Abstract
We developed a practical and easy two-step multiplex PCR assay to aid in serotyping of Streptococcus suis. The assay accurately typed almost all of the serotype reference strains and field isolates of various serotypes and also identified the genotypes of capsular polysaccharide synthesis gene clusters of some serologically nontypeable strains.
TEXT
Streptococcus suis is an important zoonotic pathogen that causes meningitis, septicemia, endocarditis, and other diseases in pigs and humans. S. suis strains have been classified into 35 serotypes (serotypes 1 to 34 and 1/2, which reacts with both serotype 1 and 2 typing sera) (1–4) on the basis of antigenic differences in their capsular polysaccharide (CP) (5). Serotyping of S. suis is one of the most useful methods to understand the epidemiology of a particular outbreak and monitor the prevalence of potentially hazardous strains. However, serotyping with all 35 typing antisera is time-consuming, and preparing the antisera is not easy due to the high cost and labor associated with its production. Additionally, cross-reactions in coagglutination tests with typing antisera and the presence of autoagglutinating strains increase the difficulty of serotyping in some cases. Therefore, the development of more practical and easier serotyping methods is desired.
CP synthesis (cps) genes are clustered on a single locus of the chromosome in S. suis (6, 7). We recently sequenced and analyzed the cps gene clusters of all 35 serotype reference strains (8) and reported that 31 (serotypes 3 to 13 and 15 to 34) possessed serotype-specific genes, while the cps gene clusters of serotypes 1 and 14 and serotypes 2 and 1/2 were almost identical in each pair (8). Liu et al. (9) recently developed multiplex PCR assays to target serotype-specific cps genes for the molecular serotyping of S. suis. In their method, the reference strains of 33 serotypes (serotypes 1 to 31, 33, and 1/2) were sorted into their respective serotypes by three multiplex PCR assays, although serotypes 1 and 1/2 were not distinguished from serotypes 14 and 2, respectively (9). In addition, 84 isolates from a patient and clinically healthy pigs, which covered 20 serotypes, were correctly assigned serotypes predicted by coagglutination tests, except for those of a new serotype (serotype 21/29) (9). However, these methods have not yet been validated using field isolates from diseased pigs as well as nontypeable ones by the coagglutination test. Moreover, serotypes 32 and 34 were not included as targets for typing, because the reference strains of these serotypes have been reclassified as Streptococcus orisratti (10). In addition to the serotype 32 and 34 reference strains, those of serotypes 20, 22, 26, and 33 were also recently suggested to be removed from the taxon of S. suis (11). However, whether all isolates of these serotypes actually belong to a species that is different from S. suis remains unclear. From a diagnostic point of view, the isolates of these serotypes recovered from diseased pigs are still identified as S. suis (12).
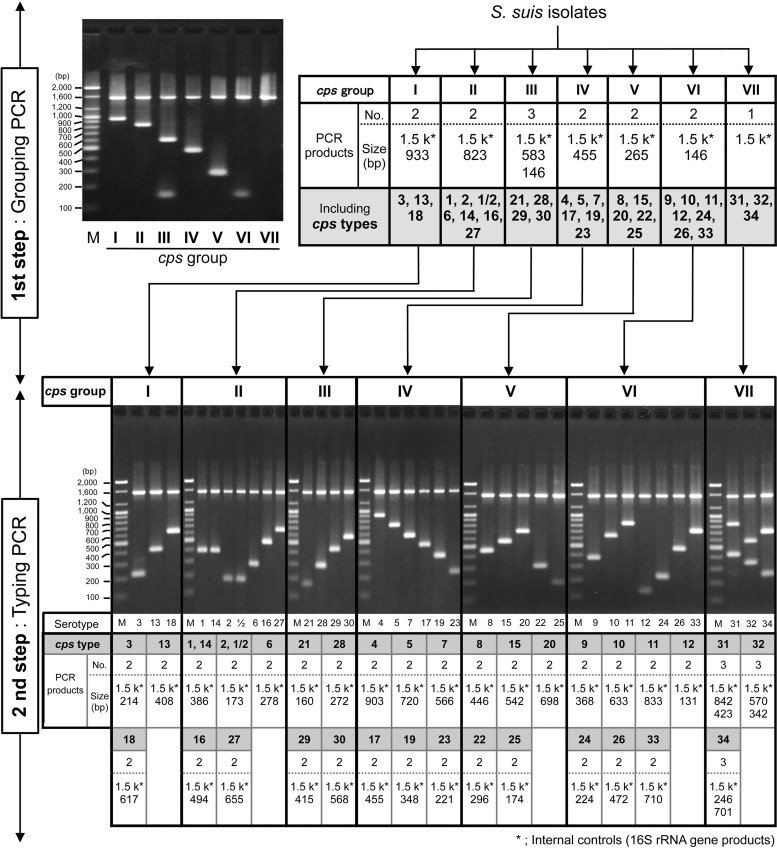
In the present study, we developed a novel method to type the 35 cps gene clusters of S. suis and validated its usefulness with 483 isolates from diseased and healthy animals and human patients, which covered 30 serotypes. With our methods (cps typing), we determined cps types, which were numbered corresponding to the serotypes, using only two multiplex PCRs (Fig. 1). The first PCR (grouping PCR) detected cps genes conserved in multiple serotypes and classified the tested strains into seven cps groups. The second PCR (typing PCR) detected cps genes specific to the respective serotypes of each group and identified the cps type of the strain.
FIG 1.
Scheme of cps typing using a two-step multiplex PCR assay and the PCR products amplified from 35 S. suis serotype reference strains. The first PCR (grouping PCR) classified the tested strains into 7 groups (cps groups I to VII). Numbers and sizes of grouping PCR products amplified from strains of each cps group and the electrophoresis image are shown in the upper panel. Approximately 1.5-kbp bands that appeared in all grouping PCR assays are the internal control products (16S rRNA gene products). Only the internal control products were amplified from cps group VII strains. The strains classified by grouping PCR were then subjected to the second PCR (typing PCR) using primers specific to the respective cps groups in order to identify the cps type of each strain. Number and size of typing PCR products amplified from all S. suis serotype reference strains and the electrophoresis images are shown in the bottom panels. Approximately 1.5-kbp bands that appeared in all typing PCR assays are the internal control products (16S rRNA gene products). The serotypes of reference strains are indicated below the lanes. All PCR products were electrophoresed on a 2% agarose gel (100 V, 40 min), stained with ethidium bromide, and photographed under UV light. Lanes M show DNA molecular weight markers (in thousands [k]) (100-bp DNA ladder; Bioneer, Daejeon, South Korea).
The target cps genes, primer sequences, and predicted product size(s) of each PCR are shown in Table 1. Target genes were selected on the basis of the clustering analysis data of all cps gene products reported previously (8). Universal primers for the amplification of 16S rRNA genes (13) were used as internal controls for all reactions. The genomic DNA of strains/isolates was extracted as described previously (for determination of the sensitivity of PCR assays) (8) or using InstaGene Matrix (Bio-Rad, Hercules, CA) following the manufacturer's instructions. All PCR mixtures contained 1× Qiagen multiplex master mix (Qiagen, Hilden, Germany) and 0.2 μM each primer. The PCR conditions were as follows: an initial activation at 95°C for 15 min followed by 30 cycles of denaturation at 94°C for 30 s, primer annealing at 60°C (for grouping PCR) or 58°C (for typing PCR) for 90 s, and extension at 72°C for 90 s, and then a final extension at 72°C for 10 min. The 483 putative S. suis isolates used to validate the developed assays were isolated from diseased pigs (394 isolates), clinically healthy pigs (60 isolates), diseased cows (2 isolates), healthy cows (17 isolates), a diseased sheep (1 isolate), and patients (9 isolates). All isolates tested were positive for the gdh-PCR assay (data not shown), which specifically detects the glutamate dehydrogenase genes of S. suis strains, including the reference strains of serotypes 20, 22, 26, and 32 to 34 (14). All isolates were also serotyped by coagglutination tests in the reference laboratory (University of Montreal) as previously described (15).
TABLE 1.
Primer sequences used in this study and the predicted sizes of the PCR products
| Target cps group or type | Primer sequences (5′ to 3′) |
Target gene(s)a | Putative gene product(s) (HGb) | Size (bp) of products | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| For grouping PCRc | |||||
| I | TGGTTCAAATATCAATGCTC | ATTGGTTGTGAGTGCATTG | cps3G, cps13G, cps18G | Aminotransferase (HG41) | 933 |
| IId | TCAAAATACGCACCTAAGGC | CACTCACCTGCCCCAAGAC | cps1Q, cps1/2P, cps2P, cps6Q, cps14P, cps16R, cps27P | N-acetylneuraminic acid synthase NeuB (HG10) | 823 |
| IIIe,f | TGATTTGGGTGAGACCATG | CTCATGCTGGATAACACGT | cps21R, cps28P, cps29Q, cps30N | N-acetylfucosamine synthase FlnC (HG26) | 583 |
| IV | ACAGTCGGTCAAGATAATCG | TCAGCTTGGGTAATATCTGG | cps4F-G, cps5F-G, cps7F-G, cps17G-H, cps19G-H, cps23F-G | Initial sugar transferase (HG21) and aminotransferase (HG22) | 455 |
| Vg | GGAAAGATGGAGGACCAGC | CCAACCAGACTCATATCCCC | cps8E, cps15E, cps20E, cps22E, cps25E | Initial sugar transferase (HG6) | 265 |
| III and VIe,h | GATGCCCCAAGCGATATGCC (F1), GACGCACCAAGTGATATGCC (F2) | GGACCAACAATGGCCATCTC (R1), GGTCCGACAATAGCCATTTC (R2) | Group III: cps21G, cps28F, cps29G, cps30F; group VI, cps9F, cps10F, cps11F, cps12F, cps24G, cps26H, cps33G | Initial sugar transferase (HG8) | 146 |
| For typing PCR | |||||
| cps group I | |||||
| 3 | GGTTTTGATTGGTCTAGTTG | CTCTAAAGCTCGATATCTAC | cps3L | Wzy polymerase (HG90) | 214 |
| 13 | TATGGTTAAAGGTGGAACTG | CCTTGTATATATTCCCTCCA | cps13L | Wzy polymerase (HG148) | 408 |
| 18 | TAATGGGATAGTTGCGTTAC | ATACATAAAGTTGTCCTGCG | cps18N | Wzy polymerase (HG170) | 617 |
| cps group II | |||||
| 2 and 1/2 | TTAGCAACGTTGCCAATAAG | AATCCTCCATTAAAACCCTG | cps2I, cps1/2I | Wzy polymerase (HG54) | 173 |
| 6 | GCTCACTATTTTTACATTACAC | TATTACTCCGCCAAATACAG | cps6I | Wzy polymerase (HG109) | 278 |
| 1 and 14 | TTAGACAGACACCTTATAGG | CTAGCTTCGTTACTTGATTC | cps14H, cps1I | Wzy polymerase (HG50) | 386 |
| 16 | AAGGTTATCCACGAAAGATG | TCCGGCAATATTCTTTCAAG | cps16I | Wzy polymerase (HG362) | 494 |
| 27 | AGACACTGCTTGCATTATTG | TCAGAATTACTTCCTGTTGC | cps27K | Wzy polymerase (HG246) | 655 |
| cps group III | |||||
| 21 | TATCATATTGAGAATCTTCCC | TTGCGTAGCATACAAAGTTC | cps21P | Wzy polymerase (HG194) | 160 |
| 28 | ATTATGTTGGTTGCAGAAGG | CGACTCAATTGTTGTAGTAG | cps28L | Wzy polymerase (HG254) | 272 |
| 29 | TTCTGGGATTTTAGGAATGC | CATGAAATACGCACTTGTAC | cps29L | Wzy polymerase (HG259) | 415 |
| 30 | TATTGCACTAGCTTCAGAAC | TGCATCCATAGTTGTATTCG | cps30I | Wzy polymerase (HG264) | 568 |
| cps group IV | |||||
| 4 | GACTATCTGTATACCCAAAC | TCCTTCCAAGTATTCTCTAG | cps4K | Wzy polymerase (HG96) | 903 |
| 5 | ATCTTAGGAATGATTCGGAC | ACCAGATATCTGAGCAAATG | cps5L | Wzy polymerase (HG103) | 720 |
| 7 | AACTACCTACCTGAACTTTG | AGTCTAAAAGTGATCGAGTC | cps7L | Wzy polymerase (HG113) | 566 |
| 17 | TAGCATCAGTTTATACGAGG | TAGTTTATCTGTGACACACC | cps17O | Wzy polymerase (HG164) | 455 |
| 19 | GTGTCGCAAATCAAGTATTG | AAGCTAGTACAACAAGCATG | cps19L | Wzy polymerase (HG174) | 348 |
| 23 | TAATGTATGCTCTGTCACTG | AACGAAACGGAATAGTTTGC | cps23J | Wzy polymerase (HG213) | 221 |
| cps group V | |||||
| 8 | AAATAAGGTAGGAGCTACTC | ATCCAACCTTAGCTTTCTGT | cps8K | Wzy polymerase (HG120) | 446 |
| 15 | ATCGTTTTGAGATTGAGTGG | TAAACGGATTCGGTTACTCA | cps15K | Wzy polymerase (HG153) | 542 |
| 20 | TGTGGATTTCTGGGATAATC | TGTGGACGAATTACTACTTG | cps20I | Wzy polymerase (HG182) | 698 |
| 22 | GCATTATCAGGATTCTTTCC | CCAATTGGGTGTTCAAAAAG | cps22K | Wzy polymerase (HG200) | 296 |
| 25 | GTTTGCTCCGATCATAATAG | CCAGTAAAAGGACTCAATAC | cps25M | Wzy polymerase (HG229) | 174 |
| cps group VI | |||||
| 9 | GAAAGTAGGTATATCTCAGC | GGGCTATTAAAACTCCTATC | cps9J | Wzy polymerase (HG123) | 368 |
| 10 | TTTCCCATTTGCTTATGGAC | GGAATAAAAACGATTGGGAG | cps10M | Wzy polymerase (HG130) | 633 |
| 11 | ATGCGATTGCAACAATTGAC | AGGCATGAGTAATACATAGG | cps11N | Wzy polymerase (HG138) | 833 |
| 12 | AACAGGTATTTCAGGATTGC | CTCGGATAAAGATAATCAGC | cps12J | Wzy polymerase (HG140) | 131 |
| 24 | TACTGAGATTTATTGGGACG | AAGCGATTGGATTACATTGC | cps24M | Wzy polymerase (HG220) | 224 |
| 26 | TTATACCGAAATTTTGTTGCC | CGTCAATCATATAAAGTGGG | cps26P | Wzy polymerase (HG240) | 472 |
| 33 | GATGTTTTCAACAGGTGTAC | CAAAGTACCTATTTTCAGCG | cps33K | Wzy polymerase (HG286) | 710 |
| cps group VII | |||||
| 31 | ACAATCGTTTCTGCAATACG | GATGAAAACATCGTTGGTAG | cps31L | Wzy polymerase (HG274) | 842 |
| ATCAGTAGTGGGAATAGTTG | TTTACTGTTTTTCGACCGTG | cps31G | Initial sugar transferase (HG269) | 423 | |
| 32 | AACCGCTGTTGAATTAAGAG | TTCGTTAGTTGAACTGTTCC | cps32H | Wzy polymerase (HG281) | 570 |
| TAGGACTATGGTTCCTAATG | TATTCTAGTTCAAGTCGCTC | cps32E | Initial sugar transferase (HG278) | 342 | |
| 34 | AAGTTTCATTCGAGGACTTC | GTATATAACACCGCAAGAAG | cps34M | Wzy polymerase (HG287) | 246 |
| ATACAGTGATGTCTTGCAAC | ATTGCTTTTTGACAATCGGC | cps34E | Initial sugar transferase (HG290) | 701 | |
| For all PCR | |||||
| Internal control | GAGTTTGATCCTGGCTCAG | AGAAAGGAGGTGATCCAGCC | 16S rRNA | 1,542–1,553 | |
Refer to our previous study (8) for gene names.
HG, homology group of putative cps gene products assigned in our previous study (8).
Only the internal control products were amplified from cps group VII strains.
Serotype 13 reference strain has a homologue of target genes for cps group II (putative N-acetylneuraminic acid synthase gene, cps13O). However, the primers were designed to not detect this gene.
In addition to the internal control products, cps group III strains produced two cps gene products by the grouping PCR (146- and 583-bp products), and the 146-bp product is common with cps group VI strains.
Serotype 17 reference strain has a homologue of target genes for cps group III (putative N-acetylfucosamine synthase gene, cps17R). However, the primers were designed to not detect this gene.
All serotype reference strains in cps-group II also possess the homologues of target genes for cps group V (genes encoding putative initial sugar transferase of HG6). However, the primers were designed not to detect these strains.
Because the genes encoding putative initial sugar transferase of HG8 have sequence variations among serotype reference strains in groups III and VI, two primers were designed for each primer site to amplify the 146-bp products. Of note, all reference strains in groups III and VI were detected only when the four primers were mixed, while with every single primer pair (forward 1 [F1]-reverse 1 [R1], F1-R2, F2-R1, or F2-R2), some of the reference strains in groups III and VI were not detected.
Our cps typing assigned all serotype reference strains, including those of serotypes 32 and 34, to the expected cps types. However, serotypes 1 and 1/2 could not be distinguished from serotypes 14 and 2, respectively, which is consistent with previous observations with the reported molecular serotyping method (9) (Fig. 1). Although the developed methods accurately assigned nearly all field isolates (341/357; 95.5%) to the cps types predicted from their serotypes, 16 of them were not typed by cps typing (Table 2). These nontypeable isolates may have mismatch sequences in their primer sites or may be novel cps types that cross-react with known typing sera. All PCR assays were repeated on three different occasions using representative strains/isolates from all serotypes and nontypeable isolates to confirm the reproducibility, and the same results were obtained (data not shown). The detection limit of the assays determined using reference strains was 1 to 100 pg of purified DNA/reaction (equivalent to approximately 4 × 102 to 4 × 104 copies of chromosome), which was comparable to or slightly better than those of the reported method (9).
TABLE 2.
Results of serotyping and cps typing of the 483 isolates tested
| Serotype using antiserum | No. of isolates | cps type(s) using PCR assays (no. of isolates) |
|---|---|---|
| 1 | 11 | 1 or 14 (9), NT (2)a |
| 1/2 | 33 | 2 or 1/2 (33) |
| 2 | 95 | 2 or 1/2 (95) |
| 3 | 19 | 3 (19) |
| 4 | 24 | 4 (21), NT (3)a |
| 5 | 8 | 5 (8) |
| 6 | 6 | 6 (6) |
| 7 | 12 | 7 (12) |
| 8 | 24 | 8 (24) |
| 9 | 29 | 9 (26), NT (3)a |
| 10 | 3 | 10 (3) |
| 11 | 3 | 11 (2), NT (1)a |
| 12 | 2 | 12 (2) |
| 13 | 1 | NT (1)a |
| 14 | 15 | 1 or 14 (15) |
| 16 | 17 | 16 (17) |
| 17 | 2 | 17 (2) |
| 19 | 3 | 19 (3) |
| 21 | 2 | 21 (2) |
| 22 | 2 | NT (2)a |
| 23 | 8 | 23 (8) |
| 24 | 4 | 24 (4) |
| 25 | 1 | 25 (1) |
| 27 | 1 | 27 (1) |
| 28 | 8 | 28 (8) |
| 29 | 4 | 29 (4) |
| 30 | 6 | 30 (2), NT (4)a |
| 31 | 6 | 31 (6) |
| 33 | 7 | 33 (7) |
| 34 | 1 | 34 (1) |
| Nonserotypeableb | 126 | 2 or 1/2 (9),c 3 (2),c 5 (2),c 7 (1),c 9 (6),c 10 (3),c 11 (3),c 12 (3),c 15 (4),c 16 (1),c 21 (1),c 23 (1),c 25 (1),c 28 (3),c 29 (1),c 30 (2),c 31 (9),c NT (74)d |
Isolates that were typeable by coagglutination tests with typing sera but nontypeable (NT) by cps typing PCR assays.
Nonserotypeable isolates, include autoagglutinating, multiagglutinating, and nonagglutinating strains.
Isolates that were typeable by cps typing PCR assays but not by the coagglutination test.
Isolates that were nontypeable by either coagglutination tests or cps typing PCR assays.
A total of 52 of the 126 nontypeable isolates by coagglutination tests (41.3%) were typed by cps typing (Table 2). Three of them (NL146, NL194, and NL290) had been confirmed to be acapsular by transmission electron microscopy analysis (16). Furthermore, we found that they had mutations in their cps2 gene clusters but still retained the target genes (cps2I and cps2P) for cps typing (reference 16 and our unpublished data). Bacterial cells of these three isolates showed autoagglutination, and their antigens extracted by autoclaving did not react with any typing antisera (data not shown). Many of the other 49 nontypeable isolates also showed similar characteristics (data not shown). Thus, these isolates might also be acapsular due to mutations in the cps genes, such as NL146, NL194, and NL290, although we cannot rule out the possibility that some of them are encapsulated and have different antigenicity from the strains of the known 35 serotypes. Although CP was believed to be essential for the virulence of S. suis (6, 17, 18), the biological significance of the acapsular phase in the pathogenesis of S. suis infection has been proposed recently (16, 19). Many acapsular S. suis isolates have been recovered from porcine cases of endocarditis (16). Therefore, the typing methods developed are also considered to be useful for epidemiological studies of acapsular strains by enabling determination of the genotypes of the cps gene clusters.
Some isolates were regarded as nonserotypeable in our validation tests due to multiagglutinating characteristics with several typing sera. Liu et al. (9) recently identified serotype 21/29 isolates that react with both serotype 21 and 29 typing sera and demonstrated that one of the isolates had a mosaic cps gene cluster consisting of serotype 21 and 29 cps gene homologues. Therefore, this may also have occurred in our multiagglutinating isolates, and novel cps gene clusters may have been generated. Furthermore, 74 isolates in this study could not be typed by either serotyping or PCR-based cps typing. These isolates may be acapsular because of a large deletion in the cps gene clusters, including the target genes. Most of them had strong hydrophobic and autoagglutinating characteristics, strongly suggesting that they are nonencapsulated (12). However, a few strains were weakly hydrophobic, implying that they have completely new cps gene clusters. So far, the CP structures have been determined only for serotypes 2 and 14 (20, 21), and cps gene clusters of most of the nonserotypeable isolates have not been analyzed yet. Further analysis, including structure determination of the CPs and cps gene clusters of S. suis strains with different antigenicity, may provide insights into the evolution of S. suis CP as well as the relationship between cps types and serotypes.
The presence of many different serotypes in the S. suis population suggests the continuous and ongoing evolution of cps gene clusters in this species. Therefore, all reported molecular serotyping methods, including ours, should lead to improvements in the typing of more field strains. A few serotyped isolates were assigned to the expected serotypes by our method, but could not be typed by the method developed by Liu and colleagues (9), and vice versa (data not shown). However, the presence of a number of nonserotypeable isolates that can be assigned to the cps types in this study, as well as the consistent results of cps typing with serotyping that were observed in most of the tested isolates, including those from many clinical cases, strongly suggest that the developed cps typing methods are practical and can further contribute to the diagnosis and epidemiological studies of S. suis infections.
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Young Scientists (B) (23780310) and a Grant-in-Aid for Exploratory Research (24659197) from the Japan Society for the Promotion of Science and a Grant-in-Aid for Scientific Research (C) (22592032, 23580420, and 24590525) from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and was supported partially by the Natural Sciences and Engineering Research Council of Canada (NSERC) (grant 154280).
Footnotes
Published ahead of print 26 February 2014
REFERENCES
- 1.Perch B, Pedersen K, Henrichsen J. 1983. Serology of capsulated streptococci pathogenic for pigs: six new serotypes of Streptococcus suis. J. Clin. Microbiol. 17:993–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. 1991. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J. Clin. Microbiol. 29:2590–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottschalk M, Higgins R, Jacques M, Mittal K, Henrichsen J. 1989. Description of 14 new capsular types of Streptococcus suis. J. Clin. Microbiol. 27:2633–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higgins R, Gottschalk M, Boudreau M, Lebrun A, Henrichsen J. 1995. Description of six new capsular types (29–34) of Streptococcus suis. J. Vet. Diagn. Invest. 7:405–406. 10.1177/104063879500700322 [DOI] [PubMed] [Google Scholar]
- 5.Staats JJ, Feder I, Okwumabua O, Chengappa MM. 1997. Streptococcus suis: past and present. Vet. Res. Commun. 21:381–407. 10.1023/A:1005870317757 [DOI] [PubMed] [Google Scholar]
- 6.Smith HE, Damman M, van der Velde J, Wagenaar F, Wisselink HJ, Stockhofe-Zurwieden N, Smits MA. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67:1750–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith HE, de Vries R, van't Slot R, Smits MA. 2000. The cps locus of Streptococcus suis serotype 2: genetic determinant for the synthesis of sialic acid. Microb. Pathog. 29:127–134. 10.1006/mpat.2000.0372 [DOI] [PubMed] [Google Scholar]
- 8.Okura M, Takamatsu D, Maruyama F, Nozawa T, Nakagawa I, Osaki M, Sekizaki T, Gottschalk M, Kumagai Y, Hamada S. 2013. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: potential mechanisms for generation of capsular variation. Appl. Environ. Microbiol. 79:2796–2806. 10.1128/AEM.03742-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Zheng H, Gottschalk M, Bai X, Lan R, Ji S, Liu H, Xu J. 2013. Development of multiplex PCR assays for the identification of the 33 serotypes of Streptococcus suis. PLoS One 8:e72070. 10.1371/journal.pone.0072070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill JE, Gottschalk M, Brousseau R, Harel J, Hemmingsen SM, Goh SH. 2005. Biochemical analysis, cpn60 and 16S rDNA sequence data indicate that Streptococcus suis serotypes 32 and 34, isolated from pigs, are Streptococcus orisratti. Vet. Microbiol. 107:63–69. 10.1016/j.vetmic.2005.01.003 [DOI] [PubMed] [Google Scholar]
- 11.Tien le HT, Nishibori T, Nishitani Y, Nomoto R, Osawa R. 2013. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogeny. Vet. Microbiol. 162:842–849. 10.1016/j.vetmic.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 12.Gottschalk M, Lacouture S, Bonifait L, Roy D, Fittipaldi N, Grenier D. 2013. Characterization of Streptococcus suis isolates recovered between 2008 and 2011 from diseased pigs in Québec, Canada. Vet. Microbiol. 162:819–825. 10.1016/j.vetmic.2012.10.028 [DOI] [PubMed] [Google Scholar]
- 13.Dorsch M, Stackebrandt E. 1992. Some modifications in the procedure of direct sequencing of PCR amplified 16S rDNA. J. Microbiol. Methods 16:271–279. 10.1016/0167-7012(92)90017-X [DOI] [Google Scholar]
- 14.Okwumabua O, O'Connor M, Shull E. 2003. A polymerase chain reaction (PCR) assay specific for Streptococcus suis based on the gene encoding the glutamate dehydrogenase. FEMS Microbiol. Lett. 218:79–84. 10.1111/j.1574-6968.2003.tb11501.x [DOI] [PubMed] [Google Scholar]
- 15.Higgins R, Gottschalk M. 1990. An update on Streptococcus suis identification. J. Vet. Diagn. Invest. 2:249–252. 10.1177/104063879000200324 [DOI] [PubMed] [Google Scholar]
- 16.Lakkitjaroen N, Takamatsu D, Okura M, Sato M, Osaki M, Sekizaki T. 2011. Loss of capsule among Streptococcus suis isolates from porcine endocarditis and its biological significance. J. Med. Microbiol. 60:1669–1676. 10.1099/jmm.0.034686-0 [DOI] [PubMed] [Google Scholar]
- 17.Chabot-Roy G, Willson P, Segura M, Lacouture S, Gottschalk M. 2006. Phagocytosis and killing of Streptococcus suis by porcine neutrophils. Microb. Pathog. 41:21–32. 10.1016/j.micpath.2006.04.001 [DOI] [PubMed] [Google Scholar]
- 18.Benga L, Fulde M, Neis C, Goethe R, Valentin-Weigand P. 2008. Polysaccharide capsule and suilysin contribute to extracellular survival of Streptococcus suis co-cultivated with primary porcine phagocytes. Vet. Microbiol. 132:211–219. 10.1016/j.vetmic.2008.05.005 [DOI] [PubMed] [Google Scholar]
- 19.Tanabe S, Bonifait L, Fittipaldi N, Grignon L, Gottschalk M, Grenier D. 2010. Pleiotropic effects of polysaccharide capsule loss on selected biological properties of Streptococcus suis. Can. J. Vet. Res. 74:65–70. 10.1128/JCM.01687-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Calsteren MR, Gagnon F, Lacouture S, Fittipaldi N, Gottschalk M. 2010. Structure determination of Streptococcus suis serotype 2 capsular polysaccharide. Biochem. Cell Biol. 88:513–525. 10.1139/O09-170 [DOI] [PubMed] [Google Scholar]
- 21.Van Calsteren MR, Gagnon F, Calzas C, Goyette-Desjardins G, Okura M, Takamatsu D, Gottschalk M, Segura M. 2013. Structure determination of Streptococcus suis serotype 14 capsular polysaccharide. Biochem. Cell Biol. 91:49–58. 10.1139/bcb-2012-0036 [DOI] [PubMed] [Google Scholar]