Abstract
Merkel cell polyomavirus (MCPyV) is linked to a cutaneous cancer mainly occurring in Caucasians. DNA from skin swabs of 255 adults, originating from the 5 continents, were subjected to MCPyV PCRs. Phylogenetic analyses demonstrate the existence of 5 major geographically related MCPyV genotypes (Europe/North America, Africa [sub-Saharan], Oceania, South America, and Asia/Japan).
TEXT
Merkel cell polyomavirus (MCPyV) is a recently discovered circular double-stranded DNA virus, etiologically linked to an uncommon and aggressive primary cutaneous cancer, Merkel cell carcinoma (MCC) (1). This neoplasm, which affects nerve-associated Merkel cells, occurs mainly on sun-exposed skin of elderly Caucasians or in immunodepressed patients (2, 3). MCPyV is one of the 12 known human polyomaviruses (4). Besides MCPyV, three other polyomaviruses (BK polyomavirus [BKPyV], JC polyomavirus [JCPyV], and trichodysplasia spinulosa-associated polyomavirus [TSPyV]) are associated with human diseases (4–6). For the JC and BK polyomaviruses, many studies have demonstrated the existence of various genotypes linked to the geographical origin of the infected individuals (7–9). Furthermore, it has been shown that JCPyV coevolved with human populations and can be used as a molecular tracer for deciphering human migrations (10). Concerning MCPyV, recent studies have suggested the existence of geographically related variants with one group composed of MCPyV strains present in Caucasians and another group comprising strains found in Asians (11–13). This hypothesis, currently based on few preliminary data, needs to be strengthened on a larger scale.
For the past 20 years, our laboratory has studied the genetic variability of human oncogenic viruses, e.g., the retrovirus human T-cell leukemia virus 1 (HTLV-1) and human herpesvirus 8 (HHV-8), in different populations. Thus, we have frequent access to healthy individuals living in villages of the western part of central Africa (14, 15) and South America (16, 17) and in patients from hospitals in Oceania (18) and Europe. In order to search for specific geographically related MCPyV genotypes, we tested many individuals originating from the five continents for MCPyV. The study was approved in France by the Comité de Protection des Personnes (approval no. CEBH 2012/02), the Comité Consultatif sur le Traitement de l'Information en Matière de Recherche dans le Domaine de la Santé (CCTIRS) (approval no. 12.541), and the Commission Nationale de l'Informatique et des Libertés (CNIL) (approval DR-2012-535) and in Cameroon by the National Ethics Committee. Detailed explanations of the study were provided to each participant. Individual written informed consent was obtained, and a short standardized questionnaire was used to collect personal epidemiological data (age, sex, location, and ethnicity). A total of 255 adult volunteers were included. The volunteers originated from South America (30 Amerindians living in Saint-Laurent du Maroni in French Guiana), Oceania (5 Polynesians and 4 Melanesians living in New Caledonia), central Africa (13 Bantus and 42 Pygmies from south Cameroon villages and settlements), and Europe (29 Caucasians living in metropolitan France or French Guiana). We also studied 81 Noir-Marrons of west African ancestry (19) and 51 Hmongs of southeast Asian origin (20) who were living in the Saint-Laurent du Maroni area of French Guiana. All of the participants were considered healthy individuals except the 9 people from New Caledonia, who were outpatients seen in a hospital dermatology unit. None of them had clinical evidence of a Merkel tumor.
As MCPyV is present on the skin, we used cutaneous swabs of the face, mainly the forehead, to study MCPyV strains as previously described (13, 21, 22) (see Supplemental Methods in the supplemental material for the methods of DNA extraction from the swabs). For each of the 255 DNA samples, we performed 3 different PCRs using specific primer pairs, generating 3 distinct MCPyV genomic fragments called A (large T antigen [LT-Ag]), B (VP1), and C (VP2 and the noncoding control region [NCCR]) (see Supplemental Methods and Table S in the supplemental material).
Out of the 255 DNA samples tested, we obtained 48 (18.8%) PCR positive for fragment A, and only 14 of these samples were also amplifiable for fragments B and C (Table 1). We have thus sequenced the 3 fragments corresponding to these 14 samples (6 from Africa, 6 from South America, and 2 from French Caucasians). We obtained a concatenated 1,468-bp-long fragment of the MCPyV genome for these 14 samples (Table 1). We also sequenced the 4 Hmong samples and the 4 samples from Oceania, despite the fact that they were only positive by PCR for fragment A, because they were the only ones representing this geographical clade in our study. Thus, we obtained 22 sequences for fragment A and 14 sequences for fragments B and C.
TABLE 1.
Geographical origin and molecular results concerning the PCR and high-throughput sequencing technologies of the 22 MCPyV strains studied
| Participant no. | Sample | Geographic origin | Age (yr) | Sexa | PCR results for fragment: |
Phylogeny (1,284-bp fragmentb) | ||
|---|---|---|---|---|---|---|---|---|
| A | B | C | ||||||
| 1 | AmePIc | South America (Amerindians) | 35 | F | + | + | + | + |
| 2 | AmePA | South America (Amerindians) | 22 | F | + | + | + | + |
| 3 | AmeKS | South America (Amerindians) | 42 | M | + | + | + | + |
| 4 | AmeKR | South America (Amerindians) | 64 | F | + | + | + | + |
| 5 | AmeJH | South America (Amerindians) | 43 | M | + | + | + | + |
| 6 | AmeKK | South America (Amerindians) | 41 | F | + | + | + | + |
| 7 | OceMelNC1 | Oceania (Melanesians) | 64 | M | + | − | − | +d |
| 8 | OceMelNC2 | Oceania (Melanesians) | 45 | M | + | − | − | +d |
| 9 | OceMelNC3 | Oceania (Melanesians) | 43 | M | + | − | − | − |
| 10 | OcePolW1c | Oceania (Polynesians) | 47 | F | + | − | − | +d |
| 11 | EurCauC1c | Europe (Caucasians) | 26 | F | + | + | + | + |
| 12 | EurCauC2 | Europe (Caucasians) | 55 | M | + | + | + | + |
| 13 | AfrBak541 | Africa (Pygmies) | 42 | M | + | + | + | + |
| 14 | AfrBad468 | Africa (Bantus) | 35 | M | + | + | + | + |
| 15 | AfrNm51 | Africa (Noir-Marrons) | 20 | F | + | + | + | + |
| 16 | AfrNm45 | Africa (Noir-Marrons) | 20 | F | + | + | + | + |
| 17 | AfrNm70 | Africa (Noir-Marrons) | 26 | F | + | + | + | + |
| 18 | AfrNm020 | Africa (Noir-Marrons) | 14 | F | + | + | + | + |
| 19 | Asi-HmLT | Asia (Hmongs) | 25 | F | + | − | − | − |
| 20 | Asi-Hm130 | Asia (Hmongs) | 28 | F | + | − | − | − |
| 21 | Asi-Hm225 | Asia (Hmongs) | 21 | F | + | − | − | − |
| 22 | Asi-HmYY | Asia (Hmongs) | 81 | F | + | − | − | − |
F, female; M, male.
The 1,284-bp fragment corresponds to the 1,468-bp fragment without the noncoding region of fragment C.
Complete genome obtained by high-throughput sequencing (see the methodologies and Fig. S1 in the supplemental material).
Importantly, in these 3 cases (samples OceMelNC1, OceMelNC2, and OcePolW1), the HTS technique generated the sequences corresponding to fragments B and C. Thus, the complete 1,468-bp fragment for these 3 cases was also obtained.
The low rate of PCR positivity is likely due to the very low MCPyV loads present on the skin of healthy individuals compared to those found on patients with MCC (12, 21).
In order to get more sequence information, we also used high-throughput DNA sequencing (HTS) (Illumina Sequencing Technology; see the HTS methodology in the supplemental material). This was performed on 2 samples (EurCauC1 and AmePI) for which the 3 PCRs were positive, suggesting a high MCPyV load in the skin DNA (Table 1), and on 8 other interesting samples (4 from Hmongs and 4 from Oceanians), even though only the PCR for fragment A was found positive (Table 1). In these 10 samples, reads (short sequences of approximately 100 nucleotides) corresponding to MCPyV were obtained. Contigs were generated, and we obtained segments of the MCPyV genome with sizes ranging from 2,288 bp to 5,388 bp (data not shown). Importantly, in 3 cases (samples OceMelNC1, OceMelNC2, and OcePolW1), the HTS technique generated the sequences corresponding to fragments B and C. Thus, the complete 1,468-bp fragment for these 3 samples was also obtained (Table 1). Unfortunately, we were not able to generate fragment B or C from the 4 Hmong samples or from the last Oceanian one. Furthermore, 3 complete MCPyV sequences were generated by HTS, one from an Amerindian (sample AmePI), one from a Polynesian (OcePolW1), and one from a European/Caucasian (sample EurCauC1) (GenBank accession numbers KF266963 through KF266965) (Table 1; see also Fig. S1 in the supplemental material).The first two sequences can be considered prototypic MCPyV strains for natives from South America (sample AmePI) and Oceania (sample OcePolW1).
Thus, a 1,468-bp-long informative genomic sequence was obtained for 17 strains by concatenation of the 3 amplicons (Table 1). In order to search for specific geographically related clades of MCPyV, we performed phylogenetic analyses using the neighbor-joining and maximum likelihood methods (Fig. 1 and data not shown, respectively). We included in these analyses the 17 novel MCPyV strains we generated and most of the available prototypic sequences. The prototypic sequences originated mainly from North America and Europe (the great majority of them purportedly from Caucasians), and a few were from Asia, including one from Japan (12, 13, 23).
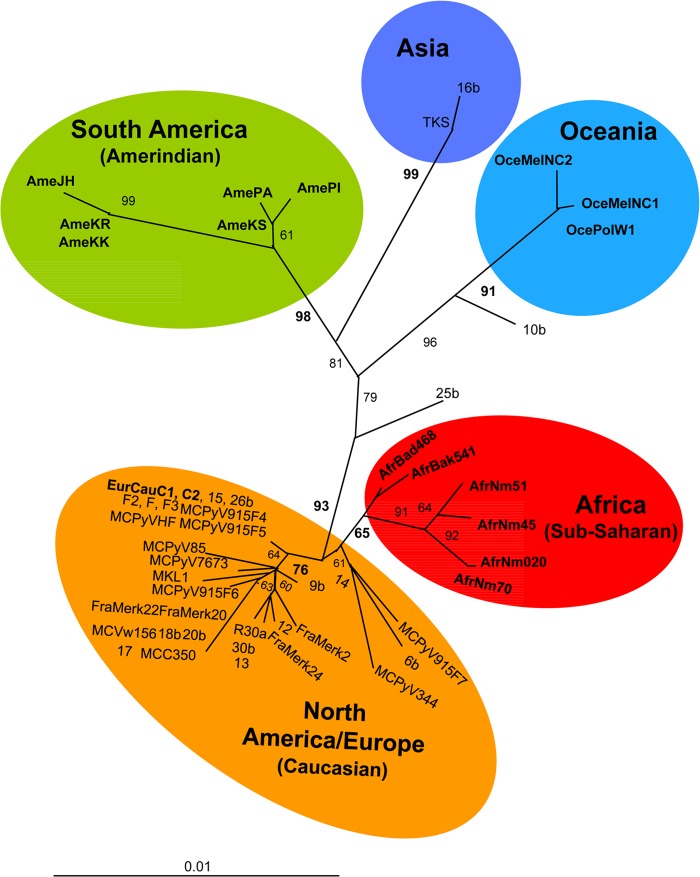
FIG 1.
Phylogenetic tree generated with the neighbor-joining method. The phylogeny was derived from the neighbor-joining method on 51 sequences of colinearized genomic fragments (1,284 bp) of MCPyV corresponding to the 1,468-bp fragment without the noncoding region of fragment C (144 bp). The phylogeny was derived by the neighbor-joining method by using the general time-reversible (GTR) model in the PAUP program version 4.0b10 (Sinauer Associates, Sunderland, MA, USA). Reliability of the inferred tree was evaluated by bootstrap analysis on 1,000 replicates. Branch lengths are drawn to scale, with the bar indicating 0.001-nucleotide replacement per site. We tentatively propose the following nomenclature for the 5 different genotypes: North America/Europe (Caucasian), Africa, Asia, Oceania, and South America (Amerindians). The 17 new colinearized sequences used in this study are in bold type (GenBank accession numbers KF266906 through KF266962).
Both analyses gave very similar results, with clear evidence of specific geographically related MCPyV genotypes. Indeed, we could identify 4 main clades, which are phylogenetically supported and correspond to the different continents (Fig. 1). The larger clade (bootstrap [BS] value, 93%) included the sequences previously generated from North America/Europe as well, as expected, the 2 new sequences (sample EurCauC1/C2) from the French Caucasian, and the 6 new sequences from individuals of African origin. The African group (BS value, 65%) included not only the sequences isolated from one Bantu (sample AfrBad468) and one Pygmy (sample AfrBak541) from central Africa (Cameroon) but also the 4 strains (samples AfrNm51, AfrNm45, AfrNm70, and AfrNm020) from Noir-Marron individuals, who were living in French Guiana but whose ancestors originated from west Africa around 2 centuries ago (19). The second clade (BS value, 91%) was composed of the 3 sequences of Oceania origin, including the sequence from a Polynesian and the two sequences from Melanesians living in New Caledonia. The third clade (BS value, 98%) included the 6 new sequences originating from Amerindians living in French Guiana, and the last clade (BS value, 99%) comprised two sequences from Asian individuals, one from a Japanese patient (strain TKS) and the other (strain 16b) from an individual born in Asia.
In order to include the 4 Hmong sequences, for which only the small A fragment was available, we created an additional phylogenetic tree using the 436-bp fragment. Interestingly, the Hmong strains were quite related to strain 10b (from an individual born in Asia) and the Oceania strains (see Fig. S2 in the supplemental material). Furthermore, a phylogenetic tree created using fragment B alone (574 bp) gave very similar results to the analysis of fragment A alone (see Fig. S3 in the supplemental material).
In conclusion, based on the analysis of the 1,284-bp-long fragment of MCPyV, we demonstrated the existence of 5 major geographically related MCPyV genotypes (Europe/North America/Caucasian, Africa [sub-Saharan], Oceania, South America/Amerindian, and Asia/Japan). Thus, our data extend to MCPyV the notion of different genotypes linked to ethnic/geographical backgrounds and demonstrated for the JC and BK polyomaviruses (9, 10, 24, 25). Further analyses based on larger fragments of MCPyV will probably strengthen our original genotype classification. Moreover, MCC is mainly reported in patients of Caucasian origin but rarely in Asia and exceptionally in Africa (2, 5, 6). The fact that the new African sequences reported here are very close to the European sequences argues against the existence of an especially carcinogenic European MCPyV strain.
Nucleotide sequence accession numbers.
The nucleotide sequences discussed above have been deposited in GenBank with accession numbers KF266906 through KF266965.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Institut de Recherche pour le Développement and the Centre Pasteur du Cameroun for their collaboration in the field work carried out in Cameroon.
This work was financially supported by the French Government's Investissement d'Avenir program Laboratoire d'Excellence, Integrative Biology of Emerging Infectious Diseases (grant no. ANR-10-LABX-62-IBEID), by the Institut Pasteur in Paris, France, by la Fondation ARC pour la Recherche sur le Cancer (A09/1/5041, 2009 and 2013), and by la Ligue Nationale contre le Cancer (RS10/75-1, RS11/75-63, and RS14/75-78).
Footnotes
Published ahead of print 12 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02348-13.
REFERENCES
- 1.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. 10.1126/science.1152586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuwamoto S, Higaki H, Kanai K, Iwasaki T, Sano H, Nagata K, Kato K, Kato M, Murakami I, Horie Y, Yamamoto O, Hayashi K. 2011. Association of Merkel cell polyomavirus infection with morphologic differences in Merkel cell carcinoma. Hum. Pathol. 42:632–640. 10.1016/j.humpath.2010.09.011 [DOI] [PubMed] [Google Scholar]
- 3.Prieto Munoz I, Pardo Masferrer J, Olivera Vegas J, Medina Montalvo MS, Jover Diaz R, Perez Casas AM. 2013. Merkel cell carcinoma from 2008 to 2012: reaching a new level of understanding. Cancer Treat Rev. 39:421–429. 10.1016/j.ctrv.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 4.DeCaprio JA, Garcea RL. 2013. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 11:264–276. 10.1038/nrmicro2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalianis T, Hirsch HH. 2013. Human polyomaviruses in disease and cancer. Virology 437:63–72. 10.1016/j.virol.2012.12.015 [DOI] [PubMed] [Google Scholar]
- 6.Feltkamp MC, Kazem S, van der Meijden E, Lauber C, Gorbalenya AE. 2013. From Stockholm to Malawi: recent developments in studying human polyomaviruses. J. Gen. Virol. 94:482–496. 10.1099/vir.0.048462-0 [DOI] [PubMed] [Google Scholar]
- 7.Agostini HT, Yanagihara R, Davis V, Ryschkewitsch CF, Stoner GL. 1997. Asian genotypes of JC virus in Native Americans and in a Pacific Island population: markers of viral evolution and human migration. Proc. Natl. Acad. Sci. U. S. A. 94:14542–14546. 10.1073/pnas.94.26.14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugimoto C, Kitamura T, Guo J, Al-Ahdal MN, Shchelkunov SN, Otova B, Ondrejka P, Chollet JY, El-Safi S, Ettayebi M, Gresenguet G, Kocagoz T, Chaiyarasamee S, Thant KZ, Thein S, Moe K, Kobayashi N, Taguchi F, Yogo Y. 1997. Typing of urinary JC virus DNA offers a novel means of tracing human migrations. Proc. Natl. Acad. Sci. U. S. A. 94:9191–9196. 10.1073/pnas.94.17.9191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng HY, Nishimoto Y, Chen Q, Hasegawa M, Zhong S, Ikegaya H, Ohno N, Sugimoto C, Takasaka T, Kitamura T, Yogo Y. 2007. Relationships between BK virus lineages and human populations. Microbes Infect. 9:204–213. 10.1016/j.micinf.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 10.Shackelton LA, Rambaut A, Pybus OG, Holmes EC. 2006. JC virus evolution and its association with human populations. J. Virol. 80:9928–9933. 10.1128/JVI.00441-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hattori T, Takeuchi Y, Takenouchi T, Hirofuji A, Tsuchida T, Kabumoto T, Fujiwara H, Ito M, Shimizu A, Okada E, Motegi S, Tamura A, Ishikawa O. 2013. The prevalence of Merkel cell polyomavirus in Japanese patients with Merkel cell carcinoma. J. Dermatol. Sci. 70:99–107. 10.1016/j.jdermsci.2013.02.010 [DOI] [PubMed] [Google Scholar]
- 12.Martel-Jantin C, Filippone C, Cassar O, Peter M, Tomasic G, Vielh P, Briere J, Petrella T, Aubriot-Lorton MH, Mortier L, Jouvion G, Sastre-Garau X, Robert C, Gessain A. 2012. Genetic variability and integration of Merkel cell polyomavirus in Merkel cell carcinoma. Virology 426:134–142. 10.1016/j.virol.2012.01.018 [DOI] [PubMed] [Google Scholar]
- 13.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515. 10.1016/j.chom.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martel-Jantin C, Pedergnana V, Nicol JT, Leblond V, Tregouet DA, Tortevoye P, Plancoulaine S, Coursaget P, Touze A, Abel L, Gessain A. 2013. Merkel cell polyomavirus infection occurs during early childhood and is transmitted between siblings. J. Clin. Virol. 58:288–291. 10.1016/j.jcv.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 15.Mauclere P, Afonso PV, Meertens L, Plancoulaine S, Calattini S, Froment A, Van Beveren M, de The G, Quintana-Murci L, Mahieux R, Gessain A. 2011. HTLV-2B strains, similar to those found in several Amerindian tribes, are endemic in central African Bakola Pygmies. J. Infect. Dis. 203:1316–1323. 10.1093/infdis/jir031 [DOI] [PubMed] [Google Scholar]
- 16.Kazanji M, Dussart P, Duprez R, Tortevoye P, Pouliquen JF, Vandekerkhove J, Couppie P, Morvan J, Talarmin A, Gessain A. 2005. Serological and molecular evidence that human herpesvirus 8 is endemic among Amerindians in French Guiana. J. Infect. Dis. 192:1525–1529. 10.1086/491744 [DOI] [PubMed] [Google Scholar]
- 17.Plancoulaine S, Gessain A, van Beveren M, Tortevoye P, Abel L. 2003. Evidence for a recessive major gene predisposing to human herpesvirus 8 (HHV-8) infection in a population in which HHV-8 is endemic. J. Infect. Dis. 187:1944–1950. 10.1086/375345 [DOI] [PubMed] [Google Scholar]
- 18.Cassar O, Charavay F, Bassot S, Plancoulaine S, Grangeon JP, Laumond-Barny S, Martin PM, Chanteau S, Gessain A. 2012. Divergent KSHV/HHV-8 subtype D strains in New Caledonia and Solomon Islands, Melanesia. J. Clin. Virol. 53:214–218. 10.1016/j.jcv.2011.12.016 [DOI] [PubMed] [Google Scholar]
- 19.Brucato N, Cassar O, Tonasso L, Tortevoye P, Migot-Nabias F, Plancoulaine S, Guitard E, Larrouy G, Gessain A, Dugoujon JM. 2010. The imprint of the Slave Trade in an African American population: mitochondrial DNA, Y chromosome and HTLV-1 analysis in the Noir Marron of French Guiana. BMC Evol. Biol. 10:314. 10.1186/1471-2148-10-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brucato N, Mazieres S, Guitard E, Giscard PH, Bois E, Larrouy G, Dugoujon JM. 2012. The Hmong diaspora: preserved south-east Asian genetic ancestry in French Guianese Asians. C. R. Biol. 335:698–707. 10.1016/j.crvi.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 21.Foulongne V, Kluger N, Dereure O, Mercier G, Moles JP, Guillot B, Segondy M. 2010. Merkel cell polyomavirus in cutaneous swabs. Emerg. Infect. Dis. 16:685–687. 10.3201/eid1604.091278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, Pariente K, Segondy M, Burguiere A, Manuguerra JC, Caro V, Eloit M. 2012. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One 7:e38499. 10.1371/journal.pone.0038499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katano H, Ito H, Suzuki Y, Nakamura T, Sato Y, Tsuji T, Matsuo K, Nakagawa H, Sata T. 2009. Detection of Merkel cell polyomavirus in Merkel cell carcinoma and Kaposi's sarcoma. J. Med. Virol. 81:1951–1958. 10.1002/jmv.21608 [DOI] [PubMed] [Google Scholar]
- 24.Sugimoto C, Hasegawa M, Zheng HY, Demenev V, Sekino Y, Kojima K, Honjo T, Kida H, Hovi T, Vesikari T, Schalken JA, Tomita K, Mitsunobu Y, Ikegaya H, Kobayashi N, Kitamura T, Yogo Y. 2002. JC virus strains indigenous to northeastern Siberians and Canadian Inuits are unique but evolutionally related to those distributed throughout Europe and Mediterranean areas. J. Mol. Evol. 55:322–335. 10.1007/s00239-001-2329-2 [DOI] [PubMed] [Google Scholar]
- 25.Yogo Y, Sugimoto C, Zheng HY, Ikegaya H, Takasaka T, Kitamura T. 2004. JC virus genotyping offers a new paradigm in the study of human populations. Rev. Med. Virol. 14:179–191. 10.1002/rmv.428 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.