Abstract
Clostridium difficile-associated diarrhea is a well-recognized complication of antibiotic use. Historically, diagnosing C. difficile has been difficult, as antigen assays are insensitive and culture-based methods require several days to yield results. Nucleic acid amplification tests (NAATs) are quickly becoming the standard of care. We compared the performance of two automated investigational/research use only (IUO/RUO) NAATs for the detection of C. difficile toxin genes, the IMDx C. difficile for Abbott m2000 Assay (IMDx) and the BD Max Cdiff Assay (Max). A prospective analysis of 111 stool specimens received in the laboratory for C. difficile testing by the laboratory's test of record (TOR), the BD GeneOhm Cdiff Assay, and a retrospective analysis of 88 specimens previously determined to be positive for C. difficile were included in the study. One prospective specimen was excluded due to loss to follow-up discrepancy analysis. Of the remaining 198 specimens, 90 were positive by all three methods, 9 were positive by TOR and Max, and 3 were positive by TOR only. One negative specimen was initially inhibitory by Max. The remaining 95 specimens were negative by all methods. Toxigenic C. difficile culture was performed on the 12 discrepant samples. True C. difficile-positive status was defined as either positive by all three amplification assays or positive by toxigenic culture. Based on this definition, the sensitivity and specificity were 96.9% and 95% for Max and 92.8% and 100% for IMDx. In summary, both highly automated systems demonstrated excellent performance, and each has individual benefits, which will ensure that they will both have a niche in clinical laboratories.
INTRODUCTION
Clostridium difficile infection (CDI) is a toxin-associated disease ranging from mild diarrhea to pseudomembranous colitis, but complications, including septic shock and death, can occur. Although the anaerobic, Gram-positive bacillus is one of the most common hospital-acquired pathogens, cases of community-acquired infections have been reported. In the past decade, the incidence of CDI has been increasing, and severe cases are becoming more common (1, 2). Furthermore, a hypervirulent strain of C. difficile (B1/NAP1/027) overproducing both toxins A and B has been associated with more severe outbreaks of the disease (reviewed in reference 3).
The diagnosis of CDI is based upon the clinical signs and symptoms and laboratory tests. Laboratory diagnosis of toxigenic C. difficile includes anaerobic culture followed by detection of toxin production by C. difficile isolates or the direct detection of toxin, a toxin gene(s), or a C. difficile-associated enzyme(s) in stool. Anaerobic toxigenic culture is the most sensitive testing method; however, it is not practical for use in most clinical laboratories, as it takes days to perform. Cell culture cytotoxicity neutralization assays (CCCNAs) were once considered the diagnostic gold standard; however, they are no longer considered acceptable reference methods because they lack standardization and they have lower sensitivities than toxigenic culture and nucleic acid amplification tests (NAATs) (3). Enzyme immunoassay (EIA), used for the detection of toxins A and B and glutamate dehydrogenase (GDH), an enzyme found in all C. difficile strains, became very popular in most clinical laboratories because the results were available the same day and the tests were easy to perform and were relatively inexpensive. However, current literature indicates that the sensitivities of these tests are suboptimal for the diagnosis of CDI (3).
Recently, rapid PCR methods for the detection of the toxin A and/or toxin B gene have been developed, demonstrating sensitivities and specificities comparable to those of toxigenic culture. Thus, the popularity of NAATs has grown, resulting in changes in the commercial market, shifting toward automated platforms. In this report, we compared two such automated systems in terms of performance and workflow.
(Part of this report was presented at the 29th Annual Clinical Virology Symposium [Daytona Beach, FL] in 2013.)
MATERIALS AND METHODS
Bacterial strains and verification panel.
A BI/NAP1/027 strain of C. difficile (12-18691), obtained from the Wadsworth Center (WC), New York State Department of Health, was cultured anaerobically, resuspended in saline to 0.5 McFarland standard (∼1 × 108 CFU/ml), and serially diluted 10-fold in RNase- and DNase-free water. The NATtrol Clostridium difficile Verification Panel (Zeptometrix Corp., Buffalo, NY), which contains two isolates of ribotype 027 and one isolate each of ribotypes 002, 078, and 17, was used for assay verification and reproducibility studies.
Clinical samples.
Prospectively analyzed liquid or loose stool specimens (n = 111) were maintained at 4°C after receipt in the laboratory and analyzed by the laboratory's test of record (TOR) (BD [Québec, Canada] GeneOhm Cdiff Assay) within 12 h of receipt, with further analysis by investigational assays within 48 h. This system was reported to have a sensitivity and specificity of 83.6% and 98.2% versus toxigenic culture (4). Retrospectively analyzed specimens (n = 88), previously determined to be positive for C. difficile by TOR, were analyzed alongside prospective samples in a blinded fashion. Retrospective samples were stored at −80°C until testing, as were all specimens after analysis. Samples were considered true positive for C. difficile if they were positive by all three amplification assays or positive by toxigenic culture. Toxigenic culture was performed only on PCR-discrepant samples.
Real-time PCR comparison.
Variations due to sample handling were minimized by performing the two investigational methods simultaneously. Two operators performed all IMDx C. difficile for Abbott m2000 Assay (IMDx) testing, and two operators performed all BD Max Cdiff Assay (Max) testing, with one common operator between the two systems. Each of the assays investigated, as well as the TOR, was performed according to the manufacturer's instructions, except for a minor variation for specimen vortexing at high speed for 15 s, as indicated for Max, rather than three times for 2 to 3 s each time, as indicated for IMDx, to ensure sufficient mixing. All retrospective samples were thawed only once and tested within 2 h of thawing. For discrepancy analysis, all samples were processed and retested as described above, and an aliquot was prepared and frozen at −80°C until transportation to the WC on dry ice for toxigenic culture. Workflow analyses to assess total time, in addition to hands-on time, were performed for 48 specimens after experience and competency were established. For the Max, subsequent specimens were placed on the instrument for DNA extraction while the first set of 24 specimens was amplifying.
IMDx.
The IMDx C. difficile for Abbott m2000 (IntelligentMDx, Waltham, MA) is a real-time PCR assay for the detection of C. difficile toxin A (tcdA), toxin B (tcdB), and toxin B variant (tcdBv) genes. Automated sample lysing and target amplification and detection are performed on the m2000 RealTime System (Abbott Laboratories, Abbott Park, IL), enabling rapid, high-throughput capabilities. Initially, sample buffer tubes were created by adding 2.5 ml Tris-EDTA (TE) buffer, pH 8.0 (Fisher Scientific, Fair Lawn, NJ), to individual tubes. A flocked swab (Puritan Medical Products Co., Guilford, ME) was dipped into the vortexed stool specimen, transferred to the sample buffer tube, swirled, and left immersed in buffer. For every 24 specimens tested, 2.45 ml proteinase K (Abbott) was mixed with 17.15 ml of molecular biology grade water (Sigma-Aldrich, St. Louis, MO). Although the system has the capacity for testing 96 samples, for our studies, a maximum of 48 uncapped samples, including the positive and negative kit controls, were loaded onto the m2000sp. In addition, Abbott reagent vessels containing either the proteinase K solution or IMDx Process Control-A and the Abbott 96-Deep-Well Plate, Abbott 96-Well Optical Reaction Plate, and Abbott Disposable Tips (200 μl and 1,000 μl) were loaded on the system. The instrument mixes 100 μl of sample with 400 μl of proteinase K and 200 μl of the process control. Amplification reagents were loaded onto the system following sample preparation completion. Amplification reaction mixtures were prepared by automated addition of 15 μl of bacterial lysates to the amplification reagents in an Abbott 96-Well Optical Reaction Plate. The plate was then manually sealed and transferred to the Abbott m2000rt instrument for amplification and detection. The results were reported as tcdA detected, tcdB detected, tcdA and tcdB detected, tcdA and tcdB not detected, or an error code indicating the results were invalid. In addition, the cycle number value (CN), a proprietary method that differs from the threshold cycle (CT) assignment method, is available in the detailed analysis (5).
Max.
The BD Max Cdiff Assay (BD Diagnostics, Québec, Canada) performed on the BD Max System (BD Diagnostics, Sparks, MD) is a real-time PCR assay for the detection of tcdB. The BD Max System automates DNA extraction and amplification/detection on a single platform for a completely hands-free system following sample addition. A 10-μl disposable inoculating loop (Copan Diagnostics Inc., Murrieta, CA) was dipped into the vortexed specimen, placed into a BD Max Sample Buffer Tube containing 1.5 ml of buffer, swirled to release the specimen, and discarded. The tube was sealed with a septum cap and vortexed for 1 min prior to being placed in the BD Max System Rack. One BD Max Cdiff Reagent Strip was placed on the System Rack for each sample tested, as were one BD Max Cdiff Extraction Tube and one BD Max Cdiff Master Mix tube, with a maximum of 24 specimens initially placed at one time. One BD Max PCR cartridge was also placed on the BD Max for every 12 specimens tested. The DNA extraction, which utilizes magnetic-bead technology (6), includes a sample-processing control (SPC). During the extraction, 475 μl of the sample is extracted and eluted into 25 μl. The eluate is neutralized and transferred to a master mix tube to rehydrate the PCR reagents, and then 4.2 μl of the mixture is amplified in the cartridge well. The results were reported as positive, negative, or invalid. C. difficile (ribotype 027) and Clostridium sordellii samples from the NATtrol Clostridium difficile Verification Panel were used as positive and negative controls, respectively. CT values were available with the research use only (RUO) data analysis software.
BI/NAP1/027 strain determination.
Most BI/NAP1/027 strains are known to contain binary toxin genes A (cdtA) and B (cdtB), as well as both a Δ117 frameshift mutation and an in-frame 18-bp deletion mutation in the tcdC gene (7). The detection of cdtA from primary specimens was performed by real-time PCR using primers (0.5 μM; Integrated DNA Technologies [IDT], Coralville, IA) and an MGB-labeled probe (0.13 μM; Life Technologies, Foster City, CA) targeting the cdtA gene, as previously described (7). Three microliters of Max extract were amplified on the BD Max in 10-μl reaction mixtures containing 1× PerfeCTa qPCR ToughMix (Quanta BioSciences, Inc., Gaithersburg, MD), 115 copies of an internal control plasmid containing a 194-bp HaeIII fragment of the ΦX174 genome, internal control primers (TGA GGA TAA ATT ATG TCT AAT ATT C and GGA GTA GTT TGA AAT GGT AA; 0.5 μM; IDT), and probe (ACC AAT CTG ACC AGC AAG GAA G; 0.25 μM; IDT) by cycling between 95°C for 15 s and 60°C for 60 s for 45 cycles after an initial hot-start activation of 95°C for 180 s. The tcdC Δ117 mutation detection assay previously described by de Boer et al. (8) was modified for amplification on a SmartCycler (Cepheid, Carlsbad, CA). Again, 3 μl of Max extract was amplified in a 25-μl reaction mixture containing 0.5 μM primers and 0.25 μM both wild-type and mutant MGB-labeled probe in TaqMan Universal PCR Master Mix (Life Technologies) by cycling between 95°C for 15 s and 60°C for 60 s for 45 cycles after an initial hot-start activation of 95°C for 10 min. Conventional PCR amplification of a 162-bp region of the tcdC gene with agarose gel electrophoresis, as described by Persson et al. (9), was used to detect large deletions in the gene. Hot-start PCR in 25-μl reaction mixtures containing 3 μl of Max extract, 0.2 mM deoxynucleoside triphosphate (dNTP) mixture, 10 mM Tris, 3 mM MgCl2, 25 μM each primer, and 1.25 U of Gold Taq (Life Technologies) was performed in a GeneAmp PCR System 9600 thermocycler (Perkin-Elmer, Norwalk, CT) by cycling between 95°C for 15 s and 60°C for 60 s for 50 cycles after an initial hot-start activation of 95°C for 10 min. Samples were considered to be presumptively positive for strain BI/NAP1/027 if they were positive for cdtA, the Δ117 tcdC mutation, and the 18-bp tcdC deletion.
Toxigenic culture.
Discrepant specimens were plated on cycloserine cefoxitin fructose agar supplemented with sodium taurocholate to a final concentration of 0.1% (TCCFA) (10, 11). The plates were incubated at 35°C for 96 h in an anaerobic atmosphere. Specimens that were initially culture negative were inoculated into cooked-meat broth and incubated for 48 h. Alcohol shock was performed by mixing 1 ml of cooked-meat broth and 1 ml of 95% alcohol for 1 h. The cooked-meat–alcohol mixture was plated on TCCFA as before.
C. difficile-like colonies were picked from TCCFA to 2 CDC anaerobe agar plates. One plate was incubated at 35°C in an anaerobic atmosphere, and 1 plate was incubated at 35°C in an aerobic atmosphere. After incubation for 48 to 72 h, the following tests were performed: Gram stain, PRO disc (Key Scientific), and UV fluorescence. Organisms were identified as C. difficile if they produced characteristic yellow colonies on TCCFA, were Gram-positive bacilli, grew only in an anaerobic atmosphere, were PRO positive, and fluoresced under UV light.
Isolates were determined to be toxigenic by multiplex real-time PCR for the presence of tcdA, tcdB, cdtA, and cdtB genes, as previously described (7). In addition, all isolates were assessed for tcdC deletion mutations by pyrosequencing (7).
RESULTS
To assess the precision of IMDx and Max, 10 μl of a NATtrol C. difficile (ribotype 027) sample was tested in triplicate in three separate runs by two different operators. The concentration of this NATtrol panel member was determined by quantification against the clinical isolate 12-18691 and determined to be 5 × 106 CFU/ml. The coefficient of variance was determined from equivalent log copies to normalize the results. Both systems demonstrated excellent reproducibility, with the intra-assay precision ranging from 0.7 to 6.3% and 0.3 to 3.9% for IMDx tcdA and tcdB targets, respectively, and 1.1 to 5.9% for Max. Similarly, interassay precision was 5.2, 5.8, and 5.0% for IMDx tcdA, IMDx tcdB, and Max, respectively.
Serial dilutions of the BI/NAP1/027 strain 12-18691 were tested in duplicate with both investigational systems. The results indicated that the analytical sensitivity for Max was 1 dilution lower than that for either IMDx or TOR. The Max reproducibly detected tcdB in samples with 225 CFU/inoculum of C. difficile (Table 1), whereas the limit of detection (LOD) was 2,250 CFU/inoculum for both IMDx and TOR. The LOD reported here is similar to that reported in the package inserts for Max and TOR, 125 to 256 CFU/loop and 4 CFU/reaction (∼700 CFU/swab), respectively, but higher than the information provided for IMDx (67 to 337 CFU/ml or 0.7 to 3.4 CFU/swab for unpreserved stool).
TABLE 1.
Analytical sensitivity
| No. of CFU/inoculuma | Mean CT/CN value |
||
|---|---|---|---|
| IMDxb | Max | TOR | |
| 22,500 | 36.6 | 27.9 | 36.6 |
| 2,250 | 39.6 | 31.2 | 39.3 |
| 225 | 0 | 35.0 | 0 |
| 23 | 0 | 0 | 0 |
Limits of detection (CFU/ml sample) were as follows: IMDx, 2.25 × 105; Max, 2.25 × 104; TOR, 2.25 × 105.
Combined CN means for tcdA and tcdB targets.
The determination of clinical performance involved both a prospective analysis of 111 stool specimens and a retrospective analysis of 88 specimens previously determined to be positive for C. difficile by the TOR. Of these 199 specimens, 1 prospective specimen that was positive by IMDx only was excluded due to loss to follow-up discrepancy analysis. One hundred two specimens were positive by TOR, of which 26 (25%) were presumptive strain NAP1, with detection of the cdtA gene and both the Δ117 frameshift mutation and the 18-bp deletion mutation in the tcdC gene. Ninety-nine specimens were positive by Max, and 90 were positive by IMDx, of which 85 were positive for both tcdA and tcdB, 2 were positive for tcdA only, and 3 were positive for tcdB only (Table 2). Max was positive for all 26 presumptive NAP1 strain-containing samples, while IMDx was positive for 25 of these samples. All Max or IMDx positive samples were positive by TOR, and all IMDx positive samples were positive by Max. One additional sample that was negative by both TOR and IMDx was initially inhibitory by Max. Upon retesting, the sample was negative with acceptable internal control recovery, albeit the CT value of the SPC was quite high (41), suggesting there was still some assay inhibition.
TABLE 2.
Assay performance with clinical samples
| Truea | Result (mean) |
No. | ||
|---|---|---|---|---|
| TORb | Maxb | IMDxc | ||
| Pos | Pos (32.6) | Pos (26.9) | Pos (33.8) | 90 |
| Pos | Pos (38.3) | Pos (30.4) | Neg | 4 |
| Pos | Pos (37.8) | Neg | Neg | 3 |
| Neg | Pos (39.3) | Pos (33.6) | Neg | 5 |
| Neg | Neg | Inhibitory | Neg | 1 |
| Neg | Neg | Neg | Neg | 95 |
| Total | 198 | |||
True result after discrepancy analysis. Pos, positive; Neg, Negative.
CT values are shown in parentheses.
Combined CN means for tcdA and tcdB targets are shown in parentheses.
A total of 12 specimens were discrepant between methods and were reanalyzed by all 3 PCRs. The high level of variability of the repeat PCR test results and the generally high CT values obtained suggest the level of C. difficile was near the limit of detection in these specimens (Table 3). In addition, toxigenic C. difficile culture was performed on the 12 discrepant samples, and indeed, many samples had very low colony counts. Interestingly, the 3 specimens that were initially positive by TOR only were all culture positive and had the highest colony counts. From these analyses, 5 out of 9 specimens, positive by both TOR and Max, were determined to be false positive, including one sample positive for the NAP1-associated molecular targets.
TABLE 3.
Discrepancy analysis
| Discrepant sample | Initial resultsa (CT/CN) |
Repeat testinga (CT/CN) |
Toxigenic culture | Colony countb | ||||
|---|---|---|---|---|---|---|---|---|
| TOR | Max | IMDx | TOR | Max | IMDx | |||
| 1 | Pos (34.2) | Neg | Neg | Pos (42.7) | Pos (32.9) | Neg | Pos | 10–12 |
| 2 | Pos (40.2) | Neg | Neg | Pos (41.2) | Pos (34.0) | Pos (41.0) | Pos | 10–12 |
| 3 | Pos (38.9) | Neg | Neg | Neg | Neg | Neg | Pos | 10–12 |
| 4 | Pos (37.9) | Pos (35.0) | Neg | Neg | Neg | Pos (39.3) | Pos | 3 |
| 5 | Pos (34.2) | Pos (31.9) | Neg | Pos (42.3) | Pos (33.1) | Neg | Pos | 1 |
| 6 | Pos (41.8) | Pos (33.2) | Neg | Pos (42.7) | Neg | Neg | Pos | 1 |
| 7 | Pos (39.1) | Pos (32.6) | Neg | Neg | Neg | Neg | Pos | 1 |
| 8 | Pos (37.2) | Pos (34.0) | Neg | Neg | Pos (34.5) | Neg | Neg | |
| 9 | Pos (44.5) | Pos (31.2) | Neg | Neg | Pos (32.2) | Neg | Neg | |
| 10 | Pos (41.8) | Pos (35.0) | Neg | Neg | Neg | Neg | Neg | |
| 11 | Pos (42.5) | Pos (32.8) | Neg | Neg | Neg | Neg | Neg | |
| 12 | Pos (39.0) | Pos (31.7) | Neg | Neg | Neg | Neg | Neg | |
Pos, positive; Neg, negative.
Colony counts from direct culture of stool samples on TCCFA.
After discrepancy analysis, samples were considered true positive for C. difficile toxin genes if they were positive by all three amplification assays or positive by toxigenic culture. Based on this definition, the sensitivity and specificity were 92.8% and 100% for IMDx and 96.9% and 95% for Max (Table 4). When evaluating the data for presumptive NAP1 versus non-NAP1 strains, it was apparent that assay sensitivity was affected by the C. difficile strain type. Whereas all assays had a sensitivity of 100% for true positives determined to be presumptive NAP1 strains, the sensitivities were lower for the non-NAP1 strains (Table 4).
TABLE 4.
Percent sensitivity and percent specificity by strain type
| Test | All strain types |
NAP1a strains |
Non-NAP1a strains |
|||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| IMDx | 92.8 | 100 | 100 | 100 | 90.3 | 100 |
| Max | 96.9 | 95.0 | 100 | 99.0 | 95.8 | 96.0 |
Presumptive NAP1.
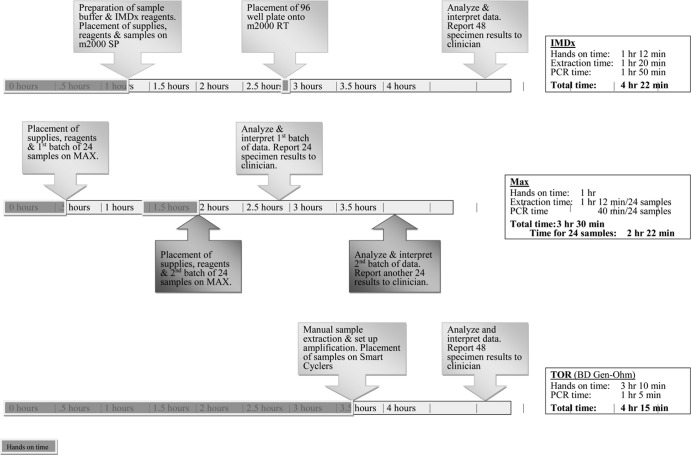
Both systems offer enhanced workflow and significantly reduced technical time compared with the TOR. For both systems, the most cumbersome step is the transfer of the stool specimen to the sample buffer tubes. The workflow for the Max is optimal for testing ≤24 samples (Table 5 and Fig. 1). Testing more than 24 samples requires more returns to the instrument. The maximal workflow benefit for IMDx is seen when testing 96 specimens, at which point the hands-on time per specimen is the shortest and there is only one return to the instrument for transfer of the amplification plate.
TABLE 5.
Processing time per sample
| No. of samples | Processing time/sample (min)a |
|||||
|---|---|---|---|---|---|---|
| Max |
IMDx |
TOR |
||||
| HOT | Total | HOT | Total | HOT | Total | |
| 24 | 1.25 | 5.92 | 1.54 | 9.88 | 4.42 | 7.15 |
| 48 | 1.25 | 4.38 | 1.50 | 5.46 | 3.94 | 5.31 |
| 96 | 1.25 | 3.44 | 1.27 | 3.25 | 3.71 | 4.39 |
HOT, hands-on time, measured with 48 samples and estimated for 24 and 96 samples.
FIG 1.
Workflow timeline for 48 samples.
DISCUSSION
CDI is a potentially life-threatening disease that causes considerable morbidity and mortality. Furthermore, CDI rates have been increasing, as has the severity of the illness. Accurate diagnosis of CDI is vital for patient management and infection control. False-negative results can lead to inappropriate therapy and loss of infection control intervention. Both EIA and NAAT are rapid methods for C. difficile testing, but only NAAT offers the level of sensitivity needed for accurate detection of this infectious agent. Although the reagent price for NAATs cannot match that of EIA systems, the added cost may be offset by savings associated with improved patient management in terms of antibiotic utilization and reduction in nosocomial transmission (12).
At the time these studies were initiated, there were nine in vitro diagnostic product (IVD)-cleared platforms in the United States; subsequently, both Max and IMDx obtained FDA clearance (all but IMDx are reviewed in reference 13). Prior to the release of the Max, the Xpert C. difficile and C. difficile/Epi (Cepheid) were the only completely automated, walk-away systems. The main purpose of this study was to compare the performances of two new automated real-time PCR systems as premarket evaluations. Not surprisingly, both systems demonstrated excellent sensitivity and specificity with our sample population, although the Max demonstrated slightly better sensitivity while IMDx demonstrated slightly better specificity. It appears that the result deviation was most likely associated with low C. difficile levels, perhaps near the detection limit, as the mean CT value of the discrepant samples was more than 6 CT values higher than the concordant samples. It is important to point out that the level of sensitivity needed for CDI diagnostic testing is not clear, as asymptomatic shedding can be detected by culture (reviewed in reference 14). Hence, some argue that toxigenic culture should not be considered the gold standard test for C. difficile disease. It is important to note that for accurate CDI diagnosis, current guidelines recommend that laboratory testing should be performed only on symptomatic patients (14, 15), as no laboratory test can differentiate between asymptomatic shedding and infection. This is particularly true for assays with greater analytical sensitivity, such as toxigenic culture and NAAT.
The enhanced sensitivity seen with the Max is most likely associated with the extraction process, where the nucleic acids in the specimen are concentrated. Indeed, the lower limit of detection with this system, as seen in the analytical sensitivity studies, supports this concept. Another potential source of false-negative results is inhibition of the enzymatic reaction, which one would expect to be more problematic with a system that tests only sample lysates, as do both TOR and IMDx. Interestingly, that was not the case in our studies, and the only inhibition that was observed occurred with Max.
A difference in assay sensitivity was more apparent in the presumptive non-NAP1 C. difficile-positive samples. This phenomenon was previously observed with EIAs (16). However, other studies have demonstrated no antigenic differences among the various ribotypes (17, 18). Perhaps this observed difference is simply due to differences in the bacterial load, as an increase in sporulation has been observed with NAP1 strains (19). In our study, the mean difference in CT values for presumptive NAP1 strain-positive versus -negative samples was approximately 3, suggesting a 1-log-unit-lower bacterial load for samples containing non-NAP1 organisms.
With regard to specificity determination, it is important to point out the limitations of these studies. First, the population was artificially skewed toward a greater percentage of positive specimens so as to have nearly equal numbers of positive and negative specimens rather than the typical 12% positivity rate we see at our institution. In addition, the study was designed as a comparison of laboratory methods rather than a clinical trial; hence, the clinical status of the patients in our study was unknown. It can only can be assumed that the patients were symptomatic based on our institutional guidelines for C. difficile testing. Lastly, the toxigenic culture discrepancy analysis was performed on stored residual specimens, which may impact organism viability. Indeed if the definition of true positivity had been based on positivity by two or more molecular assays, the specificity for the Max would have been 100%.
Both the IMDx and the Max offer enhanced workflow and significantly reduced technical time compared with the TOR. The most laborious step for both systems is the inoculation of the sample into the buffer tubes. Perhaps in the future, up front automation systems, such as the BD Kiestra, will be able to take on this functionality. Another benefit that both systems have is the availability of residual sample lysates or extracts for additional tests, such as PCR tests for mutations associated with the NAP1 strain.
Max has the added benefit of having room temperature storage for all of the test components and has the ability to combine the Max Cdiff and methicillin-resistant Staphylococcus aureus (MRSA) assays in the same run. It also enables variable batch sizes, while IMDx requires discrete batches of 24, which is not always practical in clinical laboratories. IMDx has the added benefit of additional sample types in stool samples in Cary Blair medium. In addition, the assay has redundancy in the primer design, which may provide a safeguard against false-negative results due to genetic variation in the toxin gene(s). Specifically, the assay is designed to detect a variant form of the tcdB gene, observed in some A− B+ strains (e.g., 8864 and 1470) (20, 21), as well as tcdA, which may enable the detection of the rare toxin A+ B− strain (22). Among our samples, there were five samples positive for only one of the two targets with IMDx, two for tcdA only, and three for tcdB only. However, for all five samples, the positive target had CN values of 39 or greater; hence, the target discrepancy most likely reflected the limit of detection rather than a strain variant.
It is apparent that these highly automated assays demonstrate similar performances and greatly reduce the hands-on time. Each system offers different benefits to fit different laboratory needs. For example, IMDx is more suited for higher-volume laboratories, especially those that test 96 or more samples per day. Max is especially suited for laboratories that run 24 or fewer samples per day, as discrete batch sizes are not necessary.
ACKNOWLEDGMENTS
We are grateful to BD Diagnostics for providing the Max Cdiff kits.
This study was sponsored in part by Abbott Molecular Inc.
Footnotes
Published ahead of print 19 February 2014
REFERENCES
- 1.Redelings MD, Sorvillo F, Mascola L. 2007. Increase in Clostridium difficile-related mortality rates, United States, 1999–2004. Emerg. Infect. Dis. 13:1417–1419. 10.3201/eid1309.061116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonald LC, Owings M, Jernigan DB. 2006. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996–2003. Emerg. Infect. Dis. 12:409–415. 10.3201/eid1203.051064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll KC, Bartlett JG. 2011. Biology of Clostridium difficile: implications for epidemiology and diagnosis. Annu. Rev. Microbiol. 65:501–521. 10.1146/annurev-micro-090110-102824 [DOI] [PubMed] [Google Scholar]
- 4.Stamper PD, Alcabasa R, Aird D, Babiker W, Wehrlin J, Ikpeama I, Carroll KC. 2009. Comparison of a commercial real-time PCR assay for tcdB detection to a cell culture cytotoxicity assay and toxigenic culture for direct detection of toxin-producing Clostridium difficile in clinical samples. J. Clin. Microbiol. 47:373–378. 10.1128/JCM.01613-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shain EB, Clemens JM. 2008. A new method for robust quantitative and qualitative analysis of real-time PCR. Nucleic Acids Res. 36:e91. 10.1093/nar/gkn408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wroblewski D, Hannett GE, Bopp DJ, Dumyati GK, Halse TA, Dumas NB, Musser KA. 2009. Rapid molecular characterization of Clostridium difficile and assessment of populations of C. difficile in stool specimens. J. Clin. Microbiol. 47:2142–2148. 10.1128/JCM.02498-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Boer RF, Wijma JJ, Schuurman T, Moedt J, Dijk-Alberts BG, Ott A, Kooistra-Smid AM, van Duynhoven YT. 2010. Evaluation of a rapid molecular screening approach for the detection of toxigenic Clostridium difficile in general and subsequent identification of the tcdC Delta117 mutation in human stools. J. Microbiol. Methods 83:59–65. 10.1016/j.mimet.2010.07.017 [DOI] [PubMed] [Google Scholar]
- 9.Persson S, Jensen JN, Olsen KE. 2011. Multiplex PCR method for detection of Clostridium difficile tcdA, tcdB, cdtA, and cdtB and internal in-frame deletion of tcdC. J. Clin. Microbiol. 49:4299–4300. 10.1128/JCM.05161-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George WL, Sutter VL, Citron D, Finegold SM. 1979. Selective and differential medium for isolation of Clostridium difficile. J. Clin. Microbiol. 9:214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson KH, Kennedy MJ, Fekety FR. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 15:443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sydnor ER, Lenhart A, Trollinger B, Avdic E, Maragakis LL, Carroll KC, Cosgrove SE. 2011. Antimicrobial prescribing practices in response to different Clostridium difficile diagnostic methodologies. Infect. Control Hosp. Epidemiol. 32:1133–1136. 10.1086/662381 [DOI] [PubMed] [Google Scholar]
- 13.Burnham CA, Carroll KC. 2013. Diagnosis of Clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin. Microbiol. Rev. 26:604–630. 10.1128/CMR.00016-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Planche T, Wilcox M. 2011. Reference assays for Clostridium difficile infection: one or two gold standards? J. Clin. Pathol. 64:1–5. 10.1136/jcp.2010.080135 [DOI] [PubMed] [Google Scholar]
- 15.Brecher SM, Novak-Weekley SM, Nagy E. 2013. Laboratory diagnosis of Clostridium difficile infections: there is light at the end of the colon. Clin. Infect. Dis. 57:1175–1181. 10.1093/cid/cit424 [DOI] [PubMed] [Google Scholar]
- 16.Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, Fang FC, Dascal A, Gerding DN, Nomura JH, Goering RV, Akerlund T, Weissfeld AS, Baron EJ, Wong E, Marlowe EM, Whitmore J, Persing DH. 2010. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J. Clin. Microbiol. 48:3719–3724. 10.1128/JCM.00427-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carman RJ, Wickham KN, Chen L, Lawrence AM, Boone JH, Wilkins TD, Kerkering TM, Lyerly DM. 2012. Glutamate dehydrogenase is highly conserved among Clostridium difficile ribotypes. J. Clin. Microbiol. 50:1425–1426. 10.1128/JCM.05600-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldenberg SD, Gumban M, Hall A, Patel A, French GL. 2011. Lack of effect of strain type on detection of toxigenic Clostridium difficile by glutamate dehydrogenase and polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 70:417–419. 10.1016/j.diagmicrobio.2011.03.012 [DOI] [PubMed] [Google Scholar]
- 19.Akerlund T, Persson I, Unemo M, Noren T, Svenungsson B, Wullt M, Burman LG. 2008. Increased sporulation rate of epidemic Clostridium difficile type 027/NAP1. J. Clin. Microbiol. 46:1530–1533. 10.1128/JCM.01964-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borriello SP, Wren BW, Hyde S, Seddon SV, Sibbons P, Krishna MM, Tabaqchali S, Manek S, Price AB. 1992. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4192–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Eichel-Streiber C, Zec-Pirnat I, Grabnar M, Rupnik M. 1999. A nonsense mutation abrogates production of a functional enterotoxin A in Clostridium difficile toxinotype VIII strains of serogroups F and X. FEMS Microbiol. Lett. 178:163–168 [DOI] [PubMed] [Google Scholar]
- 22.Cohen SH, Tang YJ, Hansen B, Silva J., Jr 1998. Isolation of a toxin B-deficient mutant strain of Clostridium difficile in a case of recurrent C. difficile-associated diarrhea. Clin. Infect. Dis. 26:410–412 [DOI] [PubMed] [Google Scholar]