Abstract
The erm(41) gene causes inducible macrolide resistance in Mycobacterium abscessus but not Mycobacterium chelonae. erm(41) sequencing of 285 M. abscessus and 45 M. chelonae isolates was compared to 14-day susceptibility; agreement percentages were 98.9% and 100%, respectively. Extended incubation may not be necessary for M. chelonae, and the erm(41) genotype is a useful adjunct for M. abscessus.
TEXT
Mycobacterium abscessus is a rapidly growing mycobacterium (RGM) that is increasingly responsible for chronic pulmonary and cutaneous infections (1). Whole-genome sequence analysis supports the separation of this species into 3 taxa, including: M. abscessus subsp. abscessus, M. abscessus subsp. massiliense, and M. abscessus subsp. bolletii (2).
Infections due to these pathogens are often difficult to treat, in part because of acquired and/or intrinsic antimicrobial drug resistance. Clarithromycin and azithromycin remain the cornerstones of RGM therapy (3). Acquired macrolide resistance in the M. abscessus group, as well as the closely related Mycobacterium chelonae, has been linked to the point mutations A2058 and A2059 in the rrl gene encoding the peptidyltransferase domain of 23S rRNA. rrl gene mutants rarely emerge during therapy and are readily detected by routine 72-h susceptibility testing (4). An intrinsic macrolide resistance mechanism has also been described that involves an inducible erythromycin ribosomal methylase gene [erm(41)] (5). Previous work has demonstrated that (i) M. abscessus subsp. abscessus harbors an intact erm(41) gene but a C-to-T polymorphism at the 28th nucleotide position (T28) confers inducible clarithromycin resistance while C28 isolates remain susceptible (except for rrl mutants), (ii) M. abscessus subsp. bolletii strains had erm(41) sequences and resistance patterns similar to those of T28 M. abscessus subsp. abscessus isolates, and (iii) M. abscessus subsp. massiliense contained two deletions, rendering erm(41) nonfunctional and the organism macrolide susceptible (except for rrl mutants) (5–7). M. chelonae is not thought to contain erm(41), but this conclusion is based on relatively few genotyped isolates (5) and/or strains incubated in the presence of clarithromycin for >3 days (8).
To ensure detection of inducible macrolide resistance in vitro, the Clinical and Laboratory Standards Institute (CLSI) recommends that all RGM susceptibility testing for clarithromycin be determined after 14 days of incubation, unless resistance is recognized sooner (9). An alternative approach that would decrease the turnaround time to macrolide result reporting involves genomic interrogation of the erm(41) gene. (This work was presented in part at the 113th General Meeting of the American Society for Microbiology in Denver, CO, 18 to 21 May 2013.)
The purpose of this study was to verify that erm(41) sequencing correlates with extended microbroth susceptibility testing using a large number of prospectively collected M. abscessus group and M. chelonae isolates in a clinical setting. RGM specimens were submitted to the ARUP Reference Laboratories for identification and susceptibility testing between April 2011 and April 2013. Isolates were first identified to the M. chelonae-abscessus] complex level by partial 16S rRNA gene sequencing (10). Differentiation between M. chelonae and the M. abscessus group was accomplished using real-time internal transcribed spacer PCR analysis as previously described (11, 12). Macrolide susceptibility testing was performed per the 2011 CLSI guideline using RGM Sensititre MIC plates (Trek Diagnostic Systems, Cleveland, OH). Clarithromycin results were read after 3, 5, 7, 12, and 14 days of incubation or until the time resistance was first detected.
During the 2-year study period, 427 M. chelonae and 1,025 M. abscessus group isolates were identified as a part of routine clinical care, but patient histories were not available for review. Phenotypic clarithromycin susceptibilities are displayed by day of incubation in Fig. 1. All M. chelonae isolates were clarithromycin susceptible after 3 days of incubation, which suggests an absence of rrl mutants in this sample. The majority of M. chelonae strains (426/427, 99.8%) were fully susceptible at day 14, in agreement with previous reports that clarithromycin MICs did not increase when the incubation times were extended (8). In contrast, a small proportion of M. abscessus group isolates (28/1025, 2.7%) appeared resistant after 3 days and more than half (637/1025, 62.1%) were ultimately reported as resistant after a week or more of incubation (i.e., inducibly resistant).
FIG 1.
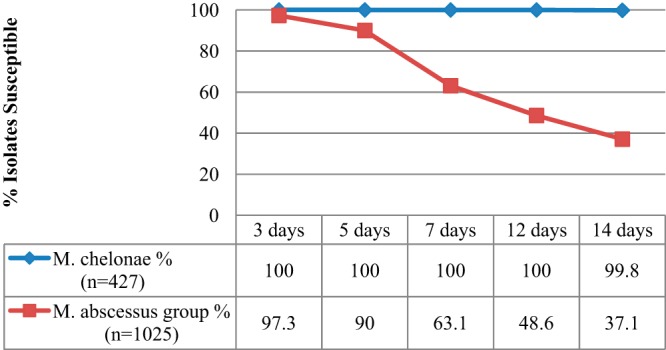
Fourteen-day clarithromycin susceptibility test results for all isolates over a 2-year period. One M. chelonae and 3 M. abscessus group isolates had intermediate clarithromycin susceptibility (MIC, 4 μg/ml) detected at day 14 on repeated testing. These organisms are classified here as nonsusceptible.
A subset of unique M. chelonae and M. abscessus group isolates from individual patients, representing all of the observed clarithromycin susceptibility profiles, were selected for erm(41) sequencing. Briefly, isolates were retrieved from liquid nitrogen storage and checked for purity. The erm(41) PCR mixture contained the following: 0.5 μM of primers (5), 1× GoTaq colorless master mix (Promega, Madison, WI) with a final MgCl concentration of 1.5 mM, 5 μl template DNA, and 200 μM of each deoxynucleoside trisphosphate (dNTP). Step-down PCR was performed on an ABI 9700 thermal cycler. Cycling conditions included a denaturation hold at 94°C for 10 min, 20 cycles at 94°C for 10 s, 65°C (with a 1° drop per cycle) for 30 s, 72°C for 60 s, 15 cycles of 94°C for 10 s, 35°C for 20 s, 72°C for 60 s, and a final primer extension at 72°C for 2 min. Amplicons were detected on a 2% agarose gel and purified using Agencourt AMPure magnetic beads (Beckman Coulter, Brea, CA). Sanger sequencing using BigDye Terminator chemistry was then performed on the ABI Prism 3730 DNA analyzer per the manufacturer's instructions. Sequences obtained for erm(41) were assembled in VectorNTI Advance 11.0 Contig Express (Life Technologies, Grand Island, NY), aligned in MEGA 5.2 (13), and compared to the NCBI Reference Sequence Database using BLAST analysis. The erm(41) sequences and predicted macrolide susceptibility were interpreted per Nash et al. (5).
In all, 330 isolates (M. chelonae [n = 45] and M. abscessus group [n = 285]) that were macrolide susceptible after 72 h of incubation had erm(41) sequencing performed (Table 1). None of the 45 M. chelonae isolates analyzed, including a single strain with intermediate susceptibility at day 14, contained the erm(41) gene. Interestingly, the erm(41) primers did amplify highly similar sequences for some isolates (21/45, 46.7%), but none matched references deposited in GenBank (data not shown). These observations, taken together with the phenotypic susceptibility results presented in Fig. 1, suggest that holding M. chelonae isolates for 14 days to exclude inducible clarithromycin resistance may not be necessary. A total of 285 M. abscessus group isolates were also sequenced. Most genotyped isolates were from respiratory (62.1%) sources, followed by skin/soft tissue (18.9%) or blood (5.6%). Initially, 5 isolates with susceptible erm(41) genotypes (C28 sequevars) were read as intermediately susceptible to clarithromycin on day 14. Sequence alignments revealed no novel erm(41) polymorphisms in these isolates. Repeat susceptibility testing reclassified 2 of the 5 as fully susceptible (MIC, ≤2 μg/ml) and 3 remained intermediate on day 14 only (MIC, 4 μg/ml). Following this discrepancy resolution testing, overall agreement between the M. abscessus group macrolide susceptibility genotype and phenotype was 98.9% (282/285). If the intermediately susceptible isolates of uncertain clinical significance were excluded, there was 100% concordance.
TABLE 1.
Correlation between erm(41) genotype and macrolide phenotypic susceptibility for selected M. chelonae-abscessus] complex isolates
| ITSa identification (n) | 14-day clarithromycin susceptibilitiesb of isolates (no. [%]) |
erm(41) genotype | ||
|---|---|---|---|---|
| Susceptible (MIC ≤ 2 μg/ml) | Intermediate (MIC 4 μg/ml) | Resistant (MIC ≥ 8 μg/ml) | ||
| M. chelonae (45) | 44 (97.8) | 1 (2.2) | 0 | Not present |
| M. abscessus group (285) | 74 (26) | 2 (0.7) | 0 | C28 sequevar |
| 0 | 0 | 94 (33.0) | T28 sequevar | |
| 114 (40) | 1 (0.3) | 0 | Deletionsc | |
ITS, internal transcribed spacer.
All sequenced isolates were clarithromycin susceptible after 72 h of incubation in an attempt to eliminate organisms with acquired macrolide resistance due to rrl mutation.
Observed deletions, which always occurred together, included deletions at nucleotides 64 and 65 and a deletion of 274 bp starting from nucleotide 159 (using the GTG start codon as nucleotide 1).
Subspecies-level identification of the M. abscessus group has been recommended based on the observation that patients with M. abscessus subsp. abscessus lung infections have poorer clinical and microbiologic responses to antimicrobial therapy than do those with M. abscessus subsp. massiliense infections (14). Outcome differences are likely related to the inducible macrolide resistance more commonly associated with M. abscessus subsp. abscessus strains. We did not attempt to differentiate members of the M. abscessus group to also determine if subspecies identification matched the expected erm(41) sequence. Procedures for identification to the species level are not trivial and require multilocus sequence analysis (15–17), a dedicated gel-based multiplex PCR targeting regions of genetic difference (18), or possibly manual analysis of individual peak differences contained within matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) spectra (19). Multilocus sequence typing and principal component cluster-based analysis of protein spectra are beyond the scope of most clinical laboratories. Gel-based PCR assays are more labor-intensive than automated real-time systems but do offer a more practical solution for this application. However, gene transfer across members of the M. abscessus group limits the inference of subspecies-specific susceptibility patterns (15). For example, 2 M. abscessus subsp. massiliense strains with full-length erm(41) gene sequences and inducible macrolide resistance were recently reported (18). Thus, predicting macrolide susceptibility based on subspecies in these cases would have been inaccurate. While identification to the species level may be useful for epidemiologic purposes, perhaps the most important information for immediate patient care is whether or not an M. abscessus group isolate harbors a wild-type erm(41) gene or rrl mutations.
In line with previous reports (5–7), we observed strong overall agreement (98.9%) between the erm(41) genotype and 14-day clarithromycin susceptibility for a large number of M. abscessus group isolates that appeared susceptible after 3 days of initial incubation. Discrepant results were derived from 3 M. abscessus isolates that had reproducibly intermediate clarithromycin results on day 14 but lacked a functional erm(41) gene. These would be reported as susceptible based on erm(41) genotype (i.e., C28 sequevar). Whether this difference would adversely affect patient treatment outcomes is not known. In addition, M. chelonae does not appear to harbor an inducible macrolide resistance mechanism.
In conclusion, determining M. chelonae and M. abscessus group macrolide susceptibility after 3 to 5 days of incubation combined with erm(41) sequence analysis is a rapid, highly accurate, and logistically feasible approach for the clinical laboratory. The elimination of 14-day clarithromycin susceptibility testing for M. chelonae and perhaps also M. abscessus group isolates could streamline clinical laboratory workflow, but additional studies are required to assess the cost-effectiveness and clinical impact of rapid molecular-based approaches. For now, erm(41) sequence analysis for the M. abscessus group may be useful as an adjunct to extended susceptibility testing.
Footnotes
Published ahead of print 19 February 2014
REFERENCES
- 1.Petrini B. 2006. Mycobacterium abscessus: an emerging rapid-growing potential pathogen. APMIS 114:319–328. 10.1111/j.1600-0463.2006.apm_390.x [DOI] [PubMed] [Google Scholar]
- 2.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. 10.1016/S0140-6736(13)60632-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175:367–416. 10.1164/rccm.200604-571ST [DOI] [PubMed] [Google Scholar]
- 4.Wallace RJ, JR, Meier A, Brown BA, Zhang Y, Sander P, Onyi GO, Böttger EC. 1996. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob. Agents Chemother. 40:1676–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nash KA, Brown-Elliott BA, Wallace RJ., Jr 2009. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 53:1367–1376. 10.1128/AAC.01275-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastian S, Veziris N, Roux AL, Brossier F, Gaillard JL, Jarlier V, Cambau E. 2011. Assessment of clarithromycin susceptibility in strains belonging to the Mycobacterium abscessus group by erm(41) and rrl sequencing. Antimicrob. Agents Chemother. 55:775–781. 10.1128/AAC.00861-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HY, Kim BJ, Kook Y, Yun YJ, Shin JH, Kim BJ, Kook YH. 2010. Mycobacterium massiliense is differentiated from Mycobacterium abscessus and Mycobacterium bolletii by erythromycin ribosome methyltransferase gene (erm) and clarithromycin susceptibility patterns. Microbiol. Immunol. 54:347–353. 10.1111/j.1348-0421.2010.00221.x [DOI] [PubMed] [Google Scholar]
- 8.Brown BA, Wallace RJ, Onyi GO, De Rosas V, Wallace RJ. 1992. Activities of four macrolides, including clarithromycin, against Mycobacterium fortuitum, Mycobacterium chelonae, and M. chelonae-like organisms. Antimicrob. Agents Chemother. 36:180–184. 10.1128/AAC.36.1.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical Laboratory Standards Institute. 2011. Susceptibility testing of Mycobacteria, Nocardiae, and other aerobic actinomycetes; approved standard—2nd ed. CLSI M24-A2 Clinical Laboratory Standards Institute, Wayne, PA: [PubMed] [Google Scholar]
- 10.Clinical Laboratory Standards Institute. 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. CLSI MM18-A Clinical Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11.Cloud JL, Hoggan K, Belousov E, Cohen S, Brown-Elliott BA, Mann L, Wilson R, Aldous W, Wallace RJ, Jr, Woods GL. 2005. Use of the MGB Eclipse system and SmartCycler PCR for differentiation of Mycobacterium chelonae and M. abscessus. J. Clin. Microbiol. 43:4205–4207. 10.1128/JCM.43.8.4205-4207.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neal H, Cloud JL, Pounder JI, Page SR, Woods GL. 2005. Sequence variant for internal transcribed spacer region of Mycobacterium abscessus. J. Clin. Microbiol. 43:6214. 10.1128/JCM.43.12.6214.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH, Lee SH, Park YK, Kim CK, Shin SJ, Huitt GA, Daley CL, Kwon OJ. 2011. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am. J. Respir. Crit. Care Med. 183:405–410. 10.1164/rccm.201003-0395OC [DOI] [PubMed] [Google Scholar]
- 15.Macheras E, Roux AL, Bastian S, Leao SC, Palaci M, Sivadon-Tardy V, Gutierrez C, Richter E, Rusch-Gerdes S, Pfyffer G, Bodmer T, Cambau E, Gaillard JL, Heym B. 2011. Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains. J. Clin. Microbiol. 49:491–499. 10.1128/JCM.01274-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macheras E, Roux AL, Ripoll F, Sivadon-Tardy V, Gutierrez C, Gaillard JL, Heym B. 2009. Inaccuracy of single-target sequencing for discriminating species of the Mycobacterium abscessus group. J. Clin. Microbiol. 47:2596–2600. 10.1128/JCM.00037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, Stock F, Conlan S, McNulty S, Brown-Elliott BA, Wallace RJ, Jr, Olivier KN, Holland SM, Sampaio EP. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J. Clin. Microbiol. 47:1985–1995. 10.1128/JCM.01688-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shallom SJ, Gardina PJ, Myers TG, Sebastian Y, Conville P, Calhoun LB, Tettelin H, Olivier KN, Uzel G, Sampaio EP, Holland SM, Zelazny AM. 2013. New rapid scheme for distinguishing the subspecies of the Mycobacterium abscessus group and identifying Mycobacterium massiliense isolates with inducible clarithromycin resistance. J. Clin. Microbiol. 51:2943–2949. 10.1128/JCM.01132-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teng SH, Chen CM, Lee MR, Lee TF, Chien KY, Teng LJ, Hsueh PR. 2013. Matrix-assisted laser desorption ionization–time of flight mass spectrometry can accurately differentiate between Mycobacterium masilliense (M. abscessus subspecies bolletti) and M. abscessus (sensu stricto). J. Clin. Microbiol. 51:3113–3116. 10.1128/JCM.01239-13 [DOI] [PMC free article] [PubMed] [Google Scholar]


