Abstract
Different antimicrobial susceptibility testing methods to detect low-level vancomycin resistance in enterococci were evaluated in a Scandinavian multicenter study (n = 28). A phenotypically and genotypically well-characterized diverse collection of Enterococcus faecalis (n = 12) and Enterococcus faecium (n = 18) strains with and without nonsusceptibility to vancomycin was examined blindly in Danish (n = 5), Norwegian (n = 13), and Swedish (n = 10) laboratories using the EUCAST disk diffusion method (n = 28) and the CLSI agar screen (n = 18) or the Vitek 2 system (bioMérieux) (n = 5). The EUCAST disk diffusion method (very major error [VME] rate, 7.0%; sensitivity, 0.93; major error [ME] rate, 2.4%; specificity, 0.98) and CLSI agar screen (VME rate, 6.6%; sensitivity, 0.93; ME rate, 5.6%; specificity, 0.94) performed significantly better (P = 0.02) than the Vitek 2 system (VME rate, 13%; sensitivity, 0.87; ME rate, 0%; specificity, 1). The performance of the EUCAST disk diffusion method was challenged by differences in vancomycin inhibition zone sizes as well as the experience of the personnel in interpreting fuzzy zone edges as an indication of vancomycin resistance. Laboratories using Oxoid agar (P < 0.0001) or Merck Mueller-Hinton (MH) agar (P = 0.027) for the disk diffusion assay performed significantly better than did laboratories using BBL MH II medium. Laboratories using Difco brain heart infusion (BHI) agar for the CLSI agar screen performed significantly better (P = 0.017) than did those using Oxoid BHI agar. In conclusion, both the EUCAST disk diffusion and CLSI agar screening methods performed acceptably (sensitivity, 0.93; specificity, 0.94 to 0.98) in the detection of VanB-type vancomycin-resistant enterococci with low-level resistance. Importantly, use of the CLSI agar screen requires careful monitoring of the vancomycin concentration in the plates. Moreover, disk diffusion methodology requires that personnel be trained in interpreting zone edges.
INTRODUCTION
Enterococci are now recognized as an important cause of hospital-acquired infections worldwide (1, 2). Notably, recent European surveys have documented pronounced yearly increases in bloodstream infections caused by multidrug-resistant (MDR) Enterococcus faecium, represented by high-risk clones (3). The relative increase in infections caused by MDR enterococci is related to several characteristics, including their intrinsic ability to withstand exposure to broad-spectrum antibiotics and environmental extremes, as well as the capacity to acquire new genetic determinants promoting gastrointestinal colonization and survival in the hospital environment (4, 5). Moreover, their remarkable ability to acquire new antimicrobial resistance determinants poses substantial therapeutic problems, and physicians are forced to use last-resort therapeutic options (4, 6). Therefore, it is important that clinical laboratories have the ability to deliver rapid accurate antimicrobial susceptibility data for enterococci, to support appropriate therapeutic and infection-control measures.
Currently, the vancomycin resistance (van) clusters in enterococci include eight acquired gene clusters, i.e., vanA, vanB, vanD, vanE, vanG, vanL (7), vanM (8), and vanN (9). The vanA genotype is the most prevalent genotype in vancomycin-resistant enterococci (VRE) worldwide, but infections with VanB-type VRE (mainly E. faecium) have shown dramatic increases in several European countries and are predominant in Australia (10–16). The vanB ligase gene has been divided into three subtypes, vanB1 to vanB3, based on phylogenetic diversity (17–19).
The VanB-type VRE have inducible resistance and express various levels of resistance to vancomycin (MICs, 4 to 1,024 mg/liter) and susceptibility to teicoplanin (MICs, ≤2 mg/liter) in vitro (7, 20). The wide range of vancomycin MICs in VanB-type enterococci is well known and has been observed within the same clone during outbreaks (12, 21) (A. Sivertsen, H. Billström, Ö. Melefors, B. Olsson Liljequist, K. Tegmark Wisell, M. Ullberg, V. Özenci, A. Sundsfjord, and K. Hegstad, submitted for publication). The MIC clinical breakpoints defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) for Enterococcus spp. are as follows: for vancomycin, susceptible, ≤4 mg/liter; resistant, >4 mg/liter; for teicoplanin, susceptible, ≤2 mg/liter; resistant, >2 mg/liter (22). The inducible phenotypes of VanB-type VRE with moderate to low vancomycin MICs challenge current phenotypic detection methods. It is important to detect these VRE isolates, as glycopeptide treatment of infections caused by such isolates may lead to treatment failure due to increased MICs or selection of constitutively expressed vanB clusters also showing resistance to teicoplanin (23–26).
The purpose of this study was to examine the ability of different antimicrobial susceptibility testing methods to detect VRE with low or medium levels of resistance. It was organized as a multicenter study in which Danish, Norwegian, and Swedish laboratories were invited to blindly examine a phenotypically and genotypically well-characterized diverse collection of Enterococcus faecalis (n = 12) and E. faecium (n = 18) isolates with and without nonsusceptibility to vancomycin. The collection was examined by the EUCAST disk diffusion method in all laboratories and by one alternative method, which could include the vancomycin-brain heart infusion (BHI) agar screening method (referred to as the Clinical and Laboratory Standards Institute [CLSI] agar screen), a commercial chromogenic agar screening method, or an automated antimicrobial susceptibility testing system.
MATERIALS AND METHODS
Bacterial strains used in this study.
All isolates were previously genotyped by multiplex identification-PCR (27), vanA/B/E/G PCRs (17, 28, 29), SmaI pulsed-field gel electrophoresis (PFGE) (30), and/or multilocus sequence typing (MLST), to ensure species identification, van genotype identification, and genetic diversity. The isolates covered the whole range of vancomycin MIC values. The vancomycin MICs of the isolates were confirmed by both gradient testing (Etest; bioMérieux, Marcy l'Etoile, France) on Mueller-Hinton (MH) agar, as described by the manufacturer, and broth microdilution testing, according to International Organization for Standardization recommendations (31).
The isolate panel consisted of well-characterized vancomycin-susceptible (n = 3; three copies of E. faecalis ATCC 29212) and vancomycin-resistant (n = 27) strains of E. faecalis and E. faecium (Table 1). The resistant isolates expressed various vancomycin MICs (Fig. 1). Twelve of the isolates were E. faecalis (vanB1, n = 3; vanB2, n = 1; vanE, n = 1; vanG, n = 1; ATCC 29212, vancomycin susceptible, n = 3; ATCC 51299, vanB, n = 3) and 18 E. faecium (vanB1, n = 1; vanB2, n = 17), of diverse geographical origins (Norway, n = 12; United States, n = 11; Sweden, n = 6; Australia, n = 1). Seven of the isolates represented Nordic outbreak strains from Bergen, Norway (1996) (MIC, 32 mg/liter) (32, 33), Örebro, Sweden (2002 to 2003) (MICs, 32 and ≥256 mg/liter) (15), Stockholm, Sweden (2007) (MIC, 8 mg/liter), and Västerås, Sweden (2008) (MIC, 16 mg/liter) (Sivertsen et al., submitted). The remaining 20 resistant strains expressed low-level (MIC, 4 to 8 mg/liter; n = 5), medium-level (MIC, 16 to 32 mg/liter; n = 13), or high-level (MIC, ≥64 mg/liter; n = 2) vancomycin resistance. The panel consisted of 23 PFGE types and two subtypes within type XII. MLST analysis of 13 isolates demonstrated 5 sequence types (STs) among the E. faecium clonal complex 17 (CC17) high-risk clones (ST16, n = 1; ST17, n = 5; ST192, n = 1; ST262, n = 1; ST313, n = 1), as well as E. faecalis ST14 (n = 1) and ST30 (ATCC 29212, n = 3) (Table 1).
TABLE 1.
Isolate identification, species, van genotype, gradient test results, broth microdilution MICs for vancomycin, and PFGE and MLST types for the blinded material used in this study
| Isolate no. | Identification | Species | Origin | van subtype | Vancomycin MIC (mg/liter) |
PFGE typeb | MLST resultsc | Reference or source | |
|---|---|---|---|---|---|---|---|---|---|
| Etest | BMDa | ||||||||
| 1 | K09-03/V1-38 | E. faecium | Norway | B2 | 16 | 16 | I | This study | |
| 2 | K25-21 | E. faecium | Norway | B2 | 16 | 8 | II | This study | |
| 3 | K26-39 | E. faecium | Norway | B2 | 16 | 8 | III | This study | |
| 4 | K30-42 | E. faecium | Norway | B2 | 8 | 4 | IV | This study | |
| 5 | K33-53 | E. faecium | Norway | B2 | 32 | 32 | V | This study | |
| 6 | K46-59 | E. faecium | Norway | B2 | 8 | 8 | VI | This study | |
| 7 | ATCC 29212 | E. faecalis | Negative | 4 | 2 | VII | E. faecalis ST30 | This study, 39 | |
| 8 | ATCC 51299 | E. faecalis | B | 16 | 128 | VIII | This study | ||
| 9 | K53-60 | E. faecalis | Norway | B1 | 8 | 16 | IX | This study | |
| 10 | K57-77 | E. faecium | Norway | B2 | 16 | 16 | X | This study | |
| 11 | K61-59 | E. faecium | Norway | B2 | 8 | 8 | XI | This study | |
| 12 | K55-34 (VRE0683) | E. faecium | Stockholm, Sweden | B2 | 8 | 32 | XIIa | ST192 (DLV ST17) | Sivertsen et al., submitted |
| 13 | K55-41 (VRE0881) | E. faecium | Västerås, Sweden | B2 | 16 | 16 | XIIb | ST17 | Sivertsen et al., submitted |
| 14 | A2-46 (BM4518) | E. faecalis | Australia | G | 16 | 16 | XIII | This study, 40 | |
| 15 | K55-41 (VRE0881) | E. faecium | Västerås, Sweden | B2 | 16 | 16 | XIIb | ST17 | Sivertsen et al., submitted |
| 16 | ATCC 29212 | E. faecalis | Negative | 4 | 2 | VII | E. faecalis ST30 | This study, 39 | |
| 17 | ATCC 51299 | E. faecalis | B | 16 | 128 | VIII | This study | ||
| 18 | A2-48 (BM4405) | E. faecalis | USA | E | 16 | 16 | XIV | This study, 28 | |
| 19 | TUH 12-1 (C68) | E. faecium | USA | B2 | ≥256 | ≥256 | XV | ST16 (SLV ST17) | 41, 42 |
| 20 | K55-41 (VRE0881) | E. faecium | Västerås, Sweden | B2 | 16 | 16 | XIIb | ST17 | Sivertsen et al., submitted |
| 21 | K45-3 (03T468) | E. faecium | Örebro, Sweden | B2 | ≥256 | ≥256 | XVI | ST262 (SLV ST18) | 15 |
| 22 | K45-12 (02T895) | E. faecium | Örebro, Sweden | B2 | 32 | 16 | XVII | ST17 | 15 |
| 23 | TUH 1-3 (V583) | E. faecalis | USA | B1 | 32 | XVIII | E. faecalis ST14 | 17, 43 | |
| 24 | TUH 1-79 | E. faecium | Norway | B2 | 16 | 16 | XIX | 17 | |
| 25 | TUH 2-18 | E. faecium | Bergen, Norway | B2 | 32 | XX | ST17 | 17, 42 | |
| 26 | TUH 4-65 | E. faecium | USA | B1 | ≥256 | ≥256 | XXI | ST313 (SLV ST18) | 17, 42 |
| 27 | TUH 7-13 | E. faecalis | USA | B1 | 32 | 16 | XXII | 17 | |
| 28 | ATCC 29212 | E. faecalis | Negative | 4 | 2 | VII | E. faecalis ST30 | This study, 39 | |
| 29 | ATCC 51299 | E. faecalis | B | 16 | 128 | VIII | This study | ||
| 30 | 50577479KRE | E. faecalis | Norway | B2 | 4 | 8 | XXIII | This study | |
BMD, broth microdilution.
Previous PFGE type designations have been changed to consecutive numbering with Roman numerals in this study, for pedagogic reasons.
SLV, single-locus variant; DLV, double-locus variant.
FIG 1.
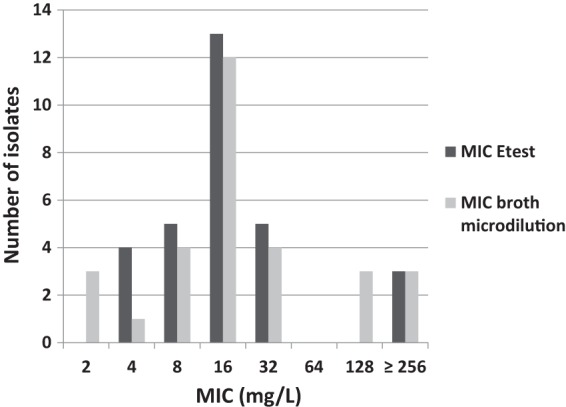
MIC distribution of the collection of blinded isolates (n = 30).
Study design.
The study was organized through the NordicAST (Nordic Committee on Antimicrobial Susceptibility Testing) network (www.nordicast.org). All Danish (n = 13), Swedish (n = 23), and Norwegian (n = 24) diagnostic laboratories for clinical microbiology were invited to participate in the study, examining the bacterial panel listed in Table 1. All laboratories were asked to perform the reference agar disk diffusion method as recommended by EUCAST (34) and at least one alternative method, i.e., the CLSI agar screening method (35) using BHI agar supplemented with vancomycin at 6 mg/liter and/or a commercial automated test available in the laboratory (Vitek 2 system). The laboratories were also welcome to test the performance of commercial VRE-selective agars. Each participating laboratory used its own supply of test reagents, disks, and agar. The laboratories were instructed to avoid any testing with a confirmatory method (van genotyping or gradient testing), as the objective was for the strains to be examined through a blinded nonbiased approach. The time frame for the laboratories to complete the susceptibility testing was a maximum of 3 weeks. A result data form was filled in for each participating laboratory for each strain and included zone diameter and zone edge quality (fuzzy or sharp zone edge) for the agar disk diffusion testing, growth/no-growth for the agar screening, results for the commercial automated antimicrobial susceptibility testing systems as reported, interpretation (susceptible or resistant) according to EUCAST clinical breakpoints (22), and a comment on whether, under normal circumstances, the laboratory would refer the sample for confirmatory testing in a reference laboratory.
Phenotypic methods used for antimicrobial susceptibility testing.
The disk diffusion test was performed with 6-mm disks with 5 μg vancomycin and MH agar plates, using the EUCAST method (34) and clinical breakpoints (22). Each laboratory participating in the study used its own supply of MH agar, which was either Oxoid agar (Oxoid Ltd./Thermo Fisher Scientific, Cambridge, United Kingdom) (n = 16), BBL MH II agar (Becton, Dickinson Diagnostics, Baltimore, MD) (n = 10), or Merck agar (Merck KGaA, Darmstadt, Germany) (n = 2).
The CLSI agar screening method was performed with BHI agar with 6 mg/liter vancomycin and results were read after 24 h, as described by Swenson et al. (35). The BHI agar used was either Difco agar (Becton, Dickinson Diagnostics) (n = 8), Oxoid agar (n = 5), BBL agar (Becton, Dickinson Diagnostics) (n = 3), Scharlau agar (Scharlab SL, Barcelona, Spain) (n = 1), or Acumedia agar (Neogen Corp., Lansing, MI) (n = 1). Growth on commercial chromogenic culture media, read after 48 h, was tested using chromID VRE agar (bioMérieux, Marcy l'Etoile, France) (n = 7) or CHROMagar VRE medium (CHROMagar, Paris, France) (n = 5). Automated susceptibility testing with the Vitek 2 system was performed using either AST-P592 (n = 2), AST-P586 (n = 2), or AST-594 (n = 1) susceptibility cards (bioMérieux).
Evaluation of results.
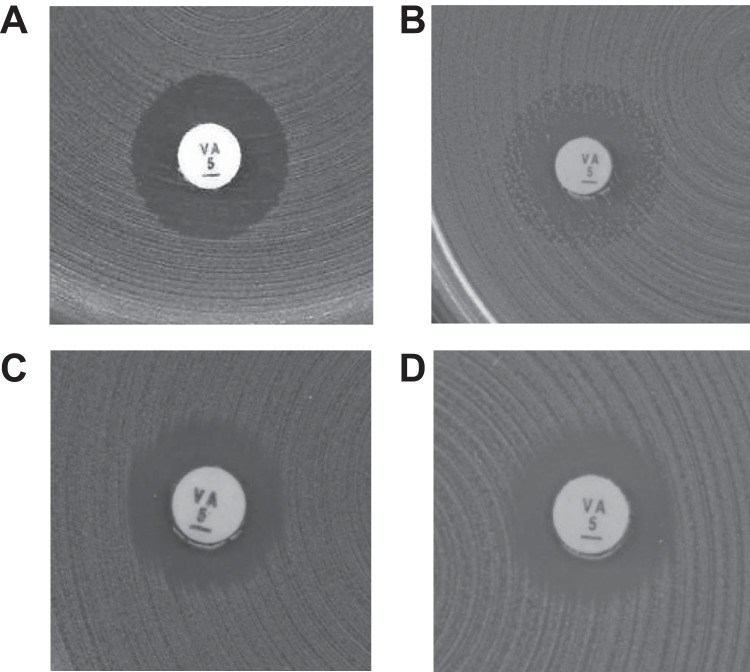
The test results were categorized as either correct (susceptible reported as susceptible and resistant reported as resistant), very major error (VME) (resistant reported as susceptible), or major error (ME) (susceptible reported as resistant), as EUCAST has not defined any intermediate category for clinical vancomycin breakpoints (thus excluding the possibility of minor errors occurring). According to the EUCAST disk diffusion method, the isolates were categorized as resistant when the zone diameter was less than 12 mm. Also, according to the method, resistance should be suspected when the vancomycin zone edge is fuzzy (examples in Fig. 2C and D) or colonies are growing within the inhibition zone (example in Fig. 2B). Thus, zone edge quality was also taken into account. The isolates were reported as vancomycin susceptible only when the zone edges were sharp and ≥12 mm (example in Fig. 2A).
FIG 2.
Examples of disk diffusion inhibition zones for Enterococcus spp. with 5-μg vancomycin disks. (A) Cultures with sharp zone edges and zone diameters of ≥12 mm should be reported as susceptible. (B to D) Cultures with fuzzy zone edges (C and D) or colonies within the zone (B) should be reported as resistant, even if the zone diameter is ≥12 mm.
Statistical methods and interpretation.
Sensitivities (conditional probabilities that resistant isolates are correctly categorized), specificities (conditional probabilities that susceptible isolates are correctly categorized), and confidence intervals (CIs) were calculated using Clinical Research Calculator 1 (vassarstats.net/index.html). Fisher's exact test with two-tailed P values, performed using an online calculator (www.graphpad.com/quickcalcs/contingency1.cfm), was used to identify statistically significant differences (P < 0.05) between pairs of methods or media.
RESULTS
A total of 34 Scandinavian laboratories agreed to participate and delivered data to the study. The results from six laboratories were excluded because the laboratories did not deliver data on the EUCAST disk diffusion method. Thus, results from 28 laboratories were included, i.e., 13 Norwegian, 10 Swedish, and five Danish laboratories. The laboratories delivered data sets on the EUCAST disk diffusion (n = 28), CLSI agar screen (n = 18), agar screen using commercial chromogenic VRE media (n = 12), and Vitek 2 (n = 5) (bioMérieux) methods.
Routine phenotypic testing for vancomycin resistance in enterococci is done by CLSI agar screening in Norwegian laboratories, while the EUCAST disk diffusion method is the preferred method in Sweden. Three of the Danish laboratories were also familiar with the EUCAST disk diffusion method for detection of VRE.
EUCAST disk diffusion and CLSI agar screen methods performed better than the Vitek 2 system.
VME and ME rates, sensitivity, and specificity were calculated for each method or agar type used (Table 2 shows calculations for n ≥ 5). None of the tested methods scored as perfect. The sensitivity (0.87 to 0.93) and specificity (0.94 to 1) values were high for the EUCAST disk diffusion method, the CLSI agar screen, and the Vitek 2 system (Table 2). Overall, the EUCAST disk diffusion and CLSI agar screen methods performed better than the Vitek 2 system (Table 2; also see Table S1 in the supplemental material). Comparisons of the methods with one another showed that both the disk diffusion method and the CLSI agar screen performed significantly better than the Vitek 2 system in identifying the isolates as resistant or susceptible (see Table S1).
TABLE 2.
Numbers of very major and major errors, sensitivity, and specificity calculated for each detection method and for each type of agar used (for n ≥ 5)a
| Method (no. of laboratories) | No. of VMEs/total no. of isolates with van genotype (%) | Sensitivity (95% CI) | No. of MEs/total no. of susceptible isolates (%) | Specificity (95% CI) |
|---|---|---|---|---|
| EUCAST disk diffusion (28) | 53/756 (7.0) | 0.93 (0.91–0.95) | 2/84 (2.4) | 0.98 (0.91–1) |
| Oxoid MH agar (16) | 14/432 (3.2) | 0.97 (0.94–0.98) | 0/48 | 1 (0.91–1) |
| BBL MH II agar (10) | 37/270 (14) | 0.86 (0.81–0.90) | 2/30 (6.7) | 0.93 (0.76–0.99) |
| CLSI agar screen (18) | 32/486 (6.6) | 0.93 (0.91–0.95) | 3/54 (5.6) | 0.94 (0.84–0.99) |
| Difco BHI agar (8) | 9/216 (4.2) | 0.96 (0.92–0.98) | 0/24 | 1 (0.83–1) |
| Oxoid BHI agar (5) | 15/135 (11) | 0.89 (0.82–0.93) | 0/15 | 1 (0.75–1) |
| Chromogenic agar screen (12) | 7/324 (2.2) | 0.98 (0.95–0.99) | 4/36 (11) | 0.89 (0.73–0.96) |
| chromID VRE agar (7) | 3/189 (1.6) | 0.98 (0.95–1) | 1/21 (4.8) | 0.95 (0.74–1) |
| VRE CHROMagar (5) | 4/135 (3.0) | 0.97 (0.92–0.99) | 3/15 (20) | 0.8 (0.51–0.95) |
| Vitek 2 system (5) | 18/135 (13) | 0.87 (0.79–92) | 0/15 (0) | 1 (0.75–1) |
In VMEs, strains were classified as susceptible when containing the van genotype. In MEs, strains were classified as resistant when containing no van genotype. According to EUCAST rules for the disk diffusion assay with vancomycin, samples were considered resistant, although the zone size suggested susceptibility, if the zone edge was fuzzy or colonies were growing within the zone.
The distribution of VMEs related to reference MIC values of the isolates (data not shown) revealed that the troublesome isolates for the disk diffusion and CLSI agar screen methods mainly expressed low MIC values (4 to 8 mg/liter), whereas the Vitek 2 system had more problems concerning detection of moderate resistance levels (MICs, 16 to 32 mg/liter). The mean, median, lowest, and highest numbers of errors per laboratory for each method are reported in Table 3. The mean and median values were similar and low for the disk diffusion and CLSI agar screen methods.
TABLE 3.
Mean, median, lowest, and highest numbers of errors per laboratory for each methoda
| Method (no. of laboratories) | No. of VMEs/laboratory |
No. of MEs/laboratory |
||||||
|---|---|---|---|---|---|---|---|---|
| Mean | Median | Lowest | Highest | Mean | Median | Lowest | Highest | |
| EUCAST disk diffusion (28) | 1.9 | 1 | 0 | 13 | 0.071 | 0 | 0 | 2 |
| CLSI agar screen (18) | 1.8 | 2 | 0 | 5 | 0.17 | 0 | 0 | 3 |
| Vitek 2 system (5) | 3.2 | 2.5 | 1 | 6 | 0 | 0 | 0 | 0 |
In VMEs, strains were classified as susceptible when containing the van genotype. In MEs, strains were classified as resistant when containing no van genotype. The median number is the middle number in a numerically sorted list of numbers.
Experience and media influence the results of the EUCAST disk diffusion method.
The observed numbers of VMEs and MEs, as well as sensitivity and specificity, for the disk diffusion method differed with laboratory country locations. Swedish laboratories, which were experienced in using the disk diffusion method to assess vancomycin resistance, in general performed better in detecting the resistant isolates with the EUCAST disk diffusion method (Table 4). Dividing the results according to the type of MH agar used for disk diffusion indicated that BBL MH II agar did not perform as well as Oxoid MH agar (Table 2) and Merck MH agar. Statistical analyses showed that laboratories using Oxoid MH agar (P < 0.0001) and Merck MH agar (P = 0.027) performed significantly better than laboratories using BBL MH II agar. The disk diffusion inhibition zones were read to be 0.8 mm larger, on average, with BBL MH II agar than with Oxoid agar (Fig. 3). Resistant isolates had zone sizes of ≥12 mm more often on BBL MH II agar than on other agars (Fig. 3).
TABLE 4.
Numbers of very major and major errors, sensitivity, and specificity calculated for EUCAST disk diffusion and CLSI agar screen methods, according to the country of the laboratories or their experience in using the detection methodsa
| Method | Country and experience (no. of laboratories) | VME rate (% [no. of VMEs/total no. resistant]) | Sensitivity (95% CI) | ME rate (% [no. of MEs/total no. susceptible]) | Specificity (95% CI) |
|---|---|---|---|---|---|
| Disk diffusion | Sweden (10) | 1.9 (5/270) | 0.98 (0.95–0.99) | 0 (0/30) | 1 (0.86–1) |
| Norway (13) | 10 (36/351) | 0.90 (0.86–0.93) | 2.6 (1/39) | 0.97 (0.85–1) | |
| Denmark (5) | 8.9 (12/135) | 0.91 (0.85–0.95) | 6.7 (1/15) | 0.93 (0.66–1) | |
| Experienced (13) | 2.6 (9/351) | 0.97 (0.95–0.99) | 0 (0/39) | 1 (0.89–1) | |
| Inexperienced (15) | 11 (44/405) | 0.89 (0.86–0.92) | 4.4 (2/45) | 0.96 (0.84–0.99) | |
| Agar screen | Sweden (1) | 3.7 (1/27) | 0.9 (0.79–1) | 0 (0/3) | 1 (0.31–1) |
| Norway (13) | 6.8 (24/351) | 0.93 (0.90–0.95) | 0 (0/39) | 1 (0.89–1) | |
| Denmark (4) | 6.5 (7/108) | 0.94 (0.87–0.97) | 25 (3/12) | 0.75 (0.43–0.93) | |
| Experienced (13) | 6.8 (24/351) | 0.93 (0.90–0.95) | 0 (0/39) | 1 (0.89–1) | |
| Inexperienced (5) | 5.9 (8/135) | 0.94 (0.88–0.97) | 20 (3/15) | 0.8 (0.51–0.95) |
In VMEs, strains were classified as susceptible when containing the van genotype. In MEs, strains were classified as resistant when containing no van genotype.
FIG 3.
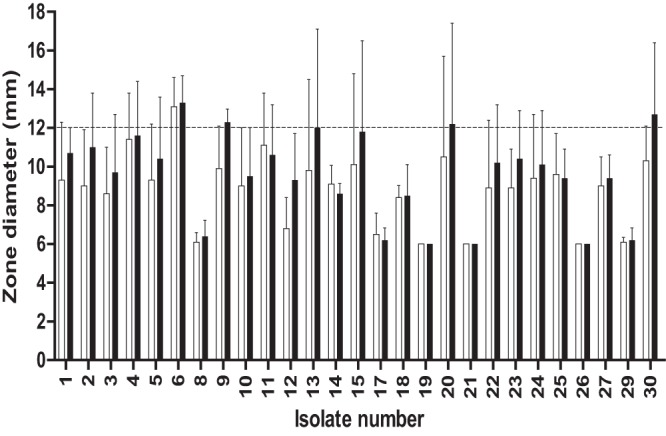
Average zone diameters and standard deviations calculated for each isolate from recorded results from laboratories using either Oxoid MH agar (□) or BBL MH II agar (■).
Nine of 10 laboratories using BBL MH II agar were inexperienced with the agar disk diffusion method for the detection of VRE. Thus, the number of experienced users who reported results with BBL MH II agar was too small to get a clear picture of whether these errors were due to inexperience or agar characteristics (see Tables S2 and S3 in the supplemental material). In line with this, the four inexperienced laboratories using Oxoid MH agar were not significantly better at identifying the isolates than the nine inexperienced laboratories using BBL MH II agar.
Three laboratories performed disk diffusion assays using Neo-Sensitabs 9-mm disks (Rosco Diagnostica A/S, Taastrup, Denmark). The VME rate (14%) and specificity of 1 for Neo-Sensitabs were higher than the values for 6-mm disks (VME rate, 6.5%; specificity, 0.98), while the ME rate (0%) and sensitivity (0.86) were lower than those for 6-mm disks (ME rate, 2.2%; sensitivity, 0.93). Agar disk diffusion assays using 6-mm disks performed significantly better (P = 0.042) than assays using Neo-Sensitabs in identifying the isolates as resistant or susceptible. However, the VME rate and sensitivity for assays with Neo-Sensitabs were slightly different from values from inexperienced personnel using 6-mm disks (VME, 11%; sensitivity, 0.89), and agar disk diffusion assays conducted by inexperienced personnel using 6-mm disks did not perform significantly better than assays with Neo-Sensitabs in identifying the isolates as resistant or susceptible (data not shown).
Media influence the results of CLSI agar screening.
The observed numbers of VMEs and MEs, sensitivity, and specificity for the CLSI agar screen revealed that the Norwegian laboratories, which were experienced in using agar screening for testing vancomycin resistance, did not appear to be better in detecting resistant isolates by this method than was the only participating Swedish laboratory that reported data for this method (Table 4). The laboratories using Oxoid agar (Table 2) and BBL BHI agar (data not shown) for agar screening performed less well in revealing the susceptibility category of isolates than did those using Difco BHI agar. Laboratories using Difco BHI agar performed significantly better (P = 0.017) than those using Oxoid BHI agar but just insignificantly (P = 0.052) better than those using BBL BHI agar (data not shown). Comparing only experienced laboratories, Difco BHI agar performed significantly better in identifying isolates correctly than did Oxoid BHI agar in the agar screen (see Table S3 in the supplemental material). The numbers of laboratories using Scharlau or Acumedia BHI agar for agar screening were too small to give any significant differences in comparison with Oxoid and Difco BHI agar.
Agar screening using commercial chromogenic VRE media.
Agar screening using commercial chromogenic VRE media (n = 12) showed a higher ME rate (11%) and sensitivity (0.98) and lower specificity (0.89) than values obtained with disk diffusion, CLSI agar screening, and Vitek 2 methods (Table 2). The high ME rates and lower specificities for commercial chromogenic VRE agars indicate that their potential use in screening of clinical strains for vancomycin resistance would result in many false-positive results. Comparison of the two different chromogenic VRE media used showed slightly better performance of chromID VRE agar than VRE CHROMagar medium (Table 2), but these results were not statistically significant.
Different performance of AST cards used in the Vitek 2 system.
The Vitek 2 system results using different types of antimicrobial susceptibility test cards showed that card AST-P592 (VME rate, 7.4%; sensitivity, 0.93) was better at identifying the isolates correctly than were cards AST-P586 (VME rate, 20%; sensitivity, 0.80) and AST-P594 (VME rate, 11%; sensitivity, 0.89). However, these results were not statistically significant.
DISCUSSION
We have examined the ability of the EUCAST disk diffusion assay, the CLSI agar screen, and the Vitek 2 automated antimicrobial susceptibility testing system to detect VanB-type VRE in a blinded panel of well-characterized E. faecalis and E. faecium strains, with or without low to medium levels of resistance to vancomycin. The study was performed with a multicenter design involving clinical laboratories in Denmark, Norway, and Sweden. All laboratories tested the EUCAST agar disk diffusion method and in addition one alternative method, i.e., the CLSI agar screen, commercial chromogenic VRE agar screens, and/or automated systems.
In projects such as this, it is possible that project samples receive more attention than routine samples and that this introduces bias in the evaluation of performance. On the other hand, the results reveal the “best achievement level,” and knowing this is valuable. Although none of the methods was perfect, the CLSI agar screen and EUCAST agar disk diffusion methods showed comparably high and acceptable sensitivity and specificity values.
Our results show that the ability to detect VRE by the agar disk diffusion method is influenced by the experience of the personnel in reading inhibition zones, while experience in the interpretation of CLSI agar screen results is not that critical. For the agar screen method, the choice of BHI agar and quality control of the vancomycin concentration seemed to be more important for correct identification of VRE. The notion that experience is more important when using the disk diffusion method was supported by the numbers calculated for the EUCAST disk diffusion and CLSI agar screen methods for experienced versus inexperienced laboratories (Table 4; see also Table S2 in the supplemental material). Laboratories experienced with the disk diffusion method performed significantly better (P < 0.0001) in identifying the isolates as resistant or susceptible by the disk diffusion method than did inexperienced personnel (data not shown). Moreover, the mean and median error values were similar and low for the EUCAST disk diffusion and CLSI agar screen methods, while the highest VME rates for the EUCAST disk diffusion method were all reported from laboratories that had no previous experience with use of the disk diffusion method to detect low-level vancomycin resistance. Comparing the different media in agar disk diffusion assays showed that the resistant isolates more often had zone sizes of ≥12 mm on BBL MH II agar (Fig. 3), which would require experienced readers to check the quality of zone edges in order to correctly categorize isolates as resistant.
In this study, chromogenic VRE media gave the lowest VME rates (Table 2) but the highest ME rates. In a recent study by Klare et al., the VME rates of the chromogenic VRE screening media were higher than those observed in our study (36). The higher VME rates observed by Klare and coworkers could be due to their selection of strains; 43/129 (33%) of their VanB-type E. faecium strains showed MICs of ≤4 mg/liter, thus pushing the detection limits more than our collection, in which only 1 of 27 VRE isolates showed a MIC of 4 mg/liter (Table 1).
It is important to have rapid routine antimicrobial susceptibility testing methods that yield results that can be easily interpreted and communicated to clinicians. We have shown that results can be influenced by the level of experience of the laboratory personnel and/or the media used. In this study, the participating laboratories were informed about the van status of the examined strain collection after they reported their results. Subsequently, many participants examined the samples again and reported that they had read the troublesome samples incorrectly the first time. This was specifically the case for the disk diffusion method. Detection of low-level vancomycin-resistant isolates with the disk diffusion method relies not only on evaluation of the zone size but also on evaluation of the zone edges, which requires experience. Hence, it is important to note that the EUCAST stresses that the zone edges should be examined with transmitted light (the plate held up to the light) and resistance suspected when the vancomycin zone edge is fuzzy (Fig. 2C and D) or when colonies are growing within the inhibition zone (Fig. 2B) and that glycopeptide susceptibility tests on enterococci should be incubated for 24 h to ensure the visibility of resistant colonies. These recommendations are specific for evaluating VRE and are different from those for reading disk diffusion results for most other antimicrobials and species. The sharp zones of susceptible isolates (Fig. 2A) are very characteristic, in comparison with fuzzy zones. Thus, positive and negative controls for comparisons facilitate the evaluation of VRE with low-level resistance. Moreover, the CLSI agar screen method requires experienced personnel who can prepare and store the agar plates correctly, so that the vancomycin concentration in the plates is accurate.
It is well known that VanB-type VRE can be difficult to detect, due to the inducible mechanism of resistance and the variable levels of vancomycin resistance expressed (12, 37). The strain collection was designed to be genetically and epidemiologically diverse and to include troublesome isolates with low vancomycin MICs in addition to ATCC reference strains (Table 1). The vanB E. faecalis ATCC 51299 and E. faecalis ATCC 29212 strains are recommended as positive and negative quality controls, respectively, for both the EUCAST disk diffusion (34) and CLSI agar screen (38) methods. Twenty-one of the 28 laboratories in this study reported using vanB E. faecalis ATCC 51299 and 5 laboratories used various CCUG VanB-positive enterococcal strains as positive controls. A previous evaluation of the vanB E. faecalis ATCC 51299 strain showed vancomycin MICs of 16 to 32 mg/liter after 24 h of incubation. After 48 h, however, a vancomycin MIC of 128 mg/liter was observed (38). A valid positive control is supposed to challenge the detection limits of the method. In this study, the vanB E. faecalis ATCC 51299 strain was included in three copies in the blinded test collection. According to the disk diffusion zone sizes recorded in the different laboratories, this reference strain does not seem to challenge the test conditions. All laboratories were able to place this isolate well within the resistant category, with zone diameters ranging from 6 to 9 mm. A more relevant reference strain with a lower inducible vancomycin MIC should be considered for quality control purposes.
In conclusion, our results demonstrate acceptable performance by both the EUCAST disk diffusion and CLSI agar screen methods. Both reliably detect VanB-type VRE with low-level resistance. The high ME rates and lower specificities of commercial chromogenic VRE agars indicate that their potential use in screening of clinical strains for vancomycin resistance would result in many false-positive results. However, confirmation of the ME rates and specificities calls for another study with more vancomycin-susceptible strains included in the strain collection. Importantly, the use of the agar disk diffusion method requires personnel trained in the interpretation of zone edges, and use of the CLSI agar screen method requires careful selection of control strains to monitor the vancomycin concentration in the plates.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bettina Aasnæs and Tracy Munthali Lunde for excellent technical assistance.
The study was performed in collaboration with the diagnostic laboratories in clinical microbiology in Norway, Sweden, and Denmark that form the NordicAST VRE Detection Study Group. Representatives of this study group included Kirsten Paulsen (Aalborg Hospital), Lars Erik Lemming (Aarhus University Hospital), Siri Haug Hänsgen (Akershus University Hospital), Marianne Bäckman (Aleris Medilab), Jenny Åhman (EUCAST Laboratory for Antimicrobial Susceptibility Testing), Thomas Ahlqvist (Central Hospital Karlstad), Elisabeth Sirnes (Central Hospital Førde), Ann Cathrine Petersson (Department of Clinical Microbiology, Laboratory Medicine Lund), Håkan Janson (Department of Clinical Microbiology, Laboratory Medicine Malmö), Ingegerd Sjögren (Hallands Hospital Halmstad), Kristin Stenhaug Kilhus (Haukeland University Hospital), Magnus Arpi (Herlev Hospital), Inger Brock (Hillerød Hospital), Pia Littauer (Hvidovre Hospital), Sara Gustavsson (Kalmar County Hospital), Hong Fang (Karolinska University Hospital Huddinge), Kirsti Jalakas Pörnull (Karolinska University Hospital Solna), Lennart E. Nilsson (Linköping University Hospital), Margreet Boer (Molde Hospital), Hege Elisabeth Larsen (Nordland Hospital Bodø), Anette Holm (Odense University Hospital), Pia Langseth (Rikshospitalet University Hospital), Ia Adlerberth (Sahlgrenska University Hospital), Ole Heltberg (Slagelse Hospital), Anette M. Hammerum (Statens Serum Institute), Anita Løvås Brekken (Stavanger University Hospital), Kjersti Wik Larssen (St. Olavs Hospital), Dagfinn Skaare (Vestfold Hospital), Anita Kanestrøm (Østfold Hospital), Ståle Tofteland (Sørlandet Hospital), Thea Bergheim (Ullevål University Hospital), Gunnar Skov Simonsen (University Hospital of North Norway), Angela Lagerqvist Vidh (Uppsala University Hospital), and Claus Østergaard (Velje Hospital).
Footnotes
Published ahead of print 5 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03544-13.
REFERENCES
- 1.Werner G, Coque TM, Franz CM, Grohmann E, Hegstad K, Jensen L, van Schaik W, Weaver K. 2013. Antibiotic resistant enterococci: tales of a drug resistance gene trafficker. Int. J. Med. Microbiol. 303:360–379. 10.1016/j.ijmm.2013.03.001 [DOI] [PubMed] [Google Scholar]
- 2.Gilmore MS, Lebreton F, van Schaik W. 2013. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr. Opin. Microbiol. 16:10–16. 10.1016/j.mib.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Kraker ME, Jarlier V, Monen JC, Heuer OE, van de Sande N, Grundmann H. 2013. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin. Microbiol. Infect. 19:860–868. 10.1111/1469-0691.12028 [DOI] [PubMed] [Google Scholar]
- 4.Gilmore MS, Ferretti JJ. 2003. Microbiology: the thin line between gut commensal and pathogen. Science 299:1999–2002. 10.1126/science.1083534 [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Top J, de Been M, Bierschenk D, Rogers M, Leendertse M, Bonten MJ, van der Poll T, Willems RJ, van Schaik W. 2013. Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J. Infect. Dis. 207:1780–1786. 10.1093/infdis/jit076 [DOI] [PubMed] [Google Scholar]
- 6.Arias CA, Contreras GA, Murray BE. 2010. Management of multi-drug resistant enterococcal infections. Clin. Microbiol. Infect. 16:555–562. 10.1111/j.1469-0691.2010.03214.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegstad K, Mikalsen T, Coque TM, Jensen LB, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and E. faecium. Clin. Microbiol. Infect. 16:541–554. 10.1111/j.1469-0691.2010.03226.x [DOI] [PubMed] [Google Scholar]
- 8.Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, Zhu D, Hu F, Zhang Y, Wang F, Jacoby GA, Wang M. 2010. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 54:4643–4647. 10.1128/AAC.01710-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lebreton F, Depardieu F, Bourdon N, Fines-Guyon M, Berger P, Camiade S, Leclercq R, Courvalin P, Cattoir V. 2011. d-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 55:4606–4612. 10.1128/AAC.00714-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinsson KG, Leclercq R, Lester CH, Lillie M, Novais C, Olsson-Liljequist B, Peixe LV, Sadowy E, Simonsen GS, Top J, Vuopio-Varkila J, Willems RJ, Witte W, Woodford N. 2008. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 13(47):pii=19046 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19046 [PubMed] [Google Scholar]
- 11.Granlund M, Carlsson C, Edebro H, Emanuelsson K, Lundholm R. 2006. Nosocomial outbreak of vanB2 vancomycin-resistant Enterococcus faecium in Sweden. J. Hosp. Infect. 62:254–256. 10.1016/j.jhin.2005.06.031 [DOI] [PubMed] [Google Scholar]
- 12.Werner G, Klare I, Fleige C, Geringer U, Witte W, Just HM, Ziegler R. 2012. Vancomycin-resistant vanB-type Enterococcus faecium isolates expressing varying levels of vancomycin resistance and being highly prevalent among neonatal patients in a single ICU. Antimicrob. Resist. Infect. Control 1:21. 10.1186/2047-2994-1-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourdon N, Fines-Guyon M, Thiolet JM, Maugat S, Coignard B, Leclercq R, Cattoir V. 2011. Changing trends in vancomycin-resistant enterococci in French hospitals, 2001–08. J. Antimicrob. Chemother. 66:713–721. 10.1093/jac/dkq524 [DOI] [PubMed] [Google Scholar]
- 14.Söderblom T, Aspevall O, Erntell M, Hedin G, Heimer D, Hökeberg I, Kidd-Ljunggren K, Melhus Å, Olsson-Liljequist B, Sjögren I, Smedjegård J, Struwe J, Sylvan S, Tegmark-Wisell K, Thore M. 2010. Alarming spread of vancomycin resistant enterococci in Sweden since 2007. Euro Surveill. 15(29):pii=19620 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19620 [DOI] [PubMed] [Google Scholar]
- 15.Bjørkeng EK, Rasmussen G, Sundsfjord A, Sjöberg L, Hegstad K, Söderquist B. 2011. Clustering of polyclonal VanB-type vancomycin-resistant Enterococcus faecium in a low-endemic area was associated with CC17-genogroup strains harbouring transferable vanB2-Tn5382 and pRUM-like repA containing plasmids with axe-txe plasmid addiction systems. APMIS 119:247–258. 10.1111/j.1600-0463.2011.02724.x [DOI] [PubMed] [Google Scholar]
- 16.Johnson PD, Ballard SA, Grabsch EA, Stinear TP, Seemann T, Young HL, Grayson ML, Howden BP. 2010. A sustained hospital outbreak of vancomycin-resistant Enterococcus faecium bacteremia due to emergence of vanB E. faecium sequence type 203. J. Infect. Dis. 202:1278–1286. 10.1086/656319 [DOI] [PubMed] [Google Scholar]
- 17.Dahl KH, Simonsen GS, Olsvik Ø, Sundsfjord A. 1999. Heterogeneity in the vanB gene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 43:1105–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gold HS, Unal S, Cercenado E, Thauvin Eliopoulos C, Eliopoulos GM, Wennersten CB, Moellering RC., Jr 1993. A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes of enterococci. Antimicrob. Agents Chemother. 37:1604–1609. 10.1128/AAC.37.8.1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel R, Uhl JR, Kohner P, Hopkins MK, Steckelberg JM, Kline B, Cockerill FR III. 1998. DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcus isolates. Antimicrob. Agents Chemother. 42:202–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Courvalin P. 2006. Vancomycin resistance in Gram-positive cocci. Clin. Infect. Dis. 42(Suppl 1):S25–S34. 10.1086/491711 [DOI] [PubMed] [Google Scholar]
- 21.Boyce JM, Opal SM, Chow JW, Zervos MJ, Potter-Bynoe G, Sherman CB, Romulo RL, Fortna S, Medeiros AA. 1994. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistance. J. Clin. Microbiol. 32:1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.European Committee on Antimicrobial Susceptibility Testing. 2013. Breakpoint tables for interpretation of MICs and zone diameters, version 3.1, 2013. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_3.1.pdf
- 23.Hayden MK, Trenholme GM, Schultz JE, Sahm DF. 1993. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J. Infect. Dis. 167:1224–1227. 10.1093/infdis/167.5.1224 [DOI] [PubMed] [Google Scholar]
- 24.Kawalec M, Gniadkowski M, Kedzierska J, Skotnicki A, Fiett J, Hryniewicz W. 2001. Selection of a teicoplanin-resistant Enterococcus faecium mutant during an outbreak caused by vancomycin-resistant enterococci with the vanB phenotype. J. Clin. Microbiol. 39:4274–4282. 10.1128/JCM.39.12.4274-4282.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.San Millan A, Depardieu F, Godreuil S, Courvalin P. 2009. VanB-type Enterococcus faecium clinical isolate successively inducibly resistant to, dependent on, and constitutively resistant to vancomycin. Antimicrob. Agents Chemother. 53:1974–1982. 10.1128/AAC.00034-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes NE, Ballard SA, Lam MM, Johnson PD, Grayson ML, Stinear TP, Howden BP. 2013. Genomic analysis of teicoplanin resistance emerging during treatment of vanB vancomycin-resistant Enterococcus faecium infections in solid organ transplant recipients including donor-derived cases. J. Antimicrob. Chemother. 68:2134–2139. 10.1093/jac/dkt130 [DOI] [PubMed] [Google Scholar]
- 27.Dutka-Malen S, Evers S, Courvalin P. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fines M, Perichon B, Reynolds P, Sahm DF, Courvalin P. 1999. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob. Agents Chemother. 43:2161–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Depardieu F, Perichon B, Courvalin P. 2004. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 42:5857–5860. 10.1128/JCM.42.12.5857-5860.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahl KH, Lundblad EW, Røkenes TP, Olsvik Ø, Sundsfjord A. 2000. Genetic linkage of the vanB2 gene cluster to Tn5382 in vancomycin-resistant enterococci and characterization of two novel insertion sequences. Microbiology 146:1469–1479 [DOI] [PubMed] [Google Scholar]
- 31.International Organization for Standardization. 2006. Clinical laboratory testing and in vitro diagnostic test systems: susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices. Part 1: reference method for testing the in vitro activity of antimicrobial agents against rapidly growing aerobic bacteria involved in infectious diseases. Publication ISO 20776–1. International Organization for Standardization, Geneva, Switzerland [Google Scholar]
- 32.Dahl KH, Røkenes TP, Lundblad EW, Sundsfjord A. 2003. Nonconjugative transposition of the vanB-containing Tn5382-like element in Enterococcus faecium. Antimicrob. Agents Chemother. 47:786–789. 10.1128/AAC.47.2.786-789.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harthug S, Digranes A, Hope O, Kristiansen BE, Allum AG, Langeland N. 2000. Vancomycin resistance emerging in a clonal outbreak caused by ampicillin-resistant Enterococcus faecium. Clin. Microbiol. Infect. 6:19–28. 10.1046/j.1469-0691.2000.00008.x [DOI] [PubMed] [Google Scholar]
- 34.European Committee on Antimicrobial Susceptibility Testing. 2013. Antimicrobial susceptibility testing: EUCAST disk diffusion method, version 3.0, 2013. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/Manual_v_3.0_EUCAST_Disk_Test.pdf
- 35.Swenson JM, Clark NC, Ferraro MJ, Sahm DF, Doern G, Pfaller MA, Reller LB, Weinstein MP, Zabransky RJ, Tenover FC. 1994. Development of a standardized screening method for detection of vancomycin-resistant enterococci. J. Clin. Microbiol. 32:1700–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klare I, Fleige C, Geringer U, Witte W, Werner G. 2012. Performance of three chromogenic VRE screening agars, two EtestR vancomycin protocols, and different microdilution methods in detecting vanB genotype Enterococcus faecium with varying vancomycin MICs. Diagn. Microbiol. Infect. Dis. 74:171–176. 10.1016/j.diagmicrobio.2012.06.020 [DOI] [PubMed] [Google Scholar]
- 37.Grabsch EA, Chua K, Xie S, Byrne J, Ballard SA, Ward PB, Grayson ML. 2008. Improved detection of vanB2-containing Enterococcus faecium with vancomycin susceptibility by Etest using oxgall supplementation. J. Clin. Microbiol. 46:1961–1964. 10.1128/JCM.01778-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swenson JM, Clark NC, Sahm DF, Ferraro MJ, Doern G, Hindler J, Jorgensen JH, Pfaller MA, Reller LB, Weinstein MP. 1995. Molecular characterization and multilaboratory evaluation of Enterococcus faecalis ATCC 51299 for quality control of screening tests for vancomycin and high-level aminoglycoside resistance in enterococci. J. Clin. Microbiol. 33:3019–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim EB, Kopit LM, Harris LJ, Marco ML. 2012. Draft genome sequence of the quality control strain Enterococcus faecalis ATCC 29212. J. Bacteriol. 194:6006–6007. 10.1128/JB.01423-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Depardieu F, Bonora MG, Reynolds PE, Courvalin P. 2003. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 50:931–948. 10.1046/j.1365-2958.2003.03737.x [DOI] [PubMed] [Google Scholar]
- 41.Carias LL, Rudin SD, Donskey CJ, Rice LB. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosvoll TC, Pedersen T, Sletvold H, Johnsen PJ, Sollid JE, Simonsen GS, Jensen LB, Nielsen KM, Sundsfjord A. 2010. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTβ-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol. Med. Microbiol. 58:254–268. 10.1111/j.1574-695X.2009.00633.x [DOI] [PubMed] [Google Scholar]
- 43.Nallapareddy SR, Wenxiang H, Weinstock GM, Murray BE. 2005. Molecular characterization of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J. Bacteriol. 187:5709–5718. 10.1128/JB.187.16.5709-5718.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.