Abstract
Our study is the first to compare the nasopharyngeal microbiota of pediatric pneumonia patients and control children by 454 pyrosequencing. A distinct microbiota was associated with different pneumonia etiologies. Viral pneumonia was associated with a high abundance of the operational taxonomic unit (OTU) corresponding to Moraxella lacunata. Patients with nonviral pneumonia showed high abundances of OTUs of three typical bacterial pathogens, Streptococcus pneumoniae complex, Haemophilus influenzae complex, and Moraxella catarrhalis. Patients classified as having no definitive etiology harbored microbiota particularly enriched in the H. influenzae complex. We did not observe a commensal taxon specifically associated with health. The microbiota of the healthy nasopharynx was more diverse and contained a wider range of less abundant taxa.
INTRODUCTION
Pneumonia is the leading cause of childhood mortality worldwide, claiming 2 million lives yearly among young children (1). The etiology of pediatric pneumonia is complex and not routinely determined in clinical practice (2). Its definitive determination remains challenging (3). Clinical research shows that Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus are the leading pathogens of bacterial pneumonia (1, 4). Respiratory syncytial virus (RSV), parainfluenza, and influenza viruses are the main causes of pediatric viral pneumonia (e.g., see references 5 and 6). S. pneumoniae and H. influenzae frequently colonize the nasopharynx of young children, and the nasopharyngeal (NP) carriage of S. pneumoniae is considered key to pneumonia and other pneumococcal diseases (7). However, the mere nasopharyngeal presence of bacterial pathogens does not necessarily lead to invasive lung infection. Various mechanisms, such as competition with resident nonpathogenic microbiota, viral coinfection, or host immune factors are likely to affect the transition from nasopharyngeal colonization to pneumonia (8). The increasing accessibility of culture-independent sequencing methods has permitted microbiota description at various body sites (9), and the potential of commensal microbiota to modify the disease process is receiving increasing attention (10).
Previous culture-independent studies of the nasopharyngeal microbiota of young children looked at healthy children (11) and compared the microbiota composition of healthy children and children with acute otitis media (12). In the present study, we analyzed the nasopharyngeal microbiota composition, including the presence of respiratory viruses, in pediatric pneumonia patients and matched healthy controls and looked for microbiota associations with the disease status.
MATERIALS AND METHODS
Patients and clinical samples.
A prospective case-control study conducted in 2008 to 2009 in 3 major hospitals in Switzerland (Geneva, Lausanne, and Sion) to investigate pediatric community-acquired pneumonia etiology was described in detail elsewhere (13). Briefly, pneumonia cases were diagnosed according to the WHO criteria (14) in children 2 months to 16 years old presenting with fever (temperature > 38°C), increased respiratory rate for age or respiratory distress, and infiltrates on chest X-ray. Controls were age matched and recruited from children undergoing minor elective surgery under anesthesia. Patients with respiratory tract infection (RTI) within the past 3 weeks and any history of chronic disease were excluded. From this case-control study population we selected 50 children with pneumonia and 50 control children who were age and sex matched. The median age of the included children was 2.7 years; 41% were girls.
Signed informed parental consent was obtained before enrollment. Ethics approval was obtained from the research ethics committees at the 3 hospitals (authorization number CER 07-269).
The nasopharynx samples were collected from all patients using swabs with liquid Amies preservation medium (Copan) and immediately frozen at −80 °C. Blood cultures were performed, and C-reactive protein (CRP) concentrations were determined for all pneumonia patients.
Detection of respiratory viruses and S. pneumoniae.
Twenty-five common respiratory viruses were detected in nasopharyngeal samples using a PCR- and microarray-based assay (Infinity RVP Plus; AutoGenomics, Inc., Vista, CA) (15) following the instructions of the manufacturer. Detection of S. pneumoniae was done by PCR targeting autolysin (lytA) and pneumolysin (ply) genes (13).
454 pyrosequencing.
Two hundred microliters of each sample was lysed in 1× Tris-EDTA (TE) buffer with 0.5% Tween and 200 μg/ml of proteinase K (Fermentas). Proteinase K was inactivated by a 10-min incubation at 95°C (16). PCR and pyrosequencing of the variable region of V1-V2 of the 16S rRNA gene was performed as previously described using Roche 454 FLX chemistry (17), except that the requirement for a minimum of 2 ng of DNA template per reaction was not followed. PCR products suitable for sequencing were obtained from samples of 27 of 50 pneumonia and 22 of 50 control patients. A total of 26,955 good-quality sequencing reads were obtained for controls and 80,022 were obtained for pneumonia patients.
Taxonomic classification.
The initial sequence reads underwent quality checks to remove sequencing artifacts and low-quality sequences, as described in reference 17. High-quality reads were classified into Bergey's taxonomy using the RDP-Classifier version 2.3 (60% confidence cutoff). Classification at the genus level was used for the analysis. Reads were also grouped into operational taxonomic units (OTUs) based on their best BLAST hit to reference type strain 16S DNAs using a 98% sequence identity cutoff. Some of the reference sequences representing type strains of distinct species were over 98% identical and hence were pooled in a single OTU. All OTUs for which the reference sequences were similar at a >98% level over the sequenced region (V1-V2) were summed up and labeled with the name of the dominant taxon. For example, the S. pneumoniae complex OTU was a cluster grouping Streptococcus pneumoniae, S. mitis, and S. pseudopneumoniae sequences. Table S1 in the supplemental material contains the list of OTUs and corresponding reference taxa.
Statistical analysis.
The Wilcoxon rank sum test was used to assess whether individual taxa/OTUs were statistically different between the groups. The 12 most abundant OTUs were used for principal component analysis (PCA). PCA biplots were used to visualize the differences between the multivariate means and the multivariate variability (dispersion of points) and which taxa contributed to this difference (arrows with corresponding label). Nonparametric multivariate analysis of variance (NP-MANOVA) was used to assess the difference between groups. All analyses were performed using R 2.12.0.
RESULTS
The nasopharyngeal microbiota of children with community-acquired pneumonia cases was compared with that of matched children undergoing minor surgery (controls). Three dominant bacterial genera, namely, Moraxella, Haemophilus, and Streptococcus, represented 92% of reads in the pneumonia patients and 76% of reads in the controls (see Fig. S1 in the supplemental material). Despite the dominance of just 3 genera, high variability among the patients was observed. Twenty samples were dominated (>50% of all reads) by Moraxella, 17 by Haemophilus, and 4 by Streptococcus. Only 2.1% of reads were unclassified at the genus level.
Richness and diversity was higher in control than in pneumonia patients.
For each sample, the number of genera present among 300 randomly chosen reads was counted, omitting all genera that were represented by ≤5 reads per sample. Control patients, with 14.7 genera, showed significantly greater genus richness than pneumonia cases, with 7.7 genera (P < 0.01, Mann-Whitney U test). Diversity, measured by the Shannon index, was also higher in controls than in patients (2.31 versus 1.71, P < 0.002, Mann-Whitney U test).
Viral diagnostics and presumed pneumonia etiology.
Eight of 27 pneumonia patients compared to only 1 of 22 control patients showed a respiratory viral pathogen in the nasopharynx (P = 0.03, Fisher exact test). The viruses detected in nasopharyngeal samples of the pneumonia patients included respiratory syncytial virus, adenovirus, human metapneumovirus, and rhinovirus (Fig. 1). The only virus-positive sample from a control patient contained a rhinovirus. We tentatively assumed that patients with respiratory virus suffered from viral pneumonia (6).
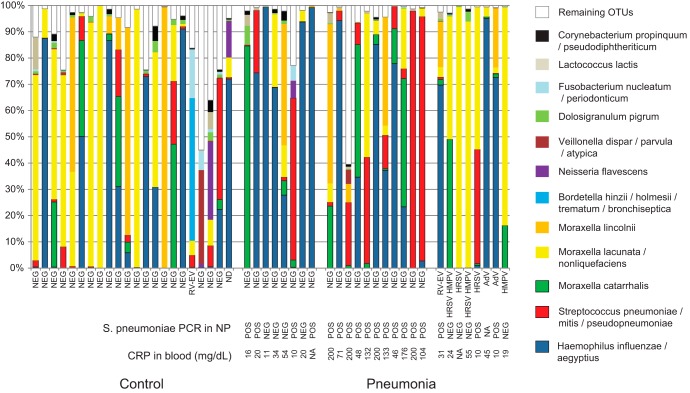
FIG 1.
The composition of nasopharyngeal microbiota of control children (left) and children with nonviral (center) or viral (right) pneumonia. Nonviral-pneumonia patients were further subdivided according to S. pneumoniae diagnosis (patients with both PCR positivity and CRP levels of >40 mg/dl were grouped apart from patients with only one or none of these features). The 12 most abundant operational taxonomic units (OTUs) are shown. Sequences belonging to OTUs which contain <0.5% of total number of sequences were pooled and are labeled “Remaining OTUs.” The samples where respiratory viruses were detected are labeled accordingly, i.e., AdV (adenovirus), RV-EV (rhinovirus or enterovirus), HRSV (human respiratory syncytial virus), and HMPV (human metapneumovirus). “NEG” along the x axis indicates negativity for virus detection. ND, not determined; NA, not available.
Blood culture was negative for all patients. Eighteen out of 26 patients had nasopharyngeal samples which were positive for S. pneumoniae by PCR. We considered patients who showed both a CRP concentration of >40 mg/dl and a positive test for S. pneumoniae in the nasopharynx by PCR to suffer from pneumococcal pneumonia (18). For a mixed infection, the two above criteria would have to be fulfilled, in addition to the detection of respiratory virus in the nasopharynx. According to these criteria, none of the patients suffered from mixed infection. The remaining patients were considered to show no definitive etiology (Fig. 1).
Presence of respiratory virus was associated with distinct microbiota.
A great majority (92%) of reads were assigned to an OTU with 98% identity cutoff, i.e., produced a BLAST hit to a reference sequence of a type strain. In total, 250 OTUs were identified (see Table S1 in the supplemental material). Out of 250 identified OTUs, only 12 showed >0.5% average relative abundance. Many samples (35 out of 49) were dominated (>50% of all reads) by a single OTU (Fig. 1).
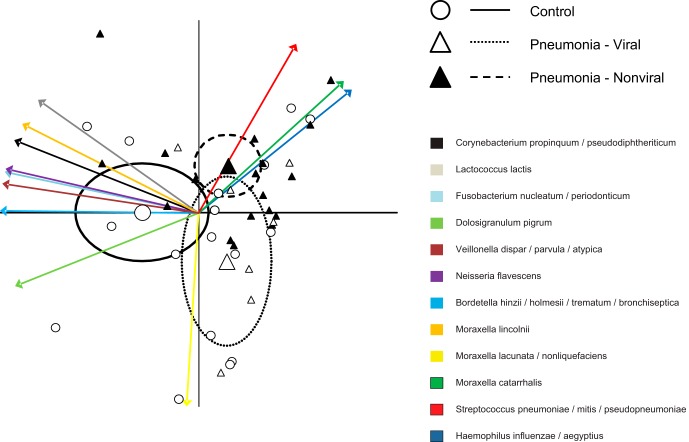
Multivariate analysis performed on the 12 most common OTUs, (Fig. 2) suggested that nonviral pneumonia patients had a high abundance of the three typical pneumonia pathogens (S. pneumoniae complex, H. influenzae complex, and M. catarrhalis), while viral pneumonia was distinguished by increased presence of the M. lacunata complex. NP-MANOVA indicated no difference between the control group and the viral pneumonia group (P = 0.86) and trends for a difference between the control group and the nonviral pneumonia group (P = 0.12) and between the bacterial and viral pneumonia groups (P = 0.11). Subsequent univariate analysis confirmed these observations. The M. lacunata complex was nearly 7-fold more abundant in viral pneumonia patients than in nonviral pneumonia patients (Fig. 3) (P = 0.005). When OTUs corresponding to the three main bacterial pathogens (S. pneumoniae complex, H. influenzae complex, and M. catarrhalis) were combined, nonviral pneumonia patients had a higher proportion of the three bacterial pathogens than did both the controls (P = 0.0005) and the viral pneumonia patients (Fig. 3) (P = 0.025). Four OTUs showed a trend toward a higher abundance in control patients than in nonviral pneumonia patients (M. lacunata complex, P = 0.075; Dolosigranulum pigrum, P = 0.058; Fusobacterium nucleatum complex, P = 0.065; Lactococcus lactis, P = 0.074). There were no statistically significant differences or trends (P < 0.15) between controls and viral pneumonia patients. We also observed that patients for whom no definitive etiology was detected showed particularly high abundances of H. influenzae complex (Fig. 1).
FIG 2.
Multivariate analysis including 12 most common OTUs. PCA-biplot representation of microbiota of control children and children with bacterial (nonviral) and viral pneumonia. Arrows represent projections of the taxa that are responsible for the differences between groups. Large points represent the group means, and ellipses represent 95% confidence limits.
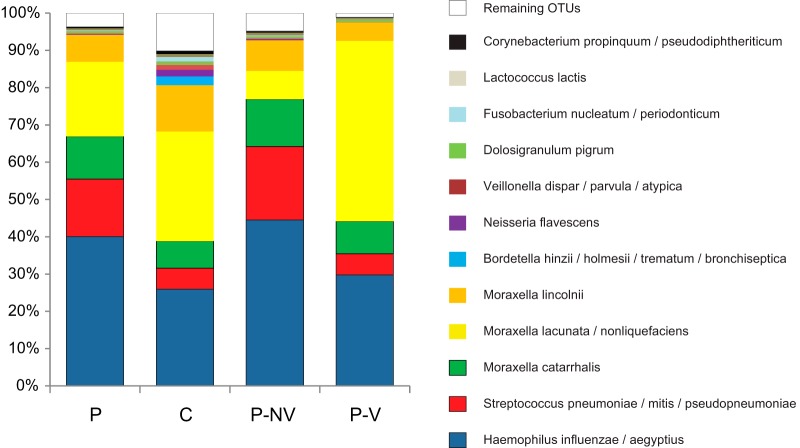
FIG 3.
Mean relative abundance of 12 most common OTUs in samples from pneumonia patients (P) and controls (C). Pneumonia patients are further differentiated as having nonviral pneumonia (P-NV) or viral pneumonia (P-V). OTUs containing <0.5% of the total number of sequences were pooled and are labeled “Remaining OTUs.”
When all pneumonia patients were compared with controls, the only significant difference between the groups was a higher relative abundance of OTUs corresponding to the combined three pathogens (S. pneumoniae complex, H. influenzae complex, and M. catarrhalis) (66% versus 36%, P = 0.005) (Fig. 3). F. nucleatum complex (P = 0.055) and D. pigrum (P = 0.084) showed trends toward higher abundances in control patients.
DISCUSSION
In this study, we used the culture-independent technique of pyrosequencing to analyze the nasopharyngeal microbiota of pediatric pneumonia patients and compare it to the microbiota of control children. Three highly dominant genera, namely, Moraxella, Streptococcus, and Haemophilus, were observed, which was in agreement with two previous studies surveying the nasopharynxes of children (11, 12). Likewise, the abundances of H. influenzae, S. pneumoniae, and M. catarrhalis in the nasopharynx in children are well known from previous culture-based studies. Here we identified other prevalent taxa, namely, M. lacunata and Moraxella lincolnii.
High dissimilarity of microbiota among patients was striking. Dominance of just one taxon in a sample was common, and more so in pneumonia patients. However, on average, the nasopharyngeal microbiota of control and pneumonia patients did not display prominent differences. In terms of abundance, the only clear difference was the increased presence of the bacterial pathogens S. pneumoniae complex, H. influenzae, and M. catarrhalis in pneumonia patients. Comparison between viral and nonviral pneumonia revealed that nonviral (and thus presumably bacterial) pneumonia patients harbored a microbiota which was particularly enriched in these three bacterial pathogens, as expected. In viral pneumonia patients the three pathogenic taxa were not more abundant than in control patients; instead, the increased presence of the little-known M. lacunata was observed. Although we think it is unlikely that M. lacunata is a pathogen on its own, its association with true respiratory pathogens, in this case respiratory viruses, should be further investigated. Different etiologies of pneumonia appear to be associated with specific patterns of microbiota dysbiosis. Patients without definitive etiology appeared to harbor a particularly high abundance of H. influenzae complex. Nontypeable (NT) H. influenzae has increasingly been considered an important pathogen of pediatric pneumonia (19). Because it is a very frequent colonizer of the nasopharynx and it does not appear to be associated with elevated CRP concentrations, definitive attribution of etiology to this species may prove difficult. Recent studies, where samples taken from the lung were investigated in cases of recurrent or severe pneumonia, showed high prevalence of NT H. influenzae. Our results suggest that this pathogen should be considered in uncomplicated pneumonia. The nasopharyngeal microbiota of children in our study showed low taxon complexity and low bacterial abundance. Although we did not perform a direct assessment of bacterial abundance on our samples, low (50%) success in amplification implies low bacterial colonization density, as previously observed (11). Moreover, higher amplification success of pneumonia samples implied denser colonization in disease than in health, as already suggested for acute otitis media (12).
The microbiota of control children was more diverse than that of pneumonia patients. The association of decreased richness and diversity of microbiota with disease appears to be a common theme, reported for a number of conditions, and also specifically for nasopharyngeal (12) and nasal (20) microbiota in children. One might speculate that the decrease in diversity is caused by the outgrowth of pathogens, which predisposes to invasive infection, but this hypothesis can be tested only in a longitudinal study. Our initial working hypothesis was that commensal bacteria keep the facultative pathogens colonizing the nasopharynx in check (21). This could prevent their descent into the lungs and invasive infection. However, we did not find evidence for a protective commensal taxon associated with health. Instead, the healthy nasopharynx was colonized with a wider range of less numerous taxa.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andreas Rytz for help with statistical analysis and Jacques Schrenzel for helpful discussions.
This work was supported by the Nestlé Research Center.
O.S., V.B.S., B.B., A.B., M.L., D.M., C.N.B., and H.B. are employees of Nestec SA. A.G. and K.K. have no conflicts to declare.
Footnotes
Published ahead of print 5 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03280-13.
REFERENCES
- 1.Rudan I, Boschi-Pinto C, Biloglav Z, Mulholland K, Campbell H. 2008. Epidemiology and etiology of childhood pneumonia. Bull. World Health Organ. 86:408–416. 10.2471/BLT.07.048769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feigin R, Cherry J, Demmler-Harrison GJ, Kaplan SL. 2009. Feigin and Cherry's textbook of pediatric infectious diseases. Saunders, Philidelphia, PA [Google Scholar]
- 3.Levine OS, O'Brien KL, Deloria-Knoll M, Murdoch DR, Feikin DR, Deluca AN, Driscoll AJ, Baggett HC, Brooks WA, Howie SRC, Kotloff KL, Madhi SA, Maloney SA, Sow S, Thea DM, Scott JA. 2012. The Pneumonia Etiology Research for Child Health Project: a 21st century childhood pneumonia etiology study. Clin. Infect. Dis. 54(Suppl 2):S93–S101. 10.1093/cid/cir1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cevey-Macherel M, Galetto-Lacour A, Gervaix A, Siegrist CA, Bille J, Bescher-Ninet B, Kaiser L, Krahenbuhl JD, Gehri M. 2009. Etiology of community-acquired pneumonia in hospitalized children based on WHO clinical guidelines. Eur. J. Pediatr. 168:1429–1436. 10.1007/s00431-009-0943-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkley JA, Munywoki P, Ngama M, Kazungu S, Abwao J, Bett A, Lassaunire R, Kresfelder T, Cane PA, Venter M, Scott JAG, Nokes DJ. 2010. Viral etiology of severe pneumonia among Kenyan infants and children. JAMA 303:2051–2057. 10.1001/jama.2010.675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Garcia ML, Calvo C, Pozo F, Villadangos PA, Perez-Brena P, Casas I. 2012. Spectrum of respiratory viruses in children with community-acquired pneumonia. Pediatr. Infect. Dis. J. 31:808–813. 10.1097/INF.0b013e3182568c67 [DOI] [PubMed] [Google Scholar]
- 7.Bogaert D, De Groot R, Hermans PWM. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4:144–154. 10.1016/S1473-3099(04)00938-7 [DOI] [PubMed] [Google Scholar]
- 8.Bosch AA, Biesbroek G, Trzcinski K, Sanders EA, Bogaert D. 2013. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 9:e1003057. 10.1371/journal.ppat.1003057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemon KP, Armitage GC, Relman DA, Fischbach MA. 2012. Microbiota-targeted therapies: an ecological perspective. Sci. Transl. Med.. 4:137rv135. 10.1126/scitranslmed.3004183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, Bruin J, Montijn R, Bonten M, Sanders E. 2011. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 6:e17035. 10.1371/journal.pone.0017035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilty M, Qi W, Brugger SD, Frei L, Agyeman P, Frey PM, Aebi S, Mühlemann K. 2012. Nasopharyngeal microbiota in infants with acute otitis media. J. Infect. Dis. 205:1048–1055. 10.1093/infdis/jis024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chappuy H, Keitel K, Gehri M, Tabin R, Robitaille L, Raymond F, Corbeil J, Maspoli V, Bouazza N, Alcoba G, Lacroix L, Manzano S, Galetto-Lacour A, Gervaix A. 2013. Nasopharyngeal carriage of individual Streptococcus pneumoniae serotypes during pediatric radiologically confirmed community acquired pneumonia following PCV7 introduction in Switzerland. BMC Infect. Dis. 13:357. 10.1186/1471-2334-13-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization Pneumonia Vaccine Trial Investigators Group. 2001. Standardization of interpretation of chest radiographs for the diagnosis of pneumonia in children. WHO document WHO/V&B/01.35. World Health Organization, Geneva, Switzerland [Google Scholar]
- 15.Raymond F, Carbonneau J, Boucher N, Robitaille L, Boisvert S, Wu WK, De Serres G, Boivin G, Corbeil J. 2009. Comparison of automated microarray detection with real-time PCR assays for detection of respiratory viruses in specimens obtained from children. J. Clin. Microbiol. 47:743–750. 10.1128/JCM.01297-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faveri M, Mayer MPA, Feres M, de Figueiredo LC, Dewhirst FE, Paster BJ. 2008. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol. Immunol. 23:112–118. 10.1111/j.1399-302X.2007.00397.x [DOI] [PubMed] [Google Scholar]
- 17.Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, de Waziers I, Cloarec O, Richards SE, Wang Y, Dumas M-E, Ross A, Rezzi S, Kochhar S, Van Bladeren P, Lindon JC, Holmes E, Nicholson JK. 2011. Colonization-induced host-gut microbial metabolic interaction. MBio 2:e00271–10. 10.1128/mBio.00271-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galetto-Lacour A, Alcoba G, Posfay-Barbe KM, Cevey-Macherel M, Gehri M, Ochs MM, Brookes RH, Siegrist CA, Gervaix A. 2013. Elevated inflammatory markers combined with positive pneumococcal urinary antigen are a good predictor of pneumococcal community acquired pneumonia in children. Pediatr. Infect. Dis. J. 32:1175–1179. 10.1097/INF.0b013e31829ba62a [DOI] [PubMed] [Google Scholar]
- 19.De Schutter I, De Wachter E, Crokaert F, Verhaegen J, Soetens O, Piérard D, Malfroot A. 2011. Microbiology of bronchoalveolar lavage fluid in children with acute nonresponding or recurrent community-acquired pneumonia: identification of nontypeable Haemophilus influenzae as a major pathogen. Clin. Infect. Dis. 52:1437–1444. 10.1093/cid/cir235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP. 2012. Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl. Environ. Microbiol. 78:6262–6270. 10.1128/AEM.01051-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shak JR, Vidal JE, Klugman KP. 2013. Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol. 3:129–135. 10.1016/j.tim.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.