Abstract
Rapid and accurate detection of multidrug resistance (MDR) in Mycobacterium tuberculosis is essential to improve treatment outcomes and reduce global transmission but remains a challenge. Rifampin (RIF) resistance is a reliable marker of MDR tuberculosis (TB) since by far the majority of RIF-resistant strains are also isoniazid (INH) resistant. We have developed a rapid, sensitive, and specific method for detecting the most common mutations associated with RIF resistance, in the RIF resistance determining region (RRDR) of rpoB, using a cocktail of six padlock probes and rolling circle amplification (RCA). We used this method to test 46 stored M. tuberculosis clinical isolates with known RIF susceptibility profiles (18 RIF resistant, 28 susceptible), a standard susceptible strain (H37Rv, ATCC 27294) and 78 M. tuberculosis culture-positive clinical (sputum) samples, 59 of which grew RIF-resistant strains. All stored clinical isolates were correctly categorized, by the padlock probe/RCA method, as RIF susceptible or resistant; the sensitivity and specificity of the method, for direct detection of phenotypically RIF-resistant M. tuberculosis in clinical specimens, were 96.6 and 89.5%, respectively. This method is rapid, simple, and inexpensive and has the potential for high-throughput routine screening of clinical specimens for MDR M. tuberculosis, particularly in high prevalence settings with limited resources.
INTRODUCTION
With one-third of the world's population infected, tuberculosis (TB) is a major public health threat worldwide (1). The two most important drugs used to treat TB are isoniazid (INH) and rifampin (RIF). Inadequate or inappropriate therapy, together with long and costly treatment courses, often result in the emergence of drug resistance (2). Multidrug-resistant (MDR) Mycobacterium tuberculosis strains—defined as bacillary resistance to at least INH and RIF—have spread rapidly and become a global health emergency (3). Identification of these strains by drug susceptibility testing (DST) allows optimization of therapeutic efficacy and improved treatment outcomes while minimizing transmission of drug-resistant strains. However, phenotypic DST involving subculture is costly, it is too slow to guide optimal management of MDR-TB, and it lacks high-throughput capability. Commercial systems for rapid molecular identification of RIF resistance are available but expensive.
RIF resistance in M. tuberculosis is associated with amino acid changes in the β-subunit of RNA polymerase, encoded by rpoB. The majority (90 to 95%) of mutations responsible for resistance are located in the 81-bp RIF resistance determining region (RRDR) of rpoB (4–6), and ca. 60 to 70% are found within two codons, 531 and 526. The fact that most mutations known to confer resistance are localized to a specific region facilitates the development of molecular methods for their detection. Sequencing and hybridization-based assays have been commonly used and can define the precise mutations involved (4, 7). Hybridization-based assays also provide a rapid and high-throughput approach (8).
Recent studies have shown that the use of padlock probes offers advantages over other techniques for detection of genetic mutations, including single-nucleotide polymorphisms (SNPs); ligation of the probe ends by DNA ligase requires perfectly matched target-probe complexes, which prevents the nonspecific amplification that can occur with conventional PCR and results in very high specificity (9). A padlock probe comprises two sequences complementary to the 5′ and 3′ termini of the target sequence, joined by a genetic linker region. When they hybridize, head to tail, with the target, the 5′ and 3′ ends of the probe are juxtaposed, forming a closed, circular molecule after incubation with a DNA ligase. The ability of padlock probes to accurately identify SNPs has been demonstrated (10, 11), and the intensity of the signal generated by the circularized probe can be increased, exponentially, by rolling circle amplification (RCA) (12).
In the present study, we developed a RCA assay using six padlock probes to recognize 12 common resistance-associated mutations in the RRDR of rpoB of M. tuberculosis. Our choice of targeted mutations was based on analysis of mutations in the RRDR of 242 M. tuberculosis isolates, by Huang et al. (13). They identified 60 different mutations in RIF-resistant isolates, of which those, at codons 511, 516, 526 and 531, accounted for almost 90% of all mutations identified. Others (14–17) have reported similar results. After validating its sensitivity and specificity, we used the assay to examine RIF resistance in M. tuberculosis isolates and sputum specimens from patients with active tuberculosis. Its diagnostic accuracy was compared to the standard rpoB PCR/sequencing and phenotypic DST.
MATERIALS AND METHODS
Clinical isolates.
A total of 46 stored M. tuberculosis clinical isolates were chosen for the present study, including five RIF-resistant from the New South Wales Mycobacterium Reference Laboratory, Centre for Infectious Diseases and Microbiology, Westmead Hospital, and 13 RIF-resistant and 28 RIF-susceptible isolates from Department of Tuberculosis, Beijing Chest Hospital, Capital Medical University, China. In addition, a standard susceptible strain, H37Rv (ATCC 27294), was included as control. Isolates were stored in 1% glycerol or 7H9 broth (without oleic acid-albumin-dextrose-catalase [OADC]) at −80°C until required, when they were thawed and subcultured on Lowenstein-Jensen (L-J) agar slants for 3 to 4 weeks at 35 to 37°C in air.
Clinical sputum samples: collection and processing.
A total of 78 nonduplicate sputum specimens were collected between May and December 2012 from pulmonary TB patients admitted to the Beijing Chest Hospital, Capital Medical University, Beijing, China. The diagnosis of pulmonary TB was based on clinical and radiological findings, together with identification of acid-fast bacilli (AFB) in sputum and subsequent isolation of M. tuberculosis in culture. All patients were HIV seronegative, and written informed consent for additional testing of specimens was obtained from all patients involved in the study.
Early morning sputum specimens were digested and decontaminated with N-acetyl-l-cysteine-sodium hydroxide (NALC-NaOH). Specimens in which AFBs were identified by microscopy were split into two aliquots. One aliquot was use for molecular testing (see below), and the other was processed for standard culture on L-J slants, followed by strain identification with 2-thiophene carboxylic acid (TCH) and paranitrobenzoic acid (PNB) and by DST.
DST.
All stored and new clinical isolates were tested for RIF susceptibility on L-J slants by the absolute concentration method. Isolates were determined to be resistant to RIF—at concentrations of either 50 μg/ml (low-level resistance) or 250 μg/ml (high-level resistance)—if the inoculum grew on one-quarter (1+) of the slant surface or there were ≥20 colonies on slants containing the corresponding RIF concentrations, after 4 weeks incubation at 35 to 37°C (18).
DNA extraction.
Scrapes of isolates grown on L-J slants were suspended in 1× Tris-EDTA (TE), and DNA was extracted by using a standard phenol-chloroform procedure (19). DNA was extracted from clinical specimens and purified using an optimized method, as described by Leung et al. (20). Briefly, 2 ml of each decontaminated sputum specimen was centrifuged at 5,000 × g for 5 min, and 1 ml of the pellet was resuspended in 1 ml of TE in a 2-ml screw-cap microcentrifuge tube containing 200 μl of 1-mm glass beads (Sigma-Aldrich, St. Louis, MO). After vortexing and centrifugation, the lysate was purified using in-house buffer with Invitrogen Dynabeads MyOne Silane (Life Technologies Corporation, Carlsbad, CA) magnetic beads. Briefly, binding buffer was prepared by dissolving 120 g of guanidium thiocyanate in 100 ml of 0.1 M Tris-HCl (pH 6.4). Subsequently, 22 ml of 0.2 M EDTA (pH 8.0) and 2.6 g of Triton X-100 were added to the solution. A wash buffer was prepared by combining 55 ml of ethanol and 45 ml of solution containing 3 M guanidium thiocyanate, 10 mM Tris-HCl, and 10 mM NaCl (pH 8.0).
PCR amplification and sequencing of rpoB region.
PCR was used to amplify a 758-bp fragment of M. tuberculosis rpoB (which included the RRDR), using the primers 5′-TGGTCCGCTTGCACGAGGGTCAGA-3′ (forward) and 5′-CAGGAAGGGAATCATCGCGG-3′ (reverse) designed for the present study. The reaction was performed in a final volume of 25 μl containing 0.5 μl of both forward and reverse primers (30 μM), 2.5 μl of 10× PCR buffer (including 1.5 mM MgCl2), 0.75 μl of deoxynucleoside triphosphates (dNTPs; 2.5 mM each of dNTP), 1 U of Qiagen Hotstart Taq polymerase, and 1 μl of purified DNA. The reaction was performed for isolates and clinical specimens under the following conditions: initial denaturation at 95°C for 15 min, followed by 40 cycles at 95°C for 30 s, 65°C for 30 s, and 72°C for 1 min, and a final elongation at 72°C for 10 min. To identify possible contamination or inhibition, appropriate negative (no-DNA) and positive (H37Rv DNA) controls were used in each step. For all clinical samples, a pair of M. tuberculosis-specific primer was designed to determine the presence of M. tuberculosis (data not shown here). PCR products were purified using a Millipore PCR purification plate (Millipore, Billerica, MA) and sequenced using the ABI Prism BigDye Terminator V3.1 ready reaction cycle sequencing kit (Applied Biosystems, Foster City, CA) on an ABI 377 automated sequencer. Multiple sequences derived from each patient were analyzed using Sequencher 3.11 software (Gene Codes Corp., Ann Arbor, MI). Chromatograms derived from both forward and reverse primers were aligned with the reference strain and carefully examined at the locations where resistance mutations have been reported.
Padlock probe design.
The padlock probes designed for the present study were based on 12 known rpoB mutations at positions 511, 514, 516, 526, and 531 in the RRDR of RIF-resistant M. tuberculosis, as previously described (3, 21).
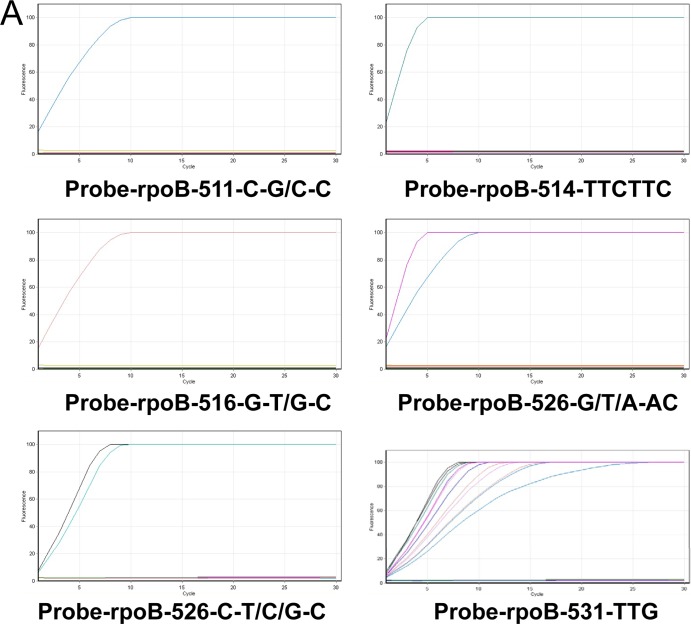
Due to the polymorphism of mutations in these locations, ambiguous positions or multiple probes were introduced. To ensure the efficiency of padlock probe binding, the padlock probes were designed with minimum secondary structure and with the Tm of the 5′ end probe binding arm close to or above the ligation temperature (65°C). To increase the discriminatory specificity, the 3′ end binding arm was designed with a Tm 10 to 15°C below ligation temperature. The genetic linker region between the two binding arms was also carefully designed to minimize any similarities to potentially cross-reacting sequences, after a BLAST search. In addition, the primers used to amplify padlock probe signals, during RCA, were designed specifically to bind to the linker regions with a Tm of about 55°C. The relevant primers and probes are given in Table 1, and the individual mutations targeted are presented in Table 2.
TABLE 1.
RCA padlock probes and primers designed for this study
| Padlock probe/primers | Sequence (5′–3′)a |
|---|---|
| Probe-rpoB-511-C-C/G-C | P-GCTGGCTGGTGCCGAAGATCATGCTTCTTCGGTGCCCATAATGCCACGTTAACAGTCAGCGCGCAGACACGATAGTCTAGGTCCATGAATTGGCTCS |
| Probe-rpoB-514-TTCTTC | P-GAATTGGCTCAGCTGGCTGGGATCATGCTTCTTCGGTGCCCATTGCGTGTTCAGTCAGACTATGCGCGCAGACACGATAGTCTAGTTGTTCTGGTCCATGAA |
| Probe-rpoB-516G-T/G-C | P-CCATGAATTGGCTCAGCTGGGATCATGCTTCTTCGGTGCCCATCCTACTAGTTGCACGCTGTTCCGCGCAGACACGATAGTCTACAGCGGGTTGTTCTGGM |
| Probe-rpoB-526-G/T/A-AC | P-GGTCAACCCCGACAGCGGGATCATGCTTCTTCGGTGCCCATTGACCGTGCTATGAATGCATCGCGCAGACACGATAGTCTAACAGTCGGCGCTTGTH |
| Probe-rpoB-526- C-T/C/G-C | P-GGGTCAACCCCGACAGCGGATCATGCTTCTTCGGTGCCCATAACGACTCCAGGTTAGCCTAGCGCGCAGACACGATAGTCTAGACAGTCGGCGCTTGV |
| Probe-rpoB-531-TTG | P-ACAGTCGGCGCTTGTGGATCATGCTTCTTCGGTGCCCATCCTAGATCAGACGTTCCTGTCCGCGCAGACACGATAGTCTACCCAGCGCCA |
| Primer-1b | ATGGGCACCGAAGAAGCA |
| Primer-2c | CGCGCAGACACGATA |
p-, 5′ phosphorylation.
RCA primer-1 binds to the padlock probe, generating a long ssDNA.
RCA primer-2 binds to the nascent ssDNA if their binding sites are available. Each bound reverse primer extends and displaces the downstream primers and their extended products.
TABLE 2.
Twelve targeted RRDR mutations in rpoB genes of M. tuberculosis from 18 rifampin-resistant isolates and 59 clinical samples
| Position | Mutation | Codon change | Amino acid change | No. of isolates or samples |
|||||
|---|---|---|---|---|---|---|---|---|---|
| RCA |
Sequencing |
Phenotype (resistance) |
|||||||
| Stored isolate | Clinical sample | Stored isolate | clinical sample | Stored isolate | Recent clinical isolate | ||||
| 514 | TTC insertion | TTC- TTCTTC | Phe→PhePhe | 1 | 0 | 1 | 0 | 1 | 0 |
| 516 | A→T | GAC→GTC | Asp→Val | 0 | 2 | 0 | 2 | 0 | 2 |
| 516 | G→T | GAC→TAC | Asp→Tyr | 0 | 3 | 0 | 3 | 0 | 3 |
| 526 | C→A | CAC→AAC | His→Asn | 0 | 5 | 0 | 5 | 0 | 5 |
| 526 | C→T | CAC→TAC | His→Tyr | 2 | 4 | 2 | 4 | 2 | 4 |
| 526 | C→G | CAC→GAC | His→Asp | 0 | 4 | 0 | 4 | 0 | 4 |
| 526 | A→T | CAC→CTC | His→Leu | 1 | 3 | 1 | 3 | 1 | 3 |
| 526 | A→C | CAC→CCC | His→Pro | 0 | 5 | 0 | 5 | 0 | 4 |
| 526 | A→G | CAC→CGC | His→Arg | 1 | 7 | 1 | 7 | 1 | 7 |
| 531 | C→T | TCG→TTG | Ser→Leu | 12 | 24 | 12 | 24 | 12 | 23 |
| 516 and 526 | A→T and A→G | GAC→GTC and CAC→CGC | Asp→Val and His→Arg | 0 | 2 | 0 | 2 | 0 | 2 |
| 511 and 516 | T→C and A→G | CTG→CCG and GAC→GGC | Leu→Ser and Asp→Gly | 1 | 0 | 1 | 0 | 1 | 0 |
| NAa | None | None | None | NA | NA | NA | NA | NA | 2 |
| Total | 18 | 59 | 18 | 59 | 18 | 59 | |||
NA, not applicable.
Preparation of standard templates.
To validate the sensitivity and specificity of padlock probes, PCR products from isolates (previously amplified from isolates and sequence, as described above), containing wild-type sequence and resistance mutations at one or more of positions 511, 514, 516, 526, and 531 were cloned into pGEM-T Easy vector system (Promega, Madison, WI) according to the manufacturer's protocol. Positive clones were resequenced to confirm the presence of the expected mutations, and plasmid DNA was further amplified to generate linear products, followed by purification using a Millipore PCR purification plate, to serve as standard template controls. DNA copy numbers of standard templates were estimated using a DNA calculator (http://cels.uri.edu/gsc/cndna.html); 1011 copies of standard DNA templates were used for testing the specificity of the system.
Probe ligation, exonucleolysis, and signal amplification by hyperbranched RCA.
Ligation of padlock probes to templates was carried out by mixing 1011 copies of each template with 1 pmol of the corresponding individual padlock probe, 2 U of Pfu DNA ligase in 20 mM Tris-HCl (pH 7.5), 20 mM KCl, 10 mM MgCl2, 0.1% igepal, 0.01 mM rATP, and 1 mM dithiothreitol in a total reaction volume of 10 μl. Multiple-cycle ligation was conducted to validate the specificity of each probe in recognizing its corresponding template (see below). The reaction conditions included one cycle of 5 min at 94°C to denature the template DNA, followed by 1 to 15 cycles of 94°C for 30 s and a 4-min ligation at 65°C. The ligation mixture was then subjected to exonucleolysis to remove unreacted padlock probe and PCR product in order to reduce subsequent ligation-independent amplification events. The exonuclease treatment was performed in a 20-μl volume by adding 10 U each of exonucleases I and III (New England BioLabs, Ipswich, MA) to the ligation mixture and incubating at 37°C for 30 min, followed by 94°C for 30 s to inactivate the exonucleases.
The amplification of circularized padlock probes was performed in a 50-μl volume by adding 8 U of Bst DNA polymerase (New England BioLabs, Ipswich, MA), 5 μl of reaction buffer, 400 μM dNTP mix, 10 pmol of each RCA primer, 5% dimethyl sulfoxide (vol/vol), and 1× Sybr green I (Sigma, St. Louis, MO) to the ligation mixture. The reaction was carried out using a Rotor-Gene 6000 real-time PCR machine (Corbett Research Pty, Ltd., Australia).
Determination of specificity and sensitivity of padlock probes and RCA system.
The specificity of padlock probes was determined by multiple-cycle ligations of each probe with the corresponding resistance template and the wild-type template to test their ability to distinguish them as described above. Testing the sensitivity of each resistance-specific padlock probe was carried out with a mixture of the resistance template with the wild-type template in various proportions to a total of 1011 copies: 100% (resistance template only), 50%, 10%, 5%, and 0% (wild-type only). The ligation reaction was performed for 15 cycles, followed by RCA of probe signal detection.
Testing the multiplex ability of padlock probes.
The use of padlock probes provides high multiplexing potential as cross-reactions between probes in the ligation step does not affect signal detection. Interactions between probes, if any, will only generate linear molecules that are easily distinguished from the circularized probes arising from “correct” reactions. To test the ability of multiplexed probes to detect any of the 12 target mutations, all padlock probes were mixed together. Ligation of the padlock probe mixture (1 pmol of each individual probe) with 1011 copies of each individual resistance or wild-type template were carried out using the reaction conditions described above, followed by RCA reactions to amplify circularized probe signals.
Padlock probe and RCA detection of resistance mutations in isolates and clinical samples.
A total of 1011 copies of PCR amplicons of rpoB region from isolates and 1010 to 1012 copies of PCR amplicons from clinical samples were subjected to individual resistance-specific padlock probe ligation or probe mixture ligation under conditions described above, followed by exonucleolysis and RCA amplification of probe signal. The signals were monitored on a Rotor-Gene 6000 real-time PCR machine.
RESULTS
DST.
Phenotypic DST of isolates from 78 clinical specimens and 46 stored isolates revealed that 59 and 18, respectively, were RIF resistant.
Sequencing of M. tuberculosis rpoB region.
Sequencing of the rpoB region from isolates and clinical samples revealed the presence of RIF resistance mutations at multiple locations in the RRDR (Table 2). Sixteen of eighteen RIF-resistant stored isolates had single mutations at codons 526 or 531; one had two mutations (at codons 511 and 516), and one, which was phenotypically RIF resistant, had the codon TTC inserted at position 514. DNA from 59 of the 78 clinical samples displayed single mutations at codon 516, 526, or 531 and two had mutations at both 516 and 526 (Table 2).
Specificity and sensitivity of RCA.
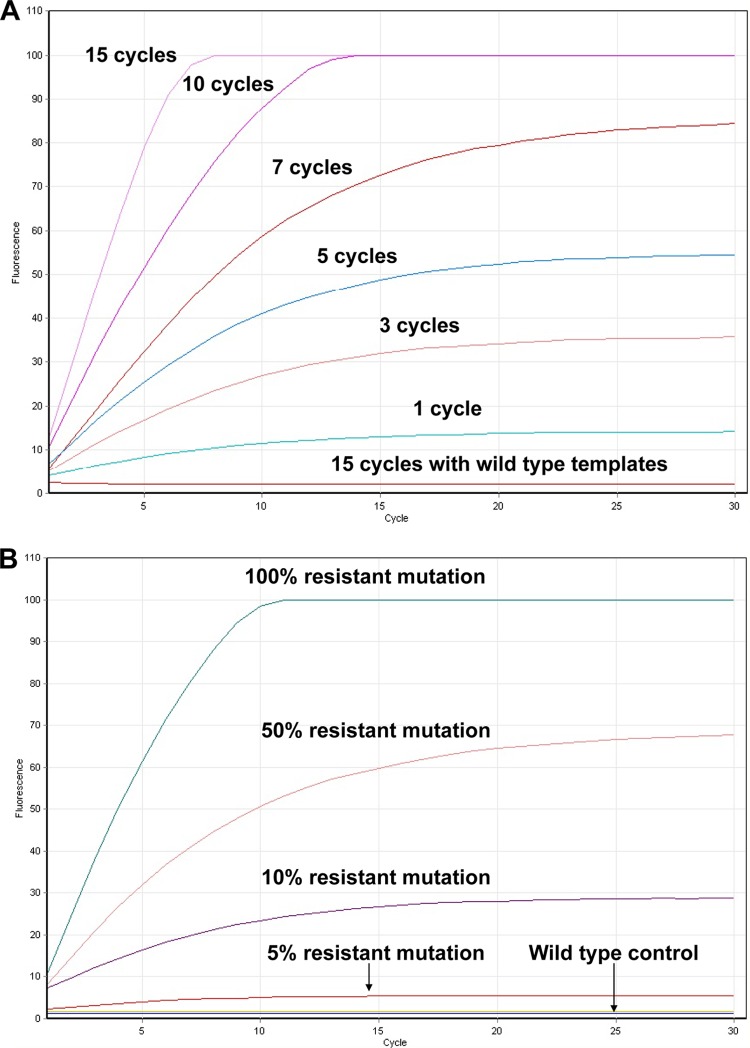
RCA of each circularized probe showed that positive signals were only detected from reaction mixtures containing the mutation-specific padlock probe and corresponding resistance template; no signals were detected from wild-type templates despite only single nucleotide differences compared to the resistance template (Fig. 1A). Increasing the number of ligation cycles resulted in earlier detection and peak of probe signal due to the increased number of circular probe molecules (Fig. 1A).
FIG 1.
(A) RCA results monitored using Rotor-Gene 6000 real-time PCR machine. Using probe-rpoB-531-TTG as an example, specificity of the probe was tested by targeting 1011 copies of 531-TTG template and 531 wild-type template with 1 pmol of the resistance mutation 531-TTG-specific probe. Ligation of the probe to the standard templates was carried out using 1, 3, 5, 7, 10, and 15 cycles, followed by RCA reaction to amplify circularized padlock probe. Specific probe signals were only detected with mutation-specific padlock probe targeting mutation template. Increasing the number of ligation cycles was associated with earlier signal detection, as more circular molecules are generated. No signal was detected in the resistance-specific padlock probe targeting wild-type template. Testing of other probes gives similar results. (B) Sensitivity testing of the probe-rpoB-531-TTG targeting 1011 copies of templates containing various proportions of the 531-TTG target. Significant elevation of signal was detected in the presence of >5% (5 × 109 copies) of the resistant template, while in wild-type control, no signal was detected. (C) Standard testing with mixture of all mutation-specific probes in mutation detection. Standard templates with resistance mutations at positions 511, 514, 516, 526, and 531, as well as the wild-type template, were ligated with the probe mixture. After 15 cycles of ligation reaction, followed by RCA, positive signals were detected only in the templates containing resistance mutations while wild-type template showed no signal. A weaker signal for two probes targeting mutation at residue 526 was observed and probably is the result of multiple 526 probes competing for the binding site.
After 15 ligation cycles, followed by RCA signal amplification, positive signals were easily detected by probes, even when resistance mutations represented only 5% of the total template tested (Fig. 1B).
Specificity of multiplexed padlock probes.
Each standard resistance template and the wild-type template were ligated with the mixture of all resistance-specific probes; after 15 cycles of each ligation reaction followed by RCA, positive signals were detected in all templates containing resistance mutations, while the wild-type template showed no signal (Fig. 1C).
Detection of rpoB mutants in RRDR of isolate DNA.
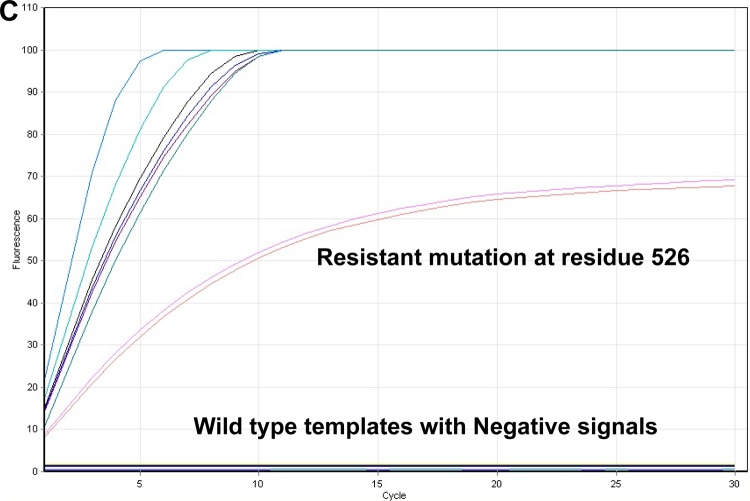
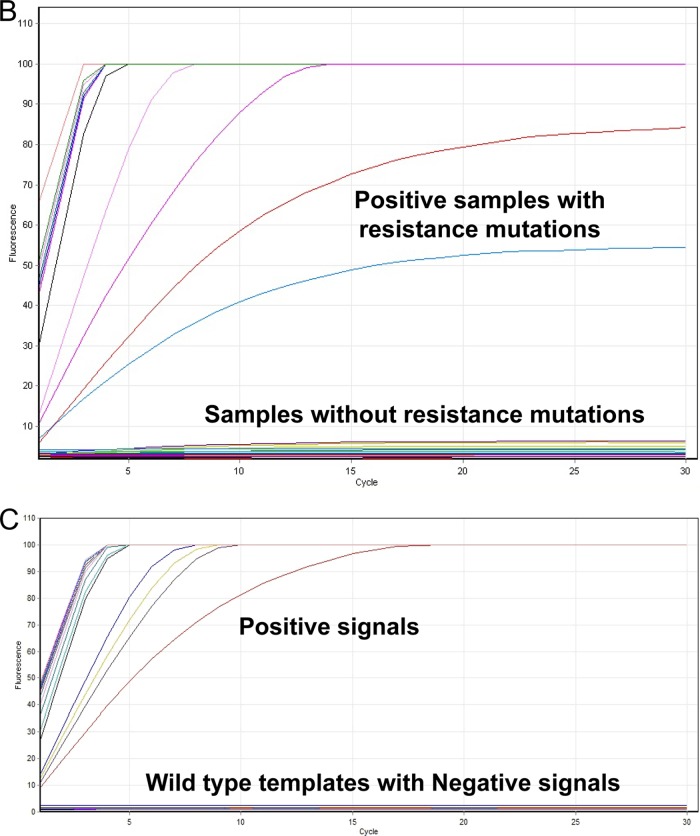
Screening of PCR amplicons from DNA extracts of all 46 stored clinical isolates and H37Rv, with individual probes, clearly showed that positive signals were obtained only from RIF-resistant isolates containing resistance mutations No signal was detected from PCR amplicons derived from RIF-susceptible isolates. To increase the mutation detection efficiency, all 47 samples were further subjected to ligation reaction using a mixture of all mutation-specific probes. Positive signals were obtained only with isolates containing one or more of the target mutations, as shown by sequencing; however, specific mutations could not be identified by this approach. No signal was produced with susceptible (wild-type) isolates (Fig. 2B).
FIG 2.
(A) Screening for resistance mutations by individual padlock probes in stored isolates. All 46 stored isolates were subjected to individual rpoB mutation-specific padlock detection. A positive signal was detected only in the matched probe-template pairs. In comparison, samples showing the absence of particular resistance mutations give no signal. (B) Using a mixture of mutation-specific probes to screen for mutations in stored isolates or clinical samples, positive signals were detected in all samples containing resistance mutations while no signals were detected in samples showing the absence of resistance mutations. (C) Screening for resistance mutations by individual padlock probes in clinical samples. All 78 clinical samples (a selection of which are shown here) were subjected to individual drug resistance-specific padlock detection, and the positive signal was detected only in the matched probe and templates pairs. No signals were produced from samples containing RIF-susceptible strains.
Detection of rpoB mutants in RRDR of clinical samples.
Screening of DNA extracts from all 78 clinical samples with individual probes clearly showed a positive signal from 59 samples and that no signals were produced from 19 patient-derived samples (a selection of samples are shown in Fig. 2C).
Comparison of genotypic and phenotypic resistance result.
Both RCA detection and rpoB sequencing revealed the presence of one or more mutations in 18 stored isolates and 59 clinical samples with 100% consistency. These results were consistent with phenotypic DST of stored isolates. Of 78 clinical samples, 4 showed discrepant results between RCA/rpoB sequencing (genotypic results) and phenotypic DST of isolates: two samples with rpoB mutations detected by both RCA and sequencing were phenotypically RIF susceptible and two, from which RIF-resistant strains were isolated, had no rpoB mutations detected by either RCA or sequencing. These discrepancies were confirmed on repeat testing.
In summary, RCA detected rpoB mutations in 57 of 59 clinical samples (i.e., two “false-negative” results) from which phenotypically RIF-resistant M. tuberculosis was isolated (96.6% sensitivity) and in 2 of 19 samples (i.e., two “false-positive” results) from which RIF-susceptible M. tuberculosis was isolated (89.5% specificity).
DISCUSSION
This study has shown that RCA using padlock probes can detect resistance mutations in RIF-resistant M. tuberculosis isolates and in template mixtures, including those in which resistance templates mutations are in a minority. This high level of sensitivity allows direct testing on clinical specimens known to contain M. tuberculosis, which reduces delay in introduction of appropriate treatment and the risk of further resistance emerging. This assay has the potential for quantitation, which would allow assessment of the proportion of resistant organisms in the specimen and provide insight into the dynamics of emergent resistance. Importantly, the ease of performance, rapid turnaround, and multiplex capacity indicate its potential for high-throughput routine screening of AFB-positive specimens or M. tuberculosis isolates, with significant clinical benefits. Formation of the circular probe molecule, mediated by DNA ligase, confers very high allele discriminatory ability (22, 23). As a result, the signal generated is highly specific and easy to interpret, in contrast to many other probe-based methods in which the result depends on a subjective assessment of spot intensity.
The specificity of the mutant-specific padlock probes, individually and when multiplexed, has been evaluated by testing both stored isolates and clinical samples. The fact that probe-probe interactions cannot give rise to closed, circular molecules means that large number of probes can be used in a single reaction tube to recognize sequences of interest. This is particularly important for the detection of rpoB mutants since multiple site substitutions can contribute to RIF resistance. When additional mutations are discovered, new probes can be easily designed and added to the panel as required.
In the present study, we included 46 clinical isolates with known RIF resistance, the standard strain (H37Rv, ATCC 27294) and 78 culture-positive clinical samples to compare the accuracy of mutation detection by PCR/sequencing and RCA using padlock probes. The use of padlock probes and RCA provided genotypic resistance data equivalent to that provided by rpoB amplification and sequencing and, with few exceptions, phenotypic RIF susceptibility testing. These results reinforced the utility of RCA, which has also been applied, successfully, to the detection of INH resistance in M. tuberculosis in our laboratory (24). The sensitivity (96.6%) and specificity (89.5%) of RCA detection for clinical samples were comparable to those of a high-resolution melting analysis (HRMA) M. tuberculosis resistance detection method previously reported by our group (25) and support the suitability of the technique for direct application of RCA detection to clinical samples.
False-positive results (rpoB mutations detected in specimens from which isolates were RIF susceptible) may be related to the threshold concentrations of RIF used for DST—which are based, historically, on clinically achievable drug levels and experience with response to therapy (18)—inhibiting growth despite low-grade RIF resistance. Moreover, it is known that not all mutations in RRDR confer phenotypic RIF resistance (26). Failure to identify rpoB mutations in samples known to contain phenotypically RIF-resistant M. tuberculosis is consistent with reports that up to 5% of RIF-resistant strains contain rpoB mutations outside of the RRDR (5) or are resistant due to other mechanisms, such as efflux pumps (5, 27).
Engstrom et al. have recently described a similar approach to detection of the most common rpoB mutations associated with RIF resistance, using multiplexed padlock probes, including an M. tuberculosis-specific probe and RCA (28). Our study has confirmed the utility of this approach and, in addition, has demonstrated its potential for routine direct screening of clinical specimens. Future studies that include samples other than sputum (blood, cerebrospinal fluid, etc.) would further extend the clinical applications of this technique in RIF resistance testing. Although we did not include an M. tuberculosis-specific probe to confirm its presence, all clinical specimens were positive for AFB on microcopy. Our intention was that RIF resistance testing would be used selectively on AFB-positive specimens, since patients with a heavy bacillary load are most infectious and most likely to fail therapy and develop further resistance if not correctly treated initially.
Addition of an M. tuberculosis-specific probe and testing of all specimens submitted for TB diagnosis would increase the cost (depending on the prevalence of TB) but allow the diagnosis of TB in patients with AFB-negative specimens. However, a multiplex testing approach to detect M. tuberculosis and RIF resistance, simultaneously, remains an option, depending on setting, need, and affordability. This would be particularly important in settings where MDR-TB is common and/or cultures are not performed routinely, where it could significantly improve TB control by optimizing treatment regimens, improving epidemiological surveillance and contributing to understanding of the global spread and transmission dynamics of MDR-TB.
The use of a commercial real-time PCR system, GeneXpert MTB/RIF (Cepheid, Sunnyvale, CA), has been endorsed by the World Health Organization and has allowed the rapid detection of RIF resistance at the point of care, directly from untreated sputum in less than 2 h with minimal hands on time. However, the cost of the instrument and the consumables remain a financial burden for patients in developing countries. Therefore, there is an urgent need for a comparable method that can be performed in resource-limited settings. RCA-based testing provides a result within 3 h, and the end product could be simply detected by agarose gel electrophoresis, which would eliminate the requirement for a real-time PCR instrument. These simple requirements and easy data interpretation make this method an attractive option.
We sought here to develop a simple, rapid, inexpensive, and accessible method of RIF resistance detection for use in high-burden, low-resource, and low-technology settings. Although the number of cases considered in the present study was limited, our findings do illustrate that the RCA assay could be utilized to achieve high sensitivity and specificity in the direct detection of rpoB mutation from clinical samples.
ACKNOWLEDGMENT
This study was supported by a grant from The National S&T Major Special Project on Major New Drug Innovation (2012ZX09301002-005-003) from the Ministry of Science and Technology of China.
Footnotes
Published ahead of print 26 February 2014
REFERENCES
- 1.Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. 1999. Consensus statement: global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA 282:677–686 [DOI] [PubMed] [Google Scholar]
- 2.Mitnick CD, Appleton SC, Shin SS. 2008. Epidemiology and treatment of multidrug resistant tuberculosis. Semin. Respir. Crit. Care Med. 29:499–524. 10.1055/s-0028-1085702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaziou P, Floyd K, Raviglione M. 2009. Global burden and epidemiology of tuberculosis. Clin. Chest Med. 30:621–636. 10.1016/j.ccm.2009.08.017 [DOI] [PubMed] [Google Scholar]
- 4.Mani C, Selvakumar N, Narayanan S, Narayanan PR. 2001. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis clinical isolates from India. J. Clin. Microbiol. 39:2987–2990. 10.1128/JCM.39.8.2987-2990.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramaswamy S, Musser JM. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuberc. Lung Dis. 79:3–29. 10.1054/tuld.1998.0002 [DOI] [PubMed] [Google Scholar]
- 6.Yue J, Shi W, Xie J, Li Y, Zeng E, Wang H. 2003. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from China. J. Clin. Microbiol. 41:2209–2212. 10.1128/JCM.41.5.2209-2212.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maschmann Rde A, Verza M, Silva MS, Sperhacke RD, Ribeiro MO, Suffys PN, Gomes HM, Tortoli E, Marcelli F, Zaha A, Rossetti ML. 2011. Detection of rifampin-resistant genotypes in Mycobacterium tuberculosis by reverse hybridization assay. Mem. Inst. Oswaldo Cruz 106:139–145 [DOI] [PubMed] [Google Scholar]
- 8.Yao C, Zhu T, Li Y, Zhang L, Zhang B, Huang J, Fu W. 2010. Detection of rpoB, katG, and inhA gene mutations in Mycobacterium tuberculosis clinical isolates from Chongqing as determined by microarray. Clin. Microbiol. Infect. 16:1639–1643. 10.1111/j.1469-0691.2010.03267.x [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Kong F, Sorrell TC, Wang B, McNicholas P, Pantarat N, Ellis D, Xiao M, Widmer F, Chen SC. 2009. Rapid detection of ERG11 gene mutations in clinical Candida albicans isolates with reduced susceptibility to fluconazole by rolling circle amplification and DNA sequencing. BMC Microbiol. 9:167. 10.1186/1471-2180-9-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antson DO, Isaksson A, Landegren U, Nilsson M. 2000. PCR-generated padlock probes detect single nucleotide variation in genomic DNA. Nucleic Acids Res. 28:E58. 10.1093/nar/28.12.e58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U. 1994. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 265:2085–2088. 10.1126/science.7522346 [DOI] [PubMed] [Google Scholar]
- 12.Tong Z, Kong F, Wang B, Zeng X, Gilbert GL. 2007. A practical method for subtyping of Streptococcus agalactiae serotype III, of human origin, using rolling circle amplification. J. Microbiol. Methods 70:39–44. 10.1016/j.mimet.2007.03.010 [DOI] [PubMed] [Google Scholar]
- 13.Huang H, Jin Q, Ma Y, Chen X, Zhuang Y. 2002. Characterization of rpoB mutations in rifampicin-resistant Mycobacterium tuberculosis isolated in China. Tuberculosis (Edinb.) 82:79–83. 10.1054/tube.2002.0326 [DOI] [PubMed] [Google Scholar]
- 14.Mokrousov I, Filliol I, Legrand E, Sola C, Otten T, Vyshnevskaya E, Limeschenko E, Vyshnevskiy B, Narvskaya O, Rastogi N. 2002. Molecular characterization of multiple-drug-resistant Mycobacterium tuberculosis isolates from northwestern Russia and analysis of rifampin resistance using RNA/RNA mismatch analysis as compared to the line probe assay and sequencing of the rpoB gene. Res. Microbiol. 153:213–219. 10.1016/S0923-2508(02)01311-6 [DOI] [PubMed] [Google Scholar]
- 15.Sharma M, Altamirano M, Prasad HK, Myneedo VP, Nand N. 2000. Characterization by single strand conformation polymorphism of mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis in strains from Vancouver, Mexico City and New Delhi. J. Assoc. Physicians India 48:568–572 [PubMed] [Google Scholar]
- 16.Suresh N, Singh UB, Arora J, Pant H, Seth P, Sola C, Rastogi N, Samantaray JC, Pande JN. 2006. rpoB gene sequencing and spoligotyping of multidrug-resistant Mycobacterium tuberculosis isolates from India. Infect. Genet. Evol. 6:474–483. 10.1016/j.meegid.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 17.Tang K, Sun H, Zhao Y, Guo J, Zhang C, Feng Q, He Y, Luo M, Li Y, Sun Q. 2013. Characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Sichuan in China. Tuberculosis (Edinb.) 93:89–95. 10.1016/j.tube.2012.10.009 [DOI] [PubMed] [Google Scholar]
- 18.Li GL, Zhao DF, Xie T, Ju HF, Mu C, Zhao H, Wang XX. 2010. Molecular characterization of drug-resistant Beijing family isolates of Mycobacterium tuberculosis from Tianjin, China. Biomed. Environ. Sci. 23:188–193. 10.1016/S0895-3988(10)60051-7 [DOI] [PubMed] [Google Scholar]
- 19.Wilson K. 2001. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. Chapter 2:Unit 24 [DOI] [PubMed] [Google Scholar]
- 20.Leung ET, Zheng L, Wong RY, Chan EW, Au TK, Chan RC, Lui G, Lee N, Ip M. 2011. Rapid and simultaneous detection of Mycobacterium tuberculosis complex and Beijing/W genotype in sputum by an optimized DNA extraction protocol and a novel multiplex real-time PCR. J. Clin. Microbiol. 49:2509–2515. 10.1128/JCM.00108-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolotin S, Alexander DC, Chedore P, Drews SJ, Jamieson F. 2009. Molecular characterization of drug-resistant Mycobacterium tuberculosis isolates from Ontario, Canada. J. Antimicrob. Chemother. 64:263–266. 10.1093/jac/dkp183 [DOI] [PubMed] [Google Scholar]
- 22.Bakht S, Qi X. 2005. Ligation-mediated rolling-circle amplification-based approaches to single nucleotide polymorphism detection. Expert Rev. Mol. Diagn. 5:111–116. 10.1586/14737159.5.1.111 [DOI] [PubMed] [Google Scholar]
- 23.Qi X, Bakht S, Devos KM, Gale MD, Osbourn A. 2001. L-RCA (ligation-rolling circle amplification): a general method for genotyping of single nucleotide polymorphisms (SNPs). Nucleic Acids Res. 29:E116. 10.1093/nar/29.22.e116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai L, Kong F, Jelfs P, Gilbert GL, Sintchenko V. 2009. Rolling circle amplification and multiplex allele-specific PCR for rapid detection of katG and inhA gene mutations in Mycobacterium tuberculosis. Int. J. Med. Microbiol. 299:574–581. 10.1016/j.ijmm.2009.05.006 [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Kong F, Wang Q, Li C, Zhang J, Gilbert GL. 2011. Rapid detection of isoniazid, rifampin, and ofloxacin resistance in Mycobacterium tuberculosis clinical isolates using high-resolution melting analysis. J. Clin. Microbiol. 49:3450–3457. 10.1128/JCM.01068-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. 2004. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48:1289–1294. 10.1128/AAC.48.4.1289-1294.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang Y, Lu J, Wang Y, Song Y, Wang S, Zhao Y. 2013. Study of the rifampin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57:893–900. 10.1128/AAC.01024-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engstrom A, Zardan Gomez de la Torre T, Stromme M, Nilsson M, Herthnek D. 2013. Detection of rifampicin resistance in Mycobacterium tuberculosis by padlock probes and magnetic nanobead-based readout. PLoS One 8:e62015. 10.1371/journal.pone.0062015 [DOI] [PMC free article] [PubMed] [Google Scholar]