Abstract
A synthesis of carolacton, a myxobacterial natural product that has profound effects on Streptococcus mutans biofilms, is reported. The synthesis proceeds via a longest linear sequence of 14 steps from an Evans β-ketoimide and enabled preliminary evaluations of the effects of late-stage intermediates on S. mutans biofilms. These studies suggest that further investigations into carolacton’s structure–function relationships are warranted.
Dental caries is a chronic health problem that impacts the majority of the global population.1 Although culture-independent sequencing has illuminated the complexity of the human oral microbiome, Streptococcus mutans remains widely regarded as a primary etiological agent in caries.2,3 This facultatively anaerobic, microaereophilic, Gram-positive, bacterium excels in the complex environment of the oral cavity where stresses including low pH and an oxidative environment are coupled to variable salivary flow and carbohydrate supply.4 The physiologic adaptations of S. mutans to these pressures provide a competitive advantage versus noncariogenic commensals that underpins the establishment and progression of caries.5
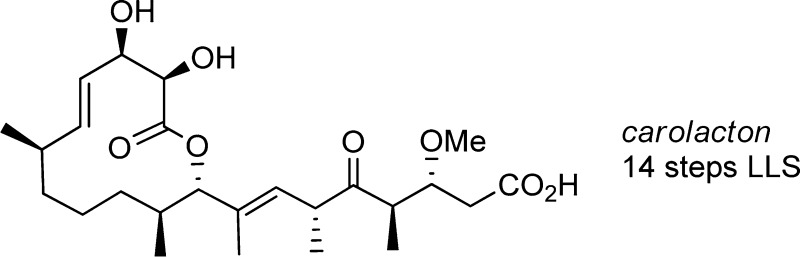
Broad-spectrum agents such as fluoride and chlorhexidine represent the state-of-the-art in anticariogenic preventative treatment, although the efficacy of chlorhexidine remains debated.6 Significant perturbation of the oral microbiome accompanies their use,7 and there is increasing evidence that such imbalances can favor pathogens versus commensals.8 Compounds that are species- or phenotype-selective would allow exploration of the hypothesis that targeting smaller elements of complex communities may be advantageous.9 In this light, we were attracted to a report describing the structure and activity of the myxobacterial metabolite carolacton (1, Figure 1).10 Initial characterizations showed that carolacton causes 35–66% cell death within S. mutans biofilms at concentrations ranging from 5 to 25 ng/mL. Remarkably, planktonic cultures of S. mutans were not affected by carolacton, nor were a variety of other bacteria.
Figure 1.
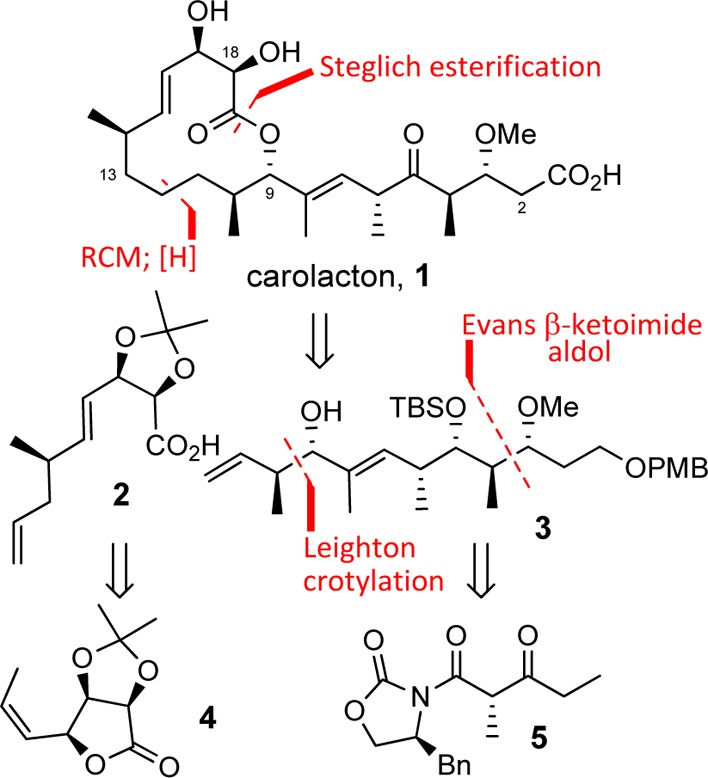
Carolacton structure and an overview of key features in the synthesis that traces to lactone 4 and β-ketoimide 5.
The structure and activity of carolacton has attracted much scientific interest. Kirschning has completed a total synthesis, and several subunit syntheses have also been reported.11 In addition, a number of investigations have pursued in more detail the biology of carolacton’s effects on S. mutans and the question of its mode-of-action.12 While there is accord on the differential activity of carolacton for biofilm versus planktonic cultures of S. mutans, coherency around mode-of-action has yet to emerge.
In this Letter, we describe a straightforward synthesis of carolacton that is amenable to an extensive exploration of structure–function relationships. Key elements of the synthesis are shown in Figure 1, including an assembly strategy that leverages macrocycle formation by esterification and a ring-closing metathesis, followed by a selective olefin hydrogenation.
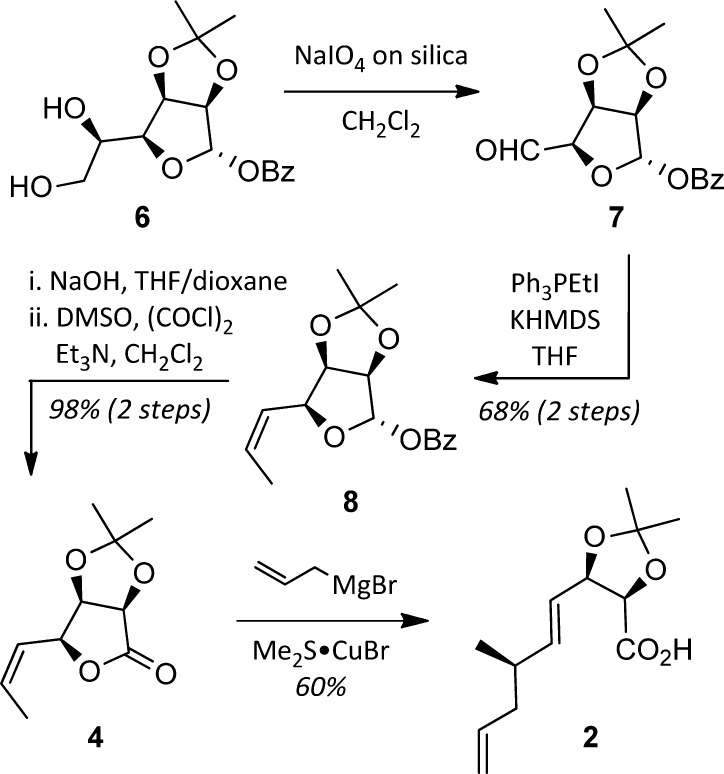
The synthesis of the C12–C19 acid 2 began with known gulonolactone-derived diol 6(13) (available in 4 steps, 62% overall from d-gulonolactone, Scheme 1). Oxidative cleavage of the diol with sodium periodate on silica14 gave aldehyde 7, which was subjected to a Z-selective Wittig olefination to yield alkene 8. Removal of the benzoate, followed by Swern oxidation, provided the key lactone 4. In accord with Trost’s studies,15 exposing 4 to allylmagnesium bromide in the presence of CuBr·SMe2 resulted in SN2′ opening of the lactone to produce acid 2 in 60% yield and as a single diastereomer.
Scheme 1. Synthesis of the C12–C19 Subunit 2 from Gulonolactone-Derived Diol 6.
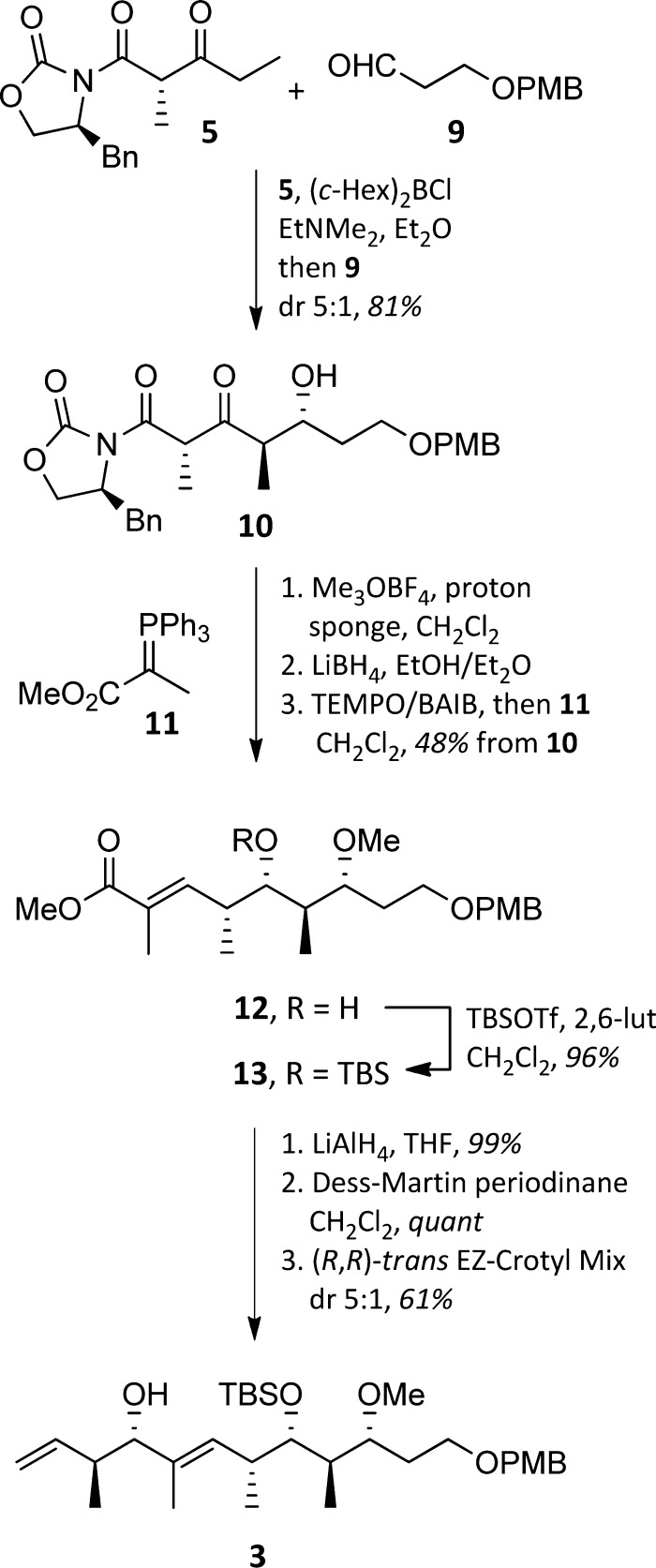
The stereochemically complex C1–C11 domain was assembled as shown in Scheme 2. Enolization of the Evans β-ketoimide 5 under conditions favoring formation of the (E)-enol borane, followed by reaction with aldehyde 9, produced 10 in 81% yield and with a dr of 5:1.16 Separation of the diastereomers was not possible. Conveniently, the subsequent three-step sequence of methylation, reduction, and oxidation–Wittig olefination not only proceeded in 48% overall yield but also allowed for isolation of enoate 12 as a single diastereomer. Silylation of the alcohol gave TBS ether 13. Introduction of the final two stereocenters was readily achieved by conversion of the ester to an aldehyde and application of Leighton’s (R,R)-trans EZ-CrotylMix to give alcohol 3 (61%, 5:1 dr).17
Scheme 2. Synthesis of the C1–C11 Subunit 3.
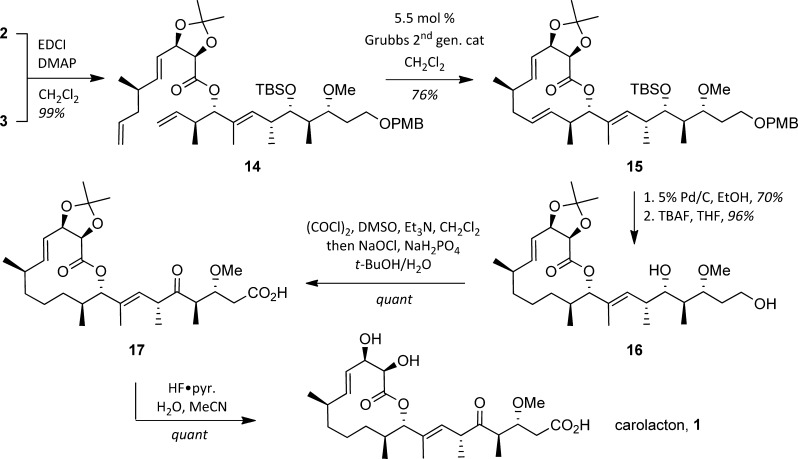
With routes to the two key subunits secured, it was possible to investigate the end-game for the synthesis, which began with a Steglich esterification to connect acid 2 and alcohol 3, producing a near-quantitative yield of 14 (Scheme 3). Reaction of 14 with 5.5 mol % of the second-generation Grubbs catalyst was unsurprisingly selective for the terminal olefins and led to lactone 15 in 76% yield. Our plans demanded a selective hydrogenation of the Δ11,12 olefin at this juncture, a strategy predicated on conformational analysis of 15 suggesting peripheral addition would favor the desired reduction.18 Pleasingly, this was borne out under operationally simple conditions: subjecting 15 to 5% Pd/C in EtOH under a hydrogen balloon resulted in selective reduction of the Δ11,12 olefin as well as removal of the PMB group (70% yield). Removal of the TBS ether with TBAF produced diol 16 in 96% yield and set the stage for a simultaneous oxidation of the primary and secondary alcohols under Swern conditions and subsequent Lindgren–Pinnick oxidation to yield keto-acid 17. Removal of the acetonide yielded synthetic carolacton (1). Analytical characteristics of this material were in full accord with data reported for the natural product.
Scheme 3. Subunit Coupling and Completion of the Synthesis.
Our initial synthetic venture provided multimilligram amounts of intermediates in the late stages of the synthesis. We capitalized on this to perform a preliminary biological evaluation of compounds 16, 17, and carolacton against S. mutans UA159 grown planktonically and also within a biofilm. None of the compounds displayed any inhibitory activity against planktonic cells at concentrations <250 μM. In the case of carolacton, this is in accord with previous reports.12b However, when the compounds were incubated with S. mutans in the presence of biofilm-inducing media, dramatic morphological and architectural changes to the biofilm matrix were observed (Figure 2). Confocal microscopy with LIVE/DEAD staining demonstrated that carolacton (>500 nM) and 16 (>62.5 μM) significantly affected both the integrity and morphology of the biofilm matrix when compared to the DMSO control. In contrast, compound 17 had no observable effect at concentrations <250 μM. While the reasons for the significant difference in activity between compound 16 and 17 remain unclear at this time, our preliminary investigations provide credence to the suggestion that a minimal set of features needed for activity in the carolacton family might be readily identified by synthesis. Future efforts will focus on expanding the understanding of structure–function relationships in the carolacton class, as well as obtaining a detailed understanding of the mechanism of action of carolacton on S. mutans biofilms.
Figure 2.
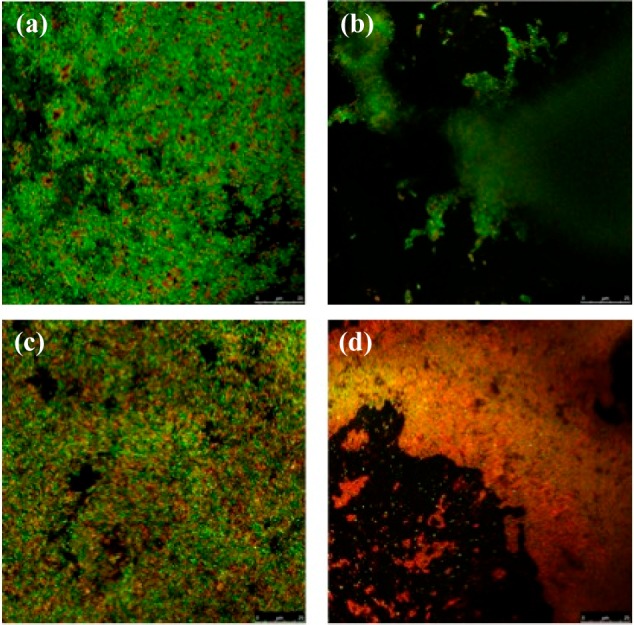
Confocal microscopy imaging of S. mutans UA159 biofilm cells treated with 250 μM (a) DMSO, (b) carolacton, (c) 17, and (d) 16.
Acknowledgments
This work was supported by Yale University (A.J.P.), in part by the NIH (CA110246, A.J.P.), and Temple University (W.M.W.). We thank Professor Bettina A. Buttaro (Temple University) for advice and assistance with confocal microscopy of S. mutans biofilms, and Dr. Jennifer A. Poole (Yale University) for mass spectrometry assistance.
Supporting Information Available
Experimental methods and data, including spectra, for new compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Bagramian R. A.; Garcia-Godoy F.; Volpe A. R. Am. J. Dent. 2009, 22, 3. [PubMed] [Google Scholar]
- Dewhirst F. E.; Chen T.; Izard J.; Paster B. J.; Tanner A. C.; Yu W. H.; Lakshmanan A.; Wade W. G. J. Bacteriol. 2010, 192, 5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kuramitsu H. K.; Wang B. Y. Am. J. Dent. 2011, 24, 153. [PMC free article] [PubMed] [Google Scholar]; b Loesche W. J. Microbiol. Rev. 1986, 50, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos J. A.; Burne R. A. Microbiology 2008, 154, 3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S. R.; Miller J. H.; Abranches J.; Zeng L.; Lefebure T.; Richards V. P.; Lemos J. A.; Stanhope M. J.; Burne R. A. PLoS One 2013, 8, e61358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Autio-Gold J. Oper. Dent. 2008, 33, 710. [DOI] [PubMed] [Google Scholar]; b Fontana M.; Young D. A.; Wolff M. S. Dent. Clin. North Am. 2009, 53, 149. [DOI] [PubMed] [Google Scholar]; c Milgrom P.; Zero D. T.; Tanzer J. M. Acad. Pediatr. 2009, 9, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Breaker R. R. Caries Res. 2012, 46, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Herrera D.; Alonso B.; Leon R.; Roldan S.; Sanz M. J. Clin. Periodontol. 2008, 35, 45. [DOI] [PubMed] [Google Scholar]; c Horner C.; Mawer D.; Wilcox M. J. Antimicrob. Chemother. 2012, 67, 2547. [DOI] [PubMed] [Google Scholar]; d Marquis R. E.; Clock S. A.; Mota-Meira M. FEMS Microbiol. Rev. 2003, 26, 493. [DOI] [PubMed] [Google Scholar]
- a Kuramitsu H. K.; He X. S.; Lux R.; Anderson M. H.; Shi W. Y. Microbiol. Mol. Biol. Rev. 2007, 71, 653. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Willing B. P.; Russell S. L.; Finlay B. B. Nat. Rev. Microbiol. 2011, 9, 233. [DOI] [PubMed] [Google Scholar]
- a Eckert R.; He J.; Yarbrough D. K.; Qi F.; Anderson M. H.; Shi W. Antimicrob. Agents Chemother. 2006, 50, 3651. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Worthington R. J.; Richards J. J.; Melander C. Org. Biomol.Chem. 2012, 10, 7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.; Irschik H.; Huch V.; Schummer D.; Steinmetz H.; Bock M.; Schmidt T.; Kirschning A.; Müller R. Eur. J. Org. Chem. 2010, 1284. [Google Scholar]
- a Rao K. S.; Ghosh S. Synthesis 2013, 45, 2745. [Google Scholar]; b Sabitha G.; Shankaraiah K.; Prasad M. N.; Yadav J. S. Synthesis 2013, 45, 251. [Google Scholar]; c Schmidt T.; Kirschning A. Angew. Chem., Int. Ed. 2012, 51, 1063. [DOI] [PubMed] [Google Scholar]; d Sharma G. V. M.; Reddy S. V. RSC Adv. 2013, 3, 21759. [Google Scholar]
- a Apel C.; Barg A.; Rheinberg A.; Conrads G.; Wagner-Döbler I. Dent. Mater. 2013, 29, 1188. [DOI] [PubMed] [Google Scholar]; b Kunze B.; Reck M.; Dotsch A.; Lemme A.; Schummer D.; Irschik H.; Steinmetz H.; Wagner-Döbler I. BMC Microbiol. 2010, 10, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Li J.; Wang W.; Wang Y.; Zeng A. P. Proteomics 2013, 13, 3470. [DOI] [PubMed] [Google Scholar]; d Reck M.; Rutz K.; Kunze B.; Tomasch J.; Surapaneni S. K.; Schulz S.; Wagner-Döbler I. J. Bacteriol. 2011, 193, 5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranathan T.; Hiremath S. V.; Reddy D. R.; Rao A. V. R. Carbohydr. Res. 1984, 134, 332. [Google Scholar]
- Zhong Y. L.; Shing T. K. M. J. Org. Chem. 1997, 62, 2622. [DOI] [PubMed] [Google Scholar]
- Trost B. M.; Klun T. P. J. Org. Chem. 1980, 45, 4256. [Google Scholar]
- Evans D. A.; Ng H. P.; Clark J. S.; Rieger D. L. Tetrahedron 1992, 48, 2127. [Google Scholar]
- Kim H.; Ho S.; Leighton J. L. J. Am. Chem. Soc. 2011, 133, 6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For pioneering applications of this concept, see:; a Still W. C.; Galynker I. Tetrahedron 1981, 37, 3981. [Google Scholar]; b Vedejs E.; Gapinski D. M. J. Am. Chem. Soc. 1983, 105, 5058. [Google Scholar]; c Schreiber S. L.; Sammakia T.; Hulin B.; Schulte G. J. Am. Chem. Soc. 1986, 108, 2106. [Google Scholar]; d Lee D.; Sello J. K.; Schreiber S. L. J. Am. Chem. Soc. 1999, 121, 10648. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.