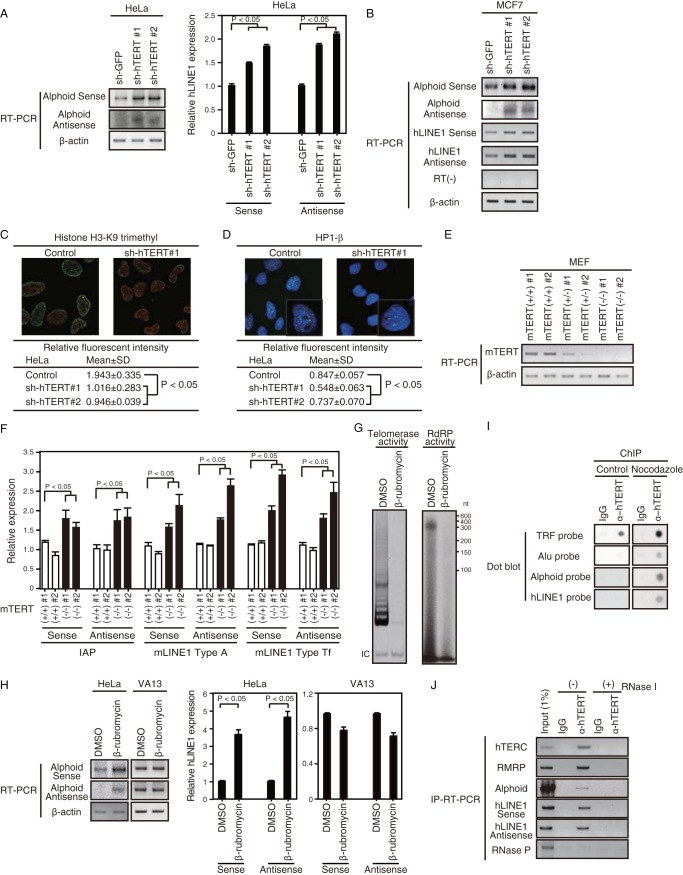
To assess the consequences of suppressing hTERT on the expression and chromatin state of these regions, we monitored the expression level of alphoid and hLINE1 in cells which we suppressed hTERT. Specifically, we monitored the alphoid and hLINE1 RNA expression levels in HeLa and MCF7 cells in which endogenous hTERT was depleted by two independent shRNAs (Fig. 1C and D). Suppressing hTERT resulted in increased transcription of both sense and antisense RNAs from alphoid and hLINE1 elements as assessed by quantitative RT-PCR in HeLa (Fig. 4A) or MCF7 cells (Fig. 4B). Moreover, when we suppressed TERT expression by TERT-specific shRNAs, we found decreased expression of the H3K9me3 and HP1-β marks, which associate with heterochromatic regions (Fig. 4C and D). These experiments show that suppression of TERT leads to alterations in chromatin.
FIG 4.
Suppression of hTERT causes heterochromatin disassembly. (A) RT-PCR of alphoid and qRT-PCR of hLINE1 RNA expression levels in HeLa cells expressing control and hTERT-specific shRNAs. β-Actin, internal control. P values for sh-hTERT 1 or sh-hTERT 2 versus sh-GFP were <0.05. (B) RT-PCR of alphoid or hLINE1 RNA in hTERT-depleted MCF7 cells. RT(−), absence of RT; β-actin, internal control. (C) Effects of hTERT suppression on trimethylation of histone H3 lysine 9 (H3-K9). Cells expressing a control shRNA (sh-GFP) or two independent hTERT-specific shRNAs were stained with anti-trimethyl H3-K9 antibody. Green represents trimethylated H3-K9 staining, and red represents DAPI staining. (D) Effects of hTERT suppression on HP1-β expression. Cells expressing a control shRNA (sh-GFP) or two independent hTERT-specific shRNAs were stained with an anti-HP1-β antibody. Green represents HP1-β staining, and blue represents DAPI staining. (E) RT-PCR of mTERT expression levels in MEF cells derived from wild-type mTERT+/+, heterozygous mTERT+/− or homozygous mTERT−/− mice. β-Actin, internal control. (F) qRT-PCR of IAP, mLINE1 Type A or mLINE1 Type Tf retrotransposons in MEF cells derived from wild-type mTERT+/+or homozygous mTERT−/− mice. β-Actin, internal control. P values for mTERT−/− versus mTERT+/+no. 1 were <0.05. (G) Inhibition of telomerase activity and RdRP activity in HeLa cells by β-rubromycin in vitro. RdRP activity was determined by IP-RdRP assay. β-Rubromycin at 15 μM and 10 μM was used for the TRAP assay and IP-RdRP assay, respectively. (H) RT-PCR of alphoid and qRT-PCR of hLINE1 RNA expression levels in HeLa or VA13 cells treated with 20 μM β-rubromycin or DMSO (Control). β-Actin and GAPDH, internal controls. P values for β-rubromycin versus DMSO were <0.05. (I) ChIP performed in HeLa cells with or without (Control) nocodazole treatment using anti-hTERT MAb. Dot blot signals were detected with the indicated γ-32P-labeled probes. (J) Detection of hTERC, RMRP, alphoid or hLINE1 RNA by RT-PCR with (+) or without (−) RNase I treatment. RNA bound to hTERT was isolated from HeLa cells by IP and subjected to RT-PCR. Antisense-strand RNAs from alphoid were below detectable levels. RNase P, negative control.
Telomeres in most inbred murine strains are longer than human telomeres, and murine TERT (mTERT) is constitutively expressed in many somatic tissues, leading to differences in telomere biology. To examine whether mTERT is also essential for heterochromatin maintenance, we investigated the transcription of transcripts from three murine transposon families: murine intracisternal A particles (IAP) (34), murine LINE1 (mLINE1) Type A and mLINE1 Type Tf (35). Using mouse embryonic fibroblast (MEF) cells derived from mTERT−/− or mTERT +/+ mice (Fig. 4E), we found that transcripts from IAP, mLINE1 Type A and mLINE1 Type Tf were upregulated in mTERT−/− MEFs compared with levels in mTERT +/+ MEFs (Fig. 4F) (P values of <0.05). mTERT−/− MEF cells, however, did not show increased binucleate formation (4.8%; 22/463 cells) compared to those of mTERT+/+ MEF cells (6.7%; 28/415 cells). We believe that the observed difference between HeLa cells with shRNAs and mTERT−/− MEFs is likely due to the difference between acute suppression and germ line deletion of the target gene. In animals that lack mTERT, the effects of loss of mTERT may be partially compensated by another factor(s) to maintain chromosome segregation. Taken together, these observations confirm that manipulation of TERT levels affects heterochromatin maintenance in both human and murine cells.
To further confirm the role of TERT in regulating the expression of these genes, we tested whether inhibiting hTERT enzymatic activity with the telomerase inhibitor β-rubromycin (36) affected the expression of transcripts from these regions. We confirmed that β-rubromycin inhibits both telomerase and RdRP activity (Fig. 4G) and found that transcription of alphoid and hLINE1 RNAs was upregulated in cells treated with β-rubromycin in HeLa cells but not in VA13 cells (Fig. 4H). Collectively, these observations suggest that TERT regulates the expression of centromeric repeated elements and transposons in a manner that correlates with the status of heterochromatin at these loci.
Single-stranded RNAs transcribed from alphoid or hLINE1 elements interact with hTERT.
TERT binds to telomeric regions through interactions with specific telomere binding proteins. To investigate whether hTERT binds directly to centromeres or hLINE1, we performed chromatin immunoprecipitation (ChIP) to determine whether we could detect alphoid or hLINE1 elements in hTERT immune complexes under conditions in which we detected telomere restriction fragments (Fig. 1I) (26). Although we failed to detect alphoid or hLINE1 elements in cycling cells, both alphoid and hLINE1 elements were observed in hTERT immune complexes isolated from cells treated with nocodazole (Fig. 4I), suggesting that hTERT associates with these regions during mitosis.
In S. pombe, siRNAs that are complementary to nascent centromeric RNA recruit the RITS complex to centromeric regions to form heterochromatin (1–3) in a manner that requires the S. pombe RdRP Rdp1. In prior work, we found that an hTERC-independent, hTERT-RMRP complex exhibited RdRP activity (10). To determine whether TERT also binds other noncoding RNAs transcribed from alphoid or hLINE elements, we isolated hTERT immune complexes and probed for RNAs corresponding to these RNAs (Fig. 4J, left panels). To confirm that hTERT interacts with single-stranded RNAs (ssRNAs), we then tested whether treatment of these hTERT-specific immune complexes with RNase I, which specifically digests ssRNAs, affected these RNAs. We found that RNase I treatment abolished the binding of all of these RNA species (Fig. 4J, right panels), suggesting that hTERT interacts with ssRNAs.
Enrichment of RdRP activity of hTERT during mitosis.
In S. pombe (28) and C. elegans (5, 6), the recruitment of RITS to heterochromatic regions requires the synthesis of dsRNAs and processing to siRNAs. To assess hTERT-associated RdRP activity, we established an IP-RdRP assay by modifying a method used to analyze the C. elegans RdRP (21). Specifically, we isolated hTERT immune complexes from cells treated with nocodazole (manipulated) or dimethyl sulfoxide (DMSO; unmanipulated) and performed the RdRP assay. We first confirmed that the observed products were dsRNAs by treating the products with RNase I. This protocol also eliminated products produced by hTERT-associated terminal transferase activity (37).
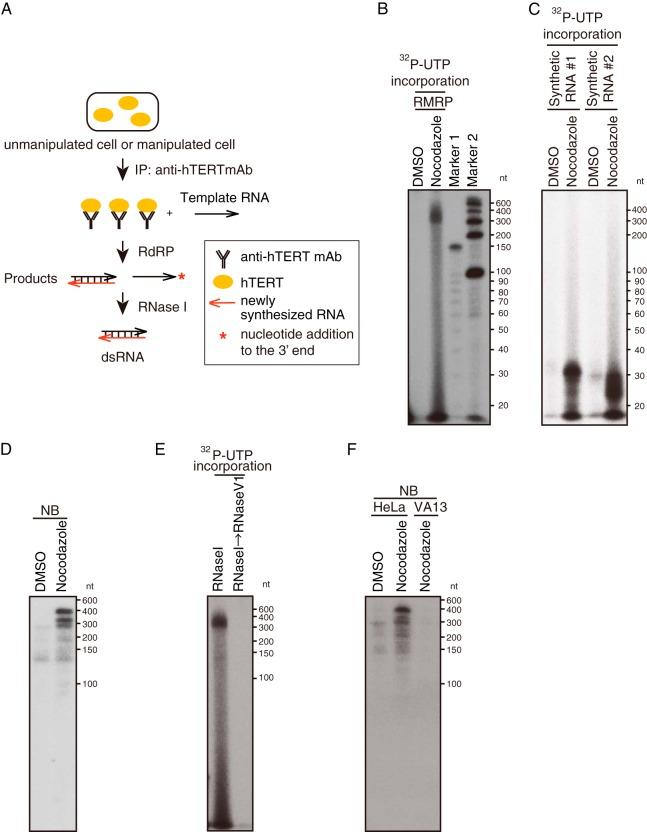
Using HeLa cells arrested in mitosis, we isolated hTERT immune complexes and performed an RdRP assay (IP-RdRP assay) using RMRP RNA (10) as a template (Fig. 5). We found 267-nucleotide (nt), RNase I-resistant products corresponding to RMRP in nocodazole-treated cells (Fig. 5B). We noted a collection of radioactive short RNA species in IP-RdRP assays using cells treated with nocodazole (Fig. 5B) that may represent intermediate products. To confirm, we performed IP-RdRP assays using synthetic RNAs as templates (34 nt). Under such conditions, the chemically synthesized templates for the reaction are pure and highly homogeneous in size. Under these conditions, we also observed a smear and confirmed the production of intermediate-sized by-products of RdRP activity in this assay (Fig. 5C). To confirm that the template-sized products were double stranded, we used the sense strand of the RMRP as a probe in a Northern blot analysis of products from this assay. As expected, we detected the antisense strand of the template RMRP RNA from the cells synchronized in mitotic phase but not in the cells that contained the complex from asynchronously dividing cells (Fig. 5D). To further confirm that the products of the RdRP assay were dsRNAs, we treated the products with RNase V1, which digests dsRNA, and found that RNase V1 treatment eliminated all of these products (Fig. 5E). In addition, since we were unable to detect any RNA products in hTERT null VA13 cells even after treatment with nocodazole to arrest the cells in mitosis, we concluded that the products are generated in a TERT-dependent manner (Fig. 5F). Taken together, these observations confirm that hTERT isolated from cells arrested in mitosis acts as an RdRP to generate dsRNA.
FIG 5.
RdRP activity in mitotic cells. (A) Scheme of the IP-RdRP assay using cell lysate and anti-hTERT MAb. hTERT immune complexes were isolated by anti-hTERT MAb and then used for an RdRP assay with RNA templates. Double-stranded RNA was detected by UTP incorporation assay followed by RNase I treatment, or antisense RNA was detected by Northern blotting (NB). (B) IP-RdRP assay using cell lysates treated with nocodazole (manipulated) or DMSO (unmanipulated) and anti-hTERT MAb. RMRP was used as the RNA template. RdRP products were treated with RNase I. (C) IP-RdRP assay for HeLa cells treated with nocodazole or DMSO using synthetic RNAs (synthetic RNA 1 and synthetic RNA 2) as templates. The RNA products were analyzed with next-generation sequencing and confirmed to be intermediate-sized products complementary to the templates. (D) Northern blotting after an IP-RdRP assay using sense probe derived from nucleotides 229 to 267 of RMRP. (E) IP-RdRP assay using cell lysates treated with nocodazole with a combination of RNase I/RNase V1 or RNase I. (F) Northern blotting after an IP-RdRP assay using HeLa cells treated with nocodazole or DMSO or using VA13 cells treated with nocodazole. A sense probe derived from nucleotides 229 to 267 of RMRP was used. IP-RdRP products were treated with RNase I.
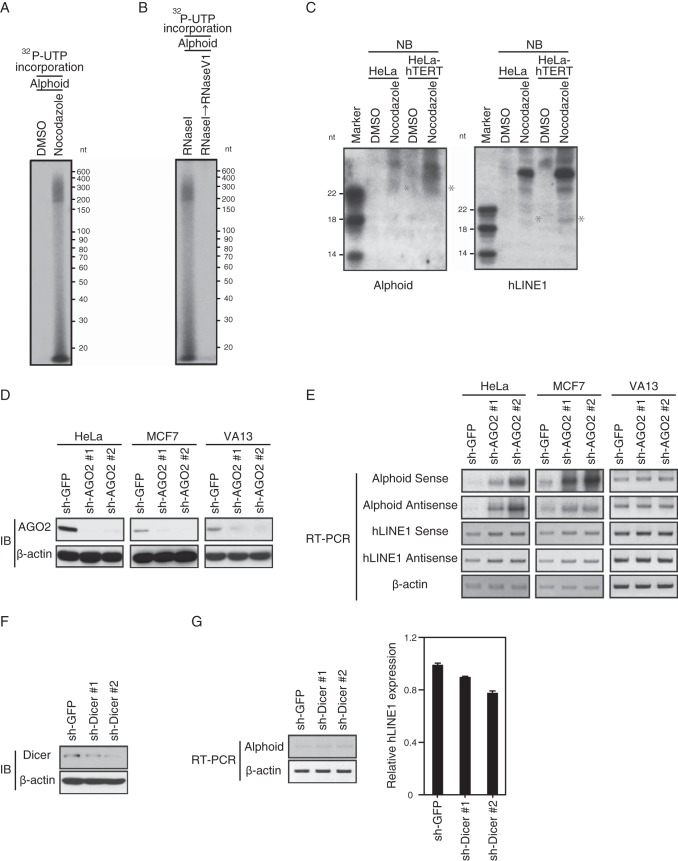
Since suppression of hTERT affected alphoid transcript expression (Fig. 4A) and since hTERT binds alphoid transcripts (Fig. 4J), we tested whether alphoid-derived RNA served as a template for an in vitro IP-RdRP reaction by hTERT. We found that mixing hTERT immune complexes and alphoid RNA transcribed in vitro produced 171-nucleotide RNA products corresponding to the length of alphoid RNA (Fig. 6A). The products were resistant to RNase I treatment but sensitive to RNase V1 treatment (Fig. 6B). These results confirm that the products are dsRNAs.
FIG 6.
hTERT has a functional role in mitotic cells. (A) IP-RdRP assay using cell lysates treated with nocodazole (manipulated) or DMSO (unmanipulated) and anti-hTERT MAb. alphoid was used as the RNA template. (B) IP-RdRP assay using cell lysates treated with nocodazole with a combination of RNase I/RNase V1 or RNase I. alphoid was used as the RNA template. (C) Detection of small RNAs in cells treated with nocodazole or DMSO. Northern blotting (NB) to detect small RNAs using antisense probe derived from nucleotides 120 to 171 of alphoid or nucleotides 1 to 70 of hLINE1. Asterisks indicate the small RNAs detected by the antisense probes. (D) Immunoblot (IB) of AGO2 expression in HeLa, MCF7, or VA13 cells expressing control or AGO2-specific shRNAs. β-Actin, internal control. (E) RT-PCR of alphoid or hLINE1 RNA expression levels in HeLa, MCF7, or VA13 cells expressing shRNAs. β-Actin, internal control. (F) IB of Dicer in Dicer-depleted HeLa cells. β-Actin, internal control. (G) RT-PCR of alphoid and qRT-PCR of hLINE1 RNA expression levels in HeLa cells expressing control or Dicer-specific shRNAs. β-Actin, internal control.
RdRPs play a central role in the synthesis of dsRNAs that are processed into siRNAs. Since the hTERT and centromeric alphoid region produce double-stranded RNA in an in vitro assay, we monitored the existence of small RNAs (19 to 23 nt) from the cells treated with nocodazole or DMSO with probes corresponding to centromeric alphoid or hLINE1 in Northern blotting. We found that these probes identified 20- to 23-nt RNAs in cells treated with nocodazole (Fig. 6C) and concluded that the RdRP activity mediated by hTERT produced small dsRNAs.
Argonaute (Ago) is essential for producing siRNAs and for heterochromatin maintenance in S. pombe (38), and an RdRP, Ago, Dicer, and nucleotidyltransferase are required for chromosome segregation in C. elegans (5, 6). We thus examined whether human AGO2 and Dicer were also essential for heterochromatin maintenance in human cells. Specifically, we examined whether suppression of human AGO2 or DICER1 affected heterochromatin formation at alphoid and hLINE1 loci. We analyzed the alphoid and hLINE1 RNA expression levels in cells in which we suppressed AGO2. We confirmed that RNAs from alphoid and hLINE1 were upregulated in AGO2-depleted HeLa and MCF7 cells compared with levels in control cells, whereas suppression of AGO2 did not affect the RNA expression levels in VA13 cells (Fig. 6D and E). In contrast, when we suppressed DICER1, we failed to observe changes in alphoid and hLINE1 expression (Fig. 6F and G). These results suggest that AGO2 but not DICER1 is essential for heterochromatin formation.
hTERT, BRG1, and NS form a functional complex during mitosis.
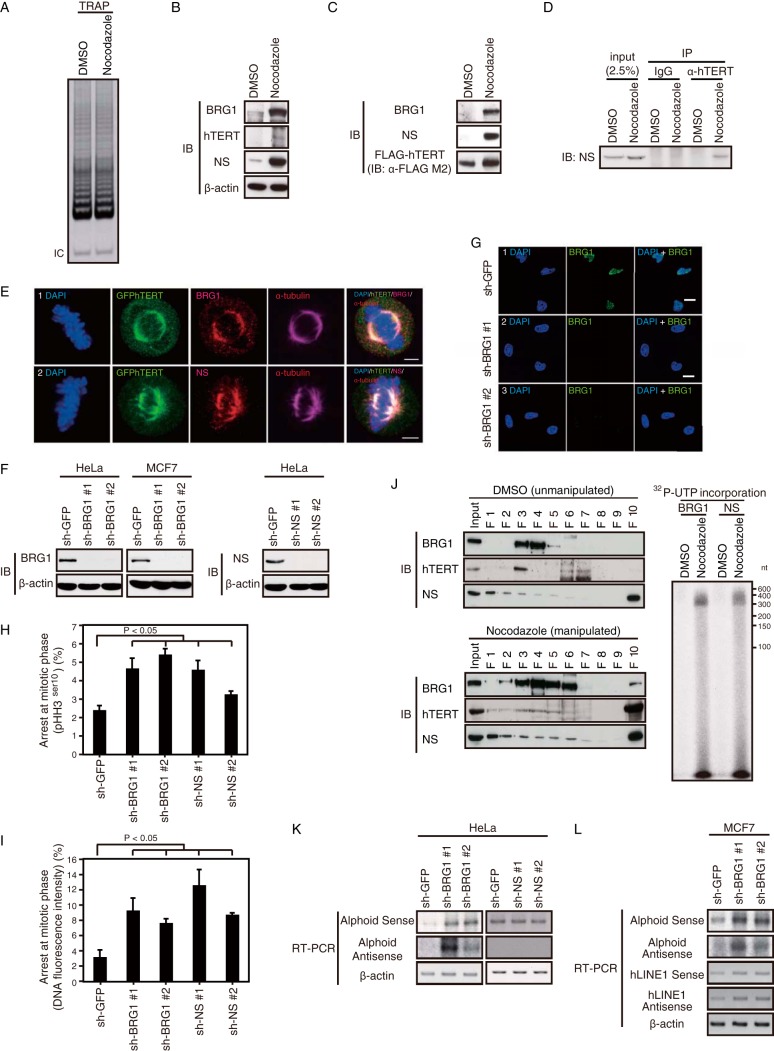
Although hTERT-associated RdRP activity was enriched in mitosis, we failed to observe increased telomerase activity (RNA-dependent DNA polymerase activity) (Fig. 7A), as described previously (27). To investigate how the hTERT RdRP activity is regulated in mitosis, we tested whether increased levels of the TBN complex were found in mitosis. We first assessed the expression levels of each of the components of the TBN complex and noted that the levels of all three components were increased in mitosis (Fig. 7B). When we isolated ectopically expressed FLAG-hTERT, we found increased hTERT associated with BRG1 and NS in cells arrested in mitosis (Fig. 7C). Similarly, we also found that the interaction between endogenous TERT and NS was increased in mitosis (Fig. 7D).
FIG 7.
hTERT, BRG1, and NS form a functional complex in mitotic cells. (A) Telomerase activity of HeLa cells with or without manipulation. Telomerase activity was detected by TRAP assay. IC, internal control. (B) Immunoblot (IB) using the indicated antibodies in HeLa cells treated with nocodazole or DMSO. β-Actin, internal control. (C) hTERT interacts with endogenous BRG1 and NS. FLAG-hTERT immune complexes in HeLa cells were precipitated with anti-FLAG M2 antibody and immunoblotted with anti-BRG1 and anti-NS antibody. (D) hTERT interacts with endogenous NS. hTERT immune complexes immunoprecipitated from HeLa cells and immunoblotted with anti-NS antibody. (E) BRG1 and NS colocalize with GFP-hTERT at mitotic spindles. HeLa-GFP-hTERT cells were immunostained with or without antibodies against BRG1, NS, and α-tubulin followed by DAPI staining. Scale bar, 5 μm. (F) IB of NS or BRG1 expression in HeLa or MCF7 cells expressing control, BRG1-specific shRNAs, or NS-specific shRNAs. β-Actin, internal control. (G) Immunofluorescence (IF) of BRG1 in HeLa cells. Suppression of BRG1 (sh-GFP as a control) was observed by IF. Representative images are shown. Scale bar, 20 μm. (H) A graph of the mitotic index according to pHH3Ser10 immunofluorescence in HeLa cells expressing shRNAs. The P value for sh-BRG1 or sh-NS versus sh-GFP was <0.05. (I) A graph of the mitotic index according to DNA fluorescence intensity and the morphology of HeLa cells expressing shRNAs. The P value for sh-BRG1 or sh-NS versus sh-GFP was <0.05. (J) hTERT, BRG1, and NS form a complex with RdRP activity. Immunoblotting (IB) of fractionated cell lysates with or without manipulation (left panels) was performed. The lysates of HeLa-FLAG-hTERT cells treated with nocodazole (manipulated) or DMSO (unmanipulated) were fractionated into 10 fractions (F1 to F10) by sucrose density gradient centrifugation. The fractions were analyzed by IB with the indicated antibodies. IP-RdRP assay of HeLa cells using cell lysates treated with nocodazole or DMSO and anti-BRG1 or anti-NS antibody for IP (right panel) was performed. (K) RT-PCR of alphoid RNA expression levels in HeLa cells expressing control, BRG1-specific shRNAs, or NS-specific shRNAs. β-Actin, internal control. (L) RT-PCR of alphoid or hLINE1 RNA in BRG1-depleted MCF7 cells. RT(−), absence of RT; β-actin, internal control.
When we investigated the subcellular location where hTERT and BRG1 interact during mitosis, we detected both TERT and BRG1 signals at spindles as marked with α-tubulin, a mitotic spindle marker (Fig. 7E, upper panel). Moreover, we found that hTERT and NS also colocalize at the mitotic spindle (Fig. 7E, lower panel). These observations suggest that increased amounts of the TBN complex are found at the mitotic spindle in mitosis.
To investigate whether manipulating the expression of the TBN complex affected mitosis, we suppressed BRG1 or NS with two independent shRNAs (Fig. 7F and G) (15) and assessed mitotic progression using pHH3Ser10 (Fig. 7H) or DNA fluorescence intensity (Fig. 7I). We found an increase in the number of mitotic cells (2- to 5-fold) after suppression of either NS or BRG1 compared to cells expressing sh-GFP (all P values of <0.05) (Fig. 7H and I). The effect of suppressing NS and BRG1 was similar to what we observed in cells lacking hTERT (Fig. 2F and G). Together, these results demonstrate that the suppression of hTERT, BRG1, or NS has an inhibitory effect on mitotic progression.
We used sucrose density gradient centrifugation and an RdRP assay to examine the correlation between TBN complex formation and its RdRP activity during mitosis using cells treated with nocodazole or DMSO. Sucrose density gradient centrifugation was used to analyze the extracts. We confirmed that hTERT, BRG1, and NS were not coseparated in any fraction in unmanipulated cells, whereas these proteins were primarily found in fraction 10 in cells treated with nocodazole, which shows that synchronization shifts all three proteins into the upper fractions (Fig. 7J, left panels). To further investigate the possibility that the TBN complex serves as an RdRP, the BRG1 immune complex or NS immune complex was isolated from cells treated with nocodazole or DMSO, and an RdRP assay was performed (Fig. 7J). We found RdRP activity in both of the immune complexes isolated from cells treated with nocodazole (Fig. 7J), as observed with the hTERT immune complex (Fig. 5B). The results indicate that both BRG1 and NS are components of the hTERT RdRP complex.
To determine whether BRG1 and NS are essential for heterochromatin assembly, we examined the consequences of suppressing BRG1 and NS on heterochromatin formation at alphoid and hLINE1 elements. Sense and antisense RNA strands from alphoid were upregulated in HeLa and MCF7 cells in which BRG1 was suppressed compared with cells expressing sh-GFP (Fig. 7K and L). However, suppression of NS did not affect alphoid RNA expression levels. These observations demonstrate that BRG1, but not NS, in addition to hTERT contributes to heterochromatin formation at alphoid and hLINE1 loci. Together with the observation that hTERT, BRG1, and NS are all essential for heterochromatin maintenance and mitotic progression, we conclude that hTERT, BRG1, and NS form a functional complex which facilitates mitotic progression by regulating centromeric heterochromatin.
TBN and AGO2 interact in human cells.
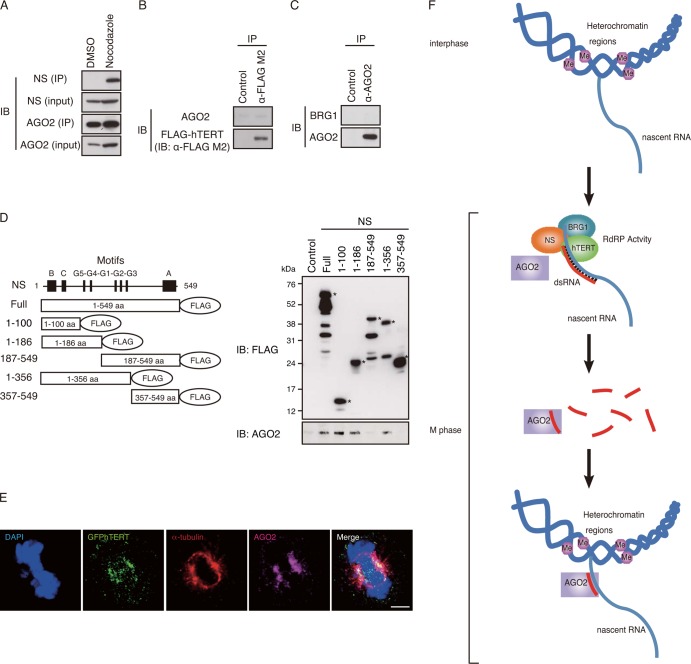
In S. pombe, Ago siRNA interacts with the RITS complex (2), raising the possibility that AGO2 interacts with the TBN complex in human cells. Since we found that expression of AGO2 is essential for heterochromatin formation at alphoid and hLINE1 loci (Fig. 6D and E), we examined whether human AGO2 is associated with the TBN complex. Specifically, we assessed whether AGO2 binds NS or hTERT and found that endogenous AGO2 binds NS but not hTERT or BRG1 (Fig. 8A to C). To further characterize the interaction of AGO2 and NS, we used NS truncation mutants (15) and found that the amino-terminal region of NS was necessary for interactions with the endogenous AGO2 (Fig. 8D). Using IF, we further confirmed that, like the TBN complex, AGO2 localized to the mitotic spindles in HeLa cells during mitosis (Fig. 8E). These observations showed that AGO and NS interact and that AGO colocalizes to the TBN complex during mitosis. These results suggest that AGO2 is essential for heterochromatin formation by interaction with the TBN complex in mitosis.
FIG 8.
AGO2 is essential for heterochromatin formation. (A) AGO2 interacts with endogenous NS. AGO2 immune complexes were isolated from HeLa cells treated with nocodazole (manipulated) or DMSO (unmanipulated) and immunoblotted with anti-NS and anti-AGO2 antibodies. IB, immunoblot. (B) hTERT does not interact with endogenous AGO2. FLAG-hTERT immune complexes were immunoblotted with anti-AGO2 antibody. (C) BRG1 does not interact with endogenous AGO2. AGO2 immune complexes were immunoblotted with anti-BRG1 antibody. (D) AGO2 interacts with NS. Schematic representation of full-length FLAG epitope-tagged NS and truncation mutants where consensus motifs are represented by boxes. 293T cells were transiently transfected with either an empty vector (Control) or FLAG-tagged NS expression vectors. Immune complexes were isolated using anti-FLAG-M2 antibody and immunoblotted with anti-FLAG M2 and anti-AGO2 antibodies. Asterisks indicate the migration of deletion mutants. Other signals in the upper panel are degradation products. (E) Immunofluorescence of GFP-hTERT, α-tubulin, and AGO2. HeLa-GFP-hTERT cells were immunostained with antibodies against α-tubulin and AGO2 followed by DAPI staining. Mitotic cells are shown. Scale bar, 5 μm. (F) A model for hTERT-mediated heterochromatin regulation in repetitive chromosomal regions. A nascent RNA transcribed from heterochromatic regions in cycling cells interacts with the TBN complex in mitosis. Antisense RNAs synthesized by the RdRP activity of hTERT are targeted to the heterochromatic regions to suppress the expression in mitosis.
DISCUSSION
Here, we show that the hTERT, BRG1, and NS complex assembles in mitosis and maintains heterochromatic repetitive elements, including those at centromeres and retrotransposons. Specifically, hTERT acting as an RdRP synthesizes dsRNA from nascent RNA from these regions, which is processed to siRNA that participates in the maintenance of heterochromatin at these same regions in a manner analogous to the RDRC complex in S. pombe. Suppressing the TBN complex disrupted heterochromatin formation at these loci and interrupted mitotic progression, suggesting that this complex participates in organizing chromatin during mitosis. Together, these observations indicate a non-telomere-directed function of hTERT in the regulation of heterochromatin.
The TBN complex.
As telomerase, TERT localizes to telomeres and protects chromosome ends. In addition, several laboratories have demonstrated that TERT forms complexes independent of the telomerase RNA hTERC (9, 11, 15). Specifically, we along with others have found that hTERT binds BRG1 and NS and that this complex regulates both gene transcription (11) and stem cell function in normal and malignant cells (15, 39, 40) in a manner independent of telomere maintenance. Here, we report that the levels of this complex are increased in mitotic cells but not in asynchronously cycling cells (41) and that suppressing each of the members of this complex arrests cells in mitosis.
TERT levels and telomerase activity have been reported to be transiently upregulated in S phase, which leads to telomere stabilization (22, 27). Using a newly generated hTERT-specific antibody, we found that during M phase hTERT localizes to mitotic spindles and that suppression of each of the components of the TBN complex arrests cells in mitosis (Fig. 2F and G and 7H and I). We previously reported that cells expressing a dominant interfering hTERT mutant (DN-hTERT) or hTERT-specific shRNA accumulated in G2/M phase (22) and exhibited slowed proliferation without telomere shortening. Suppression of hTERT or NS further induced a senescence-like phenotype in normal fibroblasts and cancer cells (15, 22). Although it remains unclear if the senescence-like proliferative arrest is related to hTERT function in M phase, these observations suggest that the distinct hTERT complexes act in different phases of the cell cycle.
We confirmed that hTERT localizes to both telomeres and Cajal bodies (Fig. 1E). In addition, we found that TERT associates with centromeres (Fig. 2D and E) and mitotic spindles (Fig. 2A to C) during mitosis. Prior work using GFP-hTERT fusion proteins showed that hTERT shuttles between subcellular compartments in both normal and malignant cells (42). We previously reported that telomerase activity is increased in S phase (22) and here confirmed that RdRP activity and the TBN complex are upregulated in mitosis. Together, these observations confirm and extend prior reports that hTERT is distributed among several complexes. Specifically, as telomerase, hTERT is shuttled between the nucleolus and the Cajal bodies during assembly and associates with telomeres during S phase while forming hTERC-independent complexes with BRG1 and NS during mitosis.
TBN and RdRP activity.
In S. pombe, RdRP localizes to centromeres and maintains heterochromatin at the centromere. Inhibition of RdRP activity leads to loss of siRNAs that are associated with the RITS complex and correlates with loss of transcriptional silencing and heterochromatin at centromeres (3). In addition, when RdRP activity is inhibited, siRNAs that are usually associated with the RITS complex are lost (4). In C. elegans, the RdRP localizes to chromosomes and is required for proper chromosome segregation (5, 6). In both S. pombe and C. elegans, the RNAi pathway is crucial for heterochromatin assembly at centromeres and is required for the accurate segregation of chromosomes during mitosis.
The S. pombe RDRC contains the RdRP Rdp1 and the RNA helicase Hrr1 (2). BRG1 has helicase (43) and ATPase (44) activity and is a component of the SNF/SWI chromatin remodeling complex (11). NS is a GTPase and is essential in pre-rRNA processing (45). In addition, we found that AGO2 interacts with NS and is required for the regulation of hLINE1 and alphoid expression. Since hTERT also exhibits RdRP activity separate from its function as telomerase (10), these observations suggest that the TBN complex contains proteins with similar functions to those found in the RdRP complexes involved in the regulation of heterochromatin in S. pombe and C. elegans.
Prior attempts to identify mammalian RdRPs based on homology to RdRP components in other organisms failed to identify homologous proteins. However, recent work suggests that proteins that are not direct homologs may serve similar functions in different organisms. For example, RNA polymerase II has been predicted to exhibit RdRP activity (46). These observations suggest that the TBN complex may function in a manner analogous to the RDRC/RITS complex in S. pombe.
Moreover, we found that AGO2 is involved in heterochromatin maintenance through its interaction with the TBN complex (Fig. 8A and D). In contrast, DICER1 is dispensable for the heterochromatin maintenance mediated by TBN-RdRP activity. Since Dicer-independent secondary siRNAs generated by RdRP in a de novo pathway have been reported in C. elegans (21, 47, 48), we speculate that the TBN complex maintains heterochromatin during mitosis using de novo-synthesized short RNAs in an AGO2-dependent but a DICER1-independent manner. Since prior reports showed that deletion of Dicer affects the maintenance of heterochromatin in human and murine cells (49, 50), other siRNA-dependent mechanisms may also regulate heterochromatin separate from hTERT.
In consonance with these observations, we found that suppression of components of the TBN complex affects the status of heterochromatin at alphoid and hLINE1 elements. Specifically, we found that hTERT binds to ssRNA transcribed from these regions and produces dsRNAs that are processed into siRNAs. Similar to what has been observed in yeast and worms, these siRNAs play a key role in regulating the status of heterochromatin in these regions. To confirm that the TBN complex is essential for the regulation of heterochromatin in these regions, we confirmed that deletion of TERT also affects the expression of RNAs transcribed from IAP, mLINE1 Type A, and mLINE1 Type Tf (Fig. 4E and F) and used the telomerase inhibitor β-rubromycin to inhibit hTERT. Together, these genetic and pharmacologic observations support the notion that hTERT has functional roles at nontelomeric regions, which are essential for heterochromatin formation. We note that suppression of NS failed to affect the transcription of alphoid repeats (Fig. 7K). NS is closely related to GNL3L, which has been reported to have redundant functions with NS (15, 51). Indeed, we have confirmed that GNL3L physically interacts with hTERT and BRG1 and forms a complex that maintains tumor initiating cells (15).
Prior work has demonstrated that the maintenance of centromeric heterochromatin is necessary for proper chromosomal segregation during mitotic progression in both S. pombe (28) and human cells (29), and regulation of expression of alphoid transcripts is required for mitosis (49). Since both centromeric alphoid regions and transposable elements are maintained in a heterochromatic state during mitosis (Fig. 3 and 4), we speculate that the regulation of heterochromatin at alphoid regions and/or hLINE1 is directly connected to proper progression of mitosis. Moreover, since these repetitive elements have been implicated in centromere maintenance (49, 52), disrupting these functions may promote genomic instability (53). Indeed, prior work suggested that TERT promotes genome stability by providing capping functions, only some of which involve the maintenance of telomeres (8, 13).
Taking these result together, we propose a model for heterochromatin regulation through the RNAi pathway mediated by the TBN complex (Fig. 8F). Specifically, the TBN complex is upregulated in M phase and binds to nascent RNA from alphoid and hLINE1 transcripts. These RNAs serve as templates for dsRNA synthesis by hTERT-RdRP and are further processed in an AGO2-dependent manner to produce siRNAs that regulate the state of heterochromatin at these loci.
Epigenetic modifications link the TBN complex with tumors or TICs.
TERT, BRG1, and NS are upregulated in malignant cells (40, 54, 55). Thus, the upregulation of TERT, BRG1, or NS may lead to aberrant heterochromatin formation due to the enhanced RdRP activity. We speculate that defective heterochromatin caused by enhanced RdRP activity may also contribute to tumor initiation and/or progression. Moreover, the TBN complex maintains the functions of tumor initiating cells (TICs) (15). These observations suggest that inhibiting these various hTERT functions may prove useful in sensitizing cancer stem cells to cytotoxic therapies such as radiation and chemotherapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Fuyuki Ishikawa for the gift of mTERT-knockout mice and Satomi Kuramochi-Miyagawa, Tokio Tani, and Hiroyuki Seimiya for providing technical assistance with RT-PCR analysis of LINE elements, satellite I RT-PCR, and TRAP assays, respectively. We thank Medical and Biological Laboratories Co., Ltd., for their assistance in creating the hTERT MAb.
This work was supported in part by a Grant-in-Aid for Young Scientists (B) (S.O.), a funding program for the Next Generation World-Leading Researchers (NEXT program) (K.M.), the Takeda Science Foundation (K.M.), the Kato Memorials Bioscience Foundation (K.M.), and National Cancer Center Research and Development Funds (23-A-8 to T.S., 23-A-7 to K.K., and 23-B-5 to K.M.). N.O. was a Research Fellow of the Japan Society for the Promotion of Science.
N.O., M.Y., S.O., K.K., Y.M., S.Y., T.K.I., T.M., and H.N. performed experiments. Y.T. and T.S. designed and carried out the bioinformatics analyses. M.Y., Y.M., and K.M. designed the experiments and discussed the interpretation of the results. K.M. and Y.M. wrote the manuscript.
Footnotes
Published ahead of print 18 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00093-14.
REFERENCES
- 1.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672–676. 10.1126/science.1093686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119:789–802. 10.1016/j.cell.2004.11.034 [DOI] [PubMed] [Google Scholar]
- 3.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. 2005. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl. Acad. Sci. U. S. A. 102:152–157. 10.1073/pnas.0407641102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wassenegger M. 2005. The role of the RNAi machinery in heterochromatin formation. Cell 122:13–16. 10.1016/j.cell.2005.06.034 [DOI] [PubMed] [Google Scholar]
- 5.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, Conte D, Jr, Mello CC. 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139:123–134. 10.1016/j.cell.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. 2009. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139:135–148. 10.1016/j.cell.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 7.Castel SE, Martienssen RA. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14:100–112. 10.1038/nrg3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masutomi K, Possemato R, Wong JM, Currier JL, Tothova Z, Manola JB, Ganesan S, Lansdorp PM, Collins K, Hahn WC. 2005. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. U. S. A. 102:8222–8227. 10.1073/pnas.0503095102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE. 2005. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 436:1048–1052. 10.1038/nature03836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. 2009. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 461:230–235. 10.1038/nature08283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, Ji H, McLaughlin M, Veenstra TD, Nusse R, McCrea PD, Artandi SE. 2009. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460:66–72. 10.1038/nature08137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart SA, Hahn WC, O'Connor BF, Banner EN, Lundberg AS, Modha P, Mizuno H, Brooks MW, Fleming M, Zimonjic DB, Popescu NC, Weinberg RA. 2002. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. U. S. A. 99:12606–12611. 10.1073/pnas.182407599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J, Wang H, Bishop JM, Blackburn EH. 1999. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc. Natl. Acad. Sci. U. S. A. 96:3723–3728. 10.1073/pnas.96.7.3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323:644–648. 10.1126/science.1165357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto N, Yasukawa M, Nguyen C, Kasim V, Maida Y, Possemato R, Shibata T, Ligon KL, Fukami K, Hahn WC, Masutomi K. 2011. Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc. Natl. Acad. Sci. U. S. A. 108:20388–20393. 10.1073/pnas.1015171108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhara M, Yoshino T, Shiokawa M, Okabe T, Mizoguchi S, Yabuhara A, Takeyama H, Matsunaga T. 2009. Magnetic separation of human podocalyxin-like protein 1 (hPCLP1)-positive cells from peripheral blood and umbilical cord blood using anti-hPCLP1 monoclonal antibody and protein A expressed on bacterial magnetic particles. Cell Struct. Funct. 34:23–30. 10.1247/csf.08043 [DOI] [PubMed] [Google Scholar]
- 17.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464–468. 10.1038/22780 [DOI] [PubMed] [Google Scholar]
- 18.Summers MK, Bothos J, Halazonetis TD. 2005. The CHFR mitotic checkpoint protein delays cell cycle progression by excluding Cyclin B1 from the nucleus. Oncogene 24:2589–2598. 10.1038/sj.onc.1208428 [DOI] [PubMed] [Google Scholar]
- 19.Hirashima K, Migita T, Sato S, Muramatsu Y, Ishikawa Y, Seimiya H. 2013. Telomere length influences cancer cell differentiation in vivo. Mol. Cell. Biol. 33:2988–2995. 10.1128/MCB.00136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thibault MM, Buschmann MD. 2006. Migration of bone marrow stromal cells in 3D: 4 color methodology reveals spatially and temporally coordinated events. Cell Motil. Cytoskeleton 63:725–740. 10.1002/cm.20160 [DOI] [PubMed] [Google Scholar]
- 21.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. 2007. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26:5007–5019. 10.1038/sj.emboj.7601910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, Weinberg RA, Stewart SA, Hahn WC. 2003. Telomerase maintains telomere structure in normal human cells. Cell 114:241–253. 10.1016/S0092-8674(03)00550-6 [DOI] [PubMed] [Google Scholar]
- 23.Zhu XD, Kuster B, Mann M, Petrini JH, de Lange T. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25:347–352. 10.1038/77139 [DOI] [PubMed] [Google Scholar]
- 24.Wu P, van Overbeek M, Rooney S, de Lange T. 2010. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol. Cell 39:606–617. 10.1016/j.molcel.2010.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T. 2011. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell 44:647–659. 10.1016/j.molcel.2011.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirai Y, Masutomi K, Ishikawa F. 2012. Kinetics of DNA replication and telomerase reaction at a single-seeded telomere in human cells. Genes Cells 17:186–204. 10.1111/j.1365-2443.2012.01581.x [DOI] [PubMed] [Google Scholar]
- 27.Nishiyama A, Muraki K, Saito M, Ohsumi K, Kishimoto T, Ishikawa F. 2006. Cell-cycle-dependent Xenopus TRF1 recruitment to telomere chromatin regulated by Polo-like kinase. EMBO J. 25:575–584. 10.1038/sj.emboj.7600964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubey RN, Nakwal N, Bisht KK, Saini A, Haldar S, Singh J. 2009. Interaction of APC/C-E3 ligase with Swi6/HP1 and Clr4/Suv39 in heterochromatin assembly in fission yeast. J. Biol. Chem. 284:7165–7176. 10.1074/jbc.M806461200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heit R, Rattner JB, Chan GK, Hendzel MJ. 2009. G2 histone methylation is required for the proper segregation of chromosomes. J. Cell Sci. 122:2957–2968. 10.1242/jcs.045351 [DOI] [PubMed] [Google Scholar]
- 30.Mitchell AR, Gosden JR, Miller DA. 1985. A cloned sequence, p82H, of the alphoid repeated DNA family found at the centromeres of all human chromosomes. Chromosoma 92:369–377. 10.1007/BF00327469 [DOI] [PubMed] [Google Scholar]
- 31.Yu F, Zingler N, Schumann G, Stratling WH. 2001. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 29:4493–4501. 10.1093/nar/29.21.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12:1577–1589. 10.1016/S1097-2765(03)00477-5 [DOI] [PubMed] [Google Scholar]
- 33.Ghoshal K, Datta J, Majumder S, Bai S, Dong X, Parthun M, Jacob ST. 2002. Inhibitors of histone deacetylase and DNA methyltransferase synergistically activate the methylated metallothionein I promoter by activating the transcription factor MTF-1 and forming an open chromatin structure. Mol. Cell. Biol. 22:8302–8319. 10.1128/MCB.22.23.8302-8319.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heidmann O, Heidmann T. 1991. Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell 64:159–170. 10.1016/0092-8674(91)90217-M [DOI] [PubMed] [Google Scholar]
- 35.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22:908–917. 10.1101/gad.1640708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueno T, Takahashi H, Oda M, Mizunuma M, Yokoyama A, Goto Y, Mizushina Y, Sakaguchi K, Hayashi H. 2000. Inhibition of human telomerase by rubromycins: implication of spiroketal system of the compounds as an active moiety. Biochemistry 39:5995–6002. 10.1021/bi992661i [DOI] [PubMed] [Google Scholar]
- 37.Lue NF, Bosoy D, Moriarty TJ, Autexier C, Altman B, Leng S. 2005. Telomerase can act as a template- and RNA-independent terminal transferase. Proc. Natl. Acad. Sci. U. S. A. 102:9778–9783. 10.1073/pnas.0502252102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837. 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- 39.Alessio N, Squillaro T, Cipollaro M, Bagella L, Giordano A, Galderisi U. 2010. The BRG1 ATPase of chromatin remodeling complexes is involved in modulation of mesenchymal stem cell senescence through RB-P53 pathways. Oncogene 29:5452–5463. 10.1038/onc.2010.285 [DOI] [PubMed] [Google Scholar]
- 40.Tsai RY, McKay RD. 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16:2991–3003. 10.1101/gad.55671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Listerman I, Gazzaniga FS, Blackburn EH. 2014. An investigation of the effects of the core protein telomerase reverse transcriptase on wnt signaling in breast cancer cells. Mol. Cell. Biol. 34:280–289. 10.1128/MCB.00844-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong JM, Kusdra L, Collins K. 2002. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat. Cell Biol. 4:731–736. 10.1038/ncb846 [DOI] [PubMed] [Google Scholar]
- 43.Muchardt C, Yaniv M. 1999. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J. Mol. Biol. 293:187–198. 10.1006/jmbi.1999.2999 [DOI] [PubMed] [Google Scholar]
- 44.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170–174. 10.1038/366170a0 [DOI] [PubMed] [Google Scholar]
- 45.Romanova L, Grand A, Zhang L, Rayner S, Katoku-Kikyo N, Kellner S, Kikyo N. 2009. Critical role of nucleostemin in pre-rRNA processing. J. Biol. Chem. 284:4968–4977. 10.1074/jbc.M804594200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lehmann E, Brueckner F, Cramer P. 2007. Molecular basis of RNA-dependent RNA polymerase II activity. Nature 450:445–449. 10.1038/nature06290 [DOI] [PubMed] [Google Scholar]
- 47.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315:244–247. 10.1126/science.1136699 [DOI] [PubMed] [Google Scholar]
- 48.Pak J, Fire A. 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315:241–244. 10.1126/science.1132839 [DOI] [PubMed] [Google Scholar]
- 49.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6:784–791. 10.1038/ncb1155 [DOI] [PubMed] [Google Scholar]
- 50.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19:489–501. 10.1101/gad.1248505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng L, Hsu JK, Tsai RY. 2011. GNL3L depletion destabilizes MDM2 and induces p53-dependent G2/M arrest. Oncogene 30:1716–1726. 10.1038/onc.2010.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH. 2009. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 5:e1000354. 10.1371/journal.pgen.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. 2002. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110:327–338. 10.1016/S0092-8674(02)00839-5 [DOI] [PubMed] [Google Scholar]
- 54.Shay JW, Wright WE. 2001. Telomeres and telomerase: implications for cancer and aging. Radiat. Res. 155:188–193. 10.1667/0033-7587(2001)155[0188:TATIFC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 55.Sentani K, Oue N, Kondo H, Kuraoka K, Motoshita J, Ito R, Yokozaki H, Yasui W. 2001. Increased expression but not genetic alteration of BRG1, a component of the SWI/SNF complex, is associated with the advanced stage of human gastric carcinomas. Pathobiology 69:315–320. 10.1159/000064638 [DOI] [PubMed] [Google Scholar]
- 56.Murray V, Monchawin C. 1994. A polymerase chain reaction on human alphoid DNA produces a characteristic ladder of bands. Biochem. Mol. Biol. Int. 34:323–327 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.