Abstract
A French national quality control study for the serological and molecular diagnosis of hepatitis delta virus (HDV) was organized. Total HDV antibodies were properly detected by all laboratories; 8/14 laboratories failed to detect low titers of IgM, and 6/11 failed to quantify and/or underestimated the RNA viral load in several samples. These discrepancies are likely related to the molecular diversity of HDV.
TEXT
Hepatitis delta virus (HDV) is a satellite of the hepatitis B virus (HBV). The HDV genome is composed of a small negative single-stranded RNA (1,679 to 1,697 nucleotides [nt]) with a rod-like structure that is related to an extensive intramolecular complementarity. This genome is closely associated to the two isoforms of the delta protein, the small (s) and the large (L) HDV antigens (Ag), and it forms a ribonucleoprotein enveloped by the HBV surface antigen (HBsAg).
Eight genotypes (HDV-1 to -8) and several subgenotypes have been described and are defined by nucleotide divergences of >20% and >10%, respectively, considering the complete genome sequence (1, 2; E. Gordien, presented at the European Association for the Study of the Liver Monothematic Conference, 24 to 26 September 2010, Istanbul, Turkey). Due to population migrations, most genotypes circulate worldwide and especially in France (4–7; S. Brichler, F. Le Gal, W. Mansour, S. Chevret, D. Roulot, and E. Gordien, presented at the 63rd Annual Meeting of the AASLD, Boston, MA, 9 to 13 November 2012).
HBV/HDV co- or superinfection leads to more severe liver disease than does HBV infection alone. HDV diagnosis relies on detection of total anti-HDV antibodies (Abs). Anti-HDV IgM Abs usually persist in patients who are chronically infected with HDV. Some authors consider anti-HDV IgM Abs a surrogate marker of HDV replication (9); however, they may be lacking in some African patients (reference 10 and E. Gordien, personal observations). Therefore, HDV RNA detection/quantification is currently the only accurate diagnostic tool for confirming HDV replication status and allowing for the optimal management of infected patients (11, 12).
Commercial tests for HDV serology have been available for many years and are used routinely worldwide. Currently, a few commercial tests are available for HDV RNA quantification, but they perform poorly, at least for non-genotype 1 HDV samples (13). Numerous in-house assays have been developed with very different protocols (11, 14–20). To our knowledge, these assays have not been evaluated on a large panel of clinical samples of various genotypes and viral loads (VL).
In 2012, the French National Reference Center for HDV (F-NRC) at Avicenne Hospital (Bobigny, France) organized an unprecedented French national quality control (FNQC) study for the diagnosis of HDV infection by serology and molecular biology. A total of 28 laboratories participated in this study, including 22 university hospital laboratories, 2 private laboratories, and 4 foreign laboratories (from Greece, Switzerland, the United Kingdom, and the United States). They performed total HDV Ab (n = 24) and/or IgM Ab (n = 14) detection and/or HDV RNA viral load (VL) quantification (n = 11), which are performed routinely for their patients, using either in-house or commercial assays. The serology panel consisted of 1 negative sample and 3 undiluted samples from blood donors, selected for their strong or weak reactivity, after 3 different experiments performed with our commercial assays (DiaSorin, Antony, France). The molecular biology panel consisted of 11 plasma samples of various genotypes and VL, including 3 samples with genotype HDV-1 Europe/Asia (Eu/As), 3 with genotype HDV-1 Africa (Afr), 2 with genotype HDV-5, 1 with genotype HDV-6, 1 with genotype HDV-7, and 1 with genotype HDV-8, and a negative control (Table 1). Quantification was performed blindly 3 times by different technicians in our lab using a consensus real-time reverse transcription-PCR (RT-PCR) assay described elsewhere (11). The mean of the 3 values was considered the expected value for each sample (Table 1). Strains were genotyped by direct sequencing of the amplicon of the R0 region (nt 889 to 1289) exactly as previously described (1).
TABLE 1.
Serological and molecular biological results
| Serological results |
Molecular biological results |
||||||
|---|---|---|---|---|---|---|---|
| Sample | HDV Ab | Expected result | Concordance (n/n [%])a | Sample | Genotype | Expected result (log10 copies/ml)b | Concordance (n/n [%])a,c |
| A | Total | Positive | 24/24 (100) | 1 | HDV-1 Eu/As | 4.18 | 6/11 (55) |
| IgM | Positive (weak) | 6/14 (43) | 2 | HDV-1 Eu/As | 5.64 | 8/11 (73) | |
| B | Total | Positive | 24/24 (100) | 3 | HDV-1 Eu/As | 7.55 | 6/11 (55) |
| IgM | Positive | 14/14 (100) | 4 | HDV-1 Afr | 5.06 | 6/11 (55) | |
| C | Total | Positive | 24/24 (100) | 5 | HDV-1 Afr | 6.48 | 6/11 (55) |
| IgM | Positive(weak) | 9/14 (64) | 6 | HDV-1 Afr | 7.52 | 6/11 (55) | |
| D | Total | Negative | 24/24 (100) | 7 | HDV-5 | 4.77 | 5/11 (45) |
| IgM | Negative | 14/14 (100) | 8 | HDV-5 | 6.33 | 7/11 (64) | |
| 9 | HDV-6 | 5.36 | 7/11 (64) | ||||
| 10 | HDV-7 | 6.42 | 6/11 (55) | ||||
| 11 | HDV-8 | 6.07 | 5/11 (45) | ||||
| 12 | Negative control | < 2 | 9/11 (82) | ||||
n/n, number of concordant results over the number of participants.
Mean of values of 3 different experiments performed by the F-NRC laboratory.
Concordance: less than ±1 log10 copy/ml compared to the expected values from the F-NRC lab.
The samples were sent in dry ice by an accredited transporter to the participating laboratories according to regulatory standards for the distribution of infected samples.
Commercial assays were used by all the labs for total and/or IgM Ab detection: DiaSorin (Antony, France) (n = 20), Adaltis (Rome, Italy) (n = 2), or Diagnostic BioProbes (Milan, Italy) (n = 2). DiaSorin (n = 14) was used for IgM detection. No discrepancies were found for total Ab detection, whatever the test used (Table 1). The negative control (sample D) and the sample with a high IgM Ab titer (sample B) were correctly identified by all the labs. However, the samples with low IgM Ab titers (samples A and C) were identified as negative by 8 and 5 labs, respectively. These results strengthen arguments for the use of HDV RNA as the sole reliable replication marker.
According to the data provided, 10 laboratories used an in-house assay based on real-time RT-PCR protocols for HDV RNA quantification, and the remaining one used the commercial Roche LightMix kit (Meylan, France). Primers and probes were located in the hepatitis delta antigen (HDAg), the ribozymes, or the interribozyme coding regions. The labs used either an RNA or a DNA standard to quantify HDV viremia. The amount of extracted sample and the declared lower limits of quantification (LLOQ), ranging from 40 to 1,000 copies/ml, were also different across the assays. It is noteworthy that 2 labs used the same assay that the F-NRC used.
Overall results are shown in Table 1. Detailed results for each sample are shown in Fig. 1. Two laboratories found a positive signal for the negative sample (sample 12), raising the problem of specificity for these assays and the interpretation of very weak signals obtained in the real-time RT-PCR assays.
FIG 1.
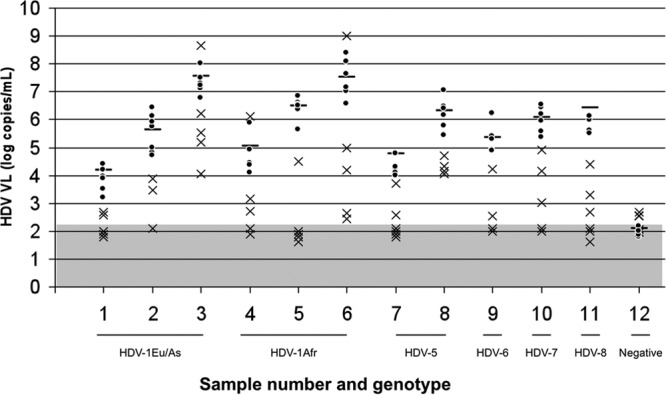
HDV RNA quantification results for the 12 samples, for all participants, expressed as log10 copies/ml. Horizontal bars represent the expected VL values obtained by the F-NRC laboratory (mean of 3 different experiments). VL values of less than ±1 log10 copies/ml or more than ±1 log10 copies/ml compared with the expected VL values are represented with dots or crosses, respectively. The gray area represents the lower limit of detection/quantification of the different assays. Genotypes of the different samples are indicated.
Moreover, 5 laboratories failed to detect one or several positive samples. Six laboratories underestimated several samples by >1 log copy/ml, and among them, 3 labs underquantified all the samples. These results were obtained whatever the genotype of the infecting strain. As described elsewhere (13), poor results were obtained with the commercial test (LightMix, Roche).
Overall, 5 laboratories found results similar to those of the F-NRC (less than ±1 log copy/ml). Interestingly, all but one lab used primers and probes designed in the same conserved ribozyme region of the HDV genome, and the other lab used primers and probes designed in the antigen-coding region.
This FNQC study provided an overview and a comparison of the performances of the different HDV tests, including those of labs outside France. Commercial total anti-HDV Ab enzyme-linked immunosorbent assay (ELISA) kits seem to be the tools of choice for HDV screening in HBsAg-positive patients. IgM anti-HDV Ab assays may fail to detect low titers of Abs and thus should not be used to evaluate HDV replication status. Therefore, HDV RNA, which provides accurate detection/quantification, should be considered the main tool for guiding patient management.
Previous work in our lab shed light on the importance of HDV genetic variability (13). In France, 40% of the spreading strains belong to HDV-1 Eu/As, 35% belong to HDV-1 Afr, and 25% belong to HDV-5 to -8; HDV-2, -3, and -4 remain very low in our cohort of patients.
This same epidemiological situation may be true for other European countries experiencing migration of populations from Africa and Asia, which are areas of endemicity, although HDV-1 remains largely predominant and ubiquitous (4–7, 17, 20–22). However, in this study, we demonstrated that even HDV-1 Eu/As strain VL could not be properly quantified by all labs.
A thorough verification of primer/probe design is necessary because extensive sequence alignments have shown mismatches that can predict quantification failure for some samples (F. Le Gal, unpublished data).
Considering available data provided by the participants, other technical points may explain some discrepant results but were not assessed in the present study; some protocols lack an internal control to validate all steps from extraction to amplification. Therefore, for some labs, we cannot exclude specific technical problems affecting particular samples. Additionally, some steps are performed manually, raising the question of the reproducibility of results, although we did not look at that issue in the present study.
Either cDNA or RNA samples were used as quantification standards; thus, precise comparisons between the tests were difficult in the absence of an international standard at the time of this study. This issue will be greatly improved with the now-available WHO HDV RNA standard (see www.who.int/biologicals/expert_committee/BS_2227_HDV_RNA.pdf). Nevertheless, this study did show that comparable results (coefficient of variation, <20%) can be obtained with different assays (data not shown).
The management of an HDV-infected patient relies on the accurate estimation of HDV replication. Indeed, a false-negative HDV VL result can lead to (i) misdiagnosis and failure to identify an HDV-replicating patient, (ii) poor management with undertreatment, and (iii) failure to detect early relapse (12, 13).
In conclusion, we advise that HDV VL be monitored only in reference virology centers using well-evaluated techniques. Also, there is a need for international comparative studies comprising samples of all genotypes and subgenotypes spreading worldwide. Indeed, all efforts to improve and standardize quantification assays should be continued, including the use of the newly available WHO HDV RNA standard, with the goal of forwarding collaborative clinical studies and, ultimately, optimizing patient management.
ACKNOWLEDGMENTS
This work was supported by grants from the Agence Nationale de Recherche sur le SIDA et les Hépatites Virales (ANRS).
We thank the heads of the virology laboratories that participated in this first French national quality control study for HDV diagnosis: Hôpital Pellegrin-Service de Virologie (Bordeaux), Laboratoire Cerba (Cergy Pontoise), Hôpital Beaujon-Service de Virologie (Clichy), Hôpital Henri Mondor-Service de Virologie (Créteil), Athens University Medical School-Department of Hygiene and Epidemiology (Greece), Laboratoire Biomnis (Ivry sur Seine), Hôpital Bicêtre-Service de Virologie (Le Kremlin-Bicêtre), Hôpital Dupuytren-Service de Virologie (Limoges), Laboratoire Biomnis (Lyon), Unité INSERM U871 (Lyon), Hôpital de la Timone-Laboratoire de Virologie (Marseille), CHU Montpellier-Laboratoire de Virologie (Montpellier), CHU Nantes-Laboratoire de Virologie (Nantes), Hôpital Archet-Service de Virologie (Nice), Hôpital Bichat-Service de Virologie (Paris), Hôpital Pitié-Salpetrière-Service de Virologie (Paris), Hôpital Saint Antoine-Service de Virologie (Paris), Institut National de Transfusion Sanguine (Paris), CHU Poitiers-Laboratoire de Virologie (Poitiers), CHU Pontchaillou-Laboratoire de Virologie (Rennes), Hôpital Charles Nicolle-Service de Virologie (Rouen), CHU Saint Etienne-Laboratoire de Virologie (Saint-Etienne), CHU Strasbourg-Laboratoire de Virologie (Strasbourg), CHUV Lausanne (Switzerland), Hôpital Purpan-Service de Virologie (Toulouse), Faculté de Médecine de Tours, Clinical Microbiology-Virology UCLH NHS Foundation of London (United Kingdom), Laboratory Branch Division of Viral Hepatitis, NCHHSTP, Centers for Disease Control and Prevention (Atlanta, GA).
We declare no conflicts of interest.
Footnotes
Published ahead of print 12 February 2014
REFERENCES
- 1.Radjef N, Gordien E, Ivaniushina V, Gault E, Anais P, Drugan T, Trinchet JC, Roulot D, Tamby M, Milinkovitch MC, Deny P. 2004. Molecular phylogenetic analyses indicate a wide and ancient radiation of African hepatitis delta virus, suggesting a deltavirus genus of at least seven major clades. J. Virol. 78:2537–2544. 10.1128/JVI.78.5.2537-2544.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Gal F, Gault E, Ripault MP, Serpaggi J, Trinchet JC, Gordien E, Deny P. 2006. Eighth major clade for hepatitis delta virus. Emerg. Infect. Dis. 12:1447–1450. 10.3201/eid1209.060112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reference deleted.
- 4.Cross TJ, Rizzi P, Horner M, Jolly A, Hussain MJ, Smith HM, Vergani D, Harrison PM. 2008. The increasing prevalence of hepatitis delta virus (HDV) infection in south London. J. Med. Virol. 80:277–282. 10.1002/jmv.21078 [DOI] [PubMed] [Google Scholar]
- 5.De Paschale M, Manco MT, Belvisi L, Magnani C, Re T, Vigano P, Biagiotti S, Capelli F, Mazzone A, Baldacci MP, Ferrara A, Neri AL, Guastoni CM, Bonazzina RA, Brando B, Clerici P. 2012. Epidemiology of hepatitis D virus (HDV) infection in an urban area of northern Italy. Infection 40:485–491. 10.1007/s15010-012-0247-4 [DOI] [PubMed] [Google Scholar]
- 6.Heidrich B, Deterding K, Tillmann HL, Raupach R, Manns MP, Wedemeyer H. 2009. Virological and clinical characteristics of delta hepatitis in central Europe. J. Viral Hepat. 16:883–894. 10.1111/j.1365-2893.2009.01144.x [DOI] [PubMed] [Google Scholar]
- 7.Reinheimer C, Doerr HW, Berger A. 2012. Hepatitis delta: on soft paws across Germany. Infection 40:621–625. 10.1007/s15010-012-0287-9 [DOI] [PubMed] [Google Scholar]
- 8. Reference deleted.
- 9.Mederacke I, Yurdaydin C, Dalekos GN, Bremer B, Erhardt A, Cakaloglu Y, Yalcin K, Gurel S, Zeuzem S, Zachou K, Bozkaya H, Dienes HP, Manns MP, Wedemeyer H. 2012. Anti-HDV immunoglobulin M testing in hepatitis delta revisited: correlations with disease activity and response to pegylated interferon-alpha2a treatment. Antivir. Ther. 17:305–312. 10.3851/IMP1926 [DOI] [PubMed] [Google Scholar]
- 10.Lunel-Fabiani F, Mansour W, Amar AO, Aye M, Le Gal F, Malick FZ, Baidy L, Brichler S, Veillon P, Ducancelle A, Gordien E, Rosenheim M. 2013. Impact of hepatitis B and delta virus co-infection on liver disease in Mauritania: a cross sectional study. J. Infect. 67:448–457. 10.1016/j.jinf.2013.06.008 [DOI] [PubMed] [Google Scholar]
- 11.Le Gal F, Gordien E, Affolabi D, Hanslik T, Alloui C, Deny P, Gault E. 2005. Quantification of hepatitis delta virus RNA in serum by consensus real-time PCR indicates different patterns of virological response to interferon therapy in chronically infected patients. J. Clin. Microbiol. 43:2363–2369. 10.1128/JCM.43.5.2363-2369.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castelnau C, Le Gal F, Ripault MP, Gordien E, Martinot-Peignoux M, Boyer N, Pham BN, Maylin S, Bedossa P, Deny P, Marcellin P, Gault E. 2006. Efficacy of peginterferon alpha-2b in chronic hepatitis delta: relevance of quantitative RT-PCR for follow-up. Hepatology 44:728–735. 10.1002/hep.21325 [DOI] [PubMed] [Google Scholar]
- 13.Brichler S, Le Gal F, Butt A, Chevret S, Gordien E. 2013. Commercial real-time reverse transcriptase PCR assays can underestimate or fail to quantify hepatitis delta virus viremia. Clin. Gastroenterol. Hepatol. 11:734–740. 10.1016/j.cgh.2013.01.025 [DOI] [PubMed] [Google Scholar]
- 14.Hofmann J, Frenzel K, Minh BQ, von Haeseler A, Edelmann A, Ross SR, Berg T, Kruger DH, Meisel H. 2010. Quantitative detection and typing of hepatitis D virus in human serum by real-time polymerase chain reaction and melting curve analysis. Diagn. Microbiol. Infect. Dis. 67:172–179. 10.1016/j.diagmicrobio.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 15.Mederacke I, Bremer B, Heidrich B, Kirschner J, Deterding K, Bock T, Wursthorn K, Manns MP, Wedemeyer H. 2010. Establishment of a novel quantitative hepatitis D virus (HDV) RNA assay using the Cobas TaqMan platform to study HDV RNA kinetics. J. Clin. Microbiol. 48:2022–2029. 10.1128/JCM.00084-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferns RB, Nastouli E, Garson JA. 2012. Quantitation of hepatitis delta virus using a single-step internally controlled real-time RT-qPCR and a full-length genomic RNA calibration standard. J. Virol. Methods 179:189–194. 10.1016/j.jviromet.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 17.Shang D, Hughes SA, Horner M, Bruce MJ, Dong Y, Carey I, Suddle AR, Agarwal K, Harrison PM, Atkins M. 2012. Development and validation of an efficient in-house real-time reverse transcription polymerase chain reaction assay for the quantitative detection of serum hepatitis delta virus RNA in a diverse south London population. J. Virol. Methods 184:55–62. 10.1016/j.jviromet.2012.05.008 [DOI] [PubMed] [Google Scholar]
- 18.Scholtes C, Icard V, Amiri M, Chevallier-Queyron P, Trabaud MA, Ramiere C, Zoulim F, Andre P, Deny P. 2012. Standardized one-step real-time reverse transcription-PCR assay for universal detection and quantification of hepatitis delta virus from clinical samples in the presence of a heterologous internal-control RNA. J. Clin. Microbiol. 50:2126–2128. 10.1128/JCM.06829-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kodani M, Martin A, Mixson-Hayden T, Drobeniuc J, Gish RR, Kamili S. 2013. One-step real-time PCR assay for detection and quantitation of hepatitis D virus RNA. J. Virol. Methods 193:531–535. 10.1016/j.jviromet.2013.07.033 [DOI] [PubMed] [Google Scholar]
- 20.Katsoulidou A, Manesis E, Rokka C, Issaris C, Pagoni A, Sypsa V, Hatzakis A. 2013. Development and assessment of a novel real-time PCR assay for quantitation of hepatitis D virus RNA to study viral kinetics in chronic hepatitis D. J. Viral Hepat. 20:256–262. 10.1111/jvh.12000 [DOI] [PubMed] [Google Scholar]
- 21.Le Gal F, Badur S, Hawajri NA, Akyuz F, Kaymakoglu S, Brichler S, Zoulim F, Gordien E, Gault E, Deny P. 2012. Current hepatitis delta virus type 1 (HDV1) infections in central and eastern Turkey indicate a wide genetic diversity that is probably linked to different HDV1 origins. Arch. Virol. 157:647–659. 10.1007/s00705-011-1212-8 [DOI] [PubMed] [Google Scholar]
- 22.Popescu GA, Otelea D, Gavriliu LC, Neaga E, Popescu C, Paraschiv S, Fratila M. 2013. Epidemiology of hepatitis D in patients infected with hepatitis B virus in Bucharest: a cross-sectional study. J. Med. Virol. 85:769–774. 10.1002/jmv.23524 [DOI] [PubMed] [Google Scholar]


