Abstract
Escherichia coli sequence type 131 (ST131), a widely disseminated multidrug-resistant extraintestinal pathogen, typically exhibits serotype O25b:H4. However, certain ST131 isolates exhibit serotype O16:H5 and derive from a phylogenetic clade that is distinct from the classic O25b:H4 ST131 clade. Both clades are assigned to ST131 by the Achtman multilocus sequence typing (MLST) system and a screening PCR assay that targets ST131-specific sequence polymorphisms in the mdh and gyrB genes. However, they are classified as separate STs by the Pasteur Institute MLST system, and an ST131 PCR method that targets the O25b rfb region and an ST131-specific polymorphism in pabB detects only the O25b-associated clade. Here, we describe a novel PCR-based method that allows for rapid and specific detection of the O16-associated ST131 clade. The clade members uniformly contained allele 41 of fimH (type 1 fimbrial adhesin) and a narrow range of alleles of gyrA and parC (fluoroquinolone target genes). The virulence genotypes of the clade members resembled those of classic O25b:H4 ST131 isolates; representative isolates were variably lethal in a mouse subcutaneous sepsis model. Several pulsotypes spanned multiple sources (adults, children, pets, and human fecal samples) and locales. An analysis of recent clinical E. coli collections showed that the O16 ST131 clade is globally distributed, accounts for 1 to 5% of E. coli isolates overall, and, when compared with other ST131 isolates, it is associated with resistance to ampicillin, gentamicin, and trimethoprim-sulfamethoxazole and with susceptibility to fluoroquinolones and extended-spectrum cephalosporins. Attention to this O16-associated ST131 clade, which is facilitated by our novel PCR-based assay, is warranted in future epidemiological studies of ST131 and, conceivably, in clinical applications.
INTRODUCTION
Escherichia coli sequence type 131 (ST131), designated according to the Achtman multilocus sequence typing (MLST) system, has emerged dramatically over the past decade to become the single most prevalent human extraintestinal E. coli strain in many regions, especially among isolates resistant to fluoroquinolones and/or extended-spectrum cephalosporins (1–6). Although ST131 classically exhibits serotype O25b:H4, multiple studies have documented a minor subset of ST131 isolates with serotype O16:H5 or O-type O16 (7–12). This O16 subset within ST131 remains poorly characterized.
Among the extended-spectrum β-lactamase (ESBL)-producing E. coli isolates from Japan, the O16 ST131 subset was recently shown to represent a different Pasteur Institute sequence type (PST506) than classic O25b ST131 isolates (PST43) and to constitute a distinct phylogenetic clade (10, 11). Additionally, compared with classic O25b ST131 clade members, the studied O16 ST131 clade members exhibited a relatively greater prevalence of CTX-M-14 over (ST131-associated) CTX-M-15, were less commonly fluoroquinolone resistant but more commonly trimethoprim-sulfamethoxazole resistant, and escaped detection by a widely used PCR screening assay for ST131 that targets the O25b rfb (O lipopolysaccharide) allele and the housekeeping gene pabB (10, 11). Two O16 ST131 clade members also have been reported among 47 ESBL-producing ST131 clinical isolates from Lugo, Spain (12), and one among 12 ESBL-producing ST131 fecal isolates from children attending day care centers in southeastern France (13).
Whether these findings apply outside these three countries or to non-ESBL-producing ST131 isolates is unknown. Additional unknowns regarding O16 ST131 strains include their virulence genotypes, in vivo virulence, geographical distribution, and prevalence, both overall and in relation to the resistance phenotype. Accordingly, we extensively characterized a panel of diverse-source O16 E. coli isolates that had been presumptively identified as ST131 according to the gyrB and mdh ST131 screening PCR assay. We also surveyed several large collections of clinical E. coli isolates to assess the prevalence, geographical distribution, and antimicrobial resistance characteristics of the O16 ST131 subset.
MATERIALS AND METHODS
Isolates and genomes.
The 24 principal O16 ST131 study isolates were selected as a convenience sample based on previous PCR results showing the presence of the O16 rfb allele and ST131-specific single-nucleotide polymorphisms (SNPs) in gyrB and mdh (see below). They were taken from various collections of E. coli clinical isolates that had been assembled according to diverse selection criteria (e.g., 14, 15) and derived from 8 widely distributed cities in 5 U.S. states plus The Netherlands and Australia. The hosts included human adults and children and a dog; the dates of isolation ranged from 2006 through 2011.
For comparison, 10 other strains from subgroup I of phylogenetic group B2 (which includes ST131 [16]) were analyzed. These included 5 classic O25b ST131 isolates representing both extraintestinal pathogenic E. coli (ExPEC) and diarrheal pathotypes, 2 O-nontypeable (O-NT) ST131 isolates, and 3 non-ST131 isolates (1 each from O4, O16, and O43) (16). Also analyzed were archetypal group B2 strain E. coli CFT073 (ST73, B2 subgroup II [16]) and published genomes from ST131 strains E. coli SE15 (commensal, Japan) (17) and NA114 (extraintestinal clinical isolate, India) (18).
The prevalence and resistance characteristics of the O16 ST131 clade were assessed using two large collections of recent E. coli clinical isolates, i.e., (i) 1,475 consecutive clinical E. coli isolates from veterans at the Minneapolis Veterans Affairs (VA) Medical Center (April 2012 to January 2014) and (ii) 2,764 E. coli clinical isolates from nine geographically and demographically diverse clinical centers in the United States and Europe (2010 to 2013). The latter collection included isolates from the Group Health Cooperative, Seattle, WA (n = 956), which serves predominantly ambulatory women; Children's Hospital, Seattle, WA (n = 294), serving children only; University of Washington, Seattle, WA (n = 200); Harborview Medical Center, Seattle, WA (n = 323); Veterans Affairs Medical Center, Minneapolis, MN (n = 110); and hospitals in Münster, Germany (n = 392) and Wroclaw, Poland (n = 199).
ST131 screening and O-type PCR, gyrA and parC analysis, fumC and fimH (CH) typing, and extended virulence genotyping.
ST131 status was defined presumptively based on PCR detection of ST131-specific SNPs in mdh and gyrB (19) and, separately, of the O25b rfb variant and ST131-specific SNPs in pabB (20). The O16 rfb variant was detected by O-type-specific PCR (21). Alleles of the type-1 fimbrial adhesin gene fimH and the genes encoding DNA gyrase (gyrA) and topoisomerase (parC) were determined by sequence analysis and comparison with private sequence libraries (14, 22). Alleles of fumC were determined according to the Achtman MLST website (http://mlst.ucc.ie/mlst/dbs/Ecoli). Combined fumC and fimH alleles were used to infer STs and fimH-based subdivisions thereof, i.e., CH types (22, 23). The fimH30 (H30) ST131 subclone also was detected using allele-specific PCR (4). The presence of 23 known or suspected extraintestinal virulence genes was defined by multiplex PCR (24).
Multilocus sequence typing and phylogenetic analysis.
Extensive MLST was performed on 37 strains, including the 24 principal O16 ST131 study isolates, the 10 comparison E. coli isolates from subgroup I of phylogroup B2, 2 ST131 strains for which complete genome sequences are available (E. coli SE15 and NA114; hereafter, “genome strains”), and reference strain E. coli CFT073, by using the genes from both the Achtman (adk, fumC, gyrB, icd, mdh, purA, and recA) (http://mlst.ucc.ie/mlst/dbs/Ecoli) and Pasteur Institute (icd, pabB, polB, putP, trpA, and trpB) (http://www.pasteur.fr/recherche/genopole/PF8/mlst/) schemes. This corresponds to 12 genes, with icd being common between the 2 schemes, and 8,928 nucleotides. Phylogenetic tree reconstruction was performed using the maximum likelihood procedure (25).
PCR assay for detection of the ST131-associated O16 clade.
For the novel O16-clade-specific PCR assay described here, PCR was done using a 20-μl volume containing 2 μl of 10× buffer (supplied with Taq polymerase), 10, 15, or 20 pmol of each primer (see below) (Table 1), 200 μM each deoxynucleoside triphosphates (dNTPs), 2 U of Taq polymerase (Promega, Charbonnières-les-Bains, France), 1.5 mM MgCl2 (final concentration), and 2 μl of purified DNA (approximately 100 ng/μl) or 3 μl of bacterial lysate (prepared as follows). The bacterial lysates were prepared by suspending 10 E. coli colonies in 500 μl H2O, which was heated 10 min at 95°C, chilled in ice, and centrifuged for 3 min at 14,000 × g; the supernatant was used for PCR. PCR was performed with an Eppendorf Mastercycler with MicroAm tubes, using the steps of denaturation for 4 min at 94°C, 30 cycles of 5 s at 94°C and 20 s at 63°C or 59°C (see below), and 5 min of final extension at 72°C.
TABLE 1.
Primer sequences and sizes of the amplified fragments
| Primer designation | Primer sequence | Target | Size of PCR product (bp) | Reference or source |
|---|---|---|---|---|
| trpAST131-O16.f | 5′-AAAACCGCGCCGCGTTACCT-3′ | trpA | 145 | This study |
| trpAST131-O16.r | 5′-CCAGAAATCGCGCCCGCATT-3′ | |||
| pabBST131-O25b.f | 5′-TCCAGCAGGTGCTGGATCGT-3′ | pabB | 347 | 20 |
| pabBST131-O25b.r | 5′-GCGAAATTTTTCGCCGTACTGT-3′ | |||
| uidA.F | 5′-CATTACGGCAAAGTGTGGGTCAAT-3′ | uidA | 657 | 26 |
| uidA.R | 5′-CCATCAGCACGTTATCGAATCCTT-3′ | |||
| gndbis.f | 5′-ATACCGACGACGCCGATCTG-3′ | gnd | 21 | |
| rfbO16.r | 5′-GGATCATTTATGCTGGTACG-3′ | rfbO16 | 450 | 21 |
| rfbO25b.r | 5′-TGCTATTCATTATGCGCAGC-3′ | rfbO25b | 300 | 20 |
In this assay, we combined detection of the O16 and O25b ST131 clades. Thus, for detecting the ST131-O16 and ST131-O25b clades (Fig. 1A), the primers used were pabBST131-O25b.f and pabBST131-O25b.r (20 pmol each) (20), trpAST131-O16.f and trpAST131-O16.r (15 pmol each) (novel), and uidA.f and uidA.r (15 pmol each) (26), all in the same reaction (Table 1), with annealing at 63°C. For detecting the O16 and O25b rfb variants (Fig. 1B), the primers used were gndbis.f, rfbO16, and rfbO25b (20 pmol each) (20, 21), plus uidA.f and uidA.r (10 pmol each) (26), all in the same reaction (Table 1), with annealing at 59°C. The PCR products were loaded on 2% agarose gel with SYBR safe DNA gel stain (Invitrogen, Cergy Pontoise, France). After electrophoresis, the gels were photographed under UV light.
FIG 1.
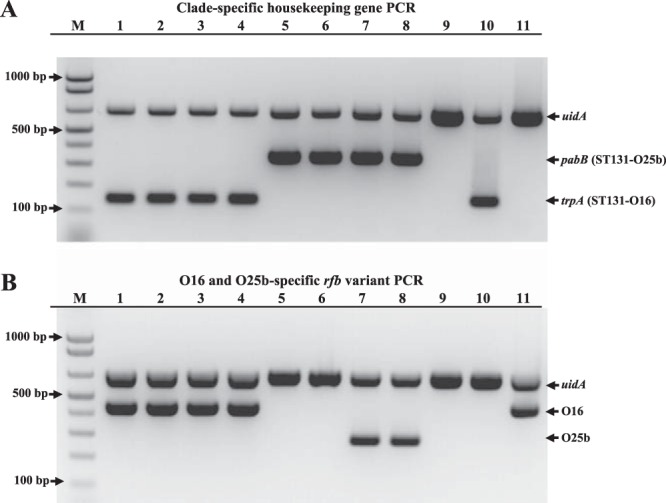
Allele-specific PCR for detection of the ST131-O16 and ST131-O25b clades. (A)), clade-specific PCR; (B), PCR O typing. M, molecular weight marker (1 Kb Plus DNA Ladder; Invitrogen). (A and B) Upper band corresponds to the amplification of the internal control uidA gene. (A) Middle band and lower bands correspond to allele-specific amplifications of the ST131-O25b clade pabB and ST131-O16 clade trpA variants, respectively. (B) Middle band and lower bands correspond to allele-specific amplifications of the O16 and O25b rfb regions, respectively. (A and B) Lanes 1 to 4, strains JJ30, JJ38, JJ59, and SE15 (all ST131-O16 clade, O16 type), respectively; lanes 5 and 6, strains JJ14 and JJ20 (both ST131-O25b clade, O nontypeable), respectively; lanes 7 and 8, strains TN03 and TU (both ST131-O25b clade, O25b type), respectively; lane 9, strain ECOR66 (non-ST131, O4 type); lane 10, strain LBC24a (non-ST131, O43 type), noting the amplification of the ST131-O16 clade trpA gene due to presumed horizontal gene transfer; lane 11, strain M1139 (non-ST131, O16 type).
This detection method for the ST131-O16 clade was validated using four sets of strains. The first encompassed 145 phylogroup B2 strains for which Pasteur Institute MLST results were available (27). This strain set, representative of the B2 phylogroup diversity (16), was composed of 15 strains from the E. coli reference (ECOR) collection (16), 22 strains from the Broad collection (http://www.broadinstitute.org/annotation/genome/escherichia_antibiotic_resistance/GenomeDescriptions.html), 14 strains with a fully sequenced genome available, and 94 strains from our collections (28). It encompasses one ST131-O16 lineage strain, i.e., strain SE15 (17), and 6 ST131-O25b strains. The second strain set encompassed 26 ST131 strains (24 of the O16 lineage, 2 of the O25b lineage) from which an extensive MLST (12 genes, 8,928 nucleotides) was available. The third strain set encompassed 23 additional ST131-O16 strains, as identified using the mdh-gyrB SNP typing and O16b rfb PCR. The fourth strain set encompassed 188 non-phylogroup B2 strains, selected randomly from the 2,764-member international collection that was used to assess the prevalence and resistance characteristics of the O16 subgroup, as described above.
Mouse subcutaneous sepsis model.
Intrinsic extraintestinal virulence was tested in a mouse model of sepsis following subcutaneous inoculation in the neck of 2 × 108 bacteria (29). Ten mice were inoculated per strain, and death was monitored as an outcome for up to 7 days after inoculation. In each experiment, two E. coli control strains were systematically tested: K-12 strain E. coli MG1655, which does not kill mice, and strain CFT073, which kills 100% of inoculated mice (24). The animal experiments were performed in compliance with the recommendations of the French Ministry of Agriculture and were approved by the French Veterinary Services (accreditation A 75-18-05).
Pulsed-field gel electrophoresis.
XbaI pulsed-field gel electrophoresis (PFGE) profiles were generated according the PulseNet protocol (30). By using BioNumerics (Bio-Rad), profiles were assigned to pulsotypes based on ≥94% similarity to the established reference ST131 profiles within a large private PFGE profile library (e.g., 15). Dendrograms based on Dice similarity coefficients were inferred according to the unweighted pair group method.
Statistical methods.
Comparisons of proportions were tested using Fisher's exact test.
RESULTS
MLST.
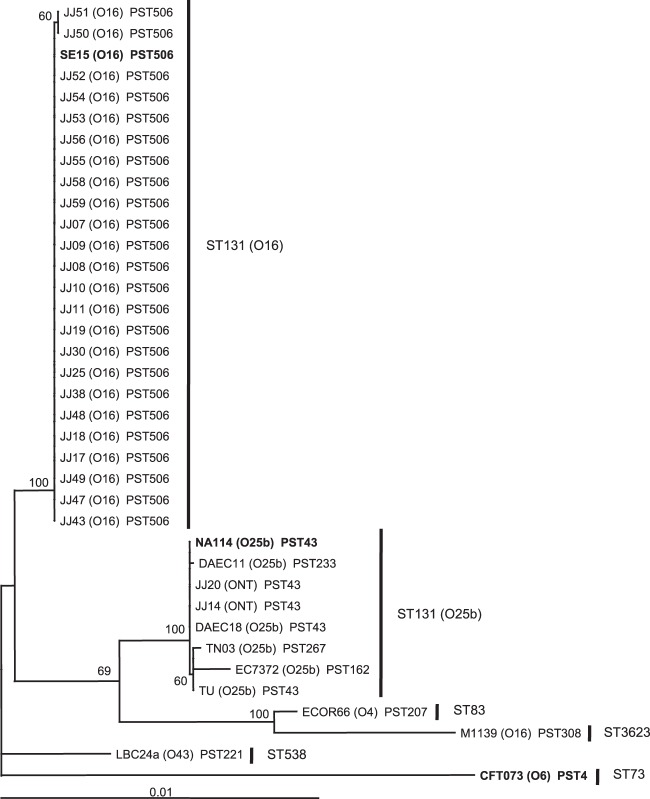
All 24 principal O16 ST131 study isolates plus genome strain SE15 (which was found to have an O16 rfb locus, not O150, as has been published) were confirmed to belong to ST131 according to Achtman MLST and to PST506 according to Pasteur Institute MLST (Fig. 2). In contrast, the seven O25b and O-nontypeable ST131 isolates, as well as genome strain NA114, although confirmed to belong to ST131 by Achtman MLST, were found by Pasteur Institute MLST to represent PST043 (n = 5), or single-locus variants thereof, i.e., PST162, PST233, or PST267, all of which fall within subgroup I of group B2 (16).
FIG 2.
Phylogenetic tree of 36 E. coli strains from phylogenetic group B2, subgroup I. Subgroup I of phylogenetic group B2 was defined as in Le Gall et al. (16). The tree was inferred from the DNA sequences of 12 housekeeping genes from the Achtman and Pasteur Institute schemes (8,928 nucleotides), using the maximum likelihood procedure. E. coli strain CFT073 (group B2, subgroup II [16]) served as an outgroup. Bootstrap values of >50% are indicated adjacent to the nodes. The strains in bold correspond to sequenced strains. PST, Pasteur Institute sequence type (ST). The other ST labels correspond to the Achtman MLST scheme.
Of the two PCR-based screening methods for ST131, the one targeting SNPs in mdh and gyrB (19) detected all ST131 study isolates, whether they belonged to O16 or O25b/O-nontypeable (NT). In contrast, the one based on pabB and the O25b rfb variant (20) detected only the O25b (both PCRs positive) and O-NT (only pabB PCR positive) ST131 isolates, giving a negative result with the O16 isolates.
Development and validation of a rapid PCR assay for detection of the O16 ST131 clade.
Primers specific for the O16 ST131 subset were derived as follows. Using MLST gene sequences of the Pasteur Institute scheme from the panel of B2 ST131 strains cited above and the 138 nonredundant B2 strains described in Materials and Methods, we searched for SNPs present only in the ST131 O16 clade strains. We choose two appropriately spaced SNPs in trpA and then positioned one at each 3′ end of two primers that together amplified a 145-bp fragment (Table 2), a size easily distinguishable from the 347-bp fragment corresponding to the allele-specific amplicon for the ST131 O25b clade-associated pabB variant (20). The PCR conditions were optimized empirically to give a robust PCR product with positive-control strains and no product with negative-control strains (Fig. 1A).
TABLE 2.
Prevalence of antimicrobial resistance in relation to ST131 subgroup among 1,475 clinical Escherichia coli isolates from U.S. veterans, Minneapolis, MN (2012 to 2013)
| Drug | No. (%) of resistant strains by sequence type |
P by subgroupa |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ST131 (n = 378) |
Non-ST131 (n = 1,097) | |||||||||
| O16 (H41) (n = 32) | O25b (n = 346) |
Within ST131 |
ST131 vs non-ST131 |
|||||||
| H30 (n = 310) | Non-H30 (n = 36)b | O16 vs H30 | O16 vs non-H30b | H30 vs non-H30 | O16 vs non-ST131 | H30 vs non-ST131 | Non-H30 vs non-ST131 | |||
| Ampicillin | 30 (94) | 253 (82) | 22 (61) | 395 (36) | 0.09 | <0.001 | 0.008 | <0.001 | <0.001 | 0.004 |
| Gentamicin | 10 (31) | 61 (20) | 6 (17) | 43 (4) | <0.001 | 0.001 | 0.009 | |||
| TMP-SMZc | 21 (66) | 129 (42) | 13 (36) | 184 (17) | 0.01 | 0.03 | <0.001 | <0.001 | 0.006 | |
| Ciprofloxacin | 6 (19) | 304 (98) | 13 (36) | 128 (12) | <0.001 | <0.001 | <0.001 | <0.001 | ||
| Ceftriaxone | 0 (0) | 52 (17) | 1 (3) | 33 (3) | 0.008 | 0.03 | <0.001 | |||
P values (by Fisher's exact test) are shown where P is <0.10.
Non-H30 refers to ST131 subclones other than the O16 and H30 ST131 subclones.
TMP-SMZ, trimethoprim-sulfamethoxazole.
The new clade-specific PCR method was validated using (i) three strain sets containing 194 total phylogroup B2 isolates, including 48 ST131-O16 isolates and 8 ST131-O25b isolates, plus (ii) a fourth strain set containing 186 non-phylogroup B2 isolates. The sensitivity of the method for detecting the O16 ST131 subset was 100%, as it detected all 48 ST131-O16 strains in the three group B2 validation sets. Its specificity was >99%, as only one non-O16 non-ST131 strain, E. coli LBC24a (from group B2, subgroup I), was misclassified into the O16 ST131 clade (Fig. 1). Indeed, this strain belongs to Achtman ST538 (PST221) and to serogroup O43; however, its trp operon sequence is of the ST131-O16 type (data not shown), presumably due to horizontal gene transfer.
To this clade-specific PCR, we added O-typing PCR, based on the principle previously described (21), using primers specific for the O16 and O25b rfb regions. With this combination of 2 PCRs, strains of both the ST131-O16 and ST131-O25b clades were easily identified, together with their characteristic O type (Fig. 1B). In both PCR assays, we included an internal control, uidA, allowing for a confirmation of the efficacy of the PCR (Fig. 1). Isolates that yielded discrepant results between the two PCRs (e.g., strain LBC24a, which gave an O16 ST131-specific trpA PCR result but was negative in the O16 O-type PCR assay) (Fig. 1) can be investigated further, e.g., with dual-SNP ST131-specific PCR (19), fumC-fimH (CH) typing (22), or full MLST.
Virulence genotypes.
The virulence genes detected in most or all of the 24 principal O16 ST131 study isolates included kpsM II, usp, ompT, malX, fyuA, and irp2 (n = 24), traT and iha (n = 23), and sat and iutA (n = 19). In contrast, afaD (n = 11), papC, hlyD, and cnf1 (n = 2), and papGII, papGIII, and hra (n = 1) appeared only sporadically. No isolate contained neuC, papGI, sfa/foc, iroN, ireA, or ibeA.
Similar results were observed with the 7 compared O25b-positive (or O25b-negative pabB allele-positive) ST131 isolates and genome strain NA114, which uniformly contained kpsM II, iha, sat, usp, ompT, malX, fyuA, and traT but lacked sfa/foc, iroN, papGI, papGIII, cnf1, hra, and ireA. Sporadically occurring virulence genes included afaD (n = 3), papC, papGII, and ibeA (n = 2), and hlyD (n = 1).
fimH, gyrA, and parC alleles.
For fimH, all 25 (100%; 95% confidence interval [CI], 87% to 100%) O16 ST131 study isolates, including the 24 principle O16 ST131 isolates and (O16) ST131 genome strain SE15, contained fimH41 or a single-nucleotide variant thereof (see Table S1 in the supplemental material). In contrast, none of the 11 other group B2 subgroup I study isolates, including 8 non-O16 ST131 study isolates, (O25b) ST131 genome strain NA114, and 3 non-ST131 subgroup I isolates, contained fimH allele 41 (0%; 95% CI, 0% to 28%: P < 0.001 versus the O16 ST131 isolates). Instead, they contained fimH30 (n = 5), fimH22 (n = 3), fimH5, fimH20, and fimH21 (n = 1 each) (see Table S1 in the supplemental material).
For gyrA and parC, among the 24 principal O16 ST131 study isolates, the three fluoroquinolone-resistant isolates all had gyrA allele 1AB, containing two replacement mutations, and parC allele 3A, a recombined variant containing one replacement mutation, whereas the 21 fluoroquinolone-susceptible isolates had gyrA allele 1 (wild type, n = 8), 1A (one replacement mutation, n = 12), or 1B (a different replacement mutation, n = 1), each combined with parC allele 1 (wild type, n = 1) or 1b (one silent mutation, n = 20). Genome strain SE15 was wild type for both gyrA and parC. In contrast, four of the non-O16 ST131 study isolates and genome strain NA114 (all fluoroquinolone resistant) contained gyrA allele 1AB and parC allele 1aAB (63%; versus none [0%] of 25 O16 ST131 isolates; P < 0.001), whereas the remaining three non-O16 ST131 isolates (all fluoroquinolone susceptible) contained wild-type gyrA and parC alleles (37%; versus 2 [8%] of 25 O16 ST131 isolates; P = 0.08).
Experimental virulence.
Five of the O16 ST131 study isolates and genome strain SE15, selected for virulence genotype diversity, were assessed for experimental virulence in the mouse sepsis model, using 10 mice per isolate. All six isolates killed 6 to 10 mice each. One isolate (E. coli JJ2591) was exceptionally rapidly lethal, killing all 10 mice within 22 h.
The proportion of mice that were killed and the rapidity of the lethality did not correspond with the assessed virulence gene content. For example, the rapidly lethal strain JJ2591 had an unexceptional ST131-like virulence gene profile (containing iha, usp, ompT, malX, irp2/fyuA, and traT genes). In contrast, the two least virulent strains, which killed only 6 mice each, contained all these genes plus iutA and afa, or all these genes except malX, plus iutA, kpsM II, and sat.
For comparison, three classic O25b ST131 isolates and two non-ST131 group B2 isolates from subgroup I were assessed in parallel for their lethal effects on mice. All five strains killed all 10 challenged mice, albeit not as quickly as did E. coli O16 ST131 strain JJ2591.
Prevalence and antimicrobial susceptibility of the O16 ST131 subgroup.
The prevalence and antimicrobial susceptibility characteristics of the O16 ST131 subgroup were assessed using two large epidemiological populations (Table 1 and 3). Within the 1,475-member Minneapolis VA-based collection (2012 to 2013), the O16 ST131 subgroup (as identified based on the mdh and gyrB ST131 PCR assay plus the O16 O-type PCR assay) accounted for 31 (2.2%) of the isolates overall, compared with 311 (21%) for the (fluoroquinolone-resistant ESBL-associated) fimH30 ST131 subclone (14, 31) and 36 (2.4%) for other ST131 subclones combined. The O16 ST131 subclone isolates exhibited distinctive resistance prevalence values, whether compared with the other ST131 subgroups (i.e., borderline or significantly higher for ampicillin, gentamicin, and trimethoprim-sulfamethoxazole (TMP-SMZ), and lower for ciprofloxacin and ceftriaxone) or the non-ST131 isolates (i.e., significantly higher for ampicillin, gentamicin, and TMP-SMZ) (Table 1).
TABLE 3.
Prevalence of antimicrobial resistance in relation to ST131 subgroup among 2,474 clinical Escherichia coli isolates from diverse centers (2010 to 2013)a
| Drug | No. (%) of resistant strains by sequence type |
P by subgroupb |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ST131 (n = 347) |
Non-ST131 (n = 2,127) | |||||||||
| O16 (H41) (n = 61) | O25b (n = 286) |
Within ST131 |
ST131 vs non-ST131 |
|||||||
| H30 (n = 233) | Non-H30 (n = 53)c | O16 vs H30 | O16 vs non-H30 | H30 vs non-H30 | O16 vs non-ST131 | H30 vs non-ST131 | Non-H30 vs non-ST131 | |||
| Ampicillin | 53 (87) | 206 (88) | 38 (72) | 1,025 (48) | 0.06 | 0.004 | <0.001 | <0.001 | <0.001 | |
| Gentamicin | 27 (44) | 61 (26) | 3 (6) | 130 (6) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |
| TMP-SMZd | 35 (57) | 119 (51) | 12 (23) | 551 (26) | <0.001 | <0.001 | <0.001 | <0.001 | ||
| Ciprofloxacin | 4 (7) | 214 (92) | 5 (9) | 306 (14) | <0.001 | <0.001 | 0.09 | <0.001 | ||
| Ceftriaxone | 8 (13) | 58 (25) | 5 (9) | 167 (8) | 0.06 | 0.016 | <0.001 | |||
The contributing centers (no. of isolates) include: Group Health Cooperative, Seattle, WA (956), Children's Hospital, Seattle, WA (294), University of Washington, Seattle, WA (200), Harborview Medical Center, Seattle, WA (323), Veterans Affairs Medical Center, Minneapolis, MN (110), Münster, Germany (392), and Wroclaw, Poland (199).
P values (by Fisher's exact test) are shown where P is <0.10.
Non-H30 refers to ST131 subclones (predominantly H22) other than the O16 and H30 ST131 subclones.
TMP-SMZ, trimethoprim-sulfamethoxazole.
Similarly, within the 2,764-member multicenter international collection (2010 to 2013), the O16 ST131 subgroup, as identified by CH typing based on presence of fumC allele 40 and fimH allele 41, accounted for 2.5% of the isolates overall (range, 0.5% to 5.6% per center), compared with 9.7% for H30 subclone ST131 isolates and 2.1% for all other (mainly H22 subclone) ST131 isolates. Susceptibility data were available for 2,474 (89.5%) of these isolates. The O16 ST131 subgroup again exhibited distinctive resistance prevalence values, whether compared with other ST131 subgroups (i.e., borderline or significantly higher for ampicillin, gentamicin, and TMP-SMZ, and lower for ciprofloxacin and ceftriaxone) or non-ST131 isolates (i.e., significantly higher for ampicillin, gentamicin, and TMP-SMZ and borderline lower for ciprofloxacin) (Table 3).
Pulsotype comparisons.
According to XbaI PFGE, the 24 principal O16 ST131 study isolates represented 12 distinct pulsotypes. These isolates, along with the index isolates from all 280 unique ST131-associated pulsotypes within a private PFGE library (containing 1,292 total ST131 isolates from diverse locales, ecological sources, and time periods) (e.g., 15), were used to construct a PFGE dendrogram. Based on the proximity within this dendrogram, 20 different O16 ST131-containing pulsotypes, with 49 isolates total, were identified. Of these pulsotypes, 7 contained multiple (i.e., 2 to 10) isolates; 2 were among the 15 most-prevalent ST131-associated pulsotypes in the library. The 6 pulsotypes with ≥3 isolates each were geographically diverse; 5 included representatives from 2 or 3 continents (North America, Europe, and/or Australia). Three of the multiple-isolate pulsotypes (with 2, 8, and 10 isolates each) included 1 to 2 clinical isolates from children, one (with 3 isolates) included a dog urine isolate, and one (with 10 isolates) included two human stool isolates.
Phylogenetic relationships.
To clarify phylogenetic relationships between and within the two main ST131 components (O16 and O25b), in comparison with non-ST131 subgroup I isolates, we used the concatenated gene sequences from Achtman and Pasteur Institute MLST schema (12 loci; 8,928 bp) to infer a maximum likelihood phylogenetic tree for the 25 principal O16 ST131 study isolates (including SE15), 8 diverse O25b and O-NT ST131 isolates (including NA114), and three non-ST131 isolates also from B2 subgroup I (one containing the O16 rfb allele, another containing the O16 ST131-associated trpA allele), rooting the tree with archetypal strain CFT073 (Fig. 2). This tree illustrated the extensive phylogenetic homogeneity among the O16 ST131 isolates, a lesser but still considerable phylogenetic homogeneity among the O25b and O-NT ST131 isolates, and the marked separation of these two clades, with the O16 clade appearing basal. Two of the 3 non-ST131 subgroup I isolates formed a separate highly supported clade.
DISCUSSION
In this molecular-epidemiological analysis of the O16 subgroup within E. coli ST131, we confirmed its considerable phylogenetic homogeneity, distinctness from the conventional O25b/O-NT ST131 isolates, and failure to be detected by two widely used screening assays for ST131, which either miss the clade (20) or fail to distinguish it from O25b ST131 strains (19), which represent mainly the H30 (fluoroquinolone-resistant) and H22 (broadly susceptible) ST131 subclones (31). We also generated novel evidence of the O16 ST131 clade's broad geographic distribution and host range (including for individual pulsotypes within the clade), associations with clinically relevant resistance phenotypes, conserved virulence genotypes, in vivo virulence in a mouse sepsis model, and tight associations with fimH41 and specific alleles of (fluoroquinolone resistance-associated) gyrA and parC. Finally, we developed a rapid and simple multiplex PCR assay that allows for simultaneous detection and differentiation of this clade from the dominant O25b subset within ST131.
Our findings regarding phylogenetic relationships within ST131 agree with those of previous analyses based on MLST (10) and whole-genome sequencing (31) that identify the O16-associated subgroup as a basal clade within the larger ST131 lineage. Our findings also confirm the close associations of this clade with the O16 rfb variant and fimH41 (14, 31), which implies that the detection of either of these traits in a known or suspected ST131 isolate reliably identifies the isolate as a clade member.
We report here the first broad survey for this clade among unselected clinical isolates, as previous studies of this clade focused on ESBL-producing isolates from a single region (10–12), diverse convenience sample isolates (14, 31), or all-comer isolates from a single locale (8, 9, 32). Our findings document that at multiple centers across the United States and Europe, the clade consistently accounts for approximately 1 to 5% of the E. coli clinical isolates overall and is associated with resistance to ampicillin, gentamicin, and TMP-SMZ but, especially in comparison with H30 ST131 isolates, with susceptibility to fluoroquinolones and extended-spectrum cephalosporins.
We likewise provide evidence that many members of the O16 ST131 clade are virulent in a mouse model of subcutaneous sepsis, and some extremely so, in agreement with a recent study on 4 Spanish O16 ST131 isolates (33). This is consistent with the extensive virulence genotypes of clade members, which closely resemble those of conventional O25b ST131 isolates. These observations suggest substantial virulence potential for humans, which is supported by the epidemiological finding of Kudinha et al. (8, 9), in both women and men, of an ascending prevalence gradient for O16 ST131 isolates in relation to clinical severity, from fecal isolates from healthy hosts, through cystitis isolates, to pyelonephritis isolates, implying enhanced virulence for this clade relative to other E. coli.
To date, although the O16 ST131 clade has not achieved the same level of clinical prominence as its (mainly H30 subclone) O25b-associated ST131 counterparts, it appears to be second in prevalence within ST131 after H30 (with its CTX-M-15 ESBL-associated H30-Rx subset [Rx, extensively resistant]) (14, 31, 34), equaling or exceeding the prevalence of the next-most-prevalent ST131 subclone, H22 (14, 23, 31). Moreover, the clade is sufficiently prevalent and widespread, and it exhibits a sufficiently distinctive resistance profile, as to make its detection potentially useful for clinical management (23). For example, within each of the two studied large clinical collections, the resistance prevalence differences between the three main subsets within ST131 (O16, H30, and others) and non-ST131 isolates were such that depending on the cut point used to determine the suitability of an agent for empirical use, knowledge of an clonal subset identity of an isolate would allow versus disallow the use of certain drugs (Tables 1 and 3). For example, for the VA-based collection (Table 1), with a 20% resistance prevalence threshold (as for cystitis), TMP-SMZ would be disallowed for O16 ST131 isolates but allowed for non-ST131 isolates, whereas ciprofloxacin would be allowed for O16 ST131 isolates and non-ST131 isolates but disallowed for H30 and other ST131 isolates. Similarly, with a 10% resistance threshold (as for pyelonephritis or urosepsis), ceftriaxone would be disallowed for H30 ST131 isolates but allowed for O16 ST131 isolates, non-H30/non-O16 ST131 isolates, and non-ST131 isolates (Table 1).
Because of the potential clinical importance and absence of a direct screening test for the O16 ST131 subset, we developed here a simple and specific PCR-based method to detect this clade and to distinguish it from the (predominantly H30 subclone) O25b ST131 subset. This novel PCR assay, which was used to screen E. coli isolates without knowledge of their phylogroup status, was highly robust, yielding strong bands with positive controls and no amplification with negative controls. Supplementation of the single-tube housekeeping gene component, which detects clade-specific SNPs in trpA and pabB, with a single-tube rfb component, which detects both the O16 and O25b rfb variants, confirms the cladal assignments or, in case of discrepancies between the two reactions, identifies isolates with possible false-positive trpA results due to horizontal gene transfer. This assay should greatly facilitate the epidemiological studies needed to better understand the geographical distribution, host range, syndrome capabilities, and transmission pathways of the O16 ST131 subset, and conceivably might help guide clinical management (23). Alternative but progressively more elaborate approaches to identifying the O16 ST131 subgroup include dual-SNP ST131 PCR (19) combined with O16 rfb detection (21), CH typing (22, 23), Pasteur Institute MLST, and whole-genome sequencing.
In summary, we found that the ST131 O16 subset, which is not reliably or specifically detected by current PCR screening assays for ST131, is clinically important (based on its prevalence, geographic distribution, host range, and distinctive resistance profiles) and virulent in mice. Although identical or highly similar to other ST131 isolates according to Achtman MLST and virulence gene profiles, it differs markedly from them according to Pasteur Institute MLST, fimH allele, pulsotypes, and resistance profiles. We describe a novel PCR screening assay for rapid and simple detection of this O16 ST131 clade for use in future epidemiological studies and, potentially, clinical applications.
Supplementary Material
ACKNOWLEDGMENTS
This material is based upon work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs, grant 1 I01 CX000192 01 (to J.R.J.) and NIH grant RC4-AI092828 (to E.S. and J.R.J.).
Isolates and associated data were provided by Ritu Banerjee, Gio J. Baracco, Roger Bedimo, David Gordon, Jan Kluytmans, JoAnn Platell, Ari Robicsek, Carl Urban, and Scott Weissman.
J.R.J. has received research grants and/or contracts from Merck, Rochester Medical, ICET, and Syntiron and has patent applications relating to diagnostic tests for ST131 and other E. coli lineages. E.S. has patent applications relating to diagnostic tests for ST131 and other E. coli lineages. The other authors report no financial conflicts of interest.
Footnotes
Published ahead of print 5 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03502-13.
REFERENCES
- 1.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14. 10.1093/jac/dkq415 [DOI] [PubMed] [Google Scholar]
- 2.Peirano G, Pitout JD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents. 35:316–321. 10.1016/j.ijantimicag.2009.11.003 [DOI] [PubMed] [Google Scholar]
- 3.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294. 10.1086/653932 [DOI] [PubMed] [Google Scholar]
- 4.Colpan A, Johnston B, Porter S, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson JR, VICTORY (Veterans Influence of Clonal Types on Resistance: Year 2011) Investigators 2013. Escherichia coli sequence type 131 (ST131) as an emergent multidrug-resistant pathogen among U.S. veterans. Clin. Infect. Dis. 57:1256–1265. 10.1093/cid/cit503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028. 10.1093/jac/dkn084 [DOI] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281. 10.1093/jac/dkm464 [DOI] [PubMed] [Google Scholar]
- 7.Platell JL, Cobbold RN, Johnson JR, Heisig A, Heisig P, Clabots C, Kuskowski MA, Trott DJ. 2011. Commonality among fluoroquinolone-resistant sequence type ST131 extraintestinal Escherichia coli isolates from humans and companion animals in Australia. Antimicrob. Agents Chemother. 55:3782–3787. 10.1128/AAC.00306-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Escherichia coli sequence type 131 (ST131) as a prominent cause of antimicrobial resistance among clinical and fecal E. coli isolates from reproductive-age women. J. Clin. Microbiol. 51:3270–3276. 10.1128/JCM.01315-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Distribution of phylogenetic groups, sequence type ST131, and virulence-associated traits among Escherichia coli isolates from men with pyelonephritis or cystitis and healthy controls. Clin. Microbiol. Infect. 19:E173–E180. 10.1111/1469-0691.12123 [DOI] [PubMed] [Google Scholar]
- 10.Matsumura Y, Yamamoto M, Nagao M, Hotta G, Matsushima A, Ito Y, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group 2012. Emergence and spread of B2-ST131-O25b, B2-ST131-O16 and D-ST405 clonal groups among extended-spectrum-β-lactamase-producing Escherichia coli in Japan. J. Antimicrob. Chemother. 67:2612–2620. 10.1093/jac/dks278 [DOI] [PubMed] [Google Scholar]
- 11.Matsumura Y, Yamamoto M, Nagao M, Ito Y, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group 2013. Association of fluoroquinolone resistance, virulence genes, and IncF plasmids with extended-spectrum-β-lactamase-producing Escherichia coli sequence type 131 (ST131) and ST405 clonal groups. Antimicrob. Agents Chemother. 57:4736–4742. 10.1128/AAC.00641-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahbi G, Mora A, López C, Alonso MP, Mamani R, Marzoa J, Coira A, García-Garrote F, Pita JM, Velasco D, Herrera A, Viso S, Blanco JE, Blanco M, Blanco J. 2013. Emergence of new variants of ST131 clonal group among extraintestinal pathogenic Escherichia coli producing extended-spectrum β-lactamases. Int. J. Antimicrob. Agents 42:347–351. 10.1016/j.ijantimicag.2013.06.017 [DOI] [PubMed] [Google Scholar]
- 13.Blanc V, Leflon-Guibout V, Blanco J, Haenni M, Madec J-Y, Rafignon G, Bruno P, Mora A, Lopez C, Dahbi G, Dunais B, Anastay M, Branger C, Moreau R, Pardier C, Nicolas-Chanoine M-H. 8 January 2014. Prevalence of day-care centre children (France) with faecal CTX-M-producing Escherichia coli comprising O25b:H4 and O16:H5 ST131 strains. J. Antimicrob. Chemother. 10.1093/jac/dkt519 [DOI] [PubMed] [Google Scholar]
- 14.Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddel K, Rogers P, Qin X, Butler-Wu S, Price LB, Aziz M, Nicolas-Chanoine MH, Debroy C, Robicsek A, Hansen G, Urban C, Platell JL, Trott DJ, Zhanel G, Weissman SJ, Cookson BT, Fang FC, Limaye AP, Scholes D, Chattopadhyay S, Hooper DC, Sokurenko EV. 2013. Abrupt emergence of a single dominant multi-drug-resistant strain of Escherichia coli. J. Infect. Dis. 207:919–928. 10.1093/infdis/jis933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JR, Nicolas-Chanoine MH, DebRoy C, Castanheira M, Robiscek A, Hansen G, Weissman SJ, Urban C, Platell J, Trott D, Zhanel G, Clabots C, Johnston BD, Kuskowski MA, MASTER Investigators 2012. Comparison of Escherichia coli sequence type ST131 pulsotypes by epidemiologic traits, 1967–2009. Emerg. Infect. Dis. 18:598–607. 10.3201/eid1804.111627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Gall T, Clermont O, Gouriou S, Picard B, Nassif X, Denamur E, Tenaillon O. 2007. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol. Biol. Evol. 24:2373–2384. 10.1093/molbev/msm172 [DOI] [PubMed] [Google Scholar]
- 17.Toh H, Oshima K, Toyoda A, Ogura Y, Ooka T, Sasamoto H, Park SH, Iyoda S, Kurokawa K, Morita H, Itoh K, Taylor TD, Hayashi T, Hattori M. 2010. Complete genome sequence of the wild-type commensal Escherichia coli strain SE15, belonging to phylogenetic group B2. J. Bacteriol. 192:1165–1166. 10.1128/JB.01543-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avasthi TS, Kumar N, Baddam R, Hussain A, Nandanwar N, Jadhav S, Ahmed N. 2011. Genome of multidrug-resistant uropathogenic Escherichia coli strain NA114 from India. J. Bacteriol. 193:4272–4273. 10.1128/JB.05413-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002–2004. Antimicrob. Agents Chemother. 53:2733–2739. 10.1128/AAC.00297-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Mulvey MR, Nordmann P, Ruppé E, Sarthou JL, Frank T, Vimont S, Arlet G, Branger C, Woodford N, Denamur E. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 64:274–277. 10.1093/jac/dkp194 [DOI] [PubMed] [Google Scholar]
- 21.Clermont O, Johnson JR, Menard M, Denamur E. 2007. Determination of Escherichia coli O types by allele-specific polymerase chain reaction: application to the O types involved in human septicemia. Diagn. Microbiol. Infect. Dis. 57:129–136. 10.1016/j.diagmicrobio.2006.08.007 [DOI] [PubMed] [Google Scholar]
- 22.Weissman SJ, Johnson J, Tchesnokova V, Billig M, Dykhuizen D, Riddel K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Chattopadhyay S, Sokurenko E. 2012. High-resolution two-locus clonal typing of extraintestinal Escherichia coli. Appl. Environ. Microbiol. 78:1353–1360. 10.1128/AEM.06663-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tchesnokova V, Billig M, Chattopadhyay S, Linardopoulou E, Aprikian P, Roberts PL, Skrivankova V, Johnston B, Gileva A, Igusheva I, Tolland A, Riddell R, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Kahl B, Price LB, Weissman SJ, Limaye A, Scholes D, Johnson JR, Sokurenko EV. 2013. Potential for predictive diagnostics of Escherichia coli infections based on the clonal association of antimicrobial resistance and clinical outcome. J. Clin. Microbiol. 51:2991–2999. 10.1128/JCM.00984-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson JR, Clermont O, Menard M, Kuskowski MA, Picard B, Denamur E. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141–1150. 10.1086/507305 [DOI] [PubMed] [Google Scholar]
- 25.Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML Online–a Web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559. 10.1093/nar/gki352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lacher DW, Steinsland H, Blank TE, Donnenberg MS, Whittam TA. 2007. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J. Bacteriol. 189:342–350. 10.1128/JB.01472-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaureguy F, Landreau L, Passet V, Diancourt L, Frapy E, Guigon G, Carbonnelle E, Lortholary O, Clermont O, Denamur E, Picard B, Nassif X, Brisse S. 2008. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9:560. 10.1186/1471-2164-9-560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clermont O, Olier M, Hoede C, Diancourt L, Brisse S, Keroudean M, Glodt J, Picard B, Oswald E, Denamur E. 2011. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11:654–662. 10.1016/j.meegid.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 29.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- 31.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-β-lactamase-producing Escherichia coli ST131 is driven by a single highly virulent subclone, H30-Rx. mBio 4(6):e00377–13. 10.1128/mBio.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudinha T, Johnson JR, Andrew SD, Kong F, Anderson P, Gilbert GL. 2013. Genotypic and phenotypic characterization of Escherichia coli isolates from children with urinary tract infection and from healthy carriers. Pediatr. Infect. Dis. J. 32:543–548. 10.1097/INF.0b013e31828ba3f1 [DOI] [PubMed] [Google Scholar]
- 33.Mora A, Dahbi G, López C, Mamani R, Marzoa J, Dion S, Picard B, Blanco M, Alonso MP, Denamur E, Blanco J. 2014. Virulence patterns in a murine sepsis model of ST131 Escherichia coli clinical isolates belonging to serotypes O25b:H4 and O16:H5 are associated to specific virotypes. PLoS One 9:e87025. 10.1371/journal.pone.0087025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee R, Robicsek A, Kuskowski M, Porter S, Johnston B, Sokurenko E, Tchesnokova V, Price L, Johnson JR. 2013. Molecular epidemiology of Escherichia coli sequence type ST131 and its H30 and H30-Rx subclones among extended-spectrum β-lactamase-positive and -negative E. coli clinical isolates from the Chicago region, 2007 to 2010. Antimicrob. Agents Chemother. 57:6385–6388. 10.1128/AAC.01604-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.