Abstract
Mycoplasma pneumoniae is an important cause of community-acquired pneumonia (CAP). In this study, M. pneumoniae strains in PCR-positive specimens collected from patients in Sydney, Australia (30 samples), and Beijing, China (83 samples), were characterized using multilocus variable-number tandem-repeat (VNTR) analysis (MLVA), P1-restriction fragment length polymorphism (RFLP) analysis, and sequencing of domain V of the 23S rRNA gene to compare genotype distribution and macrolide resistance rates between locations. Eighteen distinct MLVA types were identified in specimens from Sydney, of which 10 were known (types E, G, J, M, N, P, U, V, S, and X) and 8 previously unknown. Strains were equally distributed between P1-RFLP type 1 and type 2 variants. Among samples from Beijing, MLVA types E, G, J, P, U, X, and Z and four new types were identified. Most specimens belonged to P1-RFLP type 1. A nomenclature based on five VNTR loci is proposed to designate MLVA patterns. Macrolide resistance-associated mutations were identified in only 1 of 30 specimens (3.3%) from Sydney and 71 of 83 (85.5%) from Beijing (P < 0.05). This study demonstrated that although multiple individual M. pneumoniae strains were circulating in Beijing, the genotypes were less diverse than those in Sydney. However, the greatest regional difference was in the incidence of macrolide resistance, which may reflect differences in antibiotic use and/or measures in resistance control.
INTRODUCTION
Mycoplasma pneumoniae is an important pathogen causing community-acquired pneumonia (CAP), especially in children and young adults (1, 2). M. pneumoniae infections are increasingly recognized epidemically worldwide and as being endemic to some regions (3, 4). Previous reports showed regional differences in molecular profiles of M. pneumoniae (3, 5–7). Knowledge of these molecular characteristics is essential for outbreak investigation and to monitor the epidemiology of M. pneumoniae infections.
For the past 20 years, P1 gene restriction fragment length polymorphism (P1-RFLP) has been the most common molecular typing method for M. pneumoniae (8–10). Recombination events at two repetitive sequence loci, RepMP2/3 and RepMP4, (11) in the P1 gene contribute to gene variation, as reflected in sequence variants and subtypes, but the discriminatory power of this method is limited (12–14). Recently, a multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) method was developed by Dégrange et al. for M. pneumoniae isolates; this has a higher discriminatory power and can differentiate more than 26 distinct types (15). A culture-independent MLVA method for use directly from clinical specimens has also been described (7). Macrolide-resistant M. pneumoniae clinical isolates were first reported in the 1990s, and the incidence has been increasing ever since, with various rates in different geographic regions (16–20).
In this study, we used MLVA, P1-RFLP analysis, and detection of macrolide resistance-associated mutations to compare characteristics of M. pneumoniae detected by PCR in clinical specimens from Beijing, China, and Sydney, Australia.
MATERIALS AND METHODS
Clinical specimens.
Thirty M. pneumoniae PCR-positive clinical specimens, collected between 2008 and 2012 from patients aged between 2 and 70 years old, were obtained for further study from the diagnostic laboratory at the Centre for Infectious Diseases and Microbiology Laboratory Services (CIDMLS), Institute of Clinical Pathology and Medical Research, Westmead Hospital, Sydney, Australia. During the same 5-year period, 83 M. pneumoniae PCR-positive clinical specimens were obtained from pediatric patients in the Affiliated Children's Hospital of the Capital Institute of Pediatrics in Beijing.
Positive samples were identified using real-time PCR as described previously (21); only one sample from each patient was included. Specimen types included sputum, throat swabs, nasopharyngeal swabs or aspirates, bronchoalveolar lavage fluid (BALF), and pleural fluids. DNA was extracted using the QIAamp DNA minikit (Qiagen) or the NucliSENS easyMAG (bioMérieux) according to the manufacturer's instructions and was immediately used for PCR detection. M. pneumoniae-positive DNA extracts were stored at −20°C until required for genotyping.
MLVA typing.
MLVA was based on the five tandem repeat loci described by Dégrange et al. (15). Numbers of repeats were determined by sequencing. The MLVA loci were amplified with primers described previously (7) using a nested multiplex PCR to increase specificity. Briefly, the nested PCR was performed using 2× HotStar Taq master mix (Qiagen) in a 25-μl reaction mixture containing 12.5 μl of the 2× Qiagen master mix, 0.5 μl of each forward and reverse primer (10 μmol/liter), 1 μl extracted DNA, and 10.5 μl of molecular-grade water. The PCR was performed at 95°C for 10 min, followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min and then a final extension of 72°C for 10 min. The amplified products were purified using ExoStar PCR and a sequencing cleanup kit (GE Healthcare), as instructed by the manufacturer, and sequenced using an ABI 3730xl DNA analyzer (Applied Biosystems).
The MLVA types were assigned according to the nomenclature described by Dégrange et al. (15) using letters as MLVA types. The numbers of repeats at each of the five loci were linked together in a digital format in the order Mpn1-Mpn13-Mpn14-Mpn15-Mpn16 to designate MLVA patterns, e.g., 8-3-5-7-2.
P1 gene typing.
P1 genotyping was performed by PCR amplification of the RepMP4 and RepMP2/3 elements of the P1 gene. The PCR products were cut with restriction enzyme HaeIII. The digested fragments were separated using agarose gel electrophoresis, and patterns were interpreted as previously described (10). The P1 types were confirmed by sequencing and compared to reference sequences by performing BLAST searches in GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Detection of macrolide resistance-associated mutations.
Macrolide resistance-associated mutations in domain V of the 23S rRNA gene were detected using nested PCR-linked capillary electrophoresis and single-strand conformation polymorphisms (nPCR-CE-SSCP) as previously described by our group (20).
Data analysis.
MLVA types were analyzed based on the difference in numbers of repeats identified at each of the five loci. The repeat numbers were recorded and imported into the Bionumerics software (version 5.0; Applied Maths) to perform the dendrogram and the minimum spanning tree (MST) analysis. The dendrogram was generated based on the categorical coefficient and an unweighted-pair group method using average linkages (UPGMA) algorithm. The MST analysis was performed on the categorical coefficient with a priority rule of the first link types set as the highest number of single-locus variants (SLVs).
RESULTS
Molecular profiles of M. pneumoniae in respiratory specimens from Sydney.
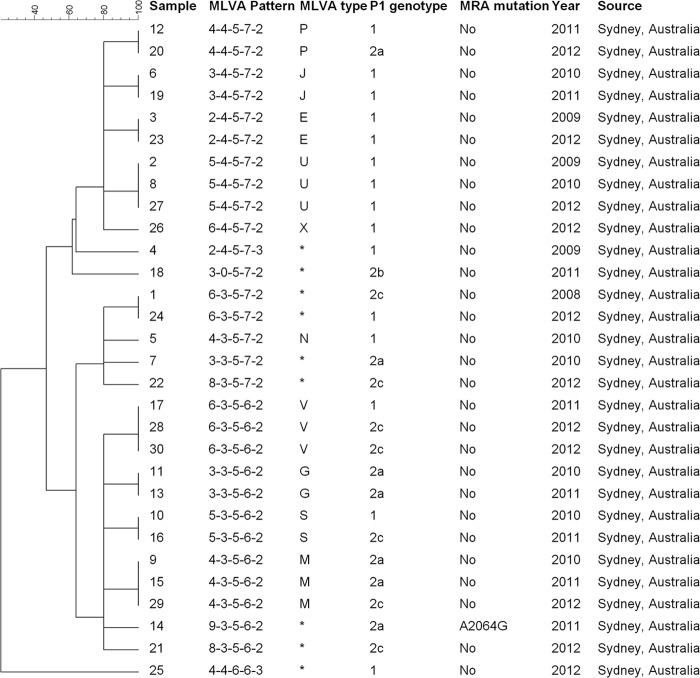
Eighteen MLVA types, each represented by one to three specimens, were identified among 30 samples from Sydney patients. Of these, 10 were known MLVA types (E, G, J, M, N, P, S, U, V, and X) based on the Dégrange nomenclature, represented by 21 (70%) specimens; eight types could not be assigned according to this nomenclature. All genotypes were represented using the proposed five-digit MLVA patterns. In one sample Mpn13 was not amplified, giving the profile 3-0-5-7-2. Mpn1 produced the most variable-repeat numbers of all loci, ranging from two to nine; the remaining four loci were less variable. When Mpn1 was excluded, three major MLVA clusters were identified: 3-5-6-2 (12 specimens, six MLVA types), 3-5-7-2 (5 specimens, four MLVA types), and 4-5-7-2 (10 specimens, five MLVA types) (Fig. 1).
FIG 1.
Molecular profiles of 30 M. pneumoniae samples collected between 2009 and 2012 from Sydney, Australia. The dendrogram was generated based on the variable-number tandem repeats according to our five-digit MLVA pattern (see the text). MLVA type was also assigned according to the Dégrange scheme (15); asterisks show MLVA types not found in the scheme. MRA, macrolide resistance associated.
There were four P1-RFLP genotypes among 30 samples: 15 of type 1, 7 each of types 2a and 2c, and 1 of type 2b. No obvious correlation between P1-RFLP and MLVA types was identified, and there was no apparent temporal clustering of either MLVA or P1-RFLP types during the period of specimen collection (Fig. 1).
Sequencing of domain V of the 23S rRNA gene identified a macrolide resistance-associated mutation (A2064G) in one specimen (3.3%).
Molecular profiles of M. pneumoniae in respiratory specimens from Beijing.
There were 11 MLVA types among 83 specimens, of which seven were previously known types (E, G, J, P, U, X, and Z), based on the Dégrange nomenclature; they accounted for 76 (91.6%) specimens; of these, the four types U (24), X (19), P (16), and J (8) accounted for 80.7% of specimens. The other four were novel MLVA types. Most specimens (78; 93.9%) shared a common pattern at the last four loci, 4-5-7-2, including seven MLVA types that clustered together in the dendrogram (Fig. 2). In one specimen, locus Mpn1was not amplified (profile 0-2-5-5-2).
FIG 2.
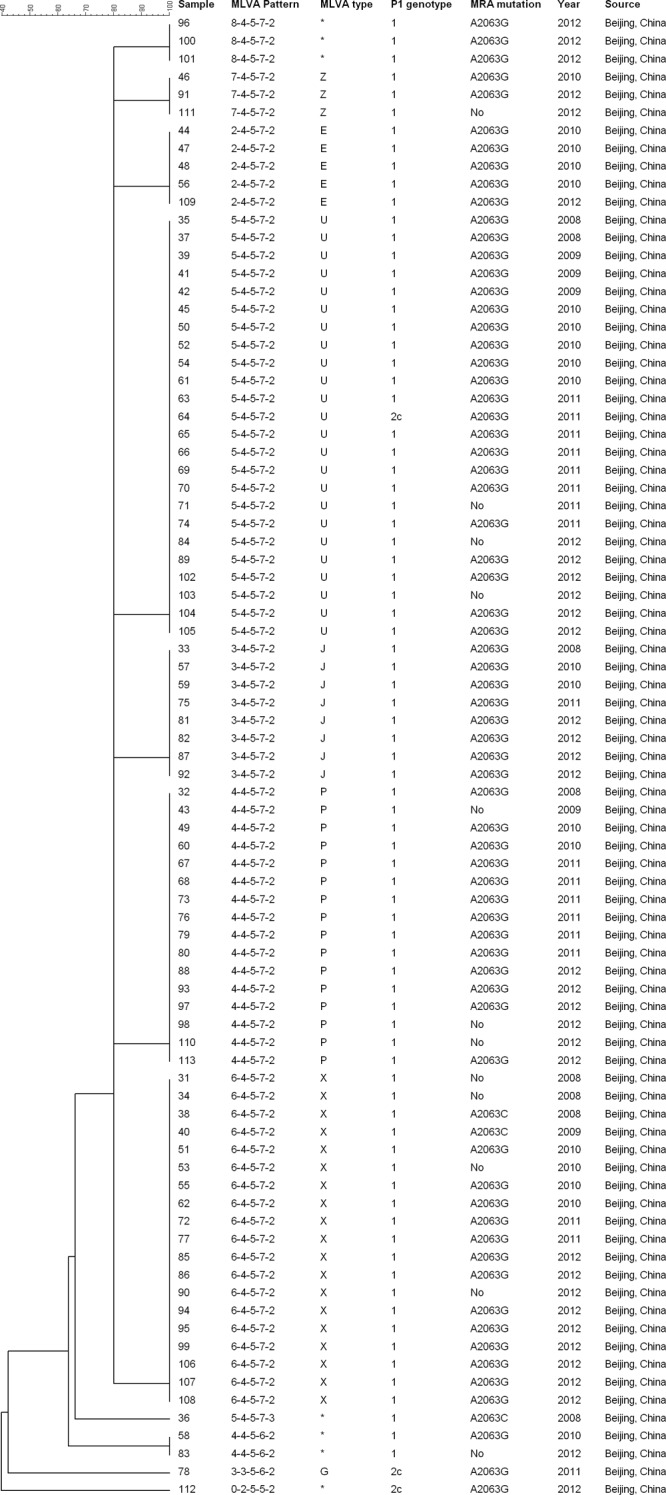
Molecular profiles of 83 M. pneumoniae samples collected between 2009 and 2012 from Beijing, China. The dendrogram was generated based on the variable-number tandem repeats according to our five-digit MLVA pattern (see the text). MLVA type was also assigned according to the Dégrange scheme (15); asterisks show MLVA types not found in the scheme. MRA, macrolide resistance associated.
In the P1-RFLP analysis, 80 (96.4%) of samples contained type 1, and the remainder contained type 2c (Fig. 2).
Macrolide resistance mutations were identified in domain V of the 23S rRNA gene in 71 specimens (85.5%), of which 68 were A2063G and three A2063C; they occurred throughout the 5-year period. There was no correlation between MLVA types and macrolide resistance (Fig. 2).
Comparison of molecular features between samples from Beijing and Sydney.
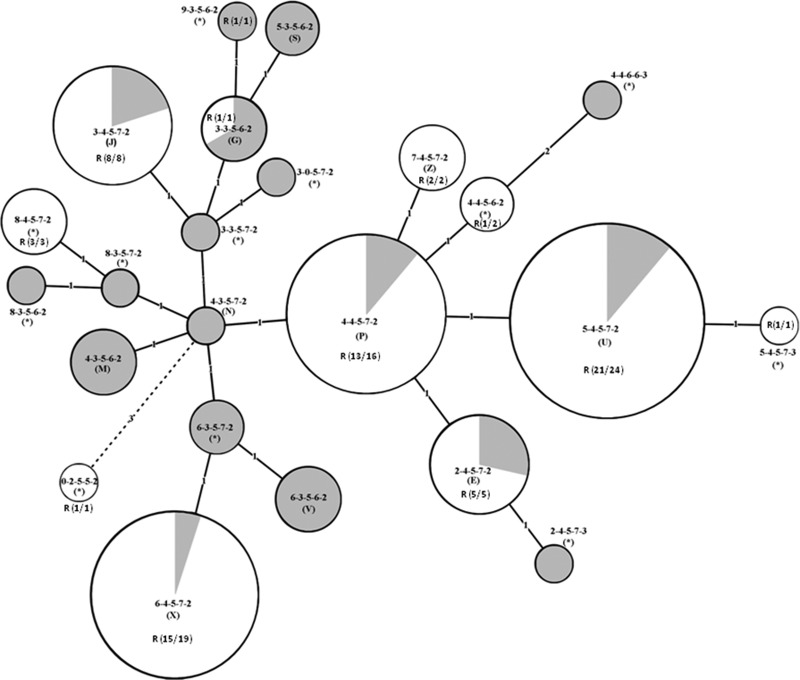
The genetic relationships of all 113 samples from Beijing, China, and Sydney, Australia, were analyzed by construction of an MST using Bionumerics software (Applied Maths) (Fig. 3). At locus three (Mpn14), there were five repeats in all samples tested. In contrast, there were major differences at the second locus (Mpn13), which, with a few exceptions, grouped MLVA patterns into clusters between and among samples from Beijing and Sydney. The majority of samples from Sydney had three repeats at Mpn13, and the most common pattern was 3-5-6(7)-2, whereas most samples from Beijing had four repeats, with the most common pattern being 4-5-7-2(3). MLVA pattern diversity among samples was highlighted by the MST, with the first locus (Mpn1) being the most diverse marker that subdivided clusters at both geographic locations. There was a marked difference in prevalence of macrolide resistance mutations between samples from Beijing and Sydney (chi square = 86.1, P < 0.05), which was found in all of Beijing MLVA patterns but in only one from Sydney (Fig. 1, 2, and 3).
FIG 3.
Minimum spanning tree (MST) analysis of molecular features of 113 M. pneumoniae samples from Beijing, China, and Sydney, Australia. Each circle represents a particular MLVA profile, designated by MLVA pattern and Dégrange type in parentheses (*, novel MLVA type). The color of the circles corresponds to sample source: white for Beijing, China, and gray for Sydney, Australia. R indicates the presence of macrolide resistance, and the number in parentheses indicates the number of the resistant samples out of the total in a particular MLVA type. The size of the circle is proportional to the number of samples belonging to the indicated MLVA type. The distance between neighboring MLVA types is expressed as the number of allelic changes and is indicated by 1, 2, and 3 for one, two, and three changes, respectively.
DISCUSSION
M. pneumoniae activity that is endemic to certain regions occurs with periodic prolonged epidemics or periods of increased activity that are separated by a few years (3, 4, 6). Molecular typing is required to determine whether periods of increased activity are true epidemics due to the spread of a predominant strain or increases in the prevalence of several strains.
In 2007, Degrange et al. described an MLVA method for M. pneumoniae based on five VNTR loci which they used to study 265 isolates, mainly from France, collected over about a decade. They identified 26 different MLVA patterns, which they designated A to Z, based on five-digit codes representing the numbers of repeats at each locus. Five MLVA types, P, U, O, J, and E, accounted for nearly 60% of isolates (15). Since then, several studies have shown different combinations of generally four to six predominant strains circulating simultaneously (3, 5–7) and changing over time (22). Most studies indicate that periods of apparent epidemic activity generally are not due to single epidemic strains (3, 6, 22). However, true epidemics can occur; Pereyre et al. reported an outbreak of M. pneumoniae infections due to clonal spread of a single strain among children attending a primary school and their household contacts (23).
Our results confirmed that Mpn1 was the most variable locus, as reported by Benitez et al., who demonstrated multiple insertions or deletions of tandem repeats within this locus (5). Excluding this locus, MLVA types in studies reported from Europe and the United States have shown two predominant patterns for the last four repeats, namely −4-5-7-2(3) and −3-5-6(7)-2. In our study, there are 11 MLVA types in pattern 4-5-7-2(3) and 17 MLVA types in pattern 3-5-6(7)-2. These MLVA types are shared by 93.3% of Sydney specimens (28/30). (Fig. 1). There was less diversity among Beijing specimens; most MLVA types shared the pattern 4-5-7-2, representing seven MLVA types and 93.9% of specimens (Fig. 2). This is consistent with the findings of another study from Beijing, in which 175 (87.1%) and 15 (7.5%) of 201 specimens contained MLVA patterns 4-5-7-2 and 3-5-6-2, respectively (24).
On the other hand, the most variable locus, Mpn1, has an important role in the investigation of local epidemics due to its high discriminatory power. A previous report showed repeat numbers ranging from one to eight (5). In this study, we identified one specimen with nine repeats at Mpn1 and one in which it failed to amplify, suggesting the potential variation at this locus is even greater. It is possible that this variation could be used to investigate outbreaks in small settings, such as schools or hospitals.
Until 2009, P1-RFLP typing was the main genotyping method. It has become a marker of gene evolution due to its low discrimination power. In this study, most specimens from China were identified as P1 type 1, and only three (3.6%) were P1 type 2c. Zhao et al. (24) also found that a majority of specimens from Beijing were P1 type 1, whereas in Sydney specimens were evenly divided between type 1 and type 2 variants, as described in European countries and the United States (4, 5, 15).
The other major difference was a much higher rate of macrolide resistance mutations in Beijing (85.5%) than in Sydney (3.3%), which has been described in other studies from Asia (18, 25, 26). In common with Zhao et al., we found no correlation between MLVA type and macrolide resistance (24). As most strains from Beijing were P1 type 1, we cannot infer that P1-RFLP type 1 strains were more likely to acquire macrolide resistance. The high rate of macrolide resistance in China more likely is related to excessive use of antibiotics. Further studies are needed to identify relatedness, if any, between macrolide resistance and genotype.
In conclusion, there were regional differences in molecular profiles of M. pneumoniae from specimens obtained in Sydney and Beijing, but these should be interpreted with caution as the patient populations were different (a wide age range among Sydney patients but only children in Beijing). MLVA typing is far more discriminatory than P1 gene subtyping, which makes it more useful for epidemiological studies and comparisons between regions. Alternative molecular typing methods, including whole-genome sequencing, may provide a better understanding of the molecular characteristics of M. pneumoniae.
ACKNOWLEDGMENTS
This work was supported by the Beijing Natural Science Foundation (7112019) and the Beijing City talent training project fund (20071A0303200118).
Footnotes
Published ahead of print 26 February 2014
REFERENCES
- 1.Hammerschlag MR. 2001. Mycoplasma pneumoniae infections. Curr. Opin. Infect. Dis. 14:181–186. 10.1097/00001432-200104000-00012 [DOI] [PubMed] [Google Scholar]
- 2.Waites KB, Atkinson TP. 2009. The role of Mycoplasma in upper respiratory infections. Curr. Infect. Dis. Rep. 11:198–206. 10.1007/s11908-009-0030-6 [DOI] [PubMed] [Google Scholar]
- 3.Pereyre S, Charron A, Hidalgo-Grass C, Touati A, Moses AE, Nir-Paz R, Bébéar C. 2012. The spread of Mycoplasma pneumoniae is polyclonal in both an endemic setting in France and in an epidemic setting in Israel. PLoS One 7:e38585. 10.1371/journal.pone.0038585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenglet A, Herrador Z, Magiorakos AP, Leitmeyer K, Coulombier D. 2012. Surveillance status and recent data for Mycoplasma pneumoniae infections in the European Union and European Economic Area, January 2012. Euro Surveill. 17:20075 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20075 [DOI] [PubMed] [Google Scholar]
- 5.Benitez AJ, Diaz MH, Wolff BJ, Pimentel G, Njenga MK, Estevez A, Winchell JM. 2012. Multilocus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical isolates from 1962 to the present: a retrospective study. J. Clin. Microbiol. 50:3620–3626. 10.1128/JCM.01755-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalker V, Stocki T, Litt D, Bermingham A, Watson J, Fleming D, Harrison T. 2012. Increased detection of Mycoplasma pneumoniae infection in children in England and Wales, October 2011 to January 2012. Euro Surveill. 17:20081 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20081 [PubMed] [Google Scholar]
- 7.Dumke R, Jacobs E. 2011. Culture-independent multi-locus variable-number tandem-repeat analysis (MLVA) of Mycoplasma pneumoniae. J. Microbiol. Methods 86:393–396. 10.1016/j.mimet.2011.06.008 [DOI] [PubMed] [Google Scholar]
- 8.Cousin-Allery A, Charron A, de Barbeyrac B, Fremy G, Skov Jensen J, Renaudin H, Bebear C. 2000. Molecular typing of Mycoplasma pneumoniae strains by PCR-based methods and pulsed-field gel electrophoresis. Application to French and Danish isolates. Epidemiol. Infect. 124:103–111 (http://journals.cambridge.org/action/displayAbstract?fromPage=online&aid=40019&fulltextType=RA&fileId=S0950268899003313). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenri T, Okazaki N, Yamazaki T, Narita M, Izumikawa K, Matsuoka M, Suzuki S, Horino A, Sasaki T. 2008. Genotyping analysis of Mycoplasma pneumoniae clinical strains in Japan between 1995 and 2005: type shift phenomenon of M. pneumoniae clinical strains. J. Med. Microbiol. 57:469–475. 10.1099/jmm.0.47634-0 [DOI] [PubMed] [Google Scholar]
- 10.Sasaki T, Kenri T, Okazaki N, Iseki M, Yamashita R, Shintani M, Sasaki Y, Yayoshi M. 1996. Epidemiological study of Mycoplasma pneumoniae infections in Japan based on PCR-restriction fragment length polymorphism of the P1 cytadhesin gene. J. Clin. Microbiol. 34:447–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenri T, Taniguchi R, Sasaki Y, Okazaki N, Narita M, Izumikawa K, Umetsu M, Sasaki T. 1999. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect. Immun. 67:4557–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumke R, Lück PC, Noppen C, Schaefer C, von Baum H, Marre R, Jacobs E. 2006. Culture-independent molecular subtyping of Mycoplasma pneumoniae in clinical samples. J. Clin. Microbiol. 44:2567–2570. 10.1128/JCM.00495-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorigo-Zetsma JW, Wilbrink B, Dankert J, Zaat SA. 2001. Mycoplasma pneumoniae P1 type 1- and type 2-specific sequences within the P1 cytadhesin gene of individual strains. Infect. Immun. 69:5612–5618. 10.1128/IAI.69.9.5612-5618.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao F, Cao B, Li J, Song S, Tao X, Yin Y, He L, Zhang J. 2011. Sequence analysis of the P1 adhesin gene of Mycoplasma pneumoniae in clinical isolates collected in Beijing in 2008 to 2009. J. Clin. Microbiol. 49:3000–3003. 10.1128/JCM.00105-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dégrange S, Cazanave C, Charron A, Renaudin H, Bébéar C, Bébéar CM. 2009. Development of multiple-locus variable-number tandem-repeat analysis for molecular typing of Mycoplasma pneumoniae. J. Clin. Microbiol. 47:914–923. 10.1128/JCM.01935-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okazaki N, Narita M, Yamada S, Izumikawa K, Umetsu M, Kenri T, Sasaki Y, Arakawa Y, Sasaki T. 2001. Characteristics of macrolide-resistant Mycoplasma pneumoniae strains isolated from patients and induced with erythromycin in vitro. Microbiol. Immunol. 45:617–620. 10.1111/j.1348-0421.2001.tb01293.x [DOI] [PubMed] [Google Scholar]
- 17.Peuchant O, Ménard A, Renaudin H, Morozumi M, Ubukata K, Bébéar CM, Pereyre S. 2009. Increased macrolide resistance of Mycoplasma pneumoniae in France directly detected in clinical specimens by real-time PCR and melting curve analysis. J. Antimicrob. Chemother. 64:52–58. 10.1093/jac/dkp160 [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Ye X, Zhang H, Xu X, Wang M. 2012. Multiclonal origin of macrolide-resistant Mycoplasma pneumoniae isolates as determined by multilocus variable-number tandem-repeat analysis. J. Clin. Microbiol. 50:2793–2795. 10.1128/JCM.00678-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuoka M, Narita M, Okazaki N, Ohya H, Yamazaki T, Ouchi K, Suzuki I, Andoh T, Kenri T, Sasaki Y, Horino A, Shintani M, Arakawa Y, Sasaki T. 2004. Characterization and molecular analysis of macrolide-resistant Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob. Agents Chemother. 48:4624–4630. 10.1128/AAC.48.12.4624-4630.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin C, Li S, Sun H, Zhao H, Feng Y, Cao L, Yuan Y, Zhang T. 2010. Nested PCR-linked capillary electrophoresis and single-strand conformation polymorphisms for detection of macrolide-resistant Mycoplasma pneumoniae in Beijing. J. Clin. Microbiol. 48:4567–4572. 10.1128/JCM.00400-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumke R, Schurwanz N, Lenz M, Schuppler M, Lück C, Jacobs E. 2007. Sensitive detection of Mycoplasma pneumoniae in human respiratory tract samples by optimized real-time PCR approach. J. Clin. Microbiol. 45:2726–2730. 10.1128/JCM.00321-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereyre S, Touati A, Petitjean-Lecherbonnier J, Charron A, Vabret A, Bébéar C. 2013. The increased incidence of Mycoplasma pneumoniae in France in 2011 was polyclonal, mainly involving M. pneumoniae type 1 strains. Clin. Microbiol. Infect. 19:E212–E217. 10.1111/1469-0691.12107 [DOI] [PubMed] [Google Scholar]
- 23.Pereyre S, Renaudin H, Charron A, Bébéar C. 2012. Clonal spread of Mycoplasma pneumoniae in primary school, Bordeaux, France. Emerg. Infect. Dis. 18:343–345. 10.3201/eid1802.111379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao F, Liu G, Cao B, Wu J, Gu Y, He L, Meng F, Zhu L, Yin Y, Lv M, Zhang J. 2013. Multiple-locus variable-number tandem-repeat analysis of 201 Mycoplasma pneumoniae isolates from Beijing in 2008 to 2011. J. Clin. Microbiol. 51:636–639. 10.1128/JCM.02567-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun H, Xue G, Yan C, Li S, Cao L, Yuan Y, Zhao H, Feng Y, Wang L, Fan Z. 2013. Multiple-locus variable-number tandem-repeat analysis of Mycoplasma pneumonia clinical specimens and proposal for amendment of MLVA nomenclature. PLoS One 8:e64607. 10.1371/journal.pone.0064607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki Y, Itagaki T, Seto J, Kaneko A, Abiko C, Mizuta K, Matsuzaki Y. 2013. Community outbreak of macrolide-resistant Mycoplasma pneumoniae in Yamagata, Japan in 2009. Pediatr. Infect. Dis. J. 32:237–240. 10.1097/INF.0b013e31827aa7bd [DOI] [PubMed] [Google Scholar]