Abstract
Protein kinases are thought to mediate their biological effects through their catalytic activity. The large number of pseudokinases in the kinome and an increasing appreciation that they have critical roles in signaling pathways, however, suggest that catalyzing protein phosphorylation may not be the only function of protein kinases. Using the principle of hydrophobic spine assembly, we interpret how kinases are capable of performing a dual function in signaling. Its first role is that of a signaling enzyme (classical kinases; canonical), while its second role is that of an allosteric activator of other kinases or as a scaffold protein for signaling in a manner that is independent of phosphoryl transfer (classical pseudokinases; noncanonical). As the hydrophobic spines are a conserved feature of the kinase domain itself, all kinases carry an inherent potential to play both roles in signaling. This review focuses on the recent lessons from the RAF kinases that effectively toggle between these roles and can be “frozen” by introducing mutations at their hydrophobic spines.
INTRODUCTION
Protein kinases play important roles in the regulation of most biological processes in eukaryotic cells, including cell growth, differentiation, cell death, and metabolism, and their dysfunction is associated with many diseases. They function by catalyzing the transfer of phosphate from ATP to protein substrates. Protein phosphorylation, in general, can modulate the activity of a wide variety of enzymes as well as control specific protein-protein interactions. Their importance is attested to by the fact that the mammalian genome contains over 500 different protein kinases, constituting about 2% of all of the proteins encoded by the genome (1).
Questions about whether protein kinases function solely as catalytic enzymes are raised by the presence of large numbers of pseudokinases in the genome. About 10% of the 518 members of the mammalian kinome are considered pseudokinases because they lack one or more critical conserved kinase residues, suggesting that they lack or have impaired kinase activity (1). An even larger percentage of pseudokinases are present in pathogens such as Giardia and Toxoplasma (2–4). Some protein kinases originally considered to be pseudokinases, such as WNK, CASK, and Haspin, are probably bona fide protein kinases that use variant motifs to substitute for the absence of conventional catalytic residues (5–7) and are probably committed to very specific substrates. However, the large number of remaining pseudokinases suggests that they may have a more general essential noncatalytic role in signaling pathways. Several classification schemes have been proposed for pseudokinases and suggest that they may have distinct mechanistic functions, with some being true catalysts with specific dedicated substrates, others being inert scaffolds that cannot even bind ATP, and others that bind ATP to induce a conformational change but do not require transfer of the phosphate for their function (8, 9). A classification scheme based on differences in nucleotide binding has recently been proposed (8). Since excellent reviews on pseudokinases are available that focus on their noncatalytic roles in signaling pathways (9–12), we will focus here on a new way of thinking about kinase and pseudokinase regulation as driven by hydrophobic spine assembly and the potential noncatalytic functions for bona fide kinases. Specifically, we wish to extend ideas about pseudokinases by asking (i) do all kinases, in fact, have bifunctional properties with both catalytic and noncatalytic functions, (ii) can we uncouple and independently explore these two functions, and (iii) are both functions essential for signaling or in some cases is the noncatalytic function sufficient?
An important insight into the function of pseudokinases was recently provided by a unifying model showing how all kinases are dynamic molecular switches that toggle between inactive and active states (13). This switch mechanism is driven by the assembly and disassembly of hydrophobic spines that define the core fold and architecture of the kinase domain itself. While the specific mechanisms underlying kinase activation are diverse and kinase specific, assembly and disassembly of the hydrophobic spines function as the common underlying mechanisms that drive most regulatory mechanisms. Therefore, mutations that stabilize the spines can be used to decipher both the catalytic and noncatalytic functions of protein kinases. Although still largely unexplored, we propose that the function of pseudokinases is also likely to correlate with the assembly and disassembly of their hydrophobic spines. Our current knowledge from the RAF kinases as a model system shows how both pseudokinases and constitutively active kinases can be created by simple manipulation of the hydrophobic spines. This indicates how the kinase domain itself carries an ability to function as a signaling enzyme or as an inert signaling scaffold where the actual transfer of the phosphate may not be essential.
In this review, we explore the possibility that kinases, both conventional active kinases and catalytically inactive pseudokinases, have important noncatalytic roles as allosteric regulators and that both the catalytic and noncatalytic functions can be interrogated independently by manipulation of the hydrophobic spines. Analogous to the G-protein family of signal transduction regulators, we propose that kinases are highly regulated and dynamic molecular switches that use ATP binding and assembly of the regulatory spine (R-spine) to induce conformational changes that can engage the catalytic machinery and regulate interactions with downstream effectors. These are likely to be important, previously unrecognized mechanisms for kinase function and regulation and have great relevance not only for understanding oncogenic mutations but also for the design of kinase inhibitors.
KINASES TOGGLE BETWEEN INACTIVE AND ACTIVE CONFORMATIONS
The protein kinases have evolved to be highly dynamic molecular switches that are turned off and on rapidly in response to intracellular and extracellular cues. Typically, protein kinases are thought to be maintained in a basal, inactive state and recruited to action only transiently by stimuli that include binding of a growth factor to its receptor, binding of a chemokine to a receptor, a photon of light, or generation of a variety of second messengers. Often, activation is associated with release of an inhibitory domain. The discovery of dynamically assembled hydrophobic regulatory spines (R-spines) introduced a new concept for how these dynamic molecular switches are assembled.
The first protein kinase structures of cyclic AMP (cAMP)-dependent protein kinase (PKA) revealed the conserved kinase fold and provided functional insight for how the conserved sequence motifs contribute to both structure and function (14). By comparing many kinase structures, in both the inactive and active conformation, Kornev et al. were able to identify two key structural elements that are assembled in every active kinase and not assembled in every inactive kinase (13) (Fig. 1). The first structural element that they identified was called the regulatory spine (15). This is a stack of four hydrophobic residues that are aligned in active kinases but broken in inactive kinases. Two residues (RS1 and RS2) come from the C-terminal lobe (C-lobe) and two (RS3/RS4) from the N-terminal lobe (N-lobe). RS1 comes from the important HRD sequence that precedes the catalytic loop, RS2 from the DFG sequence that marks the beginning of the activation loop, RS3 from the αC-helix, and RS4 from the β4-strand (16). In PKA, these residues are Y164, F185, L95, and L106, respectively. The assembly of this R-spine illustrates how kinase activation involves the integration of many separate components of the kinase. This includes not only the conserved kinase core but also, more importantly, the regions that flank the core that are often conserved in family-specific ways.
FIG 1.
Hydrophobic catalytic and regulatory spines define the active protein kinase conformation. Using PKA as a prototypical model for the protein kinase family, we show how the active kinase is assembled around two hydrophobic spines (left). The catalytic cycle is initiated by the binding of ATP, which completes the C-spine and connects the γ-phosphate of ATP with the catalytic machinery (center). This “commits” the kinase to catalysis by completing the C-spine, whereas in the active kinase the C-spine is broken in the absence of ATP and in this case the kinase is “uncommitted” to catalysis (Apo structure, right). In the case of pseudokinases, there is no commitment to catalysis. Simple stability of the R-spine to allow for dimerization is the endpoint. The C-spine is yellow; the R-spine is red.
Later, a second hydrophobic spine was identified and is referred to as the catalytic spine (C-spine) (17). Assembly of the C-spine is mediated by the binding of ATP. The adenine ring of ATP interacts with residues in both the N-lobe and the C-lobe and functions to appose the two lobes and stabilize the kinase in a relatively “closed” position, where the γ-PO4 is positioned for transfer to a protein substrate (Fig. 1). Binding of ATP thus engages all of the catalytic machinery and, based on nuclear magnetic resonance (NMR), creates an enzyme that is now “committed” to catalysis (18). Thus, catalysis for all active kinases involves the binding of ATP that completes the C-spine, while regulatory mechanisms specific to each kinase function to assemble the R-spine. Both spines are anchored to a highly unusual hydrophobic F-helix, which horizontally spans the C-lobe (17).
While the R-spine can be broken in many ways, a common mechanism, as seen in BRAF, is that the DFG phenylalanine actually occupies the ATP binding site (Fig. 2) (19). In this scenario, not only is the R-spine broken but ATP binding is also blocked. This inhibited state may be even more stable in the full-length proteins when inhibitor domains are present. In fact, the entire N-lobe of the kinase could be significantly rearranged in many full-length proteins, but we have very few kinase structures that represent full-length proteins. By studying only the kinase domain, the kinase is likely to be much more dynamic in comparison to the fully inhibited state that exists in the full-length kinase. Understanding the structure and dynamics of full-length kinases is one of the next significant challenges for the structural biology community.
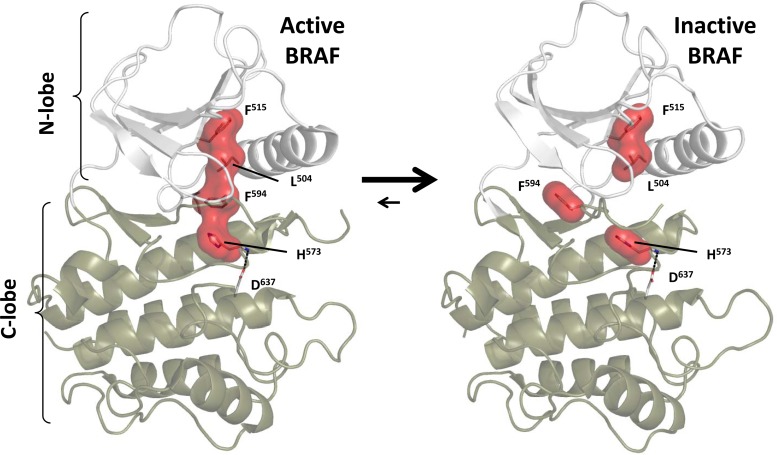
FIG 2.
Activation of BRAF is mediated by assembly of the regulatory spine. In the inactive BRAF (right), the R-spine is broken, whereas the R-spine is assembled in the active BRAF (left). The BRAF numbering here does not count the initiator methionine. Note the movement of F594, part of the DFG motif, which is displaced in the inactive kinase (right). In the absence of the inhibitory N-terminal domain, the kinase will toggle between these two states, with equilibrium most likely favoring the disassembled spine.
Once inhibition is released, the active and inactive conformations that correlate with the assembled and disassembled R-spine are typically in dynamic equilibrium (Fig. 3). Release of inhibition also allows for dimerization, which is an essential step in the activation of many protein kinases, including the RAF family of kinases. Assembly of the active R-spine is typically facilitated and/or further stabilized by phosphorylation of the activation loop (AL) or by binding of accessory molecules such as cyclins (20, 21). Since the DFG motif defines the beginning of the AL, repositioning of the AL by phosphorylation would function to reposition the DFG into its active position in the R-spine and would also establish communication with the C-helix, allowing it to correctly align the next R-spine residue (RS3).
FIG 3.
Activation of Raf is initiated by the release of the inhibitory N-terminal domain. The initial step in the activation of Raf is binding of Ras(GTP) to the N-terminal domain, which “unleashes” the kinase domain. In the absence of Ras(GTP), the kinase domain is locked into an inhibited state where the R-spine is assumed to be broken and, we suspect, ATP cannot bind. Unleashing of the kinase domain allows for the dynamic toggling of the R-spine between its assembled and disassembled state, although equilibrium most likely favors the broken spine.
The assembly of the R-spine in conjunction with C-spine assembly then results in a kinase that is primed for catalysis (Fig. 4). While the “primed” conformation is generally thought to enhance the catalytic activity of the kinase by facilitating the transfer of the γ-phosphate from ATP to the substrate, the stable, “primed” conformation might also have enhanced interactions between the kinase and binding partners like substrates, other kinases, or other scaffold proteins. The increased affinity for a substrate may enhance the efficiency of the kinase reaction, but interactions with other cellular proteins may also have important or even essential functions for downstream signaling. These may not be related to a traditional kinase-substrate relationship and may not require the transfer of the phosphate. Certainly, efficiency of phosphotransfer is not a requirement for kinases. They have not evolved to be efficient enzymes but rather have evolved to be succinct regulated enzymes. Indeed, increasing kinase catalytic activity often creates an oncogene (22). Therefore, release of inhibition is likely to be far more important for regulating kinase function than catalytic efficiency.
FIG 4.
Binding of ATP to the C-spine primes the assembled hydrophobic R-spine for catalysis. Using the analogy of an oscillating pendulum, release of inhibition allows the BRAF R-spine to toggle between its inactive (broken) and active (assembled) conformations. While dimerization and AL phosphorylation shift the equilibrium to favor the assembled R-spine, it is the binding of ATP that assembles the C-spine, allowing interactions between the R-spine and C-spine to prime the kinase for catalysis. The primed active conformation of the kinase is important for both bona fide kinases and pseudokinases, but in the pseudokinase this is the endpoint; there is no subsequent catalysis.
KINASES ARE SLOW ENZYMES
The efficiency of enzymes can be judged by their turnover rate (kcat), the maximum number of substrate molecules they can convert to product per catalytic cycle per unit of time. The fastest known enzymes, so-called perfect enzymes, have turnover rates close to 600,000/s. This means they can turn over 600,000 substrate molecules to product in 1 s. The majority of enzymes, however, are much slower, with average turnover rates of about 10 to 20/s (23). Among the slowest of the enzymes are the protein kinases that have turnover rates that range from 0.05 to 1.0/s. This is in the same range as the GTPases, which are typically thought of as switches and not enzymes (24, 25). Since the normal time scale of signaling reactions is in the seconds to minutes range, the ability of kinases to phosphorylate more than one downstream substrate may also be very limited.
The ability of protein kinases to phosphorylate more than one substrate is based on in vitro solution studies where substrate, typically a small peptide, is not limiting. However, protein kinases in cells worked on tethered proteins, not peptides, and do not typically function under these Michaelis-Menten conditions. Our greater understanding of the role for scaffolds that function to tether substrates close to kinases and phosphatases suggests, for example, that, in vivo, access to substrate might not be diffusion limited (26). Targeted access to specific substrates and slow turnover suggest that a single turnover (a single perfect catalytic cycle) may be more important in kinase function than turning over large amounts of substrate. Certainly, in some cases this will be true. The efficiency of kinase-inactivating mechanisms like dephosphorylation by phosphatases further limits the ability of kinases to phosphorylate more than one substrate. The slow enzymatic rate for kinases is another reason to consider that noncatalytic functions of kinases could also be important in signaling pathways and suggests that we need to reevaluate the idea that kinase cascades function mainly for signal amplification.
VRK, MLKL, STRAD, AND ERBB3 PSEUDOKINASES AS ALLOSTERIC ACTIVATORS
Another reason to think about the noncatalytic roles of kinases in signaling is suggested by recent data regarding the role of pseudokinases in signaling pathways. As noted above, kinases lacking conserved residues required for catalytic activity are classified as pseudokinases. Several pseudokinases have now been shown to function noncatalytically as allosteric activators.
VRK3.
Vaccinia-related kinase (VRK) is a family of kinases that contain two active kinases, VRK1 and VRK2, and a pseudokinase, VRK3. VRK1 and VRK3 are found in the nucleus, while VRK2 is found in the endoplasmic reticulum. VRK3 is structurally similar to VRK2 but lacks the ability to bind to ATP, because hydrophobic side chains fill the adenine binding pocket. It is thus a naturally occurring bona fide pseudokinase in which the C-spine is fused by hydrophobic residues that fill the adenine binding pocket; it is nevertheless fixed in an active-like conformation (27). In addition, the N-terminal linker is docked onto the glycine loop and further restricts binding of ATP. The crystal structure of VRK3 shows that both spines are assembled in an active-like but inert conformation (Fig. 5) (27, 28). It is thus a truly dead pseudokinase. VRK3 functions by binding of a mitogen-activated protein kinase (MAPK) phosphatase, vaccinia H1 related (VHR), and enhancing its phosphatase activity, and this function does not require catalytic activity.
FIG 5.

Vrk3, an example of a true pseudokinase. The structure of Vrk3 reveals that it is a true pseudokinase (catalytically inactive), because it cannot bind ATP but is locked into an “active-like” conformation where the R-spine is assembled. Several of the residues near the ATP binding pocket (shown in tan on the left), including two C-spine residues, are replaced with larger hydrophobic residues, and these fill the adenine binding site, causing the C-spine to fuse. ATP, drawn in black, is sterically blocked from binding. The two assembled spines (C-spine in yellow and R-spine in red) are shown on the right. The overall Vrk3 structure in the middle shows several reasons why it is an inert pseudokinase: the adenine binding site is filled with hydrophobic side chains (1), the glycine loop is shorter than other protein kinases (2), and the N-terminal tail, shown in teal, is locked onto the glycine loop (3).
MLKL.
Mixed-lineage kinase domain-like (MLKL) is a pseudokinase that functions to regulate programmed necrosis mediated by the inflammatory cytokine tumor necrosis factor alpha (TNF-α). Normally, signaling by TNF receptor 1 (TNFR1) functions to activate caspase 8 to stimulate apoptosis. Caspase 8 also cleaves the RIP kinases (RIPK1 and RIPK3) to block the regulated necrosis pathway. When caspase 8 activation is blocked by microbial proteins like viral FLIP proteins, RIP3 kinase phosphorylates MLKL to stimulate necrosis. The structure of full-length mouse MLKL confirms that it is a pseudokinase and shows that the catalytic lysine (K219) coordinates with a glutamine residue (Q343) that is in an unusual helix in the activation loop preventing the lysine from coordinating with the conserved glutamate in the αC-helix (29). Certain mutated catalytically inactive forms of MLKL induced constitutive cell death, suggesting that RIP3 kinase phosphorylation of MLKL functions to induce a conformational change that allows it to induce cell necrosis (29). The recently solved structure of human MLKL shows an active-like conformation, with assembly of the R-spine mediated by coordination of the catalytic lysine with a glutamic acid in the αC-helix (30). It is interesting to speculate that the two structures represent active and inactive forms of MKLK, but functional studies are necessary to confirm this. Nevertheless, the evidence supports the idea that MLKL toggles between an active and inactive conformation.
STRADα.
STE20-related adapter (STRADα) is a pseudokinase regulator of the kinase LKB1, which is an upstream activator of AMPK and the AMPK family of kinases (31). This is another example of a pseudokinase that functions through its active-like conformation. When STRAD and accessory molecule Mo25 bind to LKB1, this results in the activation of LKB1. STRAD can bind to ATP but lacks critical catalytic residues, including the aspartate in the critical DFG motif. Biochemical studies support that STRAD is a catalytically inactive pseudokinase, as it does not appear to have kinase or ATPase activity (32). Kinase-inactivating mutations do not impair the functional properties of STRAD, but ATP binding is required for STRAD function.
Structural studies demonstrate that STRAD binding to Mo25 and nucleotide binding help it to be stabilized in an active-like conformation (33). In its “active-like” conformation, STRAD binds to LKB1. The binding of the STRAD-Mo25 complex to LKB1, in turn, induces the active conformation of LKB1, allowing it to be active in the absence of activation loop phosphorylation; mutation of potential AL loop phosphorylation sites does not impair LKB1 activation by STRAD. STRAD is in the family of Ste20 kinases, which also include the PAK, HPK1, GCK, GLK, and HGK kinases. Interestingly, it has been proposed that the function of other Ste20-related kinases may be independent of their kinase activities (34).
ERBB3.
ERBB3 is an example of a transmembrane receptor pseudokinase that is a member of the epidermal growth factor (EGF) family of receptors that include ErbB1-4 (also known as HER1-4). The prototypical receptor in this family is the epidermal growth factor receptor (EGFR; ERB1 or HER1), which is a transmembrane protein that binds to its ligand, EGF. The EGFR family members (except for ErbB2) bind to ligands that include EGF, tumor growth factor alpha (TGF-α), and the neuregulins (35). When bound to their ligands, they form extracellular heterodimers with other EGF receptor family members. ErbB2 is thought to function in a dimerization competent state, and its kinase activity is stimulated when the extracellular EGF binding domain is dimerized with another member of the EGF receptor family. The detailed mechanism whereby dimerization of the extracellular ligand binding domain then mediates dimerization and activation of the cytoplasmic kinase domain in the full-length EGF receptor is still a great challenge for the signaling community (36).
ERBB3 is a kinase-impaired pseudokinase, but when heterodimerized, it is a potent activator of signaling (37, 38). Dimerization of another EGFR family member with ERBB3 can allosterically activate the kinase activity of the catalytically active partner (39). The current model is that ligand binding to any of the receptors of this family generate homo- and heterodimers, and dimerization then allows for allosteric activation (39, 40). Attesting to its importance, ERBB3 mutations are present in over 10% of tumors (41). While many of the mutations are in the extracellular domain and presumably promote dimerization of the extracellular EGF binding domain, some of the mutations are in the kinase domain, suggesting that the allosteric function of ERBB3 is also regulated. The ligand independence of all of these mutants suggests that conformational changes in the ERBB3 kinase domain promote dimerization and allosteric activation. How the spines specifically contribute to these conformational switch mechanisms has not been addressed. Interestingly, certain ATP analogs based on quinazoline can induce dimerization of the EGFR kinase domain in cells, suggesting that ATP binding and other conformation changes may be involved in promoting dimerization (42–44).
RAF KINASES DEMONSTRATE THE CATALYTIC AND NONCATALYTIC FUNCTIONS OF A PROTEIN KINASE
It is now becoming clear that even bona fide kinases have functions that are independent of their catalytic activity. The RAF kinases are good examples that have been clearly shown to have noncatalytic signaling activities (45). The surprising finding that drug inhibitors of the BRAF serine/threonine kinase could, in certain situations, activate RAF in cells led to new insights that showed that inhibited BRAF kinases can dimerize and allosterically activate other members of the RAF family of kinases (46–48). For the remainder of this review, we will summarize the evidence for noncatalytic functions for RAF and then discuss the potential implications of this work to our general understanding of kinase function.
The RAF family of kinases includes ARAF, BRAF, and CRAF. Mutations in BRAF but not other forms of RAF are important in cancer. Thus, there has been great clinical interest in understanding their function and regulation. While BRAF-specific drugs efficiently inhibit the kinase activity of a mutated form of BRAF that is constitutively active, these drugs also paradoxically activate RAF in cells containing wild-type (WT) BRAF and constitutively active RAS (46–48).
As indicated in Fig. 3 and 6, the activation of the RAFs includes several steps (49). The small GTPase, RAS, initiates the process that leads to the activation of the MAP kinase extracellular signal-regulated kinase (ERK) through an upstream kinase cascade that includes RAF and MEK. RAF kinases are ∼75- to 90-kDa proteins with a regulatory domain at the N terminus and the kinase domain at the C terminus. The N-terminal domain (NTD) contains an RAS binding domain (RBD), and binding to RAS at the membrane leads to the release of the inhibitory NTD from the kinase domain (Fig. 3). Supporting a role of the NTD as an inhibitory domain, removal of the NTD can generate oncogenic forms of RAF (49). Release of the NTD initiates the activation process by “unleashing” the kinase domain and allowing it to dynamically toggle between inactive and active conformations, a process that is driven by the assembly and disassembly of the R-spine. “Unleashing” of the kinase domain also allows for dimerization, which shifts the equilibrium in favor of the assembled R-spine. The released kinase domain can also bind ATP, whereas in its inhibited state, we predict that ATP will not be bound to the full-length RAF. This will need to be tested. In the case of the RAFs, two additional steps are required after the kinase domain is “unleashed,” dimerization with another RAF molecule and phosphorylation of the activation loop (AL) (Fig. 6).
FIG 6.
The active conformation of RAF is facilitated by dimerization and activation loop phosphorylation. Ras binding releases the N-terminal inhibitory domain, allowing the R-spine to toggle between the inactive and inactive conformations (as shown by the stacked red balls). Dimerization helps to stabilize the active conformation of the R-spine and facilitates cis autophosphorylation of its activation loop. Once activation loop phosphorylation occurs, the kinase can maintain the active conformation of the R-spines as a monomer, but this is rapidly reversed by active phosphatase.
Mutation of a C-spine residue in BRAF creates a pseudokinase that functions as an allosteric activator of WT BRAF and CRAF.
Previously, it was shown that certain oncogenic forms of BRAF were either kinase inactive or kinase impaired (45). Also we had previously shown that mutation of a C-spine residue in BRAF (A481F) blocked catalytic activity but resulted in a form of BRAF that constitutively dimerized and activated ERK (50). In this case, because Ala481 is a C-spine residue that lies directly on top of the adenine ring, the phenylalanine ring most likely fills the adenine pocket, similar to what is seen in VRK3 (Fig. 5 and 7), and this is sufficient to unleash the N-terminal inhibitory domain. Because downstream activation of MEK and ERK still required dimerization, we concluded that the A481F BRAF mutant was functioning as a kinase-inactive allosteric activator. Formation of the dimer with a wild-type BRAF did indeed induce autophosphorylation of the partner RAF, and because the activator could not bind ATP, we surmised that this must occur by cis autophosphorylation. Our next challenge was to understand why the same mutation in CRAF also induced dimerization but did not activate ERK. Truncation experiments with BRAF revealed that the major difference was in a segment that just precedes the beginning of the kinase domain, and this was confirmed by mutations of this region in both BRAF and CRAF (51).
FIG 7.
The C- and R-spines can be manipulated to control the active conformation of kinases. Replacing the conserved C-spine residues, Ala70 (PKA), with Phe fuses the C-spine so that ATP cannot bind. Ala70 (PKA) normally lies on top of the adenine ring. Substitution with Ala70 with Phe (shown on the left in teal) abolishes ATP binding and by locking the kinase in the active conformation can lead to dimerization and allosteric activation of wild-type RAF. Replacing the equivalent R-spine residue (L95-PKA) of BRAF or CRAF (L505 and L397, respectively) with Phe (shown in teal on the right) helps to stabilize the R-spine, resulting in a constitutively active form of RAF that is independent of Ras, dimerization, and activation loop phosphorylation. This diagram also shows how assembly of the R-spine positions the αC-helix and the activation loop to achieve the active conformation. The arrows in the middle panel show the positions of the two spine residues in the assembled spines.
Requirements for dimerization.
Just N-terminal to the kinase domain is a conserved acidic motif (NtA) that is part of the dimer interface. A functional equivalent to this motif may be conserved in other kinases, including the EGF receptor and the Src and ITK family of kinases (39, 52–54). These flanking segments, which are typically quite dynamic, can be thought of as “capping” motifs that help to organize and activate the kinase core. In BRAF, the NtA (SSDDW) has two serine residues that are constitutively phosphorylated plus two acidic aspartic acids, so that this sequence is always acidic (55). When BRAF dimerizes with another RAF molecule, the presence of BRAF's acidic NtA is required to induce formation of the R-spine and phosphorylation of the activation loop on the partner kinase. If the NtA motif in BRAF is mutated to Ala, it no longer functions as an activator even though it can still dimerize (51). Thus, this “capping motif,” the acidic NtA, is a key feature that allows for allosteric activation.
Phosphorylation of the equivalent NtA on CRAF (SSYYW) and ARAF (SGYYW) occurs only during or after RAF is activated. This explains why the C-spine mutation in CRAF is not sufficient to generate an allosteric activator. However, substituting acidic residues for the NtA motif in CRAF does allow the catalytically inert form of CRAF to function as an allosteric activator. These studies demonstrate convincingly that the RAF kinases have an important noncatalytic, activating function.
The allosteric function of the kinase is distinct from its catalytic activity. A mutated form of BRAF or CRAF with alanine residues substituted for the NtA, for example, can still be allosterically activated by activator forms of BRAF and CRAF (51). In the resting cell, since BRAF is the only RAF kinase with a constitutively phosphorylated NtA, when it dimerizes, it will transactivate the partner RAF kinase regardless of whether the partner kinase has an acidic NtA or not. Thus, BRAF, even inactive BRAF, can activate CRAF; however, CRAF cannot activate BRAF until it is phosphorylated on its NtA motif. This explains why pharmacologic inhibitors that preferentially target BRAF allow for CRAF activation and suggests that drugs that inhibit CRAF in addition to BRAF may minimize problems with paradoxical activation. It also potentially explains the function of the pseudokinase KSR in the regulation of this pathway (51).
Mutation of the R-spine can hijack the entire regulatory pathway.
The essential step in activation of any kinase is the assembly of the R-spine, which in turn positions the αC-helix to facilitate transfer of the γ-phosphate from ATP to a protein substrate. Because it activates the kinase, allosteric activation of RAF via dimerization must function by assembling the R-spine in the receiver kinase. Can we replicate this by directly mutating the R-spine? Constitutive assembly of the BRAF R-spine could indeed be induced by mutating an R-spine residue (Leu505) to Phe, which generated a constitutively active kinase that no longer required dimerization (51) (Fig. 7). Unlike the WT kinase where the spine is destabilized by dephosphorylation of the AL, this mutant does not require AL phosphorylation and is thus also phosphatase resistant. As the mutant also does not require binding to RAS, formation of the active conformation is sufficient to “unleash” the inhibitory domain, similar to the C-spine mutation. This R-spine mutant thus hijacks the entire regulatory pathway and creates a stable active kinase that is capable of downstream activation of MEK and ERK.
cis autophosphorylation of kinases—a function of allostery.
Most kinases require phosphorylation of the activation loop to achieve full activation, and in many cases activation loop phosphorylation is achieved by kinase dimerization. The simplest model for activation loop phosphorylation is trans phosphorylation, with one kinase phosphorylating the activation loop of the dimerization partner when the two kinases are brought together. This could occur between two different kinases, for example, MEK phosphorylation of ERK, or could be mediated by two identical kinases (SRC autophosphorylation).
Experimental data used to support models of trans autophosphorylation are of two types. The first is based on the assumption that trans phosphorylation requires dimerization and utilizes dilution as an argument for validation. If the autophosphorylation reaction attenuates with dilution, this is used as evidence in favor of trans autophosphorylation. The other assumption here is that that cis autophosphorylation occurs only with monomeric kinases, which would not attenuate with dilution. The second, more definitive proof uses a catalytically inactive form of the kinase as the substrate. If phosphorylation of the activation loop of the “dead” kinase occurs, this is strong proof for trans autophosphorylation. Most of the evidence in the literature, however, is based on dilution experiments. A dilution experiment is not sufficient to distinguish between cis and trans phosphorylation.
Until now, cis autophosphorylation of the activation loop has been thought to be relatively rare and reported for only a few kinases, like DYRK (56), glycogen synthase kinase 3β (GSK-3β) (57, 58), p38α (59), and Fus3 (60), although the precise mechanism for cis autophosphorylation is not well understood. For some kinases, like the DYRK family of kinases, cis autophosphorylation is thought to occur during biosynthesis. In other cases, it is binding of an accessory molecule that induces AL autophosphorylation. Hsp90 binding to GSK3-β (57), TAB1 binding to p38α (59), and Ste5 binding to Fus3 are required for cis autophosphorylation to occur (60). Importantly, the requirement of an accessory protein demonstrates that dimerization (in this case with a nonkinase partner) is still a requirement for cis autophosphorylation. In the case of RAF, the accessory protein is RAF itself. The presence of a phosphorylated NtA domain allows the activator molecule to induce the active conformation of the receiver and in the process allows the receiver to cis autophosphorylate its activation loop. Whether all of these examples use accessory proteins to drive R-spine assembly and subsequent AL phosphorylation remains to be established. However, a dilution experiment would also attenuate cis autophosphorylation if binding of a secondary accessory protein was required. Many of our current assumptions about cis and trans autophosphorylation thus need to be reconsidered.
The mechanism for allosteric activation by the C-spine mutants of BRAF requires dimerization and results in phosphorylation of the activation loop of the receiver kinase. Since the activator kinase with the C-spine mutation is catalytically inactive, autophosphorylation of the activation loop by RAF dimers most likely occurs by cis autophosphorylation. When a kinase-inactive form of BRAF dimerizes with a WT CRAF molecule, we could detect induced activation loop phosphorylation only on CRAF and not on the kinase-inactive BRAF. While it is conceivable that the formation of higher-order clusters such as tetramers or octamers could allow for trans autophosphorylation, it is difficult to explain why in these structures only the activation loop of the active kinase would be phosphorylated. We favor the simplest explanation, that allosteric activation induces cis autophosphorylation of the partner.
This has implications for the function of other predicted pseudokinases, which could function as allosteric activators even though they are inactive as catalysts. For example, the ErbB2/EGFR dimer (39, 40, 61), the JAK kinases where the pseudokinase domain of JAK interacts with and regulates the bona fide kinase domain of JAK (62), or the STRAD/Mo25 binding to AMPKK (32, 33) could potentially induce cis autophosphorylation, helping to stabilize the active conformation.
CONCLUSION
The large number of pseudokinases in the kinome seems to be telling us that the enzymatic activity of protein kinases is not the only thing that kinases can do. Clearly, in some cases the catalytic activity is not essential for function. Here, we suggest that the assembly of the active R-spine functions in the regulation of both pseudokinases and catalytic kinases. The fact that the R-spine residues can toggle between active and inactive states and that kinases in their active state can bind to some substrates with higher affinity suggests that protein kinases may function in a manner that is similar to how we currently view small GTPases. Small GTPases toggle between their GTP-bound and their GDP-bound states. They are most stable in their inactive GDP-bound state, while in their active GTP-bound state, they can bind, recruit, and activate downstream effector molecules. The half-life of the active GTP-bound protein is controlled by the intrinsic GTPase activity of the G-protein that is potentiated by interactions with GTPase-activating proteins (GAPs). In the absence of GAPs, the GTP turnover is <1/s, while this is accelerated in the presence of GAPs (up to 15/s). Thus, the time scale of G-protein activation is similar to the time scale of kinase activity. In the case of the kinase, the GAP would be the allosteric activator and the terminating signal would be the phosphatase; however, the essential features of a switch are conserved. Another difference is that the substrate for the GTP-bound enzyme is water, whereas the substrate for a protein kinase is usually another protein.
Because kinases have catalytic activity and bona fide substrates, the focus has always been on their enzymatic activity. The dimer activation mechanisms described here show that there are at least two functionally important states for any kinase. First are the substrate complexes where the active kinase is bound to and phosphorylates a substrate. The dimer activation mechanism provides a new way of thinking about allosteric activator-receiver complexes, where a kinase could function as an allosteric activator. Both are likely to be biologically relevant. Here, we suggest that the mechanisms of kinase regulation are also likely to be relevant in the regulation of pseudokinases. The discovery of the hydrophobic spines and the mechanisms that regulate their assembly will likely be useful as tools to probe the functional consequences of the inactive and active states of both kinases and pseudokinases. It also suggests that stabilization of the inactive conformation of a pseudokinase or targeting the dimer interface might allow us to think of pseudokinases as viable drug targets.
Footnotes
Published ahead of print 24 February 2014
REFERENCES
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934. 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 2.Manning G, Reiner DS, Lauwaet T, Dacre M, Smith A, Zhai Y, Svard S, Gillin FD. 2011. The minimal kinome of Giardia lamblia illuminates early kinase evolution and unique parasite biology. Genome Biol. 12:R66. 10.1186/gb-2011-12-7-r66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reese ML, Zeiner GM, Saeij JP, Boothroyd JC, Boyle JP. 2011. Polymorphic family of injected pseudokinases is paramount in toxoplasma virulence. Proc. Natl. Acad. Sci. U. S. A. 108:9625–9630. 10.1073/pnas.1015980108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. 2007. Structural and functional diversity of the microbial kinome. PLoS Biol. 5:e17. 10.1371/journal.pbio.0050017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Min X, Lee BH, Cobb MH, Goldsmith EJ. 2004. Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure 12:1303–1311. 10.1016/j.str.2004.04.014 [DOI] [PubMed] [Google Scholar]
- 6.Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. 2000. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J. Biol. Chem. 275:16795–16801. 10.1074/jbc.275.22.16795 [DOI] [PubMed] [Google Scholar]
- 7.Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, Higgins JM, Knapp S. 2009. Structure and functional characterization of the atypical human kinase haspin. Proc. Natl. Acad. Sci. U. S. A. 106:20198–20203. 10.1073/pnas.0901989106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy JM, Zhang Q, Young SN, Reese ML, Bailey FP, Eyers PA, Ungureanu D, Hammaren H, Silvennoinen O, Varghese LN, Chen K, Tripaydonis A, Jura N, Fukuda K, Qin J, Nimchuk Z, Mudgett MB, Elowe S, Gee CL, Liu L, Daly RJ, Manning G, Babon JJ, Lucet IS. 2014. A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J. 457:323–334. 10.1042/BJ20131174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeqiraj E, van Aalten DM. 2010. Pseudokinases—remnants of evolution or key allosteric regulators? Curr. Opin. Struct. Biol. 20:772–781. 10.1016/j.sbi.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. 2006. Emerging roles of pseudokinases. Trends Cell Biol. 16:443–452. 10.1016/j.tcb.2006.07.003 [DOI] [PubMed] [Google Scholar]
- 11.Kannan N, Taylor SS. 2008. Rethinking pseudokinases. Cell 133:204–205. 10.1016/j.cell.2008.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyers PA, Murphy JM. 2013. Dawn of the dead: protein pseudokinases signal new adventures in cell biology. Biochem. Soc. Trans. 41:969–974. 10.1042/BST20130115 [DOI] [PubMed] [Google Scholar]
- 13.Taylor SS, Kornev AP. 2010. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 10.1016/j.tibs.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. 1991. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253:407–414. 10.1126/science.1862342 [DOI] [PubMed] [Google Scholar]
- 15.Kornev AP, Haste NM, Taylor SS, Eyck LF. 2006. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. U. S. A. 103:17783–17788. 10.1073/pnas.0607656103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meharena HS, Chang P, Keshwani MM, Oruganty K, Nene AK, Kannan N, Taylor SS, Kornev AP. 2013. Deciphering the structural basis of eukaryotic protein kinase regulation. PLoS Biol. 11:e1001680. 10.1371/journal.pbio.1001680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kornev AP, Taylor SS, Ten Eyck LF. 2008. A helix scaffold for the assembly of active protein kinases. Proc. Natl. Acad. Sci. U. S. A. 105:14377–14382. 10.1073/pnas.0807988105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masterson LR, Shi L, Metcalfe E, Gao J, Taylor SS, Veglia G. 2011. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 108:6969–6974. 10.1073/pnas.1102701108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huse M, Kuriyan J. 2002. The conformational plasticity of protein kinases. Cell 109:275–282. 10.1016/S0092-8674(02)00741-9 [DOI] [PubMed] [Google Scholar]
- 20.Pavletich NP. 1999. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J. Mol. Biol. 287:821–828. 10.1006/jmbi.1999.2640 [DOI] [PubMed] [Google Scholar]
- 21.Endicott JA, Noble ME, Johnson LN. 2012. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 81:587–613. 10.1146/annurev-biochem-052410-090317 [DOI] [PubMed] [Google Scholar]
- 22.Blume-Jensen P, Hunter T. 2001. Oncogenic kinase signalling. Nature 411:355–365. 10.1038/35077225 [DOI] [PubMed] [Google Scholar]
- 23.Berg JM, Tymoczko JL, Stryer L. 2002. Biochemistry, 5th ed. W. H. Freeman, New York, NY [Google Scholar]
- 24.Berman DM, Wilkie TM, Gilman AG. 1996. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell 86:445–452. 10.1016/S0092-8674(00)80117-8 [DOI] [PubMed] [Google Scholar]
- 25.Watson N, Linder ME, Druey KM, Kehrl JH, Blumer KJ. 1996. RGS family members: GTPase-activating proteins for heterotrimeric G-protein alpha-subunits. Nature 383:172–175. 10.1038/383172a0 [DOI] [PubMed] [Google Scholar]
- 26.Pawson T, Scott JD. 1997. Signaling through scaffold, anchoring, and adaptor proteins. Science 278:2075–2080. 10.1126/science.278.5346.2075 [DOI] [PubMed] [Google Scholar]
- 27.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G. 2009. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure 17:128–138. 10.1016/j.str.2008.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornev AP, Taylor SS. 2009. Pseudokinases: functional insights gleaned from structure. Structure 17:5–7. 10.1016/j.str.2008.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S, Lewis R, Lalaoui N, Metcalf D, Webb AI, Young SN, Varghese LN, Tannahill GM, Hatchell EC, Majewski IJ, Okamoto T, Dobson RC, Hilton DJ, Babon JJ, Nicola NA, Strasser A, Silke J, Alexander WS. 2013. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39:443–453. 10.1016/j.immuni.2013.06.018 [DOI] [PubMed] [Google Scholar]
- 30.Murphy JM, Lucet IS, Hildebrand JM, Tanzer MC, Young SN, Sharma P, Lessene G, Alexander WS, Babon JJ, Silke J, Czabotar PE. 2013. Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL. Biochem. J. 457:369–377. 10.1042/BJ20131270 [DOI] [PubMed] [Google Scholar]
- 31.Baas AF, Boudeau J, Sapkota GP, Smit L, Medema R, Morrice NA, Alessi DR, Clevers HC. 2003. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 22:3062–3072. 10.1093/emboj/cdg292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeqiraj E, Filippi BM, Goldie S, Navratilova I, Boudeau J, Deak M, Alessi DR, van Aalten DM. 2009. ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 7:e1000126. 10.1371/journal.pbio.1000126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM. 2009. Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science 326:1707–1711. 10.1126/science.1178377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chadee DN, Yuasa T, Kyriakis JM. 2002. Direct activation of mitogen-activated protein kinase kinase kinase MEKK1 by the Ste20p homologue GCK and the adapter protein TRAF2. Mol. Cell. Biol. 22:737–749. 10.1128/MCB.22.3.737-749.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yarden Y, Sliwkowski MX. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2:127–137. 10.1038/35052073 [DOI] [PubMed] [Google Scholar]
- 36.Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. 2013. Architecture and membrane interactions of the EGF receptor. Cell 152:557–569. 10.1016/j.cell.2012.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. 2009. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U. S. A. 106:21608–21613. 10.1073/pnas.0912101106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. 2011. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol. Cell 42:9–22. 10.1016/j.molcel.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. 2006. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125:1137–1149. 10.1016/j.cell.2006.05.013 [DOI] [PubMed] [Google Scholar]
- 40.Lemmon MA, Schlessinger J. 2010. Cell signaling by receptor tyrosine kinases. Cell 141:1117–1134. 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaiswal BS, Kljavin NM, Stawiski EW, Chan E, Parikh C, Durinck S, Chaudhuri S, Pujara K, Guillory J, Edgar KA, Janakiraman V, Scholz RP, Bowman KK, Lorenzo M, Li H, Wu J, Yuan W, Peters BA, Kan Z, Stinson J, Mak M, Modrusan Z, Eigenbrot C, Firestein R, Stern HM, Rajalingam K, Schaefer G, Merchant MA, Sliwkowski MX, de Sauvage FJ, Seshagiri S. 2013. Oncogenic ERBB3 mutations in human cancers. Cancer Cell 23:603–617. 10.1016/j.ccr.2013.04.012 [DOI] [PubMed] [Google Scholar]
- 42.Arteaga CL, Ramsey TT, Shawver LK, Guyer CA. 1997. Unliganded epidermal growth factor receptor dimerization induced by direct interaction of quinazolines with the ATP binding site. J. Biol. Chem. 272:23247–23254. 10.1074/jbc.272.37.23247 [DOI] [PubMed] [Google Scholar]
- 43.Gan HK, Walker F, Burgess AW, Rigopoulos A, Scott AM, Johns TG. 2007. The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor AG1478 increases the formation of inactive untethered EGFR dimers. Implications for combination therapy with monoclonal antibody 806. J. Biol. Chem. 282:2840–2850. 10.1074/jbc.M605136200 [DOI] [PubMed] [Google Scholar]
- 44.Lichtner RB, Menrad A, Sommer A, Klar U, Schneider MR. 2001. Signaling-inactive epidermal growth factor receptor/ligand complexes in intact carcinoma cells by quinazoline tyrosine kinase inhibitors. Cancer Res. 61:5790–5795 [PubMed] [Google Scholar]
- 45.Garnett MJ, Rana S, Paterson H, Barford D, Marais R. 2005. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol. Cell 20:963–969. 10.1016/j.molcel.2005.10.022 [DOI] [PubMed] [Google Scholar]
- 46.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. 2010. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464:427–430. 10.1038/nature08902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. 2010. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140:209–221. 10.1016/j.cell.2009.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S. 2010. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464:431–435. 10.1038/nature08833 [DOI] [PubMed] [Google Scholar]
- 49.Morrison DK, Cutler RE. 1997. The complexity of Raf-1 regulation. Curr. Opin. Cell Biol. 9:174–179. 10.1016/S0955-0674(97)80060-9 [DOI] [PubMed] [Google Scholar]
- 50.Hu J, Yu H, Kornev AP, Zhao J, Filbert EL, Taylor SS, Shaw AS. 2011. Mutation that blocks ATP binding creates a pseudokinase stabilizing the scaffolding function of kinase suppressor of Ras, CRAF and BRAF. Proc. Natl. Acad. Sci. U. S. A. 108:6067–6072. 10.1073/pnas.1102554108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu J, Stites EC, Yu H, Germino EA, Meharena HS, Stork PJ, Kornev AP, Taylor SS, Shaw AS. 2013. Allosteric activation of functionally asymmetric RAF kinase dimers. Cell 154:1036–1046. 10.1016/j.cell.2013.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joseph RE, Min L, Andreotti AH. 2007. The linker between SH2 and kinase domains positively regulates catalysis of the Tec family kinases. Biochemistry 46:5455–5462. 10.1021/bi602512e [DOI] [PubMed] [Google Scholar]
- 53.Cowan-Jacob SW, Fendrich G, Manley PW, Jahnke W, Fabbro D, Liebetanz J, Meyer T. 2005. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure 13:861–871. 10.1016/j.str.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 54.LaFevre-Bernt M, Sicheri F, Pico A, Porter M, Kuriyan J, Miller WT. 1998. Intramolecular regulatory interactions in the Src family kinase Hck probed by mutagenesis of a conserved tryptophan residue. J. Biol. Chem. 273:32129–32134. 10.1074/jbc.273.48.32129 [DOI] [PubMed] [Google Scholar]
- 55.Mason CS, Springer CJ, Cooper RG, Superti-Furga G, Marshall CJ, Marais R. 1999. Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. EMBO J. 18:2137–2148. 10.1093/emboj/18.8.2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lochhead PA, Sibbet G, Morrice N, Cleghon V. 2005. Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell 121:925–936. 10.1016/j.cell.2005.03.034 [DOI] [PubMed] [Google Scholar]
- 57.Lochhead PA, Kinstrie R, Sibbet G, Rawjee T, Morrice N, Cleghon V. 2006. A chaperone-dependent GSK3beta transitional intermediate mediates activation-loop autophosphorylation. Mol. Cell 24:627–633. 10.1016/j.molcel.2006.10.009 [DOI] [PubMed] [Google Scholar]
- 58.Cole A, Frame S, Cohen P. 2004. Further evidence that the tyrosine phosphorylation of glycogen synthase kinase-3 (GSK3) in mammalian cells is an autophosphorylation event. Biochem. J. 377:249–255. 10.1042/BJ20031259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ge B, Gram H, Di Padova F, Huang B, New L, Ulevitch RJ, Luo Y, Han J. 2002. MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38alpha. Science 295:1291–1294. 10.1126/science.1067289 [DOI] [PubMed] [Google Scholar]
- 60.Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. 2006. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 311:822–826. 10.1126/science.1120941 [DOI] [PubMed] [Google Scholar]
- 61.Mendrola JM, Shi F, Park JH, Lemmon MA. 2013. Receptor tyrosine kinases with intracellular pseudokinase domains. Biochem. Soc. Trans. 41:1029–1036. 10.1042/BST20130104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bandaranayake RM, Ungureanu D, Shan Y, Shaw DE, Silvennoinen O, Hubbard SR. 2012. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat. Struct. Mol. Biol. 19:754–759. 10.1038/nsmb.2348 [DOI] [PMC free article] [PubMed] [Google Scholar]