Abstract
The comparison of 16S rRNA gene sequences is widely used to differentiate bacteria; however, this gene can lack resolution among closely related but distinct members of the same genus. This is a problem in clinical situations in those genera, such as Neisseria, where some species are associated with disease while others are not. Here, we identified and validated an alternative genetic target common to all Neisseria species which can be readily sequenced to provide an assay that rapidly and accurately discriminates among members of the genus. Ribosomal multilocus sequence typing (rMLST) using ribosomal protein genes has been shown to unambiguously identify these bacteria. The PubMLST Neisseria database (http://pubmlst.org/neisseria/) was queried to extract the 53 ribosomal protein gene sequences from 44 genomes from diverse species. Phylogenies reconstructed from these genes were examined, and a single 413-bp fragment of the 50S ribosomal protein L6 (rplF) gene was identified which produced a phylogeny that was congruent with the phylogeny reconstructed from concatenated ribosomal protein genes. Primers that enabled the amplification and direct sequencing of the rplF gene fragment were designed to validate the assay in vitro and in silico. Allele sequences were defined for the gene fragment, associated with particular species names, and stored on the PubMLST Neisseria database, providing a curated electronic resource. This approach provides an alternative to 16S rRNA gene sequencing, which can be readily replicated for other organisms for which more resolution is required, and it has potential applications in high-resolution metagenomic studies.
INTRODUCTION
Rapid and reliable identification of bacteria is fundamental to experimental microbiology, particularly in clinical settings where it is frequently necessary to distinguish organisms which are genetically very closely related but which have stable and distinct disease phenotypes. A good example is the genus Neisseria, which comprises mostly commensal inhabitants of the mucosal surfaces of humans and animals but includes two significant pathogens, Neisseria gonorrhoeae, the gonococcus, which causes gonorrhea, and Neisseria meningitidis, the meningococcus, which can cause meningitis and septicemia. As the meningococcus is an “accidental pathogen,” which is frequently carried but rarely invasive, species identification is particularly important in community studies, where meningococcal carriage rates are estimated in the presence of related species which are not easily distinguished using conventional methods. This is especially important when vaccines are being introduced, such as the recently developed protein-polysaccharide conjugate serogroup A vaccine (PsA-TT; MenAfriVac) (1). Conventional phenotypic identification of bacteria is time-consuming and difficult to deploy, especially in resource-limited settings, and may suffer from errors in interpretation leading to misidentification.
For isolate characterization purposes, approaches based on DNA sequencing offer accuracy and reproducibility with the additional advantage that the data generated can be transferred electronically and stored on public databases. For many years, sequence analysis of 16S rDNA, encoding 16S rRNA (ribosomal DNA sequencing), has played a principal role in this endeavor. In this approach, part or all of the 16S rRNA gene is sequenced, and identification is achieved by comparison of this sequence to curated sequences on web-accessible databases (for example http://www.ridom.de/rdna/ [2] and http://eztaxon-e.ezbiocloud.net/ [3]). The 16S rRNA gene has been a valuable target as it is ubiquitous and composed of both conserved and variable regions, allowing the design of universal PCR primers to generate nucleotide sequences that can be used to differentiate among isolates. The 16S rRNA molecule is so conserved, however, that very similar or identical sequences are frequently present in more than one species which have distinct and stable phenotypic properties (4–6).
Recently, ribosomal multilocus sequence typing (rMLST) (7) has been proposed as a method which provides an additional rational and universal approach to species classification. This approach exploits the availability of whole-genome sequence (WGS) data by indexing variation at the 53 genetic loci encoding the bacterial ribosomal protein genes. This method has been shown to unambiguously determine the species identity of Neisseria isolates, demonstrating good congruence with both whole-genome analyses and more conventional approaches (4). These data indicated that some species had been misidentified using conventional methods, and that minor changes in nomenclature were required (8). The rMLST approach, however, requires nucleotide sequence variation data at 53 loci and, although these are readily extracted from WGS data, such information is not always economically or practically available from all specimens. Therefore, the loci in the rMLST scheme were examined to identify a gene fragment from a single locus that can be used to rapidly identify Neisseria species in both the diagnostic and research settings. The target identified, a 413-bp fragment of the 50S ribosomal protein L6 (rplF) gene, includes both conserved regions suitable for primer design and variable regions to distinguish sequences from different Neisseria species. Comparison of the rplF gene fragments provided sufficient discrimination to identify most species within the genus accurately, rapidly, and inexpensively.
MATERIALS AND METHODS
Isolates and genome sequences.
Nucleotide sequences were obtained from 44 genomes which were part of the data set used to validate rMLST in Neisseria (4); a different set of 44 Neisseria DNA samples (a gift from Bachra Rokbi, Sanofi Pasteur, Marcy l'Etoile, France), which were used to validate the assay using Sanger sequencing (see Table S1 in the supplemental material); and 839 publicly available genome sequences downloaded from the PubMLST Neisseria database (http://pubmlst.org/neisseria/) (9), including those deposited as part of the MRF Meningococcus Genome Library (www.meningitis.org/research/genome). All isolates analyzed are listed in Table S2 in the supplemental material, including culture collection isolates and the type strains of Neisseria polysaccharea, Neisseria cinerea, Neisseria lactamica, Neisseria subflava, Neisseria mucosa, Neisseria oralis, Neisseria weaveri, Neisseria bacilliformis, Neisseria dentiae, Neisseria shayeganii, Neisseria canis, Neisseria wadsworthii, Neisseria animalis, and Neisseria elongata and the type strains of the previous species, Neisseria sicca, Neisseria macacae, and Neisseria flavescens (8).
Extracting and analyzing sequence data from the PubMLST Neisseria database.
Nucleotide sequences from the 53 concatenated ribosomal protein genes used in rMLST, the seven housekeeping gene fragments used in MLST, individual ribosomal protein genes, and the 16S rRNA gene were extracted from the PubMLST Neisseria database (9). Individual allele designations were also extracted from the database. Sequences were aligned with Muscle version 3.7 (10), and Mega5 (11) was employed to reconstruct phylogenies using the neighbor-joining method. Genetic distances were determined according to the Kimura two-parameter model (12), with all ambiguous positions removed from each pairwise sequence comparison and bootstrap values (13) based on 1,000 replications. DNA divergence between sequences was calculated using DnaSP5 (14), with fixed nucleotide sequence differences defined as sites at which all of the sequences in one sample are different from all the sequences in a second sample.
Nucleotide sequence determination.
The rplF fragment was amplified using the PCR primers rplF-F (5′-CAGTGACTGTTCCCGCTGGTGT-3′) and rplF-R (5′-AGGYTCAGGAGKWCGGAAHG-3′), which were designed using the primer-BLAST tool (15) available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and MEGA5 (11). For PCR amplification of the rplF gene fragment, reaction mixes were incubated for 35 cycles; each cycle consisted of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. PCR products were purified using a precipitation method (16) and the nucleotide sequences of the purified PCR products were determined on each DNA strand using the primers described above by cycle sequencing with Applied Biosystems BigDye ready reaction mix (Life Technologies), used in accordance with the manufacturer's instructions. Sequence termination reaction products were separated and the sequence data collected using an Applied Biosystems 3730 DNA analyzer (Life Technologies). Nucleotide sequence data from forward- and reverse-strand electropherograms were assembled into single contiguous sequences using SeqSphere (http://www.ridom.de/seqsphere/) and checked using the Staden suite of programs (17).
Defining rplF fragment alleles and associating with species.
The database was seeded with the first rplF fragment allele identified (arbitrarily assigned allele 1), and all genomes in the PubMLST Neisseria database were searched against this sequence (scanned) for the rplF fragment allele within the BIGSdb software using the BLAST algorithm (18). All variants with distinct nucleotide sequences were assigned unique allele designations. Each allele was also assigned a genospecies association, based on rMLST species designations (4), with type strains used to confirm these designations, where available. If genome sequences for type strains were unavailable, seven locus MLST data were used to confirm species identity (19). A reference table of alleles with associated genospecies was constructed within the PubMLST Neisseria database, which can be used to compare rplF fragment sequences to aid species identification. If an allele was obtained from a type strain or had an associated rMLST profile, the genospecies was considered confirmed; if not, it was considered provisional and labeled as such within the database.
RESULTS
Phylogenetic analysis of ribosomal protein genes.
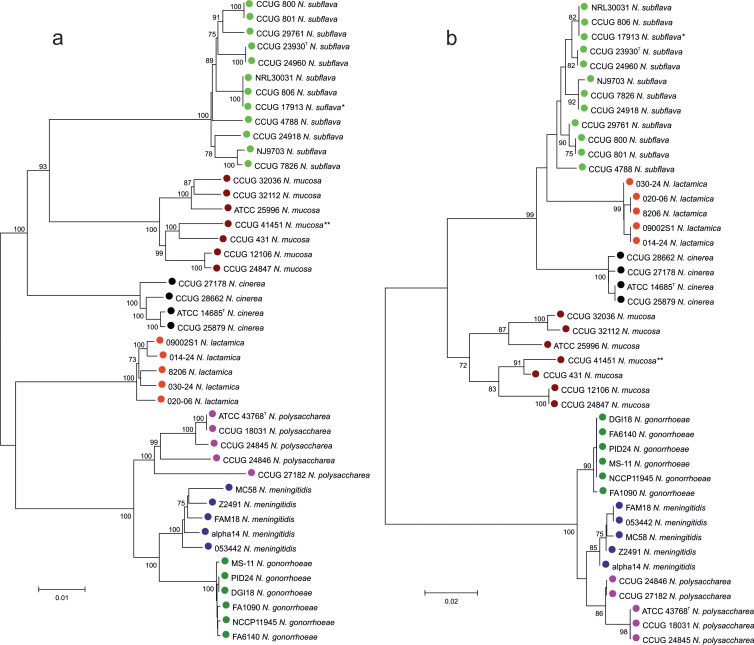
For the 44 Neisseria isolates for which WGS were available, phylogenies were generated from the 53 concatenated whole-ribosomal protein gene sequences used in rMLST and for each of the 53 ribosomal protein genes individually. These were compared to identify the single-locus tree that was most congruent with the 53-locus tree in terms of clustering the different taxa. The rplF gene phylogeny clustered the sequences consistently with the rMLST tree, and this locus was chosen for further analyses as it was of sufficient length and variability, with conserved flanking regions suitable for primer design. Sanger sequencing of the rplF gene using two primers designed from sequences extracted from the WGS data produced a nucleotide fragment of 413 bp, and this determined the length of the rplF fragment alleles for the assay. A phylogeny reconstructed from the rplF fragment alleles exhibited the same species clusters as the phylogeny produced from the 53 concatenated ribosomal protein gene sequences used in rMLST (Fig. 1).
FIG 1.
Evolutionary relationships among 44 Neisseria species based on concatenated sequences from 53 whole ribosomal protein genes and single rplF gene fragments. (a) Concatenated sequences from 53 ribosomal protein genes. (b) rplF gene fragments. Type strains of previous species: *, N. flavescens; **, N. macacae. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The unit of measure for the scale bars is the number of nucleotide substitutions per site.
rplF allele fragment variability.
A total of 27 rplF fragment alleles were identified among the set of 44 isolates used to validate the rplF assay in vitro, which included 10 Neisseria species (see Table S1 in the supplemental material). An examination of the allele sequences from these samples suggested that some isolates had been misidentified. For example, ATCC 19243, originally classified as N. subflava, has been identified as N. mucosa using rMLST. For some isolates, WGS data were unavailable and discrepancies were resolved by examining MLST loci. Of five isolates with rplF fragment allele 40, one had been previously identified as N. subflava, whereas four had been identified as N. sicca; however, they had almost identical MLST profiles, differing at only one or two loci and clustered with N. subflava when a phylogeny was reconstructed using concatenated MLST nucleotide sequences (data not shown). With the use of the rplF fragment alleles, an isolate identified previously as N. sicca with rplF fragment allele 58 was clustered with N. subflava. With the use of concatenated MLST sequences, this isolate also clustered with N. subflava, supporting the species designation identified by the rplF assay.
A total of 65 unique alleles of the rplF fragment were identified among 926 isolates present in the PubMLST Neisseria database at the time of analysis. Each allele was assigned to a genospecies as described previously (Table 1). N. mucosa, N. sicca, and N. macacae are now considered one species (N. mucosa), as they clustered as one group using rMLST (8). These organisms exhibit either indistinguishable (2) or highly similar 16S rRNA sequences (21). N. flavescens is now considered to be the same species as N. subflava, as these two species were indistinguishable using rMLST (8). The rplF fragment alleles were specific for each species group, except for allele 21, which was present in N. mucosa as well as a species previously defined as “Neisseria mucosa var. heidelbergensis” (22), now renamed N. oralis (23). Among WGS data for 804 N. meningitidis and 17 N. gonorrhoeae isolates, there were 6 and 2 unique rplF fragment alleles, respectively. The rplF fragment alleles from N. polysaccharea and N. meningitidis, the two species most closely related to the type species N. gonorrhoeae (4), were most similar to N. gonorrhoeae allele 7, with 10 and 12 nucleotide differences, respectively. Fixed nucleotide sequence differences were present among all species groups examined, with N. polysaccharea and N. meningitidis alleles having four and seven fixed differences, respectively, from allele 7. Although the sequences from N. polysaccharea and N. meningitidis were similar, there were 15 polymorphisms and 5 fixed differences that differentiated these two species. Compared to allele 7, the rplF fragment alleles from the other species of Neisseria were more distantly related, with the allele from a novel Neisseria species (isolate CCUG 21444), originally defined as N. cinerea, having 120 nucleotide differences.
TABLE 1.
Species associations of rplF fragment alleles among 926 Neisseria isolates
| Species | No. of isolates | No. of alleles | Allele(s)d | No. of polymorphic sites | No. of fixed differences |
|---|---|---|---|---|---|
| N. gonorrhoeae | 17 | 2 | 5, 7 | 1 | 1 |
| N. polysaccharea | 12 | 3 | 9, 39, 44 | 10 | 4 |
| N. meningitidis | 804 | 6 | 1, 2, 3, 4, 8, 18 | 12 | 7 |
| N. oralis | 4 | 4 | 21, 26, 36, 68 | 49 | 42 |
| N. elongata | 4 | 4 | 15, 37, 60, 75 | 50 | 45 |
| “N. bergeri”a | 1 | 1 | 16 | 63 | 63 |
| N. lactamica | 15 | 6 | 6, 32, 33, 34, 51, 52 | 65 | 56 |
| N. cinerea | 8 | 5 | 10, 19, 20, 45, 74 | 70 | 51 |
| N. subflavab | 34 | 14 | 11, 12, 23, 25, 31, 38, 40, 42, 43, 53, 56, 57, 58, 59 | 72 | 49 |
| N. animalis | 1 | 1 | 76 | 72 | 72 |
| N. mucosac | 16 | 12 | 13, 14, 17, 21, 22, 27, 28, 29, 30, 35, 41, 54 | 76 | 36 |
| N. dentiae | 1 | 1 | 47 | 82 | 82 |
| N. canis | 1 | 1 | 48 | 102 | 102 |
| N. wadsworthii | 1 | 1 | 77 | 102 | 102 |
| N. weaveri | 1 | 1 | 24 | 102 | 102 |
| N. bacilliformis | 4 | 2 | 46, 49 | 115 | 111 |
| N. shayeganii | 1 | 1 | 78 | 116 | 116 |
| Neisseria sp. (novel) | 1 | 1 | 50 | 120 | 120 |
Strain originally defined as N. polysaccharea (20), but rMLST shows that it is a distinct novel species (4) which has yet to be validly published.
Includes the previous species N. flavescens.
Includes the previous species N. sicca and N. macacae.
All alleles are compared to allele 7 from type species N. gonorrhoeae.
Comparison with 16S rRNA species identification.
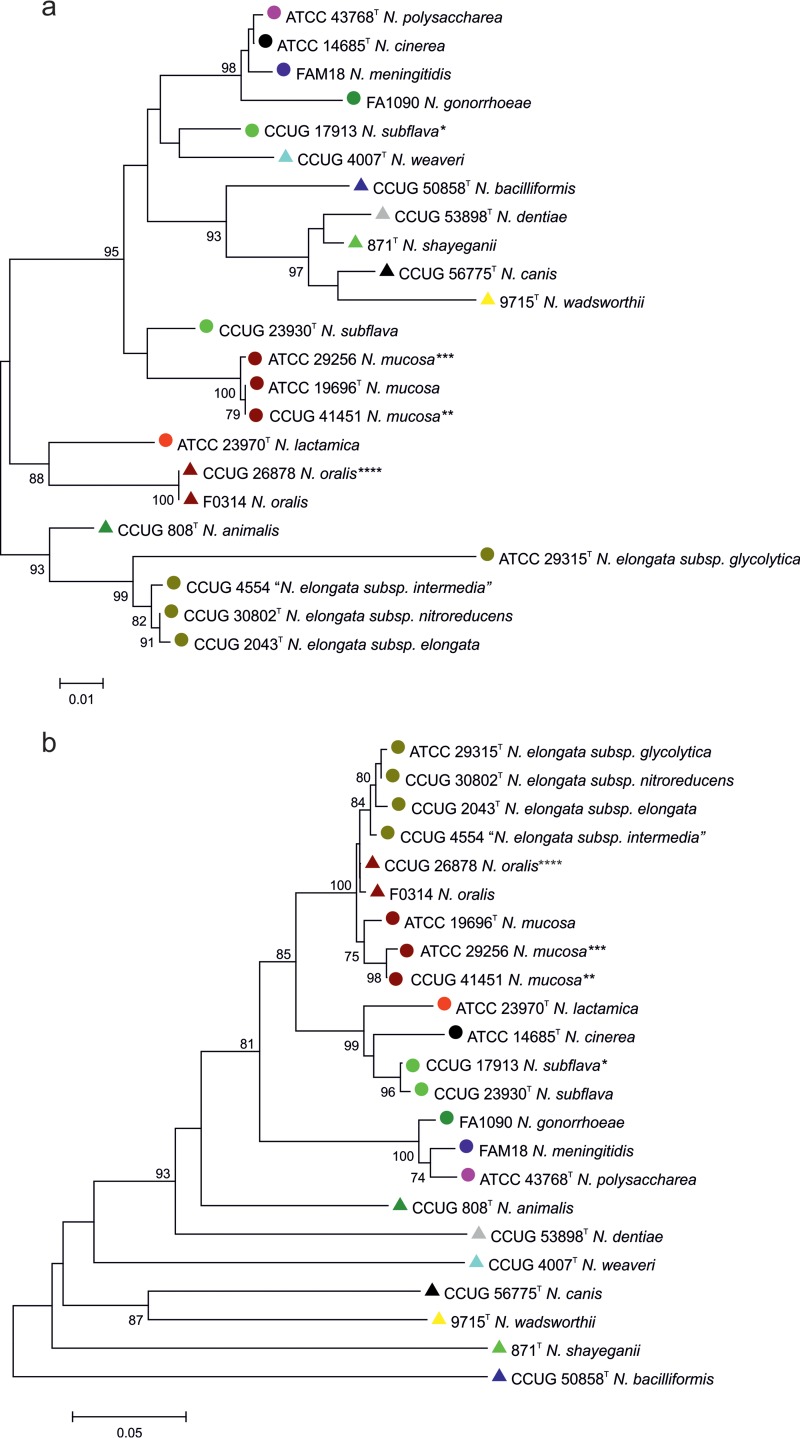
Comparison of a phylogeny reconstructed from the rplF fragment alleles from Neisseria type strains with a phylogeny reconstructed using 16S rRNA gene allele fragments (5) demonstrated improved resolution of members of the genus achieved with the rplF fragment phylogeny. Species relationships determined using rplF fragment alleles were more consistent with rMLST species identification and DNA-DNA hybridization studies (24) than relationships inferred from 16S rRNA gene phylogenies (Fig. 2). The rplF fragment allele phylogeny also clustered the more closely related species that are often found in the human oropharynx separately from the more distantly related species that are not associated with humans. A search of the PubMLST Neisseria database also revealed that some 16S rRNA gene sequences are present in both commensals and meningococci. For example, 16S rRNA gene fragment allele 5, originally identified in isolates belonging to the species N. polysaccharea and N. cinerea, including the type strain of N. cinerea, was harbored by three pathogenic serogroup W, meningococcal isolates. Allele 46 has also been found in both an N. polysaccharea isolate and a serogroup B, invasive meningococcus.
FIG 2.
Evolutionary relationships among Neisseria species based on fragments from 16S rRNA and rplF genes. (a) 16S rRNA gene fragments. (b) rplF gene fragments. T, type strain. Type strains of previous species: *, N. flavescens; **, N. macacae; ***, N. sicca; ****, N. mucosa var. heidelbergensis. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The unit of measure for the scale bars is the number of nucleotide substitutions per site.
Identifying Neisseria rplF fragment alleles using the PubMLST Neisseria database.
To identify a species using an rplF fragment, the PubMLST Neisseria database can be queried using the sequence query interface. Users should choose “rplF species” and then paste in their nucleotide sequence. If there is an exact match, an rplF genospecies designation is returned. If there are polymorphisms present, the closest match is shown and any nucleotide differences are identified and shown in an alignment, which can then be translated. All known rplF fragment alleles can be downloaded from the Neisseria locus/sequence definitions database in PubMLST, as can the rplF profiles. The Isolate database can also be searched for any related provenance data. In order to assign a new allele, novel rplF sequences can be submitted via PubMLST and a curator will then assign a provisional species identity by comparing the percentage identity to known species-specific alleles within the database and reconstructing a phylogeny using all known rplF fragment alleles and the novel allele.
DISCUSSION
The human body hosts a complex microbiota that is important in both health and disease (25). In the case of the genus Neisseria, for example, a variety of species colonize the mouth and oropharynx, with co-colonization providing a reservoir for horizontal genetic exchange (26). While most Neisseria species are harmless commensals, the meningococci and gonococci are important pathogens, and understanding the transition from commensal to pathogen is important in understanding their disease epidemiology (27). Phenotypic characteristics, such as nutritional requirements and biochemical tests, have provided the basis of diagnostic microbiology for many years; however, there are limitations with these methods and the results obtained can be ambiguous, with N. cinerea isolates, for example, being misidentified as gonococci (28, 29). Misidentification of Neisseria can have serious medicolegal consequences (28), as well as distorting the results of epidemiological studies.
Molecular techniques have increasingly replaced phenotypic approaches for characterizing commensal and pathogenic bacteria, with the sequencing of 16S rRNA gene fragments widely employed in diagnostic applications and studies of the microbiome (25, 30, 31). Limitations of this target, due to the similarity of 16S rRNA genes present in different species, are exemplified by Neisseria. For example, there are indistinguishable 16S rRNA gene sequences in N. polysaccharea, N. cinerea, and the meningococci, and some meningococci contain a 16S rRNA gene sequence identical to that found in gonococci (6). Further, public 16S rRNA databases, such as the Human Oral Microbiome database (32) and the EzTaxon-e database (3), can provide misleading results. The closest match to the 16S rRNA gene sequence from N. lactamica 020–06 (33) in both databases is a meningococcal sequence.
A variety of other approaches have been investigated to address this problem, for example, the phylogenetic analysis of the nucleotide sequences of the seven MLST loci, sometimes referred to as multilocus sequence analysis (MLSA) (34). This approach was very effective in distinguishing N. meningitidis, N. gonorrhoeae, and N. lactamica (19) but did not group all members of the genus into species-specific clusters (4). Another method with promise is matrix-assisted laser desorption–ionization time of flight mass spectrometry (MALDI-TOF); however, this method requires optimization, as it has been shown only to separate Neisseria into three groups, N. meningitis, N. gonorrhoeae, and other species (35). The availability of rapid and inexpensive whole-genome sequencing and the gene-by-gene approach (36), as implemented in the BIGSdb software (9), has allowed techniques to be developed such as rMLST, which unambiguously identifies species and accurately determines relationships among Neisseria species (4, 7); however, rMLST requires WGS data or the analysis of multiple sequences which, while definitive, is not necessarily feasible or cost-effective for clinical specimens.
A short (413-bp) fragment of the rplF gene which encodes the 50S ribosomal protein L6 was found to be a suitable genetic target for rapid differentiation within Neisseria species, as phylogenies reconstructed from rplF fragment alleles were consistent with a phylogeny reconstructed from the concatenated sequences of 53 whole-ribosomal protein genes. The rplF gene variable region is flanked by conserved regions, a characteristic that enables this fragment to be sequenced on both DNA strands with two primers. Among 65 distinct alleles of this gene fragment identified among 926 isolates, none were shared among commensals and pathogens or between the meningococci and the gonococci, confirming the suitability of the rplF fragment assay in differentiating pathogenic and commensal Neisseria species. Only one fragment allele (rplF 21) was found in more than one species (N. oralis and N. mucosa), neither of which have been known to cause disease. Although the sequence clusters obtained with the rplF fragment alleles were the same as those obtained with concatenated ribosomal protein gene sequences, the phylogeny reconstructed from them was not identical. Consequently, this single genetic target should not be used on its own to define a species or used as a replacement for rMLST. The rplF assay is, however, a practical, rapid and inexpensive single-locus tool to differentiate among species within the genus Neisseria which can be combined with additional single-locus tests, such as porA sequencing (37) and capsule gene sequencing (38), for example, to confirm meningococcal identity. The assay was specifically developed to identify Neisseria species as part of the MenAfriCar study and has been successfully used to characterize thousands of samples from heat-killed cell suspensions, assisting in determining the impact of serogroup A polysaccharide conjugate vaccines on meningococcal carriage (1, 39).
The rplF fragment allele sequences and associated metadata are stored in the PubMLST Neisseria database. It is curated and continually updated, providing an extensive library of genomes and DNA sequences along with the tools to analyze these data. Although the majority of the isolates are meningococci, it contains a number of representative strains from most species, including culture collection strains, as well as isolates from population studies, which can be used to query sequences to provide a species identity. While the rplF gene fragment assay is specific for Neisseria, the general approach can be adapted to identify other bacterial species, as the rp genes are universal (7). However, the rplF gene fragment assay has not, at the time of this writing, been adapted to identify species within other genera. In addition to species identification, ribosomal genes have potential applications in the investigation of noncultured samples and in metagenomic studies, where resolution finer than that provided by the 16S rRNA gene is required. (20).
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by The Wellcome Trust. M. C. J. Maiden is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science. This publication made use of the Meningitis Research Foundation Meningococcus Genome Library (www.meningitis.org/research/genome) developed by Public Health England, the Wellcome Trust Sanger Institute, and the University of Oxford as a collaboration. The project is funded by the Meningitis Research Foundation.
We thank Bachra Rokbi at Sanofi Pasteur, Marcy l'Etoile, France, for the provision of Neisseria DNA samples.
Footnotes
Published ahead of print 12 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03529-13.
REFERENCES
- 1.Doumagoum Moto D, Gami JP, Gamougam K, Naibei N, Mbainadji L, Narbé M, Toralta J, Kodbesse B, Ngadoua C, Coldiron M, Ferman F, Page A-L, Djingarey M, Hugonnet S, Harrison OB, Rebbetts LS, Tekletsion Y, Watkins ER, Caugant DA, Chandramohan D, Hassan-King M, Manigart O, Nascimento M, Woukeu A, Trotter C, Stuart JM, Maiden MCJ, Greenwood B. 2013. Effect of a serogroup A meningococcal conjugate vaccine (PsA-TT) on serogroup A meningococcal meningitis and carriage in Chad: a community study [corrected]. Lancet 83:40–47. 10.1016/S0140-6736(13)61612-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harmsen D, Singer C, Rothganger J, Tonjum T, de Hoog GS, Shah H, Albert J, Frosch M. 2001. Diagnostics of Neisseriaceae and Moraxellaceae by ribosomal DNA sequencing: ribosomal differentiation of medical microorganisms. J. Clin. Microbiol. 39:936–942. 10.1128/JCM.39.3.936-942.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62:716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- 4.Bennett JS, Jolley KA, Earle SG, Corton C, Bentley SD, Parkhill J, Maiden MC. 2012. A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiology 158:1570–1580. 10.1099/mic.0.056077-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harmsen D, Rothganger J, Frosch M, Albert J. 2002. RIDOM: ribosomal differentiation of medical microorganisms database. Nucleic Acids Res. 30:416–417. 10.1093/nar/30.1.416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walcher M, Skvoretz R, Montgomery-Fullerton M, Jonas V, Brentano S. 2013. Description of an unusual N. meningitidis isolate containing and expressing N. gonorrhoeae-specific 16S rRNA gene sequences. J. Clin. Microbiol. 51:3199–3206. 10.1128/JCM.00309-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jolley KA, Bliss CM, Bennett JS, Bratcher HB, Brehony CM, Colles FM, Wimalarathna HM, Harrison OB, Sheppard SK, Cody AJ, Maiden MC. 2012. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology 158:1005–1015. 10.1099/mic.0.055459-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett JS, Jolley KA, Maiden MC. 2013. Genome sequence analyses show that Neisseria oralis is the same species as “Neisseria mucosa var. heidelbergensis.” Int. J. Syst. Evol. Microbiol. 63:3920–3926. 10.1099/ijs.0.052431-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- 14.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 15.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Embley TM. 1991. The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett. Appl. Microbiol. 13:171–174. 10.1111/j.1472-765X.1991.tb00600.x [DOI] [PubMed] [Google Scholar]
- 17.Staden R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233–241. 10.1007/BF02900361 [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- 19.Bennett JS, Jolley KA, Sparling PF, Saunders NJ, Hart CA, Feavers IM, Maiden MC. 2007. Species status of Neisseria gonorrhoeae: evolutionary and epidemiological inferences from MLST. BMC Biol. 5:35. 10.1186/1741-7007-5-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger U. 1985. First isolation of Neisseria polysacchareae species nova in the Federal Republic of Germany. Eur. J. Clin. Microbiol. 4:431–433. 10.1007/BF02148705 [DOI] [PubMed] [Google Scholar]
- 21.Tonjum T. 2005. Genus I. Neisseria, p 777–798 In Garrity GM, Brenner DJ, Krieg NR, Staley JR. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer-Verlag, New York, NY [Google Scholar]
- 22.Berger U. 1971. Neisseria mucosa var. heidelbergensis. Z Med. Mikrobiol. Immunol. 156:154–158 (In German.) 10.1007/BF02124646 [DOI] [PubMed] [Google Scholar]
- 23.Wolfgang WJ, Passaretti TV, Jose R, Cole J, Coorevits A, Carpenter AN, Jose S, Van Landschoot A, Izard J, Kohlerschmidt DJ, Vandamme P, Dewhirst FE, Fisher MA, Musser KA. 2013. Neisseria oralis sp. nov. isolated from healthy gingival plaque and clinical samples. Int. J. Syst. Evol. Microbiol. 63(Pt 4):1323–1328. 10.1099/ijs.0.041731-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guibourdenche M, Popoff MY, Riou JY. 1986. Deoxyribonucleic acid relatedness among Neisseria gonorrhoeae, N. meningitidis, N. lactamica, N. cinerea and “Neisseria polysaccharea.” Ann. Inst. Pasteur. Microbiol. 137B:177–185 [DOI] [PubMed] [Google Scholar]
- 25.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J. Bacteriol. 192:1422–1431. 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong Y, Ma JH, Warren K, Tsang RS, Low DE, Jamieson FB, Alexander DC, Hao W. 2013. Homologous recombination drives both sequence diversity and gene content variation in Neisseria meningitidis. Genome Biol. Evol. 5:1611–1627. 10.1093/gbe/evt116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maiden MC. 2008. Population genomics: diversity and virulence in the Neisseria. Curr. Opin. Microbiol. 11:467–471. 10.1016/j.mib.2008.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dossett JH, Appelbaum PC, Knapp JS, Totten PA. 1985. Proctitis associated with Neisseria cinerea misidentified as Neisseria gonorrhoeae in a child. J. Clin. Microbiol. 21:575–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knapp JS, Totten PA, Mulks MH, Minshew BH. 1984. Characterization of Neisseria cinerea, a nonpathogenic species isolated on Martin-Lewis medium selective for pathogenic Neisseria spp. J. Clin. Microbiol. 19:63–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 8:e1002606. 10.1371/journal.pcbi.1002606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huse SM, Ye Y, Zhou Y, Fodor AA. 2012. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 7:e34242. 10.1371/journal.pone.0034242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010:baq013. 10.1093/database/baq013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett JS, Bentley SD, Vernikos GS, Quail MA, Cherevach I, White B, Parkhill J, Maiden MCJ. 2010. Independent evolution of the core and accessory gene sets in the genus Neisseria: insights gained from the genome of Neisseria lactamica isolate 020–06. BMC Genomics 11:652. 10.1186/1471-2164-11-652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de Peer Y, Vandamme P, Thompson FL, Swings J. 2005. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3:733–739. 10.1038/nrmicro1236 [DOI] [PubMed] [Google Scholar]
- 35.Ilina EN, Borovskaya AD, Malakhova MM, Vereshchagin VA, Kubanova AA, Kruglov AN, Svistunova TS, Gazarian AO, Maier T, Kostrzewa M, Govorun VM. 2009. Direct bacterial profiling by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for identification of pathogenic Neisseria. J. Mol. Diagn. 11:75–86. 10.2353/jmoldx.2009.080079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maiden MC, van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11:728–736. 10.1038/nrmicro3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell JE, Jolley KA, Feavers IM, Maiden MC, Suker J. 2004. PorA variable regions of Neisseria meningitidis. Emerg. Infect. Dis. 10:674–678. 10.3201/eid1004.030247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison OB, Claus H, Jiang Y, Bennett JS, Bratcher HB, Jolley KA, Corton C, Care R, Poolman JT, Zollinger WD, Frasch CE, Stephens DS, Feavers I, Frosch M, Parkhill J, Vogel U, Quail MA, Bentley SD, Maiden MCJ. 2013. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg. Infect. Dis. 19:566–573. 10.3201/eid1904.111799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ali O, Aseffa A, Bedru A, Lemma T, Moti T, Workhu A, Guebre Xabher H, Yamuah L, Boukary RM, Collard JM, Dano ID, Habiboulaye I, Issaka B, Jusot JF, Ousmane S, Rabe I, Gami JP, Gamougam K, Kodbesse B, Naibei N, Ngadoua C, Mbainadji L, Moto DD, Narbe M, Toralta J, Berthe A, Keita M, Diallo K, Onwuchekwa U, Sow SO, Tamboura B, Traore A, Toure A, Amodu M, Beida O, Gadzama G, Omotara B, Sambo Z, Yahya S, Chandramohan D, Greenwood B, Hassan-King M, Manigart O, Nascimento M, Stuart J, Woukeu A, Bai X, Borrow R, Findlow H, Avalo S, Bassene H, Diallo A, Dieng M, Doucoure S, Gomis JF, Ndiaye A, Sokhna C, Trape JF, Akalifa B, Forgor A, Hodgson A, Osei I, Quaye S, Williams J, Wontuo P, Basta N, Irving T, Trotter C, Bennett J, Hill D, Harrison O, Rebbetts L, Maiden M, Tekletsion Y, Watkins E. 2013. Meningococcal carriage in the African meningitis belt. Trop. Med. Int. Health 18:968–978. 10.1111/tmi.12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.