Abstract
Pneumocystis jirovecii is a symbiotic respiratory fungus that causes pneumonia (PcP) in immunosuppressed patients. Because P. jirovecii cannot be reliably cultured in vitro, it has proven difficult to study and gaps in our understanding of the organism persist. The release of a draft genome for the organism opens the door for the development of new genotyping approaches for studying its molecular epidemiology and global population structure. We identified and validated 8 putatively neutral microsatellite markers and 1 microsatellite marker linked to the dihydropteroate synthase gene (dhps), the enzymatic target of sulfa drugs used for PcP prevention and treatment. Using these tools, we analyzed P. jirovecii isolates from HIV-infected patients from three geographically distant populations: Uganda, the United States, and Spain. Among the 8 neutral markers, we observed high levels of allelic heterozygosity (average He, 0.586 to 0.842). Consistent with past reports, we observed limited global population structuring, with only the Ugandan isolates showing minor differentiation from the other two populations. In Ugandan isolates that harbored mutations in dhps, the microsatellite locus linked to dhps demonstrated a depressed He, consistent with positive directional selection for sulfa resistance mutations. Using a subset of these microsatellites, analyses of individual and paired samples from infections in San Francisco, CA, showed reliable typeability within a single infection and high discriminatory power between infections. These features suggest that this novel microsatellite typing approach will be an effective tool for molecular-epidemiological investigations into P. jirovecii population structure, transmission, and drug resistance.
INTRODUCTION
Pneumocystis jirovecii is a symbiotic respiratory fungus that causes pneumonia (PcP) in immunocompromised patients, including those with AIDS. In the United States, the incidence of PcP in HIV-infected populations has significantly declined owing to PcP prophylaxis and initiation of combined antiretroviral therapy (cART) for HIV infection (1–3). However, PcP remains a leading cause of death among those who do not receive cART or PcP prophylaxis (4, 5). Among patients with HIV infection, mortality rates for PcP remain high even in the cART era, ranging from 10% to nearly 40% (6–8). Concerningly, trends have suggested that PcP is an increasingly important disease among HIV-infected patients in low-income countries, where the majority of persons with HIV infection reside (9). Despite advances in preventing PcP, Pneumocystis is still cause for ample concern because (i) prophylaxis and treatment with sulfa drugs may be selecting for resistance mutations in key P. jirovecii metabolic enzymes, including dihydrofolate reductase (encoded by dhfr) and dihydropteroate synthase (encoded by dhps) (10–12); (ii) PcP incidence is increasing in some parts of the world (13); and (iii) PcP outbreaks periodically occur in immunocompromised patient populations, causing significant morbidity and mortality (13–20).
Because Pneumocystis cannot be reliably cultured in vitro (21), fundamental questions about P. jirovecii biology, ecology, and epidemiology remain unanswered. For example, we do not possess a clear understanding of the Pneumocystis life cycle, including sexual and clonal reproduction, transmission dynamics, the propensity for Pneumocystis strains to persist in unaffected, immunocompetent carriers, or the true risk of putatively sulfa drug-resistant Pneumocystis strains (reviewed in reference 22). Molecular epidemiology studies involving strain typing can help elucidate many of these issues and, in particular, population structure, the evolution of drug resistance, and transmission dynamics.
The current P. jirovecii strain-typing options have revealed much about Pneumocystis population structure and outbreak dynamics (23, 24). These typing methods include single- and multilocus genotyping using karyotypes (25), multilocus enzyme electrophoresis (25), Sanger sequence analysis (25, 26), type-specific oligonucleotide hybridization (25), single-strand conformation polymorphism (25), and multiplex-PCR/single-base extension (MPCR/SBE) (27, 28). More recently, a four-locus scheme has emerged, which includes internal transcribed spacer 1 (ITS1), 26S, mt26S, and beta-tubulin (β-TUB) (29, 30). Though each of these approaches has contributed significantly to our understanding of Pneumocystis biology, many questions remain unresolved (27, 31). A genotyping approach with neutrally evolving markers, a high discriminatory index, and a stepwise model of allele adaptation would be a useful addition for studying P. jirovecii.
The publication of the draft P. jirovecii genome (32) enables the development of such an approach. Using this information, we have identified and validated assays for 8 putatively neutral microsatellites in order to develop a more robust multilocus strain typing tool. Additionally, we have identified 1 microsatellite linked to dhps, a gene in which mutations are thought to confer reduced drug susceptibility to sulfa medications. Microsatellites are short tandem repeats in coding and noncoding regions of the genome that vary in length between strains and offer reproducible genotype calling for individual strains (typeability) and high resolution to distinguish between strains (discriminatory power) (33). This type of marker has been used extensively and has proven to be robust in humans and in pathogens such as Plasmodium falciparum for “molecular fingerprinting” and for studying the evolution of drug resistance (34–36). Here, we describe our multilocus microsatellite genotyping method and use it to investigate the global population structure of P. jirovecii, to study the evolution of sulfa antibiotic resistance, and to evaluate typeability and discriminatory power within one cohort.
MATERIALS AND METHODS
Sample collection and ethics statement.
Molecular analyses were performed on deidentified clinical specimens from Uganda (2008 to 2009, from Mulago Hospital, Makerere University), the United States (2005 to 2011, from San Francisco General Hospital, University of California, San Francisco), and Spain (Barcelona, 2001 to 2004). Patients from Uganda and San Francisco were enrolled on the basis of HIV infection and suspected PcP, as described before (12). Patients from Spain were enrolled on the basis of slide-positive microscopy, as described before (37). Among the samples from Spain, 47% of cases were late presenters and 94% of cases had not received previous sulfa or sulfone prophylaxis. Sample collection and molecular analysis of isolates from each site were approved by the appropriate Institutional Review Boards (IRBs) as described previously (12, 37, 38). As part of these prior studies, all specimens had been genotyped for dihydropteroate synthase gene (dhps) mutations (12, 37, 39). Molecular analyses detailed in this study were approved by the University of North Carolina at Chapel Hill IRB (study no. 12-1783).
Identifying and validating microsatellites.
In order to identify microsatellites, we scanned the published P. jirovecii genome (32) using Tandem Repeat Finder (40), which identified approximately 150 di- or trinucleotide tandem repeats with a repeat size of ≥8. For these, we were able to design primers to amplify 50 tandem repeats using WebSat (41, 42). PCRs were optimized on five total isolates from Kampala, Uganda, San Francisco, CA, and Chapel Hill, NC, and amplicons were sequenced using ABI BigDye Terminator Chemistry (Applied Biosystems, Grand Island, NY). Chromatograms were analyzed in Sequencher v5.0 (Gene Codes Corp., Ann Arbor, MI) to determine repeat unit counts, and sequence data were deposited in GenBank (see below). Loci that showed evidence of ≥1 repeat-unit variation in pairwise comparisons among sequenced isolates were deemed true microsatellites.
We assessed the functional genic context (i.e., intergenic, intragenic, exonic, and intronic) of each true microsatellite using the annotations at the European Nucleotide Archive (accession no. PRJEA68827), which were created using Maker v2.10 (32, 43). This annotation pipeline utilized de novo, homology, protein, and transcriptomic evidence (from 5 × 107 RNA-Seq reads) to predict the genic features. Physical linkage with the dhps locus was determined using BLAST alignment (44) of published P. jirovecii genome contigs against P. murina supercontigs (Liang Ma, personal communication, December, 2012; genome downloaded from the Broad Institute [http://www.broadinstitute.org/annotation/genome/Pneumocystis_group.2/MultiHome.html]).
Amplification and fragment sizing.
Fluorescently tagged forward primers for each variable microsatellite locus were designed (Applied Biosystems, Foster City, CA). Reverse primers carried a 5′ CTGTCTT “pigtail” to promote full adenylation and reduce stutter peaks (Table 1) (45). PCR was performed using HotStarTaq (Qiagen, Valencia, CA) according to the following generalized cycling parameters: 95°C for 15 min; 40 cycles of 95°C for 45 s, primer-specific annealing temperature (Ta) for 45 s, 72°C extension temperature for 60 s; 72°C extension temperature for 10 min; 12°C hold. To increase throughput, thermocycling was performed on Bio-Rad T100 (Bio-Rad, Hercules, CA) or ABI 2720 (Applied Biosystems, Foster City, CA) machines. For primer-specific sequences and Ta, see Table 1.
TABLE 1.
Primers and primer annealing temperatures for the P. jirovecii microsatellite loci
| Primer name | Primer sequence (5′→3′) |
Ta (°C)b | |
|---|---|---|---|
| Forward | Reversea | ||
| PjMS1 | AAGATGACAACGAGAATTGGCT | CTGTCTTAAAAGGCGATAAATGTGTGTCC | 53.3 |
| PjMS2 | TCATATACCGATCCTTTGGGAG | CTGTCTTGCCTGGACAATCCTGTCTCTAT | 53.3 |
| PjMS3 | AATAGGCGGAATCTCACTAGCA | CTGTCTTGACGGCAAAGAGTTGTTTCCT | 53.3 |
| PjMS4 | ATCGTAGAAGGATGGAAAGAAG | CTGTCTTGCAAATCGCATGTTCAAT | 50.5 |
| PjMS5 | ACTGTACCTAATCTTTCATCGG | CTGTCTTAAGGTTTTGGACGTTTGA | 50.5 |
| PjMS6 | TGCTCGAATTGCAGTAGAGATT | CTGTCTTCATCAGCAAGACGCTTAACTTG | 55.2 |
| PjMS7 | GATCTGGGTTGAATATAAGCGT | CTGTCTTTCTTTGGTTTCACAACAGCA | 55.2 |
| PjMS8 | CTTTGATTGCTCACGATATGGA | CTGTCTTATACGTTCACGGGCATAAGAGT | 53.3 |
| PjMS9 | GAACTTTGTCTTTAACAGGACG | CTGTCTTCTCAGTAGTAGCACCAATTCTTT | 47.0 |
Reverse primers have the 5′ addition of a CTGTCTT “pigtail” to decrease stutter.
Ta, temperature of primer annealing.
PCR fragment length was determined on an ABI 310 genetic analyzer and analyzed using ABI GeneMapper v4.1 software. A genomic DNA (gDNA) standard from a P. jirovecii-positive clinical specimen isolated at the University of North Carolina was used as an interrun standard to adjust for batch variability in fragment sizes. Capillary electrophoresis was performed using a denaturing polymer (POP-4; Applied Biosystems) in a 40-mm capillary at 60°C.
Excepting the possibility of diploidy, multiple microsatellite peaks likely indicate a multiclone infection (22). In the case of multiple peaks, the 2nd, 3rd, and 4th highest peaks were recorded if they were greater than one-third of the maximum peak height and also greater than 100 fluorescence units. For analyses, haplotypes were built from dominant peaks at each marker. This method of haplotype construction is reasonable, as peak height is relatively quantitative, and this has become a standard approach in other organisms in which polyclonal infections are common (46, 47). However, there is the potential that this method does not necessarily produce the single correct haplotype due to biases such as overamplification of shorter fragments during PCR. A further limitation to our method, which is common to all multilocus genotyping schemes, is the difficulty of assigning the correct phase of typed alleles in mixed populations. Fragment sizes were binned to the nearest repeat multiple. For most analyses, data points 3 standard deviations beyond the overall mean repeat size for that marker were considered missing data, and any individual with four or more missing data points was excluded from analysis.
Data analysis.
Summary statistics and genetic linkage were determined per the following methods. Heterozygosity (He) was determined using GenAlEx v6.5 for Microsoft Excel (48). He, a commonly used measure to quantify the diversity at a genetic locus in haploid organisms, is defined as , where pi is the frequency of the ith of k alleles. Linkage between genetic loci was determined in Arlequin v3.11 using an exact test of linkage disequilibrium (49), in which haplotypic data were arranged into contingency tables, and 105 Markov chains were used to explore simulated data with 104 burn-in (dememorization) steps. Bonferroni corrections were used to adjust significance levels for multiple comparisons.
Geographic population structure was determined per the following methods. Distance matrices and phylogenetic trees were generated in Populations v1.2.31 (http://bioinformatics.org/populations/) using the Cavalli-Sforza and Edwards chord distance (51). RST (52) and FST (53) measures of genetic differentiation between populations were calculated in SPAGeDi v1.4 (54) as follows: RST = 1 − (SW/S), where S is twice the estimated total allele size variance and SW is twice the estimated allele size variance within subpopulations; and FST = 1 − (HS/HT), where HT is total heterozygosity and HS is the mean heterozygosity within subpopulations. Ninety-five percent confidence intervals were generated in SPAGeDi using jackknifing over loci with 1,000 permutations.
To further investigate population structure, we used a combination of statistical and visual approaches. Fast UniFrac, a program for microbial population genetic analysis, was used to measure the reproducibility of the estimates of genetic distance between geographic populations (55). The reproducibility of genetic splits between populations was assessed using Monte Carlo simulations and jackknifing with 1,000 replicates. Principal coordinates analysis (PCoA), a multidimensional scaling analysis, was carried out in the Fast UniFrac browser window. PCoA identifies the main axes through a matrix using an eigenanalysis to quantitate dissimilarity between populations. A median-joining network was calculated and visualized in Network v4.611 (Fluxus Engineering, Suffolk, England). These networks allow for a visual representation of the mutational paths that may have led to the observed data and assume that mutations are more likely to derive from a more frequent haplotype and proceed to a less frequent haplotype.
In order to determine an optimal set of microsatellite loci for use in transmission studies, we used Simpson's index of diversity (D) to seek the minimum number of microsatellite loci necessary to fully explain the diversity of our data set (56). A Perl script was used to calculate Simpson's D for combinations of the microsatellite markers, using the following equation: , where N is the total number of parasite multilocus genotypes, s is the number of unique multilocus genotypes, and nj is the count frequency of the jth multilocus genotype variant (56). We added microsatellites to our scheme until D was ≥0.999 for our data set.
A set of six microsatellites was then used to investigate the relatedness between clinical isolates and transmission using two approaches. A neighbor-joining phylogeny (57) was generated in Populations v1.2.31, as outlined above, and visualized in the APE package for R (58). All samples from San Francisco with genotype calls for at least one-half of the markers (n = 40) were used in this analysis. In addition, hypotheses of evolutionary descent and transmission were generated in eBURST v3 (59). In this analysis, only samples with complete haplotypes for the selected markers were used due to the design of the program (n = 28). These analyses were performed in the San Francisco cohort alone, as this was the only sample set that contained paired samples from the same individuals.
Nucleotide sequence accession numbers.
Data for the sequences determined in this study were deposited in GenBank under accession numbers KF499042 to KF499075).
RESULTS
Of the 50 putative microsatellites identified and tested for interstrain length variability in the P. jirovecii genome, 9 were variable and thus carried forward into analyses (Table 1). Based upon predicted genome annotations, one microsatellite locus was intergenic, seven loci were intragenic noncoding, and one was intragenic coding (see Table S1 in the supplemental material). P. jirovecii isolates from 91 clinical specimens from Uganda, San Francisco, and Spain were typed at all 9 loci, and complete haplotypes were obtained for 63 (70%) isolates (Table 2). Amplification efficacy for each marker ranged from 78.0% for MS8 to 97.8% for MS1 (see Table S1 in the supplemental material). Each of the 9 markers was polyclonal in at least one sample. Similar to what was observed in other studies (60, 61), a high proportion of the clinical specimens were multiclonal: of the 91 BAL specimens, 63 (70%) contained a minimum of two P. jirovecii strains (at least 1 microsatellite with two peaks) and 14 isolates (15%) contained a minimum of three P. jirovecii strains (at least 1 microsatellite with three peaks).
TABLE 2.
Per-population summary statistics for P. jirovecii isolates
| Population source | No. of isolates | No. of complete haplotypesa | No. of paired samplesb | % of isolates with dhps mutant |
|---|---|---|---|---|
| Uganda | 13 | 10 | 0 | 100 (13/13) |
| San Francisco | 49 | 28 | 5 | 65.3 (32/49) |
| Spain | 29 | 25 | 0 | 10.3 (3/29) |
| All sources | 91 | 63 | 5 | 52.7 (48/91) |
Complete haplotypes are isolates for which fragment length was determined for all nine microsatellite markers.
Paired samples represent two bronchoalveolar lavage samples taken from a single patient, during either the same or different PcP episodes.
We assessed the variability of microsatellite repeats in order to quantify which loci were most informative. To do this, we computed summary statistics including heterozygosity (He) for each marker, both overall and on a per-population basis (Fig. 1; see Table S1 in the supplemental material). MS1 through MS8 had high mean He values (>0.5), indicating that they are diverse and likely informative. In contrast, MS9 had a reduced He value (<0.5). Interestingly, MS9 showed highly depressed He values relative to MS1 to 8 in Uganda but had similar He values relative to MS1 to 8 in Spain. Notably, all 13 Uganda samples contained mutations in dhps (3 Thr55Ala/Pro57Ser double mutants and 10 Pro57Ser single mutants), but very few of the Spain samples (3/29) contained mutations at dhps. Upon performing a scaffolded reconstruction of the P. jirovecii dhps region using Pneumocystis murina supercontigs, we found that MS9 was located ca. 50 kb upstream of the dhps locus. In pairwise tests of linkage disequilibrium, dhps was in significant linkage disequilibrium with MS9 (exact test, P < 0.05); no other markers were near the dhps locus.
FIG 1.
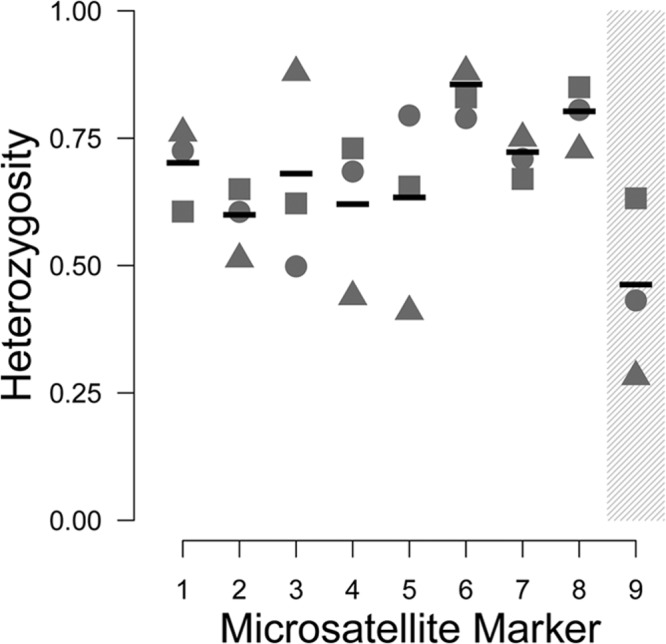
Per-population diversity index for each microsatellite studied. Nine microsatellites were typed in multiple samples from Uganda (triangles), San Francisco (circles), and Spain (squares), and heterozygosity (He) values were calculated for each population. Mean He values are indicated by horizontal bars. A higher He value indicates that a marker is more variable and thus more informative for population and transmission studies. Marker MS9 (shaded) was determined to be in linkage disequilibrium with the dhps locus.
The fact that the dhps locus and MS9 are in linkage disequilibrium with one another enables studies to investigate the genetic background of dhps mutations and to acquire evidence for a selective sweep of dhps mutant alleles. Eleven dhps mutant samples from Uganda and 14 dhps mutant samples from San Francisco with unambiguous MS9 genotype calls were used to construct dhps-MS9 haplotypes (see Table S2 in the supplemental material). In Uganda samples, we observed two unique dhps mutant genotypes (Thr-Arg-Ser and Ala-Arg-Ser) and two MS9 alleles, (TA)9 and (TA)10. Of the four possible combinations of dhps and MS9 alleles, we identified three with an excess of Thr-Arg-Ser/(TA)10 haplotypes. In San Francisco samples, we observed three unique dhps mutant genotypes (Ala-Arg-Pro, Thr-Arg-Ser, and Ala-Arg-Ser) and 5 MS9 alleles, (TA)8 to (TA)12. Of the 15 possible combinations of dhps and MS9 alleles, we identified 7 with an excess of Ala-Arg-Ser/(TA)10.
Intercontinental population structure.
In order to minimize bias in analyses of population structure, we tested for linkage between microsatellite loci (62). Because the Pneumocystis jirovecii draft genome is in 356 contigs (32, 63) rather than in chromosomes, we could not rely on genetic maps and instead tested linkage disequilibrium between markers. Bonferroni-corrected per-population pairwise tests of linkage disequilibrium showed no significant linkage disequilibrium between any locus pairs in any of the three populations (results not shown).
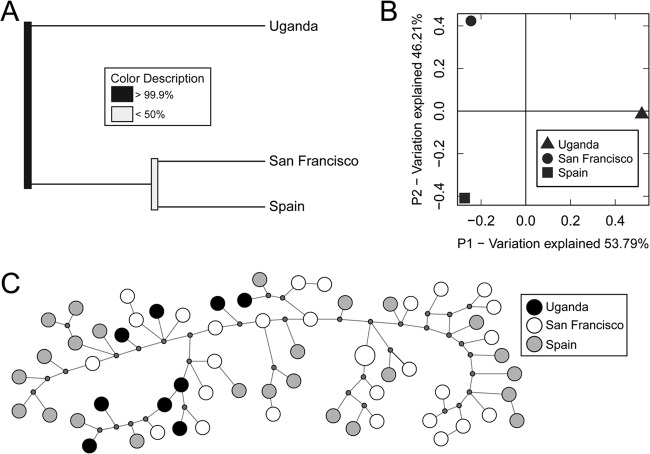
Using the 8 unlinked microsatellites, we examined the extent of genetic differentiation between P. jirovecii populations from Uganda, San Francisco, and Spain. We employed UniFrac, a widely used method for probing the genetic diversity between microbial communities, which can draw conclusions about the relative distances between multiple populations. In this analysis, P. jirovecii isolates from Uganda were genetically distinct from those in San Francisco and Spain (Fig. 2A). Principal coordinates analysis revealed that the first coordinate explained approximately one-half (53.8%) of the observed genetic difference between Uganda isolates and the other populations (Fig. 2B). Finally, we constructed a median-joining network analysis charting potential mutational intermediates between observed P. jirovecii isolates. This showed little structure between populations, with the exception being that Uganda occupied a restricted range of genetic diversity (Fig. 2C).
FIG 2.
Limited population structure and genetic differentiation between disease-causing P. jirovecii populations. (A) UniFrac revealed a distinct genetic division between Uganda and San Francisco/Spain but little genetic difference between P. jirovecii isolates from San Francisco and Spain. (B) Principal coordinates analysis demonstrated that approximately one-half of the genetic difference between population clusters was associated with geographic differences. (C) A median-joining network predicts potential mutational paths between haplotypes. The samples from Uganda (black circles) show clustering on one side of the network, likely leading to the differences seen in the other analyses.
We also evaluated genetic distances between these populations using RST (52) and FST (53), two classic measures of population demographic history (64). Similar to our previous analyses, RST and FST suggested that while Ugandan samples were significantly divergent from the San Francisco and Spain populations, there was limited genetic differentiation between the three populations overall (Table 3) (65). Specifically, RST, which assumes a stepwise mutation model of microsatellite evolution, revealed no genetic distance between the populations from San Francisco and Spain and limited distances in the Uganda-San Francisco and Uganda-Spain comparisons (RST range, −0.017 to 0.125). FST also revealed limited genetic distances between populations (FST range, 0.022 to 0.067).
TABLE 3.
Pairwise RST and FST statistics for between-population pairwise comparisons
| Source | RST or FST (95% CI)a |
||
|---|---|---|---|
| Uganda | San Francisco | Spain | |
| Uganda | 0.125 (0.074–0.220) | 0.115 (0.060–0.233) | |
| San Francisco | 0.048 (0.020–0.090) | −0.017 (−0.044–0.044) | |
| Spain | 0.067 (0.036–0.109) | 0.022 (0.006–0.043) | |
Data from all 8 neutral microsatellite loci were used to calculate RST (above the diagonal) and FST (below the diagonal) in SPAGeDi v1.4. In general, an FST or RST value of 0 to 0.05 indicates little genetic differentiation, 0.05 to 0.15 indicates moderate differentiation, 0.15 to 0.25 indicates substantial differentiation, and 0.25 to 1.00 indicates very great genetic differentiation. CI, confidence interval.
Relatedness between isolates within individuals and within a single population.
To maximize the efficiency of a potential microsatellite genotyping scheme, we determined the minimum number of microsatellite loci necessary for discriminating between strains, using both the loci with the greatest heterozygosity and those with the greatest amplification efficiency (see Table S3 in the supplemental material). We achieved high resolution (Simpson's D, ≥0.999) across all specimens tested by using the 6 most heterozygous markers (MS6, MS8, MS7, MS1, MS3, and MS5) (see Table S3 in the supplemental material). This extensive diversity is similar to the diversity described in both Pneumocystis (30) and other pathogens (66–69), making this typing method a powerful tool for P. jirovecii population genetic studies (56).
In order to evaluate the suitability of this array to study the transmission of P. jirovecii in humans, we investigated paired specimens collected from the same patient. This analysis was restricted to isolates from San Francisco, which included five isolate pairs (10 isolates) in which two samples were taken from the same individual. The genotypes of each pair are shown in Table S4 in the supplemental material. In a neighbor-joining phylogeny of all San Francisco samples, all paired isolates grouped closely together, with differing extents of genetic similarity (Fig. 3). The extent of genetic similarity between paired isolates was reflective of the time elapsed between collection of the two isolates (see Table S4 in the supplemental material).
FIG 3.
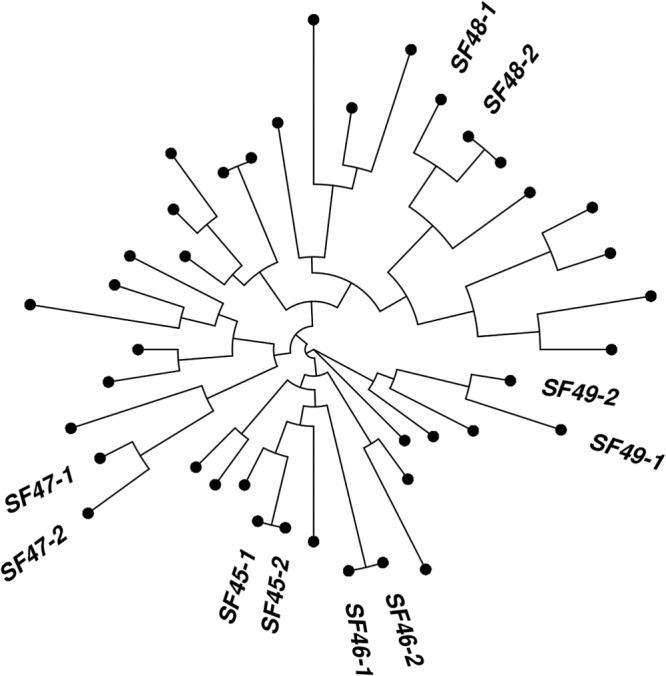
Neighbor-joining phylogenetic tree of P. jirovecii clinical isolates from San Francisco. Repeat samples from the same clinical episode produced an identical genotype (SF45-1 and -2 and SF46-1 and -2). The other paired samples were genetically similar between samples but not identical. The other points represent isolates from patients from which a single sample is available. Very few samples had identical genotypes, suggesting that the six-microsatellite array will have a high discriminatory power between isolates.
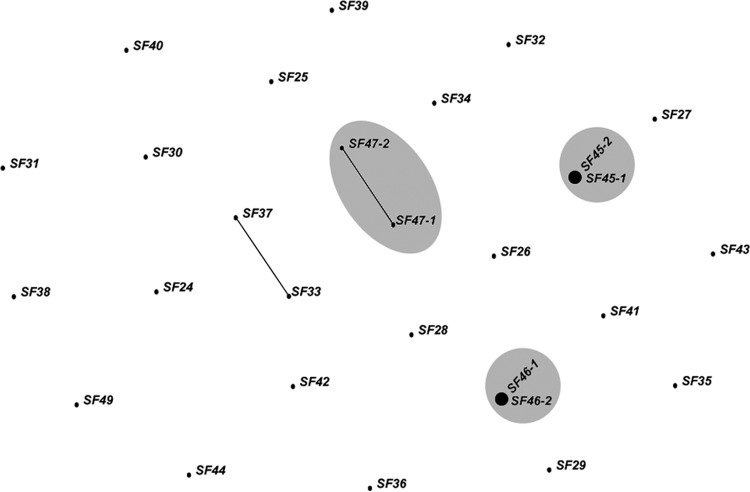
To more rigorously investigate this finding, we employed eBurst, a tool developed to study outbreaks and explore pathogen transmission dynamics (59, 70). eBurst visualizes relationships (edges) between individual parasite isolates (nodes) to generate hypotheses about pathogen relatedness. We investigated all San Francisco isolates from the years 2008 to 2011 for which there were complete haplotypic data at the 6 most informative markers (including three isolate pairs). eBurst analysis showed that the two isolate pairs that were temporally closest to one another (SF45 and SF46 in Table S4 in the supplemental material) were identical at these six markers, while the third isolate pair (SF47) was closely related, differing at only one locus (Fig. 4). The other paired isolates (SF48 and SF49) contained missing alleles and could not be included in this analysis (see Table S4 in the supplemental material).
FIG 4.
Related P. jirovecii genotypes in the San Francisco population. eBURST outbreak analysis on San Francisco samples in which a complete six-microsatellite haplotype was available. Paired samples are highlighted in gray. Two pairs (SF45-1 and -2 and SF46-1 and -2) were identical to one another and occurred in the same clinical episode of PcP. Another pair of isolates (SF47-1 and -2) were related, with only one microsatellite difference.
DISCUSSION
Understanding the population genetics and transmission dynamics of fungal pathogens is key for understanding their life history and for designing new prevention and treatment strategies. Appropriate strain typing methods are crucial to achieve this understanding (71). Here, we introduce the first multilocus microsatellite genotyping scheme for Pneumocystis jirovecii. In contrast to past genotyping schemes, which rely in large part on genic coding regions, including potential drug resistance (DHFR, DHPS, and SOD) genes and housekeeping (mtLSU-rRNA and mt26S) genes (25, 27, 31), we employ microsatellites, most of which are noncoding (see Table S1 in the supplemental material). Thus, this genotyping method carries a high discriminatory index that may be relatively free of selective pressure or functional constraints, making it a useful technique for studying population structure, sulfonamide resistance, and transmission dynamics. Microsatellites are widely employed for genotyping individuals owing to their robustness, scalability, and high information content. Given these advantages, this genotyping approach will enable studies that improve our understanding of P. jirovecii biology.
Using these microsatellite markers, we observed limited genetic differentiation between P. jirovecii populations from Uganda, the United States, and Spain. This observation was supported by an ecological clustering algorithm (Fig. 2A) and classical measures of genetic distance (Table 3), both of which indicate a slight divergence of the Ugandan isolates but relatively little divergence overall. Principal coordinates analysis revealed that approximately one-half of the genetic differences between all three populations could be explained by geography (Fig. 2B), with the maximum distance occurring in coordinate one, which separated Uganda from San Francisco and Spain. Furthermore, a median-joining network showed that isolates from all three sites were thoroughly admixed, with the only notable trend being that Ugandan isolates clustered loosely in one part of the network (Fig. 2C).
The relative lack of genetic divergence that we observed between sites, and over time, is similar to what has been reported before (72–74). This has several potential explanations, including the following: (i) P. jirovecii underwent a recent global spread, (ii) P. jirovecii has a low mutation rate, or (iii) intercontinental gene flow between P. jirovecii populations occurs frequently. Our understanding of Pneumocystis populations will be further enhanced as the P. jirovecii genome takes a more complete form and as additional samples from other regions are analyzed.
Interestingly, we identified one microsatellite linked to the dhps gene, which encodes the enzymatic target of sulfonamide antibiotics that are used to treat and prevent PcP. Polymorphisms in dhps are associated with exposure to antifolate drugs (10, 75) and, in some studies, clinical outcomes (76–79). All 13 isolates from Uganda harbored mutations in dhps: three with both the Thr55Ala and Pro57Ser substitutions and 10 with the single Pro57Ser substitution. These mutant haplotypes of dhps likely reflect directional selection by sulfonamide antimicrobials used to treat infections; consistent with this hypothesis, we observed very low heterogeneity in the microsatellite locus linked to dhps in Ugandan isolates (He = 0.282). In contrast, the isolates from Spain, which were largely sulfa naive and which harbored primarily wild-type dhps haplotypes, demonstrated a higher heterozygosity at this dhps-linked locus (He = 0.632). Taken together with evidence that antibiotic pressure causes changes in dhps mutant genotype frequency (73), this pattern is consistent with a selective sweep occurring around the dhps locus due to drug pressure, as has been observed in pathogens such as Plasmodium falciparum (36). In order to confirm this, further studies will require additional markers near the dhps locus and greater numbers of isolates bearing wild-type and mutant dhps haplotypes.
In addition to understanding population structure and drug selection, microsatellite analysis has potential as a tool for molecular epidemiology studies evaluating transmission. The currently used multilocus genotyping schemes have revealed that (i) specific P. jirovecii strains are associated with failure of prophylaxis (80) and can cause more-virulent PcP episodes (81), (ii) specific strains persist in hospitals for weeks at a time (80), and (iii) P. jirovecii strains can be acquired later in life by immunocompromised and immunocompetent patients (22, 82). Despite this wealth of knowledge, the current multilocus genotyping strategies have left many questions regarding transmission of P. jirovecii unanswered. For example, while it is clear that infants and immunocompetent adults are environmental reservoirs for P. jirovecii (83–87), there is not yet molecular evidence that these reservoirs are an important source of disease-causing P. jirovecii transmission to immunocompromised individuals. Because there is compelling evidence that transiently colonized immunocompetent mice can transmit P. jirovecii to immunosuppressed mice and vice versa (88, 89), studies of P. jirovecii transmission among immunocompetent and immunosuppressed humans may help uncover a mechanism for disease prevention. A highly discriminating genotyping method with a stepwise mutation model would prove useful in addressing this and other questions.
The microsatellite genotyping method described here could help reveal details of P. jirovecii reservoirs and transmission. By assessing five paired clinical samples (10 isolates), we showed that this approach has high discriminatory power, making it ideal for molecular epidemiological studies (56). Among our five paired isolates, we observed two cases (SF45 and SF46) in which the two samples in the pair had identical genotypes in a clinical scenario consistent with a single infection. In the case of SF45, both samples were collected by a single bronchoscopy in two different lobes of the lung. In the case of SF46, the first sample was collected by bronchoscopy upon initial presentation, and the second sample was collected 15 days later after clinical deterioration despite antifolate therapy. The last three cases (SF47, SF48, and SF49) all represent PcP patients who received PcP therapy but experienced a recurrence weeks to months later. In all three cases, the pairs were genetically close, but not identical, similar to past observations (61). This finding could be explained in multiple ways, including the following: (i) these patients may have been infected with multiple P. jirovecii strains, which were differentially detected at different time points (90), or (ii) after clinical resolution of their initial PcP case, these patients returned home and were subsequently colonized by genetically related—though not identical—organisms. Importantly, these data show that this method provides high discriminatory power between infections: other than those isolate pairs expected to be related, there were no identical isolates and only two related isolates (Fig. 3 and 4). This observation suggests that this genotyping method will prove useful for future studies of P. jirovecii outbreaks and transmission dynamics. Additionally, work from others suggests that PCR-based P. jirovecii genotyping approaches may prove successful even in immunocompetent individuals (91), enabling future studies of transmission between immunocompromised patients and immunocompetent health care workers or close contacts.
Pneumocystis jirovecii remains an important opportunistic pathogen and causes significant disease in immunocompromised individuals. Many questions about the biology of the organism remain unanswered, and the PcP disease burden is incompletely understood. Molecular epidemiology studies have the potential to provide critical information concerning organismal biology, and insights from these studies could impact health policy and influence medication use in patients. While high-resolution genotyping methods, such as this one, should be applied with care to some study types (61, 73), our approach provides a stepwise mutational model, high typeability, and high discriminatory power that will be valuable for future molecular epidemiological studies of P. jirovecii.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by ULTR000083, NIAID R01 AI089819, and the University of North Carolina at Chapel Hill Center for AIDS Research (CFAR), an NIH-funded program (P30 AI50410). Subject enrollment and specimen collection in Uganda and San Francisco were funded by NHLBI R01 HL090335 (L.H.). Subject enrollment and specimen collection in Spain were supported by the “Fundación para la Investigación y la Prevención del SIDA en España” (FIPSE, Madrid, Spain), grant 24298/01. C.M.P. was supported by GM008719, GM007092, and a grant from the IDSA Medical Scholars Program. L.H. was supported by NHLBI K24 HL087713, R01 HL090335, and U01 HL098964. S.M.T. is supported by NIAID K08 AI100924.
The opinions expressed here are the authors' own and do not necessarily reflect the opinions of these funding organizations.
Footnotes
Published ahead of print 12 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02531-13.
REFERENCES
- 1.Hoover DR, Saah AJ, Bacellar H, Phair J, Detels R, Anderson R, Kaslow RA. 1993. Clinical manifestations of AIDS in the era of pneumocystis prophylaxis. Multicenter AIDS Cohort Study. N. Engl. J. Med. 329:1922–1926 [DOI] [PubMed] [Google Scholar]
- 2.Wolff AJ, O'Donnell AE. 2001. Pulmonary manifestations of HIV infection in the era of highly active antiretroviral therapy. Chest 120:1888–1893. 10.1378/chest.120.6.1888 [DOI] [PubMed] [Google Scholar]
- 3.San-Andrés F-J, Rubio R, Castilla J, Pulido F, Palao G, de Pedro I, Costa J-R, del Palacio A. 2003. Incidence of acquired immunodeficiency syndrome-associated opportunistic diseases and the effect of treatment on a cohort of 1115 patients infected with human immunodeficiency virus, 1989-1997. Clin. Infect. Dis. 36:1177–1185. 10.1086/374358 [DOI] [PubMed] [Google Scholar]
- 4.Jain MK, Skiest DJ, Cloud JW, Jain CL, Burns D, Berggren RE. 2003. Changes in mortality related to human immunodeficiency virus infection: comparative analysis of inpatient deaths in 1995 and in 1999-2000. Clin. Infect. Dis. 36:1030–1038. 10.1086/368186 [DOI] [PubMed] [Google Scholar]
- 5.Pulvirenti J, Herrera P, Venkataraman P, Ahmed N. 2003. Pneumocystis carinii pneumonia in HIV-infected patients in the HAART era. AIDS Patient Care STDs 17:261–265. 10.1089/108729103322108139 [DOI] [PubMed] [Google Scholar]
- 6.Mansharamani NG, Garland R, Delaney D, Koziel H. 2000. Management and outcome patterns for adult Pneumocystis carinii pneumonia, 1985 to 1995: comparison of HIV-associated cases to other immunocompromised states. Chest 118:704–711. 10.1378/chest.118.3.704 [DOI] [PubMed] [Google Scholar]
- 7.Walzer PD, Evans HER, Copas AJ, Edwards SG, Grant AD, Miller RF. 2008. Early predictors of mortality from Pneumocystis jirovecii pneumonia in HIV-infected patients: 1985-2006. Clin. Infect. Dis. 46:625–633. 10.1086/526778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fei MW, Kim EJ, Sant CA, Jarlsberg LG, Davis JL, Swartzman A, Huang L. 2009. Predicting mortality from HIV-associated Pneumocystis pneumonia at illness presentation: an observational cohort study. Thorax 64:1070–1076. 10.1136/thx.2009.117846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisk DT, Meshnick S, Kazanjian PH. 2003. Pneumocystis carinii pneumonia in patients in the developing world who have acquired immunodeficiency syndrome. Clin. Infect. Dis. 36:70–78. 10.1086/344951 [DOI] [PubMed] [Google Scholar]
- 10.Armstrong W, Meshnick S, Kazanjian P. 2000. Pneumocystis carinii mutations associated with sulfa and sulfone prophylaxis failures in immunocompromised patients. Microbes Infect. 2:61–67. 10.1016/S1286-4579(00)00284-7 [DOI] [PubMed] [Google Scholar]
- 11.Huang L, Beard CB, Creasman J, Levy D, Duchin JS, Lee S, Pieniazek N, Carter JL, del Rio C, Rimland D, Navin TR. 2000. Sulfa or sulfone prophylaxis and geographic region predict mutations in the Pneumocystis carinii dihydropteroate synthase gene. J. Infect. Dis. 182:1192–1198. 10.1086/315824 [DOI] [PubMed] [Google Scholar]
- 12.Taylor SM, Meshnick SR, Worodria W, Andama A, Cattamanchi A, Davis JL, Yoo SD, Byanyima P, Kaswabuli S, Goodman CD, Huang L, International HIV-associated Opportunistic Pneumonias Study 2012. Low prevalence of Pneumocystis pneumonia (PCP) but high prevalence of pneumocystis dihydropteroate synthase (dhps) gene mutations in HIV-infected persons in Uganda. PLoS One 7:e49991. 10.1371/journal.pone.0049991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coyle PV, McCaughey C, Nager A, McKenna J, O'Neill H, Feeney SA, Fairley D, Watt A, Cox C, Curran T. 2012. Rising incidence of Pneumocystis jirovecii pneumonia suggests iatrogenic exposure of immune-compromised patients may be becoming a significant problem. J. Med. Microbiol. 61:1009–1015. 10.1099/jmm.0.043984-0 [DOI] [PubMed] [Google Scholar]
- 14.Branten AJ, Beckers PJ, Tiggeler RG, Hoitsma AJ. 1995. Pneumocystis carinii pneumonia in renal transplant recipients. Nephrol. Dial. Transplant. 10:1194–1197 [PubMed] [Google Scholar]
- 15.Hennequin C, Page B, Roux P, Legendre C, Kreis H. 1995. Outbreak of Pneumocystis carinii pneumonia in a renal transplant unit. Eur. J. Clin. Microbiol. 14:122–126. 10.1007/BF02111870 [DOI] [PubMed] [Google Scholar]
- 16.de Boer MGJ, Bruijnesteijn van Coppenraet LES, Gaasbeek A, Berger SP, Gelinck LBS, van Houwelingen HC, van den Broek P, Kuijper EJ, Kroon FP, Vandenbroucke JP. 2007. An outbreak of Pneumocystis jiroveci pneumonia with 1 predominant genotype among renal transplant recipients: interhuman transmission or a common environmental source? Clin. Infect. Dis. 44:1143–1149. 10.1086/513198 [DOI] [PubMed] [Google Scholar]
- 17.Arichi N, Kishikawa H, Mitsui Y, Kato T, Nishimura K, Tachikawa R, Tomii K, Shiina H, Igawa M, Ichikawa Y. 2009. Cluster outbreak of Pneumocystis pneumonia among kidney transplant patients within a single center. Transplant. Proc. 41:170–172. 10.1016/j.transproceed.2008.10.027 [DOI] [PubMed] [Google Scholar]
- 18.Sassi M, Ripamonti C, Mueller NJ, Yazaki H, Kutty G, Ma L, Huber C, Gogineni E, Oka S, Goto N, Fehr T, Gianella S, Konrad R, Sing A, Kovacs JA. 2012. Outbreaks of Pneumocystis pneumonia in 2 renal transplant centers linked to a single strain of Pneumocystis: implications for transmission and virulence. Clin. Infect. Dis. 54:1437–1444. 10.1093/cid/cis217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka M, Sakai R, Koike R, Komano Y, Nanki T, Sakai F, Sugiyama H, Matsushima H, Kojima T, Ohta S, Ishibe Y, Sawabe T, Ota Y, Ohishi K, Miyazato H, Nonomura Y, Saito K, Tanaka Y, Nagasawa H, Takeuchi T, Nakajima A, Ohtsubo H, Onishi M, Goto Y, Dobashi H, Miyasaka N, Harigai M. 2012. Pneumocystis jirovecii pneumonia associated with etanercept treatment in patients with rheumatoid arthritis: a retrospective review of 15 cases and analysis of risk factors. Mod. Rheumatol. 22:849–858. 10.1007/s10165-012-0615-z [DOI] [PubMed] [Google Scholar]
- 20.Goto N, Oka S. 2011. Pneumocystis jirovecii pneumonia in kidney transplantation. Transpl. Infect. Dis. 13:551–558. 10.1111/j.1399-3062.2011.00691.x [DOI] [PubMed] [Google Scholar]
- 21.Huang L, Cattamanchi A, Davis JL, den Boon S, Kovacs J, Meshnick S, Miller RF, Walzer PD, Worodria W, Masur H, International HIV-associated Opportunistic Pneumonias (IHOP) Study Lung HIV Study. 2011. HIV-associated Pneumocystis pneumonia. Proc. Am. Thorac. Soc. 8:294–300. 10.1513/pats.201009-062WR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matos O, Esteves F. 2010. Pneumocystis jirovecii multilocus gene sequencing: findings and implications. Future Microbiol. 5:1257–1267. 10.2217/fmb.10.75 [DOI] [PubMed] [Google Scholar]
- 23.Wakefield AE. 1998. Genetic heterogeneity in Pneumocystis carinii: an introduction. FEMS Immunol. Med. Microbiol. 22:5–13. 10.1111/j.1574-695X.1998.tb01182.x [DOI] [PubMed] [Google Scholar]
- 24.de Boer MGJ, de Fijter JW, Kroon FP. 2011. Outbreaks and clustering of Pneumocystis pneumonia in kidney transplant recipients: a systematic review. Med. Mycol. 49:673–680 [DOI] [PubMed] [Google Scholar]
- 25.Hauser PM, Blanc DS, Bille J, Francioli P. 1998. Typing methods to approach Pneumocystis carinii genetic heterogeneity. FEMS Immunol. Med. Microbiol. 22:27–35. 10.1111/j.1574-695X.1998.tb01184.x [DOI] [PubMed] [Google Scholar]
- 26.Keely SP, Cushion MT, Stringer JR. 2003. Diversity at the locus associated with transcription of a variable surface antigen of Pneumocystis carinii as an index of population structure and dynamics in infected rats. Infect. Immun. 71:47–60. 10.1128/IAI.71.1.47-60.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esteves F, Gaspar J, de Sousa B, Antunes F, Mansinho K, Matos O. 2012. Pneumocystis jirovecii multilocus genotyping in pooled DNA samples: a new approach for clinical and epidemiological studies. Clin. Microbiol. Infect. 18:E177–E184. 10.1111/j.1469-0691.2012.03828.x [DOI] [PubMed] [Google Scholar]
- 28.Esteves F, Gaspar J, De Sousa B, Antunes F, Mansinho K, Matos O. 2011. Clinical relevance of multiple single-nucleotide polymorphisms in Pneumocystis jirovecii pneumonia: development of a multiplex PCR-single-base-extension methodology. J. Clin. Microbiol. 49:1810–1815. 10.1128/JCM.02303-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hauser PM, Blanc DS, Sudre P, Senggen Manoloff E, Nahimana A, Bille J, Weber R, Francioli P. 2001. Genetic diversity of Pneumocystis carinii in HIV-positive and -negative patients as revealed by PCR-SSCP typing. AIDS 15:461–466. 10.1097/00002030-200103090-00004 [DOI] [PubMed] [Google Scholar]
- 30.Maitte C, Leterrier M, Pape PL, Miegeville M, Morio F. 2013. Multilocus sequence typing of Pneumocystis jirovecii from clinical samples: how many and which loci should be used? J. Clin. Microbiol. 51:2843–2849. 10.1128/JCM.01073-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esteves F, Montes-Cano MA, de la Horra C, Costa MC, Calderón EJ, Antunes F, Matos O. 2008. Pneumocystis jirovecii multilocus genotyping profiles in patients from Portugal and Spain. Clin. Microbiol. Infect. 14:356–362. 10.1111/j.1469-0691.2007.01944.x [DOI] [PubMed] [Google Scholar]
- 32.Cissé OH, Pagni M, Hauser PM. 2012. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio 4:e00428–00412. 10.1128/mBio.00428-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson KE, Williams CM, Graham NM. 2004. Infectious disease epidemiology: theory and practice. Jones and Bartlett, Burlington, MA [Google Scholar]
- 34.Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. 1996. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154. 10.1038/380152a0 [DOI] [PubMed] [Google Scholar]
- 35.Cortese JF, Caraballo A, Contreras CE, Plowe CV. 2002. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J. Infect. Dis. 186:999–1006. 10.1086/342946 [DOI] [PubMed] [Google Scholar]
- 36.Taylor SM, Antonia AL, Parobek CM, Juliano JJ, Janko M, Emch M, Alam MT, Udhayakumar V, Tshefu AK, Meshnick SR. 2013. Plasmodium falciparum sulfadoxine resistance is geographically and genetically clustered within the DR Congo. Sci. Rep. 3:1165. 10.1038/srep01165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alvarez-Martínez MJ, Moreno A, Miró JM, Valls ME, Rivas PV, de Lazzari E, Sued O, Benito N, Domingo P, Ribera E, Santín M, Sirera G, Segura F, Vidal F, Rodríguez F, Riera M, Cordero ME, Arribas JR, Jiménez de Anta MT, Gatell JM, Wilson PE, Meshnick SR, Spanish Working Group PCP 2008. Pneumocystis jirovecii pneumonia in Spanish HIV-infected patients in the combined antiretroviral therapy era: prevalence of dihydropteroate synthase mutations and prognostic factors of mortality. Diagn. Microbiol. Infect. Dis. 62:34–43. 10.1016/j.diagmicrobio.2008.04.016 [DOI] [PubMed] [Google Scholar]
- 38.Yoon C, Subramanian A, Chi A, Crothers K, Meshnick SR, Taylor SM, Beard CB, Jarlsberg LG, Lawrence GG, Avery M, Swartzman A, Fong S, Roth B, Huang L, International HIV-Associated Opportunistic Pneumonias (IHOP) Study 2013. Dihydropteroate synthase mutations in Pneumocystis pneumonia: impact of applying different definitions of prophylaxis, mortality endpoints and mutant in a single cohort. Med. Mycol. 51:568–575. 10.3109/13693786.2013.770604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor SM, Meshnick SR, Worodria W, Andama A, Davis JL, Cattamanchi A, den Boon S, Yoo SD, Goodman CD, Huang L. 2012. Low prevalence of Pneumocystis jirovecii lung colonization in Ugandan HIV-infected patients hospitalized with non-Pneumocystis pneumonia. Diagn. Microbiol. Infect. Dis. 72:139–143. 10.1016/j.diagmicrobio.2011.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martins WS, Lucas DCS, Neves KF, de S, Bertioli DJ. 2009. WebSat—a web software for microsatellite marker development. Bioinformation 3:282–283. 10.6026/97320630003282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 43.Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18:188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 45.Brownstein MJ, Carpten JD, Smith JR. 1996. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20:1004–1006, 1008–1010 [DOI] [PubMed] [Google Scholar]
- 46.Ford AF, Schall JJ. 2011. Relative clonal proportions over time in mixed-genotype infections of the lizard malaria parasite Plasmodium mexicanum. Int. J. Parasitol. 41:731–738. 10.1016/j.ijpara.2011.01.010 [DOI] [PubMed] [Google Scholar]
- 47.Van Den Eede P, Erhart A, der Auwera GV, Overmeir CV, Thang ND, Hung LX, Anne J, D'Alessandro U. 2010. High complexity of Plasmodium vivax infections in symptomatic patients from a rural community in central Vietnam detected by microsatellite genotyping. Am. J. Trop. Med. Hyg. 82:223–227. 10.4269/ajtmh.2010.09-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10:564–567. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 50. Reference deleted.
- 51.Cavalli-Sforza LL, Edwards AW. 1967. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 19:233–257 [PMC free article] [PubMed] [Google Scholar]
- 52.Slatkin M. 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright S. 1950. Genetical structure of populations. Nature 166:247–249. 10.1038/166247a0 [DOI] [PubMed] [Google Scholar]
- 54.Hardy O, Vekemans X. 2002. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2:618–620. 10.1046/j.1471-8286.2002.00305.x [DOI] [Google Scholar]
- 55.Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27. 10.1038/ismej.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 58.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 59.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530. 10.1128/JB.186.5.1518-1530.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hauser PM. 2004. The development of a typing method for an uncultivable microorganism: the example of Pneumocystis jirovecii. Infect. Genet. Evol. 4:199–203. 10.1016/j.meegid.2004.01.011 [DOI] [PubMed] [Google Scholar]
- 61.Esteves F, Gaspar J, Tavares A, Moser I, Antunes F, Mansinho K, Matos O. 2010. Population structure of Pneumocystis jirovecii isolated from immunodeficiency virus-positive patients. Infect. Genet. Evol. 10:192–199. 10.1016/j.meegid.2009.12.007 [DOI] [PubMed] [Google Scholar]
- 62.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959 http://www.genetics.org/content/155/2/945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cushion MT, Keely SP. 2013. Assembly and annotation of Pneumocystis jirovecii from the human lung microbiome. mBio 4:e00224. 10.1128/mBio.00224-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holsinger KE, Weir BS. 2009. Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat. Rev. Genet. 10:639–650. 10.1038/nrg2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hartl DL, Clark AG. 2006. Principles of population genetics, 4th ed. Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 66.Weisser M, Oostdijk EA, Willems RJL, Bonten MJM, Frei R, Elzi L, Halter J, Widmer AF, Top J. 2012. Dynamics of ampicillin-resistant Enterococcus faecium clones colonizing hospitalized patients: data from a prospective observational study. BMC Infect. Dis. 12:68. 10.1186/1471-2334-12-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guerrero C, Bernasconi C, Burki D, Bodmer T, Telenti A. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin IMC, Ison CA, Aanensen DM, Fenton KA, Spratt BG. 2004. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J. Infect. Dis. 189:1497–1505. 10.1086/383047 [DOI] [PubMed] [Google Scholar]
- 69.Marais BJ, Mlambo CK, Rastogi N, Zozio T, Duse AG, Victor TC, Marais E, Warren RM. 2013. Epidemic spread of multidrug-resistant tuberculosis in Johannesburg, South Africa. J. Clin. Microbiol. 51:1818–1825. 10.1128/JCM.00200-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner KME, Hanage WP, Fraser C, Connor TR, Spratt BG. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 7:30. 10.1186/1471-2180-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taylor JW, Geiser DM, Burt A, Koufopanou V. 1999. The evolutionary biology and population genetics underlying fungal strain typing. Clin. Microbiol. Rev. 12:126–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wakefield AE, Fritscher CC, Malin AS, Gwanzura L, Hughes WT, Miller RF. 1994. Genetic diversity in human-derived Pneumocystis carinii isolates from four geographical locations shown by analysis of mitochondrial rRNA gene sequences. J. Clin. Microbiol. 32:2959–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller RF, Lindley AR, Malin AS, Ambrose HE, Wakefield AE. 2005. Isolates of Pneumocystis jirovecii from Harare show high genotypic similarity to isolates from London at the superoxide dismutase locus. Trans. R. Soc. Trop. Med. Hyg. 99:202–206. 10.1016/j.trstmh.2004.09.005 [DOI] [PubMed] [Google Scholar]
- 74.Tsolaki AG, Beckers P, Wakefield AE. 1998. Pre-AIDS era isolates of Pneumocystis carinii f. sp. hominis: high genotype similarity with contemporary isolates. J. Clin. Microbiol. 36:90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kazanjian P, Armstrong W, Hossler PA, Burman W, Richardson J, Lee CH, Crane L, Katz J, Meshnick SR. 2000. Pneumocystis carinii mutations are associated with duration of sulfa or sulfone prophylaxis exposure in AIDS patients. J. Infect. Dis. 182:551–557. 10.1086/315719 [DOI] [PubMed] [Google Scholar]
- 76.Meshnick SR. 1999. Drug-resistant Pneumocystis carinii. Lancet 354:1318–1319. 10.1016/S0140-6736(99)00240-8 [DOI] [PubMed] [Google Scholar]
- 77.Helweg-Larsen J, Benfield TL, Eugen-Olsen J, Lundgren JD, Lundgren B. 1999. Effects of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of AIDS-associated P carinii pneumonia. Lancet 354:1347–1351. 10.1016/S0140-6736(99)03320-6 [DOI] [PubMed] [Google Scholar]
- 78.Navin TR, Beard CB, Huang L, del Rio C, Lee S, Pieniazek NJ, Carter JL, Le T, Hightower A, Rimland D. 2001. Effect of mutations in Pneumocystis carinii dihydropteroate synthase gene on outcome of P. carinii pneumonia in patients with HIV-1: a prospective study. Lancet 358:545–549. 10.1016/S0140-6736(01)05705-1 [DOI] [PubMed] [Google Scholar]
- 79.Crothers K, Beard CB, Turner J, Groner G, Fox M, Morris A, Eiser S, Huang L. 2005. Severity and outcome of HIV-associated Pneumocystis pneumonia containing Pneumocystis jirovecii dihydropteroate synthase gene mutations. AIDS 19:801–805. 10.1097/01.aids.0000168974.67090.70 [DOI] [PubMed] [Google Scholar]
- 80.Hauser PM, Sudre P, Nahimana A, Francioli P, Study Group 2001. Prophylaxis failure is associated with a specific Pneumocystis carinii genotype. Clin. Infect. Dis. 33:1080–1082. 10.1086/322659 [DOI] [PubMed] [Google Scholar]
- 81.Esteves F, Gaspar J, Marques T, Leite R, Antunes F, Mansinho K, Matos O. 2010. Identification of relevant single-nucleotide polymorphisms in Pneumocystis jirovecii: relationship with clinical data. Clin. Microbiol. Infect. 16:878–884. 10.1111/j.1469-0691.2009.03030.x [DOI] [PubMed] [Google Scholar]
- 82.Beard CB, Carter JL, Keely SP, Huang L, Pieniazek NJ, Moura IN, Roberts JM, Hightower AW, Bens MS, Freeman AR, Lee S, Stringer JR, Duchin JS, del Rio C, Rimland D, Baughman RP, Levy DA, Dietz VJ, Simon P, Navin TR. 2000. Genetic variation in Pneumocystis carinii isolates from different geographic regions: implications for transmission. Emerg. Infect. Dis. 6:265–272. 10.3201/eid0603.000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gigliotti F, Wright TW. 2012. Pneumocystis: where does it live? PLoS Pathog. 8:e1003025. 10.1371/journal.ppat.1003025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chabé M, Dei-Cas E, Creusy C, Fleurisse L, Respaldiza N, Camus D, Durand-Joly I. 2004. Immunocompetent hosts as a reservoir of pneumocystis organisms: histological and RT-PCR data demonstrate active replication. Eur. J. Clin. Microbiol. Infect. Dis. 23:89–97. 10.1007/s10096-003-1092-2 [DOI] [PubMed] [Google Scholar]
- 85.Totet A, Meliani L, Lacube P, Pautard JC, Raccurt C, Roux P, Nevez G. 2003. Immunocompetent infants as a human reservoir for Pneumocystis jirovecii: rapid screening by non-invasive sampling and real-time PCR at the mitochondria1 large subunit rRNA gene. J. Eukaryot. Microbiol. 50:668–669. 10.1111/j.1550-7408.2003.tb00678.x [DOI] [PubMed] [Google Scholar]
- 86.Cushion MT. 2010. Are members of the fungal genus pneumocystis (a) commensals; (b) opportunists; (c) pathogens; or (d) all of the above? PLoS Pathog. 6:e1001009. 10.1371/journal.ppat.1001009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rivero L, de la Horra C, Montes-Cano MA, Rodriguez-Herrera A, Respaldiza N, Friaza V, Morilla R, Gutierrez S, Varela JM, Medrano FJ, Calderon EJ. 2008. Pneumocystis jirovecii transmission from immunocompetent carriers to infant. Emerg. Infect. Dis. 14:1116–1118. 10.3201/eid1407.071431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dumoulin A, Mazars E, Seguy N, Gargallo-Viola D, Vargas S, Cailliez JC, Aliouat EM, Wakefield AE, Dei-Cas E. 2000. Transmission of Pneumocystis carinii disease from immunocompetent contacts of infected hosts to susceptible hosts. Eur. J. Clin. Microbiol. Infect. Dis. 19:671–678. 10.1007/s100960000354 [DOI] [PubMed] [Google Scholar]
- 89.Gigliotti F, Harmsen AG, Wright TW. 2003. Characterization of transmission of Pneumocystis carinii f. sp. muris through immunocompetent BALB/c mice. Infect. Immun. 71:3852–3856. 10.1128/IAI.71.7.3852-3856.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ambrose HE, Ponce CA, Wakefield AE, Miller RF, Vargas SL. 2001. Distribution of Pneumocystis carinii f. sp. hominis types in the lung of a child dying of Pneumocystis pneumonia. Clin. Infect. Dis. 33:e100–e102. 10.1086/322690 [DOI] [PubMed] [Google Scholar]
- 91.Dimonte S, Berrilli F, D'Orazi C, D'Alfonso R, Placco F, Bordi E, Perno CF, Di Cave D. 2013. Molecular analysis based on mtLSU-rRNA and DHPS sequences of Pneumocystis jirovecii from immunocompromised and immunocompetent patients in Italy. Infect. Genet. Evol. 14:68–72. 10.1016/j.meegid.2012.11.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.