Abstract
An outbreak of Newcastle disease (ND) in poultry was reported in Belize in 2008. The characteristics of three virulent Newcastle disease virus (NDV) isolates from this outbreak (NDV-Belize-3/08, NDV-Belize-4/08, and NDV-Belize-12/08) were assessed by genomic analysis and by clinicopathological characterization in specific-pathogen-free (SPF) chickens. The results showed that all three strains belong to NDV genotype V and are virulent, as assessed by the intracerebral pathogenicity index and the polybasic amino acid sequence at the fusion protein cleavage site. In 4-week-old SPF chickens, NDV-Belize-3/08 behaved as a typical velogenic viscerotropic NDV strain, causing severe necrohemorrhagic lesions in the lymphoid organs, with systemic virus distribution. Phylogenetic analysis of multiple NDV genotype V representatives revealed that genotype V can be divided into three subgenotypes, namely, Va, Vb, and Vc, and that all tested Belizean isolates belong to subgenotype Vb. Furthermore, these isolates are nearly identical to a 2007 isolate from Honduras and appear to have evolved separately from other contemporary viruses circulating in Mexico, clustering into a new clade within NDV subgenotype Vb.
INTRODUCTION
Newcastle disease (ND), caused by virulent strains of Newcastle disease virus (NDV), is one of the most severe diseases of poultry worldwide, claiming major economic losses in developing countries due to mortality, disease containment measures, outbreak eradication, and trade restrictions (1). In underdeveloped and developing countries, outbreaks of ND can be a limiting factor for backyard poultry production, therefore decreasing the amount of protein available for human consumption, especially as egg products (1–3). NDV is synonymous with avian paramyxovirus serotype 1 (APMV-1) and belongs to the Avulavirus genus, Paramyxoviridae family (4, 5). NDV has a genome composed of single-stranded negative-sense RNA approximately 15.2 kb in length, which encodes six structural proteins in the 3′ to 5′ sense (nucleoprotein [NP], phosphoprotein [P], matrix [M], hemagglutinin neuraminidase [HN], fusion [F], and RNA-dependent RNA polymerase [L] [4, 5]), and at least one nonstructural protein (V protein, encoded by a posttranscriptional editing of the P gene mRNA [6]). Based on the severity of disease produced in chickens, NDV strains have been historically classified into five pathotypes: asymptomatic enteric, lentogenic, mesogenic, velogenic neurotropic, and velogenic viscerotropic (1, 7). Enteric strains do not cause disease in birds, lentogenic strains cause very mild clinical signs, and mesogenic strains have intermediate virulence and produce respiratory and/or neurological signs (1, 7). Velogenic viscerotropic NDV strains (VVNDV) cause severe necrosis and hemorrhages in the intestines and lymphoid organs, whereas velogenic neurotropic NDV (VNNDV) strains induce neurological disease (7). The main molecular determinant of NDV pathogenesis is the amino acid sequence of the F protein cleavage site, which determines its ability to be cleaved (activated) by ubiquitous proteases and to produce a systemic infection (8–10). The internationally accepted definition of a virulent NDV strain is based on the intracerebral pathogenicity index (ICPI) and the sequence of the fusion (F) cleavage site. According to the World Organization for Animal Health (former Office International des Epizooties [OIE]), virulent strains are those that have an ICPI of ≥0.7 or whose deduced amino acid sequence of the F protein has at least three basic amino acids between residues 113 and 116 and a phenylalanine at residue 117 (11). Strains considered virulent by these criteria are notifiable to the international community (11).
All NDV isolates are part of a single serotype; however, they display marked genetic diversity (12, 13). Based on genome length and phylogenetic relationships, NDV isolates are classified into two major groups, class I and class II (12–14). Class I viruses have worldwide distribution, are isolated mainly from waterfowl and shorebirds, and include nonvirulent strains, with the exception of a single isolate from the Republic of Ireland in 1990 (12–15). Class II viruses are both virulent and nonvirulent and are recovered from poultry, pet birds, and waterfowl (12–14). Based on the mean nucleotide distance, class I viruses were recently shown to be included into a single genotype (genotype I) (16), whereas class II viruses have been divided into 18 genotypes (I to XVIII) (16–18). Genotype V is most commonly isolated in North and Central America (12, 14, 19, 20) and was recently divided into two subgenotypes (Va and Vb) (16). Subgenotype Va includes isolates from the United States and Canada, almost exclusively sampled from cormorants (16, 21), and more rarely from other wild birds, such as gulls and pelicans (22). In Mexico and Central America, the most commonly isolated NDV strains belong to the recently assigned subgenotype Vb (20, 23). The last Mexican ND outbreak that occurred in poultry in 2011 was caused by a strain that is closely related to other NDV strains isolated from 2004 to 2006 in Mexico (24) (http://www.oie.int/wahis_2/public/wahid.php/Wahidhome/Home). Phylogenetic and epidemiological studies of Mexican NDV isolates have shown that they can be differentiated, within subgenotype Vb, into “recent” (2004 to 2006) and “old” (1998 to 2001) evolutionary groups, which diverged after an intensive vaccination campaign started in Mexico around 2001 (20). Although another report recently described NDV evolution in Mexico (19), there are few epidemiological and evolutionary data regarding NDV in neighboring regions, such as Central America. Monitoring NDV evolution and epidemiology in this region is important for the United States, as the geographical proximity increases the possibility of NDV importation through illegal trade or the migration of neotropical birds harboring the virus (25). Since 2008, there have been six ND outbreaks reported in Central America: one in Belize (2008), three in Honduras (one in 2009 and two in 2010), and two in Nicaragua (one in 2011 and one in 2012) (http://www.oie.int/wahis_2/public/wahid.php/Wahidhome/Home). However, no studies characterize the NDV strains responsible for these outbreaks in Central America. The aim of the present study is to characterize three NDV isolates (NDV-Belize-3/08, NDV-Belize-4/08, and NDV-Belize-12/08) by clinicopathological assessment and phylogenetic analysis, which caused an ND outbreak in commercial poultry in Belize in November 2008.
MATERIALS AND METHODS
Viruses.
The three viruses (APMV1/chicken/Belize [Spanish Lookout]/4224-03/2008 [NDV-Belize-3/08], APMV1/chicken/Belize/4224-12/2008 [NDV-Belize-12/08], and APMV1/turkey/Belize/4438-4/2008 [NDV-Belize-4/08]) were received from the National Veterinary Service Laboratories (NVSL) (Ames, IA). The viruses were isolated in specific-pathogen-free (SPF) chicken embryos at the NVSL from swab samples received through the U.S. Embassy (USDA-APHIS-U.S. Embassy), upon authorization by the Belize Agricultural Health Authority Animal Health Department (BAHA). The samples were collected in Belize during an outbreak of ND affecting domestic poultry and turkey flocks in 2008. NDV-Belize-3/08 was collected from the proventriculus of a 5-week-old male broiler and NDV-Belize-12/08 from the lung of a 14-week-old layer from a poultry farm in Spanish Lookout, Cayo District, on 15 November 2008. The submission form listed the flock size as 19,900, with 2,145 birds being affected or dead (10.78%). NDV-Belize-4/08 was collected from a turkey as a tracheal/oropharyngeal swab on 19 November 2008 from a turkey farm in the same district as the others. The turkey flock size was listed as 8,000, with 245 birds being affected or dead (3%). After initial isolation at the NVSL, the three NDV isolates were propagated and used for RNA extraction, pathotyping, and pathogenesis experiments by the Southeast Poultry Research Laboratory (SEPRL).
Eggs and chickens.
Embryonated chicken eggs and chickens for NVSL were obtained from the Charles River Avian Vaccine Service or from the SEPRL SPF white Leghorn flock. Birds used in the ICPI and pathogenesis experiments were housed in negative-pressure isolators under biosafety level 3 (BSL3) enhanced (E) containment at SEPRL. Water and food were provided ad libitum.
ICPI test.
The intracerebral pathogenicity index (ICPI) test was performed to assess the virulence of NDV-Belize-3/08, NDV-Belize-4/08, and NDV-Belize-12/08, as described previously (4, 11). Briefly, day-old SPF chickens were inoculated intracerebrally with 0.05 ml of a 1:10 dilution of infective allantoic fluid. The chickens were observed every day for 8 consecutive days, and each bird was scored as normal (score of 0), sick or paralyzed (score of 1), or dead (score of 2). The final ICPI score was tallied using the mean daily scores for each bird (4, 11).
Clinicopathological assessment in chickens.
Due to the high genetic similarity of the three strains, clinicopathological assessment was conducted with NDV-Belize-3/08 only. Twenty (n = 20) 4-week-old SPF white Leghorn chickens were randomly placed into one of 2 experimental groups of 10 birds each (n = 10). One group consisted of birds infected with NDV-Belize-3/08 and the other of mock-infected birds. The birds were inoculated with a virus suspension (0.1 ml) in phosphate-buffered saline (PBS) containing 105× 50% embryo infectious dose (EID50)/0.1 ml. Half of the inoculum (0.05 ml) was applied to the right conjunctival sac, and the other half (0.05 ml) to the choanal cleft. Mock-infected control birds were inoculated in the same manner with 0.1 ml of PBS. The birds were monitored daily for observation of clinical signs, and two birds from each group were euthanized and necropsied at 2, 5, 10, and 14 days postinfection (dpi). Before the collection of samples for histology, complete postmortem examinations were carried out to assess macroscopic lesions. Birds with a severe clinical condition were euthanized regardless of the sampling schedule. Organs (n = 25) were collected for histopathology and immunohistochemistry: eyelid, spleen, bursa of Fabricius, thymus, Harderian gland, proventriculus, small intestine (duodenum and Meckel's diverticulum), cecal tonsil, large intestine, air sacs, trachea, lung, heart, tongue, esophagus, pharynx, crop, brain, liver, pancreas, kidney, comb, head of the femur, and nasal turbinate. The tissue samples were fixed in 10% neutral buffered formalin for 52 h and subsequently processed for histological and immunohistochemical examinations.
Microscopic pathology and immunohistochemistry.
Evaluations of microscopic lesions and immunohistochemistry were performed as previously described (17, 26, 27).
RNA isolation and sequencing.
RNA was extracted from the allantoic fluids of infected eggs using TRIzol LS (Invitrogen, Carlsbad, CA). Reverse transcription-PCR (RT-PCR) amplification and sequencing of the entire F gene coding sequence for NDV-Belize-4/08 and NDV-Belize-12/08 were conducted as previously published (27). The sequence of the full genome of NDV-Belize-3/08 was determined by using a shotgun RT-PCR sequencing approach, as previously described (17, 27). Editing and assembly of the multiple sequences was carried out using the Lasergene sequence analysis software package (Lasergene, version 5.07; DNAStar, Inc., Madison, WI).
Sequence alignment.
The sequences were aligned with MEGA (version 6) software (28), using both MUSCLE alignment algorithm and manual editing.
Phylogenetic analysis.
The concatenated aligned sequences of all six structural protein-coding regions were used to construct a phylogenetic tree in order to infer the evolutionary history between NDV-Belize-3/08 and other representatives of class II NDV genotypes (total n = 150). In addition, the sequence of the entire coding region of the F gene (“full fusion”) was used to determine the phylogenetic relationships of NDV-Belize-3/08, NDV-Belize-4/08, and NDV-Belize-12/08 with other representatives of NDV genotype V (total n = 54). The sequences for both phylogenetic trees were retrieved from GenBank, and the numbers used to identify the sequences are the NCBI (National Center for Biotechnology Information) accession numbers or GI (Gene Identifier) numbers. Evolutionary histories were inferred using the maximum likelihood method based on the general time reversible model (29) using the MEGA software (version 6) (28). The initial tree was inferred by the maximum parsimony method, and a discrete gamma distribution was used to model evolutionary rate differences among the sites (5 categories). The analysis allowed for some sites to be invariable (G+I). The codon positions included in the analysis were the 1st, 2nd, 3rd, and noncoding. The positions in the final alignment that contained gaps were eliminated from the data set (“complete deletion” option).
To test the presence of subgenotypes within genotype V, the evolutionary distances were computed between putative subgenotypes Va, Vb, and Vc, and the clade that included the 2008 Belize plus the 2007 Honduras strains (putative clade Vb.1). The distances were shown as the average (per each group pair) number of base substitutions per site and inferred using the maximum likelihood method (30),with a gamma distribution (shape parameter, 1) of rate variation among the sites, as implemented in MEGA6 (28). The criteria for the assignment of subgenotypes and genotypes were based on the recently proposed nomenclature by Diel et al. (16).
Nucleotide sequence accession numbers.
The full open reading frame of the F gene for NDV-Belize-12/08 was successfully sequenced and assigned the GenBank accession no. KF767467. The full F sequences for NDV-Belize-3/08 and NDV-Belize-4/08 were submitted to GenBank by the NVSL with the accession no. JN872163 and JN942045, respectively. The complete genome of NDV-Belize-3/08 was successfully sequenced and assigned the GenBank accession no. KF767466. Additionally, the complete coding sequences of the F gene from three additional genotype V strains were included in the phylogenetic analysis. These samples were received from the NVSL and sequenced at SEPRL: (i) APMV-1/745 FL-5682-1985, isolated from a green-cheeked parakeet in Florida in 1985 (submitted to GenBank with accession no. KF767468); (ii) APMV-1/747 Mexico-17741-1976, isolated from Mexico in 1976 from a yellow-cheeked Amazon (submitted to GenBank with accession no. KF767469); and (iii) APMV-1/750 Argentina-12567-1976, isolated from Argentina in 1976 from a blue-fronted parrot (submitted to GenBank with accession no. KF767470).
RESULTS
Biological characterization.
The virulence of the three strains was assessed by the deduced amino acid sequence of the F protein cleavage site and by the ICPI test. The deduced amino acid sequence of the F protein cleavage site for all isolates was identical. All three isolates had three basic amino acid residues at positions 113, 115, and 116, plus a phenylalanine at position 117 (112R-R-Q-K-R-F117). The ICPI scores in day-old SPF chickens were 1.75, 1.66, and 1.69 for NDV-Belize-3/08, NDV-Belize-4/08, and NDV-Belize-12/08, respectively. Both the amino acid configuration of the F cleavage site and the ICPI scores are typical of virulent NDV strains (7, 11).
Genomic and phylogenetic characterization of NDV-Belize/03.
The complete genome arrangement of NDV-Belize-3/08 is summarized in Table 1. NDV-Belize-3/08 has an RNA genome of 15,192 nucleotides. The G+C content is 46%. Compared to representatives of earlier genotypes (I to IV), there is a 6-nucleotide (nt) insertion at the 5′ noncoding region of the NP gene from nucleotide 1738 to nucleotide 1743.
TABLE 1.
Genomic features of Newcastle disease isolate NDV-Belize-3/08
| Gene | Gene start positions/lengtha | Length of 3′-UTRb | Coding sequence positions/length | Length of 5′-UTR | Gene end positions/length | Length of intergenic regions | Length of whole gene | Protein size (aa)c |
|---|---|---|---|---|---|---|---|---|
| NP | 56–65/10 | 56 | 122–1591/1,470 | 206 | 1798–1808/11 | 1 | 1,753 | 489 |
| P | 1810–1819/10 | 73 | 1893–3080/1,188 | 169 | 3250–3260/11 | 1 | 1,451 | 395 |
| M | 3262–3271/10 | 24 | 3296–4390/1,095 | 102 | 4493–4502/10 | 1 | 1,241 | 364 |
| F | 4504–4513/10 | 36 | 4550–6211/1,662 | 73 | 6285–6295/11 | 31 | 1,792 | 553 |
| HN | 6327–6336/10 | 81 | 6418–8133/1,716 | 185 | 8319–8328/10 | 47 | 2,002 | 571 |
| L | 8376–8385/10 | 1 | 8387–14001/6,615 | 67 | 15069–15078/10 | NA | 6,703 | 2,204 |
| Leader | 1–55/55 | NAd | NA | NA | NA | NA | 55 | NA |
| Trailer | 15079–15192/114 | NA | NA | NA | NA | NA | 114 | NA |
| Total length | 15,192 |
All lengths and genomic positions are expressed in nucleotides.
UTR, untranslated region.
aa, amino acids.
NA, not applicable.
In order to localize NDV-Belize-3/08 among other NDV isolates, a phylogenetic tree was inferred using the concatenated complete coding sequences for all six genes of NDV-Belize-3/08 and the representatives of other NDV genotypes (total n = 150; class I, n = 12; genotype I, n = 22; II, n = 22; III n = 4; IV, n = 4; V, n = 6; VI, n = 24; VII, n = 39; VIII, n = 3; IX, n = 6; XI, n = 2; XII, n = 1; XIII, n = 2; XVI, n = 1; unassigned, n = 1). The inferred tree showed that NDV-Belize-3/08 belongs to genotype V, with the closest related taxa being Gamefowl/USA(CA)/2002 (GI 45511218) (see Fig. S1 in the supplemental material).
NDV genotype V can be divided into three subgenotypes, with NDV-Belize-3/08, NDV-Belize-4/08, and NDV-Belize-12/08 included in a new clade of subgenotype Vb.
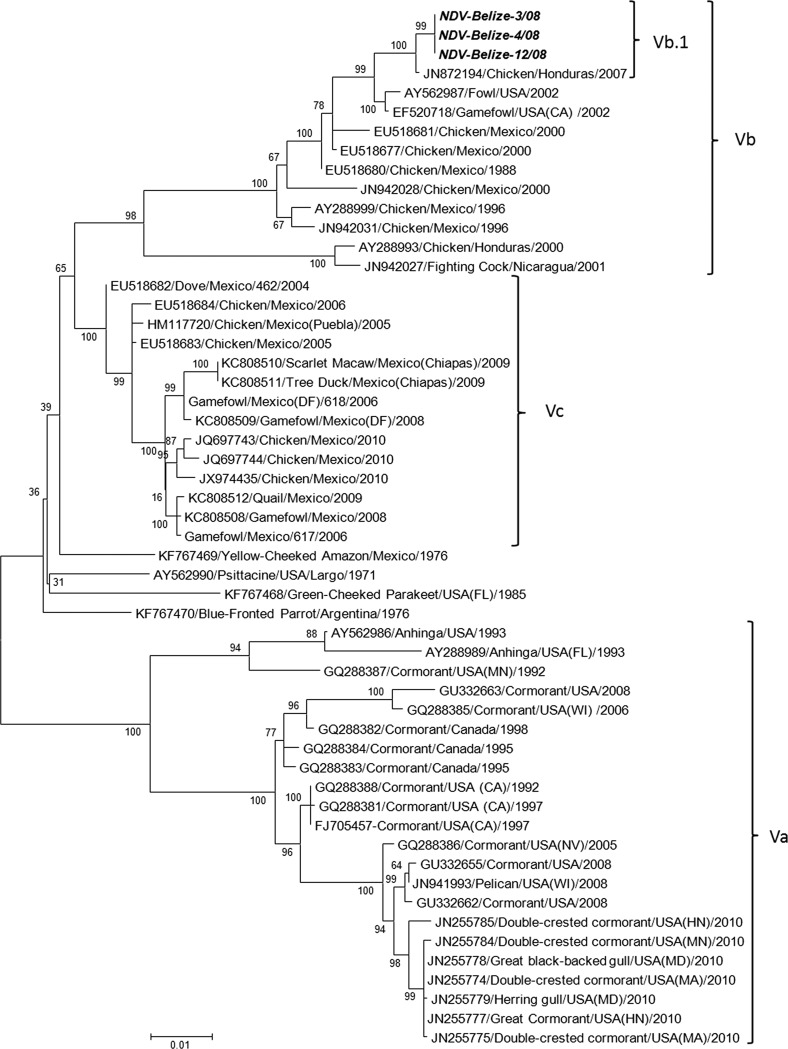
Phylogenetic analysis based on the complete F gene coding sequence of NDV genotype V representatives showed that the previously assigned subgenotype Vb is composed of two main outgroups (Fig. 1). One includes late (2004 to 2010) isolates, while the other is composed of earlier isolates (1988 to 2002), with the exception of the more recent taxa Belize (2008) and Honduras (2007). The defining node for these two groups has a bootstrap value of 65%, and their mean interpopulational evolutionary distance is 6.6% (Table 2), fulfilling the requirements for subgenotype assignment (16), here designated subgenotype Vb and subgenotype Vc (Fig. 1). Subgenotype Va is defined by a very high bootstrap value at the defining node (100%) and by an interpopulational evolutionary distance with subgenotypes Vb and Vc of 10.9% and 8.7%, respectively. The intrapopulational evolutionary distance within the whole genotype V (Va, Vb, and Vc) is 6.87%. Taken together, these data suggest that NDV genotype V can be divided evolutionarily into three subgenotypes: Va, Vb, and Vc (16). Subgenotype Va includes mostly North American isolates from cormorant species, with the exception of rare isolates from anhinga, pelican, and gull species. Subgenotype Vb includes mostly poultry isolates from Mexico and Central America that span from 1988 to 2008, as well as the isolates from the 2002 California NDV outbreak. Subgenotype Vc includes only Mexican isolates from different species isolated from 2004 to 2010. In addition, the phylogenetic analysis shows that subgenotypes Va, Vb, and Vc originated from evolutionarily close strains isolated during the 1970 and 1980s. The division of NDV genotype V into subgenotypes Va, Vb, and Vc is confirmed also by Bayesian inference (data not shown).
FIG 1.
Molecular phylogenetic analysis by maximum likelihood method using the F gene complete coding sequence of NDV-Belize-3/08, NDV-Belize-4/08, and NDV-Belize-12/08, among other genotype V NDV isolates. The evolutionary history was inferred by using the maximum likelihood method based on the general time reversible model (29). The tree with the highest log likelihood (−6389.9289) is shown. The percentage of trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches (number of replicates, 500). The accession numbers are shown to the left of the strain names. The initial tree(s) for the heuristic search were obtained automatically by applying the maximum parsimony method. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories; [+G] parameter, 0.7850). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 18.8989% of sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 54 nucleotide sequences. The codon positions included are 1st + 2nd + 3rd+ noncoding. All positions containing gaps and missing data were eliminated. There were a total of 1,652 positions in the final data set. Evolutionary analyses were conducted in MEGA6 (28).
TABLE 2.
Estimates of evolutionary distances between subgenotypes in genotype Va
| Subgenotype or clade | Evolutionary distance for indicated subgenotype or clade |
|||
|---|---|---|---|---|
| Vb.1 | Va | Vb | Vc | |
| Va | (0.009) | (0.008) | ||
| Vb (including Vb.1) | 0.109 | (0.006) | ||
| Vc | 0.087 | 0.066 | ||
| Vb.1 | (0.011) | (0.004) | (0.008) | |
| Va | 0.116 | (0.009) | (0.007) | |
| Vb (not including Vb.1) | 0.033 | 0.106 | (0.006) | |
| Vc | 0.074 | 0.087 | 0.062 | |
The number of base substitutions per site from averaging over all sequence pairs between groups within genotype V are shown. The upper three rows indicate the evolutionary distances found when clade Vb.1 is included within subgenotype Vb. The lower four rows indicate the evolutionary distances found when clade Vb.1 is considered by itself. The total number of sequences was 50. The numbers of sequences analyzed per group were as follows: clade Vb.1, n = 4; subgenotype Va, n = 22; subgenotype Vb, n = 10 (or n = 14 when including clade Vb.1); subgenotype Vc, n = 14. The values in parentheses are standard errors, calculated by bootstrap procedure (500 replicates). Analyses were conducted using the maximum composite likelihood model (30), as implemented in MEGA6 (28). The rate variation among the sites was modeled with a gamma distribution (shape parameter, 1). Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 1,652 positions in the final data set.
The full fusion sequences from NDV-Belize-3/08, NDV-Belize-4/08, and NDV-Belize-12/08 have 100% homology and grouped very closely with a 2007 isolate from Honduras (JN872194), forming a distinct clade branching from subgenotype Vb, with a 100% bootstrap value at the defining node (putative clade Vb.1; Fig. 1). The mean evolutionary distances between this clade and subgenotypes Va, Vb and Vc are 11.6%, 3.3%, and 7.4%, respectively (Table 2).
Clinicopathological characteristics of NDV-Belize-3/08.
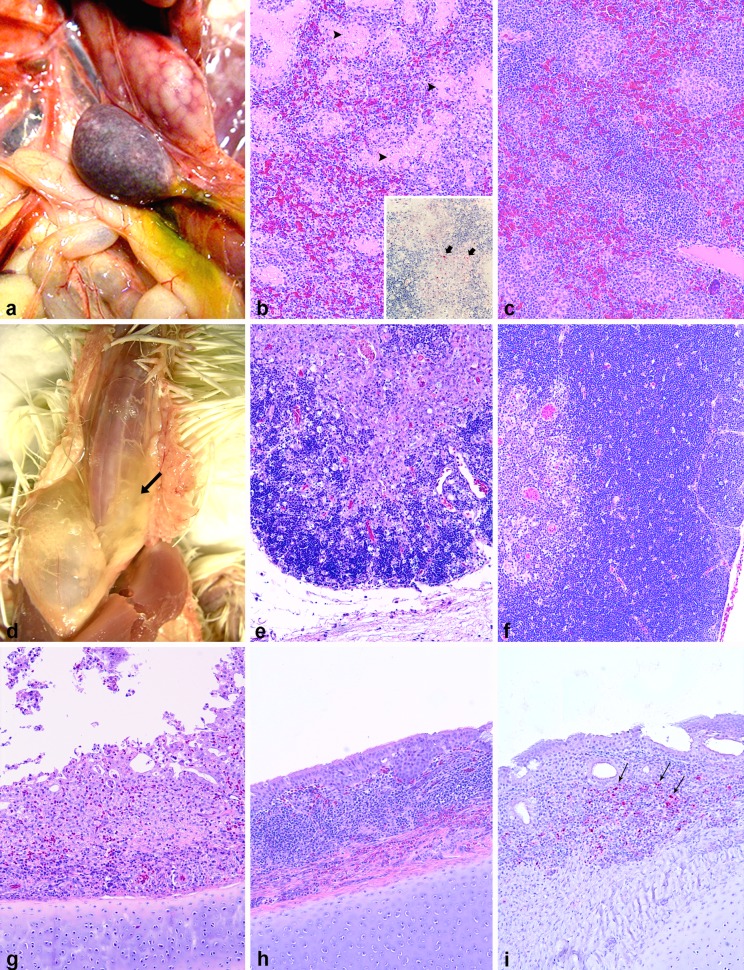
Due to the close genetic relationship between the three Belizean NDV isolates, only NDV-Belize-3/08 was used to conduct the clinicopathological experiment. All chickens inoculated with NDV-Belize-3/08 were successfully infected and exhibited depression, ruffled plumage, cyanotic comb, and mucous diarrhea, which peaked in severity at 6 dpi, when all animals where dead or euthanized in extremis. Gross lesions consisted of severe bilateral conjunctivitis (first observed by 2 dpi), multifocal necrosis in the spleen (day 3) (Fig. 2a), multifocal necrosis in the cecal tonsil, proventricular hemorrhages, scaling and erythema of the comb (day 4), multifocal areas of necrosis in the small and large intestine (day 5), and severe atrophy of the thymus characterized by a marked reduction in size associated with perithymic hemorrhages and edema (day 6) (Fig. 2d).
FIG 2.
Pathological changes in 4-week-old chickens inoculated with NDV-Belize-3/08. (a) Gross lesions in the spleen at 3 dpi. There are multifocal white specks (necrosis) that confer the organ a mottled appearance. (b) Microscopically, the spleen shows multifocal areas of necrosis and fibrin exudation (arrowheads; hematoxylin and eosin [HE], magnification ×10). Inset, positive immunolabeling for NDV nucleoprotein in the splenic ellipsoids (arrows; immunohistochemistry [IHC], magnification ×20). (c) Spleen from a control bird inoculated with PBS. There is the typical perivascular accumulation of lymphocytes eccentric to the splenic ellipsoids. (HE, magnification ×10). (d) Gross lesions of the thymus at 4 dpi. There is diffuse severe serous atrophy of the thymus (arrow). (e) Microscopically, the thymic cortex is diffusely and markedly thinned, with loss of lymphocytes and typical “starry sky” appearance (HE, magnification ×10). (f) Thymus from a control bird inoculated with PBS; note the thick cortex (HE, magnification ×10). (g) Laryngeal tonsil of an infected bird at 3 dpi. There is lymphocytic depletion, multifocal coalescing necrosis, and mucosal infiltration with macrophages and heterophils. The overlaying epithelium is eroded, and rafts of sloughed material are free in the lumen (HE, magnification ×10). (h) Normal laryngeal tonsil showing organized lymphoid tissue (arranged in follicles) and a well-organized pseudostratified ciliated epithelium (HE, magnification ×10). (i) There is multifocal immunolabeling for NDV nucleoprotein (arrows) in the laryngeal tonsil, associated with areas of necrosis and lymphoid depletion (IHC, magnification ×10).
The severities of the microscopic lesions in selected organs are presented in Table 3. The lesions peaked in severity at 6 dpi. Overall, the virus principally and bilaterally targeted the eyelids, the lymphoid organs (spleen [Fig. 2b], thymus [Fig. 2e], and bursa), and the mucosa-associated lymphoid tissues (MALT) (such as cecal tonsil). The lesions consisted of severe lymphoid depletion and necrosis, resulting in the accumulation of prominent macrophages, necrotic debris, and scattered heterophils. In the intestines, destruction of the MALT resulted in necrosis and ulceration of the overlying epithelium. The respiratory tract had severe lesions in the laryngeal tonsils (Fig. 2g), and a certain degree of lymphocyte depletion and necrosis was observed within the bronchus-associated lymphoid tissue (BALT) of the secondary and tertiary bronchi, occasionally resulting in a loss of overlaying respiratory epithelium. The myocardium showed areas of multifocal necrosis associated with the accumulation of macrophages.
TABLE 3.
Severity of microscopic lesions and intensity of immunolabeling for NDV NP in selected organs
| Organ | Staining method | Scoring value by dpi: |
||||
|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | ||
| Eyelid | HEa | +/− | +/− | ++ | +++ | +++ |
| IHCb | − | ++++ | ++ | ++ | + | |
| Spleen | HE | + | ++ | ++++ | ++++ | ++++ |
| IHC | + | +++ | +++ | + | − | |
| Thymus | HE | +/− | + | ++ | +++ | +++ |
| IHC | +/− | ++ | +++ | + | ++ | |
| Bursa | HE | − | + | +++ | +++ | +++ |
| IHC | − | + | ++ | − | + | |
| Harderian gland | HE | − | − | − | + | +++ |
| IHC | − | − | + | − | − | |
| Proventriculus | HE | + | + | ++ | NAc | ++ |
| IHC | − | − | ++++ | − | − | |
| Pancreas | HE | − | − | − | + | − |
| IHC | − | − | − | − | − | |
| Meckel's diverticulum | HE | ++ | − | ++ | ++ | ++ |
| IHC | − | + | ++ | + | − | |
| Cecal tonsil | HE | + | + | +++ | ++ | +++ |
| IHC | ++ | +++ | +++ | + | + | |
| Large intestine | HE | − | − | + | − | + |
| IHC | − | ++ | ++ | − | − | |
| Air sac | HE | − | − | − | − | − |
| IHC | − | − | +/− | − | − | |
| Trachea | HE | − | − | − | − | − |
| IHC | − | − | ++ | − | − | |
| Lung | HE | +BALTd | − | − | − | − |
| IHC | − | +BALT | +BALT | − | − | |
| Heart | HE | − | − | − | − | − |
| IHC | − | − | − | − | − | |
| Pharynx | HE | − | − | ++ | ++ | − |
| IHC | − | ++ | ++ | ++ | − | |
| Brain | HE | − | − | − | +/− | + |
| IHC | − | − | − | − | − | |
| Kidney | HE | − | − | − | − | − |
| IHC | − | + | + | − | + | |
| Comb | HE | − | − | − | − | − |
| IHC | − | − | ++ | − | − | |
| Femur | HE | − | − | + | − | − |
| IHC | − | − | ++ | − | − | |
| Turbinates | HE | − | − | − | − | − |
| IHC | − | − | − | − | − | |
For HE staining: spleen: +, moderate hyperplasia of lymphocytes; ++, mild lymphocytic depletion; +++, moderate (<50%) lymphocyte depletion, histiocytic accumulation, and multifocal necrosis; ++++, severe (>50%) lymphocytic depletion, histiocytosis, and necrosis. Thymus, cecal tonsil, mucosa-associated lymphoid tissue [MALT], and bursa: +, mild lymphocytic depletion, ++, moderate (<50%) lymphocytic depletion with necrosis and histiocytosis; +++, (>50%) severe lymphocytic depletion, necrosis, and histiocytosis. Bone marrow: +, mild (<20%) bone marrow necrosis; ++, mild (20–50%) bone marrow necrosis; +++, severe (>50%) bone marrow necrosis. Brain: +, vascular reactivity; ++, vascular reactivity and perivascular cuffing; +++, vascular reactivity, perivascular cuffing, and gliosis. Other organs: +, mild/focal necrosis, inflammation (or gliosis in brain), and/or acute hemorrhage; ++, moderate necrosis or inflammation; +++, severe necrosis, inflammation, or hemorrhage affecting >50% of examined sections; +/−, minimal lesions in the observed sections.
For IHC immunolabeling: −, no IHC signal present; +, rare cells in the section are positive on IHC; ++, positive cells seen, <50% of all high power fields (HPF); +++, positive signal seen in 50 to 75% of HPF; ++++, abundant positive signal in >75% of HPF; +/−, positivity for only a few scattered cells in the section.
NA, not applicable, since not enough tissue was present for HE assessment.
BALT, bronchus-associated lymphoid tissues.
The intensity and distribution of NP immunohistochemical staining of selected organs are summarized in Table 3. Immunolabeling for NDV NP revealed that NDV-Belize-3/08 had widespread distribution, with 18 positive tissues out of 25 (data not shown). Immunostaining was intracytoplasmic and finely to coarsely granular, with occasional extracellular signal (fine granular staining) observed in the necrotic areas. The highest immunohistochemical signal, both for intensity and extension in affected organs, was at 4 dpi, and it markedly decreased at 5 and 6 dpi. The organs with the strongest signal were the eyelids, the lymphoid organs, and the MALT in multiple organs. In these tissues, the positive cells consisted mainly of lymphocytes and macrophages. In the spleen, immunoreactivity was confined to the fixed-macrophage-dependent areas around the penicillary arteries (Fig. 2b, inset), while the lymphocyte-dependent areas were devoid of signal. In the respiratory system, the positive signal was confined to the lymphoid areas of the larynx (Fig. 2i) and in scattered lymphoid aggregates closely associated with the secondary and tertiary bronchi (BALT). In the digestive tract, intense positivity for NDV was observed only within the submucosal lymphoid aggregates. Notably, NDV-Belize-3/08 did not show tropism for the pancreas.
DISCUSSION
The United States is ND free; however, the introduction of virulent NDV strains is a serious risk for the U.S. poultry economy and might occur through spillover from migratory birds or illegal importations of infected animals from neighboring countries. The last ND outbreak in the United States occurred in California during 2002 and 2003, which later spread to Nevada, Arizona, and Texas, causing the depopulation of >3 million birds and a total estimated cost of $180 to $360 million (31). The NDV strain that caused the outbreak in California had a close resemblance to Mexican and Central American isolates (23). Here, the recent 2008 outbreak of ND in Belize, a Central American country that shares a border with Mexico, represents additional evidence of the continued evolution of viruses of genotype V south of the U.S. border. The presence of a recent evolutionary group (Vb.1) emerging from the older isolates of subgenotype Vb suggests that Newcastle disease viruses belonging to this clade might have been evolving independently from the contemporary NDV strains present in the region (such as Mexico), most likely in unknown reservoirs. Since ND outbreaks were not reported in Belize from 2002 until 2008, it is possible that virulent NDV strains might have been circulating in clinically nonsusceptible birds that maintained the virus until it finally reached backyard poultry and caused disease. The presence of NDV strains closely related to the old isolates causing outbreaks in recent years has been described also in the Dominican Republic (17). The outbreak strain of 2008 from the Dominican Republic was evolutionarily close to other strains isolated from that country since 1986, suggesting that in this case, an old virus might have circulated unnoticed in an unknown reservoir before spilling again into poultry (17). Because of the existence of psittacines and other bird species that often do not shown signs of disease, it is possible that a wild bird reservoir might have played a role in maintaining the NDV strain that caused the poultry outbreak in Belize. However, the possibility of the viruses being maintained in poorly vaccinated poultry cannot be discarded.
A clinicopathological assessment of NDV-Belize-3/08 showed that this virus is a typical VVNDV, causing severe necrohemorrhagic lesions in the intestine and lymphoid organs and death of the infected birds by 6 dpi (1, 4, 7). Many VVND strains evaluated in our laboratory (in a manner similar to what is described here) caused the death/demise of infected birds by 4 dpi (7, 17, 26, 27), suggesting that NDV-Belize-3/08, by doing so by 6 dpi, might have moderately decreased pathogenicity. This is confirmed by the ICPI score of 1.75, which is lower than what is usually observed for the most virulent velogenic strains (i.e., they are commonly >1.8 [17, 26]).
NDV strains within genotype V have been divided into subgenotypes Va and Vb, with subgenotype Va including the strains isolated from cormorant species in North America (16, 22), and Vb strains isolated mainly from poultry in Mexico and Central America (12, 16, 20). The phylogenetic analysis in the present work shows that the original subgenotype Vb is composed of two main clades that fulfill the criteria for subgenotype assignment (16), and these are designated here as Vb and Vc. Therefore, NDV genotype V can be divided into three subgenotypes (Va, Vb, and Vc), and to our knowledge, this is the first description of such a classification. Subgenotype Va remains unchanged from its previous designation (16). Subgenotype Vc is composed of strains isolated in Mexico from 2004 to 2010. Subgenotype Vb is composed mostly of NDV isolates from 1988 to 2002, with the exception of the more recent isolates from Belize (2008) and Honduras (2007). This more recent evolutionary clade (tentatively called Vb.1) is supported by a very high bootstrap value in tree topology and has an average nucleotide distance of 3.3% with the rest of the subgenotype Vb isolates. These data suggest that clade Vb.1 might belong to an additional subgenotype within genotype V. According to Diel and colleagues, a new subgenotype is assigned if it includes more than four independent isolates, it shows a high bootstrap value at the defining node (>60%), and it has an average nucleotide distance between groups between 3% and 10% (16). In this specific case, there are not enough independent isolates to assign a new subgenotype, since the three Belizean isolates derive from the same outbreak and cannot be considered independent. Eventually, the addition of further isolates might lead to the assignment of a new subgenotype.
In summary, these observations highlight the importance of understanding the epidemiology of NDV and how its ability to infect multiple species, together with its high mutation rate, can affect control measures and biosecurity. The inability to identify the origin of virulent NDV in countries with recent outbreaks (the Dominican Republic, Belize, and Peru [http://www.oie.int/wahis_2/public/wahid.php/Wahidhome/Home]) hampers the application of proper control measures to prevent the infection of commercial and backyard poultry, not only in those countries, but potentially to other countries that are free of ND. Additionally, this is the first description of three subgenotypes (Va, Vb, and Vc) within NDV genotype V.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dawn Williams-Coplin and Tim Olivier for technical assistance and the SEPRL sequencing facility personnel for nucleotide sequencing.
This work was supported by USDA funding (CRIS 6612-32000-064-00D).
Footnotes
Published ahead of print 12 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00066-14.
REFERENCES
- 1.Alexander DJ, Senne DA. 2013. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections, p 89–138 In Swayne ED, Glisson JR, McDougald LR, Nolan LK, Suarez LD, Nair V. Diseases of poultry, 13th ed. Blackwell Publishing, Ames, IA [Google Scholar]
- 2.Guèye E. 1999. Poultry plays an important role in African village life. World Poultry 14:14–17 [Google Scholar]
- 3.Kitalyi AJ. 1998. Village chicken production system in rural Africa: household and food security and gender issues. FAO Animal Production and Health paper 142. Food and Agriculture Organization of the United Nations, Rome, Italy [Google Scholar]
- 4.Alexander DJ, Senne DA. 2008. Newcastle disease and other avian paramyxoviruses, p 135–141 In Dufour-Zavala L, Glisson JR, Jackwood MW, Pearson JE, Reed WM, Woolcock PR. (ed), A laboratory manual for the isolation, identification and characterization of avian pathogens, 4th ed. American Association of Avian Pathologists, Athens, GA [Google Scholar]
- 5.Lamb R, Collins PL, Kolakofsky D, Melero JA, Nagai Y, Oldstone MBA, Pringle CR, Rima BK. 2005. The negative sense single stranded RNA viruses, p 607–738 In Fauquet CM, Mayo MA, Maniloff J, Desselberg U, Ball LA. (ed), Virus taxonomy. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 6.Steward M, Vipond IB, Millar NS, Emmerson PT. 1993. RNA editing in Newcastle disease virus. J. Gen. Virol. 74:2539–2547. 10.1099/0022-1317-74-12-2539 [DOI] [PubMed] [Google Scholar]
- 7.Cattoli G, Susta L, Terregino C, Brown CC. 2012. Newcastle disease: a review of field recognition and current methods of laboratory detection. J. Vet. Diagn. Invest. 23:637–656. 10.1177/1040638711407887 [DOI] [PubMed] [Google Scholar]
- 8.de Leeuw OS, Hartog L, Koch G, Peeters BP. 2003. Effect of fusion protein cleavage site mutations on virulence of Newcastle disease virus: non-virulent cleavage site mutants revert to virulence after one passage in chicken brain. J. Gen. Virol. 84:475–484. 10.1099/vir.0.18714-0 [DOI] [PubMed] [Google Scholar]
- 9.Nagai Y, Klenk HD, Rott R. 1976. Proteolytic cleavage of the viral glycoproteins and its significance for the virulence of Newcastle disease virus. Virology 72:494–508. 10.1016/0042-6822(76)90178-1 [DOI] [PubMed] [Google Scholar]
- 10.Collins MS, Bashiruddin JB, Alexander DJ. 1993. Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity. Arch. Virol. 128:363–370 [DOI] [PubMed] [Google Scholar]
- 11.World Organization for Animal Health (OIE). 2012. Newcastle disease, p 1–19 In Manual of diagnostic tests and vaccines for terrestrial animals, 7th ed, vol 1 World Organization for Animal Health, Paris, France [Google Scholar]
- 12.Miller PJ, Decanini EL, Afonso CL. 2010. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect. Genet. Evol. 10:26–35. 10.1016/j.meegid.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 13.Miller PJ, Kim LM, Ip HS, Afonso CL. 2009. Evolutionary dynamics of Newcastle disease virus. Virology 391:64–72. 10.1016/j.virol.2009.05.033 [DOI] [PubMed] [Google Scholar]
- 14.Afonso CL, Miller PJ. 2013. Newcastle disease: progress and gaps in the development of vaccines and diagnostic Tools, p 95–106 In Roth J, Ritch JA, Morozov IA. (ed), Vaccines and diagnostics for transboundary animal diseases. Karger, Basel, Switzerland: [DOI] [PubMed] [Google Scholar]
- 15.Alexander DJ, Campbell G, Manvell RJ, Collins MS, Parsons G, McNulty MS. 1992. Characterisation of an antigenically unusual virus responsible for two outbreaks of Newcastle disease in the Republic of Ireland in 1990. Vet. Rec. 130:65–68. 10.1136/vr.130.4.65 [DOI] [PubMed] [Google Scholar]
- 16.Diel DG, da Silva LH, Liu H, Wang Z, Miller PJ, Afonso CL. 2012. Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect. Genet. Evol. 12:1770–1779. 10.1016/j.meegid.2012.07.012 [DOI] [PubMed] [Google Scholar]
- 17.Courtney SC, Susta L, Gomez D, Hines NL, Pedersen JC, Brown CC, Miller PJ, Afonso CL. 2013. Highly divergent virulent isolates of Newcastle disease virus from the Dominican Republic are members of a new genotype that may have evolved unnoticed for over 2 decades. J. Clin. Microbiol. 51:508–517. 10.1128/JCM.02393-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snoeck CJ, Owoade AA, Couacy-Hymann E, Alkali BR, Okwen MP, Adeyanju AT, Komoyo GF, Nakouné E, Le Faou A, Muller CP. 2013. High genetic diversity of Newcastle disease virus in poultry in West and Central Africa: cocirculation of genotype XIV and newly defined genotypes XVII and XVIII. J. Clin. Microbiol. 51:2250–2260. 10.1128/JCM.00684-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardenas Garcia S, Navarro Lopez R, Morales R, Olvera MA, Marquez MA, Merino R, Miller PJ, Afonso CL. 2013. Molecular epidemiology of Newcastle disease in Mexico and the potential spillover of viruses from poultry into wild bird species. Appl. Environ. Microbiol. 79:4985–4992. 10.1128/AEM.00993-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perozo F, Merino R, Afonso CL, Villegas P, Calderon N. 2008. Biological and phylogenetic characterization of virulent Newcastle disease virus circulating in Mexico. Avian Dis. 52:472–479. 10.1637/8276-022908-Reg.1 [DOI] [PubMed] [Google Scholar]
- 21.Rue CA, Susta L, Brown CC, Pasick JM, Swafford SR, Wolf PC, Killian ML, Pedersen JC, Miller PJ, Afonso CL. 2010. Evolutionary changes affecting rapid identification of 2008 Newcastle disease viruses isolated from double-crested cormorants. J. Clin. Microbiol. 48:2440–2448. 10.1128/JCM.02213-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diel DG, Miller PJ, Wolf PC, Mickley RM, Musante AR, Emanueli DC, Shively KJ, Pedersen K, Afonso CL. 2012. Characterization of Newcastle disease viruses isolated from cormorant and gull species in the United States in 2010. Avian Dis. 56:128–133. 10.1637/9886-081111-Reg.1 [DOI] [PubMed] [Google Scholar]
- 23.Pedersen JC, Senne DA, Woolcock PR, Kinde H, King DJ, Wise MG, Panigrahy B, Seal BS. 2004. 2004. Phylogenetic relationships among virulent Newcastle disease virus isolates from the 2002–2003 outbreak in California and other recent outbreaks in North America. J. Clin. Microbiol. 42:2329–2334. 10.1128/JCM.42.5.2329-2334.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao S, Paldurai A, Nayak B, Mirande A, Collins PL, Samal SK. 2013. Complete genome sequence of a highly virulent Newcastle disease virus currently circulating in Mexico. Genome Announc. 1(1):e00177–12. 10.1128/genomeA.00177-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers TD, Jr, Hicks DL, Wishusen EW, Parrish JR. 1982. Repeats, returns, and estimated flight ranges of some North American migrants in Guatemala. J. Field Ornithol. 53:133–138 [Google Scholar]
- 26.Susta L, Miller PJ, Afonso CL, Brown CC. 2011. Clinicopathological characterization in poultry of three strains of Newcastle disease virus isolated from recent outbreaks. Vet. Pathol. 48:349–360. 10.1177/0300985810375806 [DOI] [PubMed] [Google Scholar]
- 27.Diel DG, Susta L, Cardenas Garcia S, Killian ML, Brown CC, Miller PJ, Afonso CL. 2012. Complete genome and clinicopathological characterization of a virulent Newcastle disease virus isolate from South America. J. Clin. Microbiol. 50:378–387. 10.1128/JCM.06018-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY [Google Scholar]
- 30.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise MG, Suarez DL, Seal BS, Pedersen JC, Senne DA, King DJ, Kapczynski DR, Spackman E. 2004. Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 42:329–338. 10.1128/JCM.42.1.329-338.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.