Abstract
Potyviruses express most of their proteins from a long open reading frame that is translated into a large polyprotein processed by three viral proteases. To understand the constraints on potyvirus genome organization, we relocated the viral RNA-dependent RNA polymerase (NIb) cistron to all possible intercistronic positions of the Tobacco etch virus (TEV) polyprotein. Only viruses with NIb at the amino terminus of the polyprotein or in between P1 and HC-Pro were viable in tobacco plants.
TEXT
Viral genomes are compact assemblies of genes and regulatory sequences, and plant viruses are no exception (1). To compact their genomes (2, 3), plant viruses have evolved mechanisms that include overlapping open reading frames (ORFs), ambisense coding, or translational frameshift and read through (4). One of the most common strategies, however, is coding for a polyprotein that is processed into many different gene products after translation. Potyviruses (genus Potyvirus, family Potyviridae) are one of the largest groups of plant viruses and take this strategy to an extreme. Potyviral genomes are RNA molecules of positive polarity approximately 10,000 nucleotides long, consisting of a long ORF flanked by two short untranslated regions (UTRs) (5). The potyviral ORFs apparently encode 10 mature gene products: P1 proteinase; helper component proteinase (HC-Pro); P3 protein; 6K1 polypeptide; cylindrical inclusion (CI) protein; 6K2 polypeptide; nuclear inclusion a (NIa) protein, a polyprotein that is further processed to, at least, the viral protein genome-linked (VPg) and the NIa proteinase (NIaPro); nuclear inclusion b (NIb) protein, the viral RNA-dependent RNA polymerase; and coat protein (CP) (6). These products are released from the viral polyprotein through the activity of the three viral proteinases, P1, HC-Pro, and NIaPro. An additional gene product, P3N-PIPO, results from a translation frameshift in a slippery region of the P3 cistron (7).
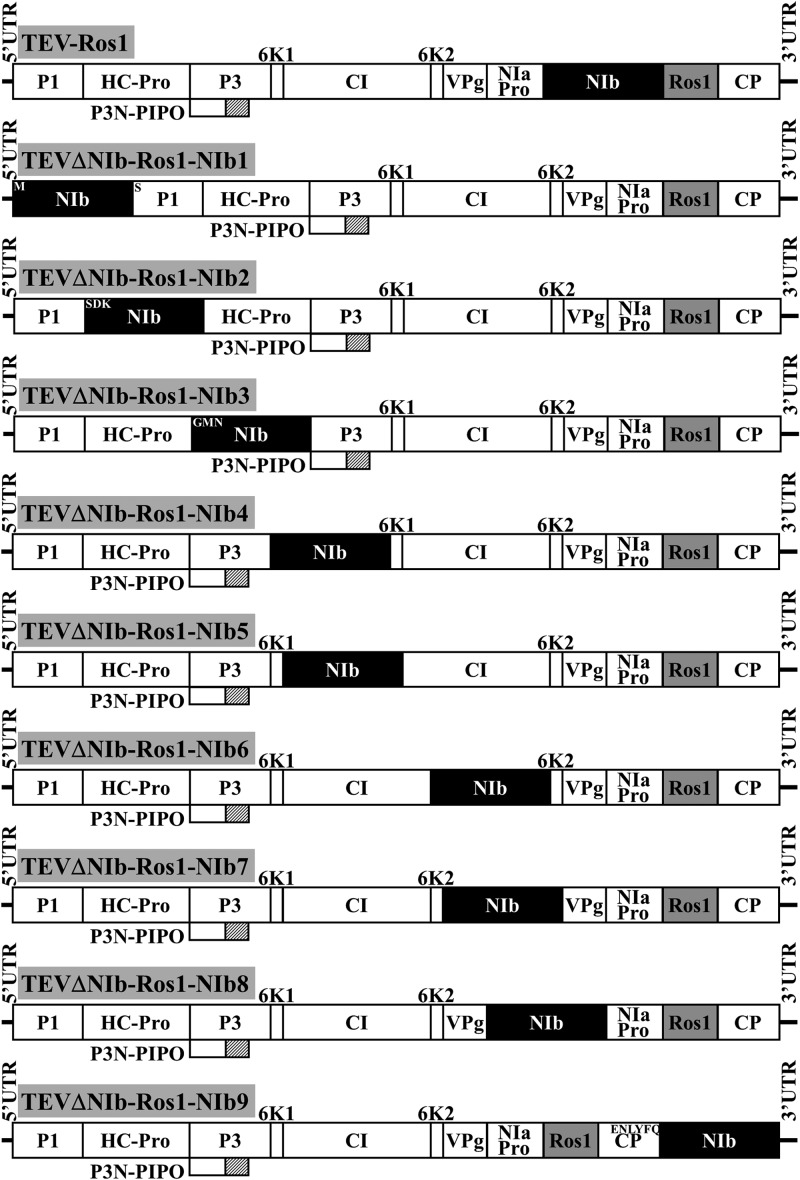
The aim of this work was to improve our understanding of the functional, structural, and evolutionary constraints that rendered the actual genome organization of potyviruses. To achieve this goal, we relocated one of the genes of Tobacco etch virus (TEV) to all possible intercistronic positions in the genome and assayed the infectivity of the resulting recombinant viruses in tobacco (Nicotiana tabacum L. cv. Xanthi nc) plants. To avoid further complexity, P3N-PIPO was excluded from this study. We chose the NIb gene because the infectivity of a TEV deletion mutant lacking the entire NIb cistron can be rescued by NIb expression in trans from a transgene (8) or a compatible viral vector (9). When relocating the NIb gene to the different intercistronic sites, in some cases we added appropriate sequences to the amino and carboxy NIb termini to create NIaPro proteolytic sites that could mediate NIb release from the viral polyprotein. To facilitate the monitoring of virus infection, spread, and accumulation, all recombinant viruses were constructed from a TEV clone carrying the Rosea1 (Ros1) visual marker (10, 11). The TEV-Ros1 load correlates with anthocyanin accumulation in tobacco tissues (10). Figure 1 outlines the genomes of the parental (TEV-Ros1) and the derived (TEVΔNIb-Ros1-NIb1 to -9) recombinant viruses. Figure S1 in the supplemental material specifies the exact nucleotide sequences of all recombinant viral clones.
FIG 1.
Schematic representation of the parental TEV clone (TEV-Ros1), including the visual Ros1 marker (gray rectangle), and the nine recombinant clones in which the NIb gene (black rectangle) was relocated to nine different intercistronic positions of the viral polyprotein (TEVΔNIb-Ros1-NIb1 to -9). Amino acid sequences that are indicated next to the NIb amino or carboxy terminus in some cases (TEVΔNIb-Ros1-NIb1, -2, -3, and -9) were inserted to complement NIb processing from the polyprotein.
The parental and recombinant TEV clones, constructed from the binary plasmid pGTEV-Ros1 (10, 11), were agroinoculated (12) into two leaves of 20 3-week-old wild-type plants and 20 transgenic plants constitutively expressing TEV NIb (8). Plants were grown in a glasshouse at 25°C with 16 h light, and Ros1 expression was visually monitored for 4 weeks. For each clone, systemic leaves from three wild-type and three transgenic plants were harvested at 15 days postinoculation (d.p.i.), photographed, and used to estimate viral load by measuring the anthocyanin accumulation induced by the Ros1 marker (10).
Most of the viral clones tested were not viable in either wild-type or transgenic plants. In other words, systemic tissue 4 weeks after agroinfiltration had no visible anthocyanin accumulation or infection symptoms. Only viruses with NIb relocated to the first two intercistronic positions were viable in wild-type plants. Ros1 activity was detected in systemic leaves of all 20 wild-type plants agroinoculated with TEVΔNIb-Ros1-NIb1 and TEVΔNIb-Ros1-NIb2, as well as the parental virus TEV-Ros1 (Fig. 2A). There is therefore a statistically significant effect of the NIb position on viability (test of equal proportions, χ2 = 180.00, 8 df, P < 0.001). An identical, statistically significant result was obtained for transgenic plants expressing NIb (χ2 = 155.077, 8 df, P < 0.001), except that approximately half of the plants agroinoculated with the virus carrying NIb in the third intercistronic position (TEVΔNIb-Ros1-NIb3) also showed systemic marker expression (Fig. 2B). The number of Ros1-expressing plants for TEVΔNIb-Ros1-NIb3 was intermediate, as it was significantly different from the results for all other clones (pairwise test of equal proportions, P < 0.001 for all comparisons).
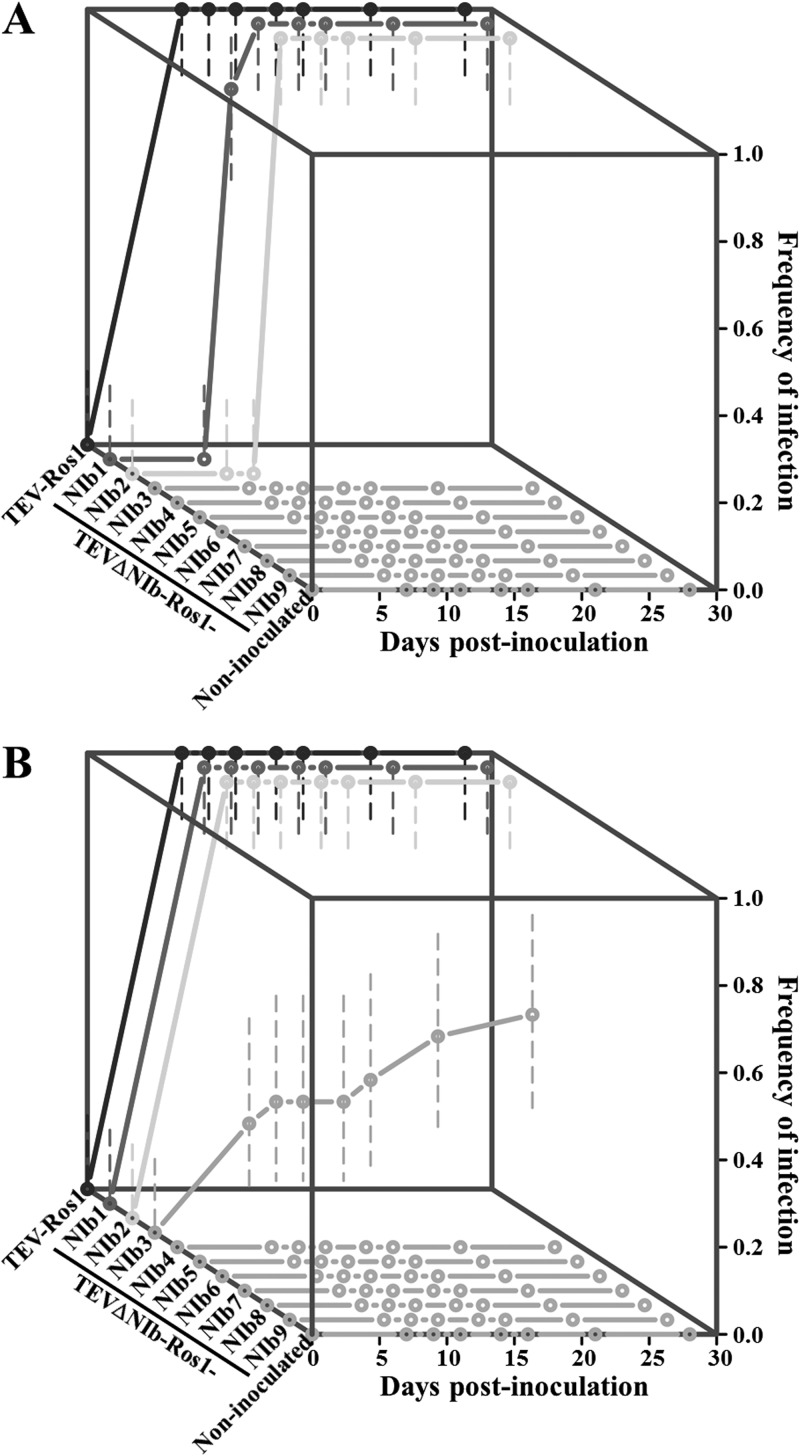
FIG 2.
Infectivities of TEV recombinant clones in which the NIb gene was relocated to different intercistronic positions in the viral polyprotein. Plots show the cumulative frequency of infection in wild-type (A) and transgenic (constitutively expressing TEV NIb) (B) tobacco plants versus days postinoculation (d.p.i.) with TEV-Ros1 and TEVΔNIb-Ros1-NIb1 to -9. Symptoms were screened at 7, 9, 11, 14, 16, 21, and 28 d.p.i. Error bars represent 95% confidence intervals of the estimated frequencies.
Next, we considered at what time after agroinfiltration anthocyanin accumulation was first apparent. In wild-type plants, there were significant differences in the median time until visual detection of Ros1 expression for all three viable viruses (Fig. 2A) (log-rank test, P < 0.001 for all three comparisons between TEV-Ros1, TEVΔNIb-Ros1-NIb1, and TEVΔNIb-Ros1-NIb2). So, TEVΔNIb-Ros1-NIb2 was significantly slower than TEVΔNIb-Ros1-NIb1, while TEVΔNIb-Ros1-NIb1 was significantly slower than TEV-Ros1. In transgenic plants, TEV-Ros1, TEVΔNIb-Ros1-NIb1, and TEVΔNIb-Ros1-NIb2 all had the exact same median time until Ros1 expression was observed visually (Fig. 2B). For TEVΔNIb-Ros1-NIb3, anthocyanin accumulation in systemic tissue was first observed significantly later than for any of the other viable viruses (pairwise log-rank test, χ2 = 11.600, 1 df, P = 0.001 for all three comparisons). In summary, TEVΔNIb-Ros1-NIb1 and TEVΔNIb-Ros1-NIb2 were viable in both plant genotypes, although infection appeared to proceed more slowly than for the ancestral virus in wild-type plants. For TEVΔNIb-Ros1-NIb3, viral spread was only seen in some transgenic plants, and when it appeared, it was significantly delayed.
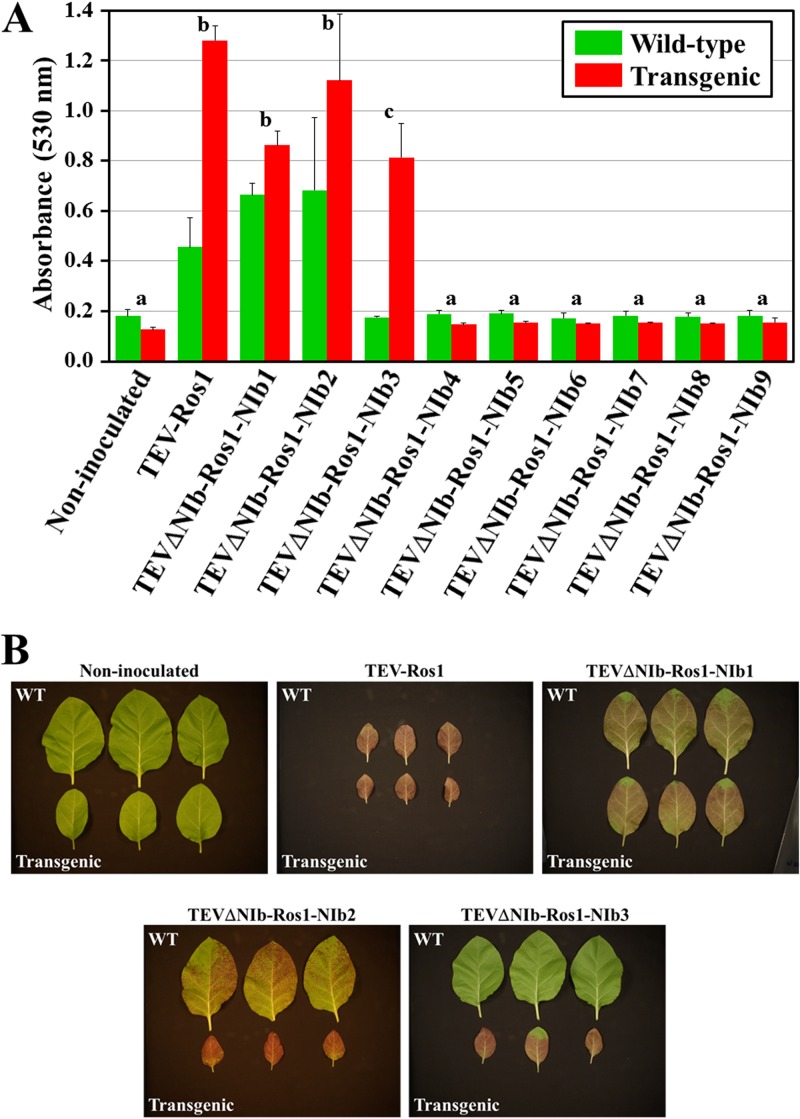
We then analyzed the stability of the relocated NIb in the progeny of the viable viruses. For TEVΔNIb-Ros1-NIb1 and TEVΔNIb-Ros1-NIb2, reverse transcription-PCR (RT-PCR) analysis of the viral progeny 15 d.p.i. confirmed the stability of the relocated NIb gene (Fig. 3A and B). For TEVΔNIb-Ros1-NIb3, which showed delayed anthocyanin accumulation in only some transgenic plants, RT-PCR analysis and sequencing confirmed that the NIb gene was lost (Fig. 3C). Anthocyanin accumulation was quantified in extracts from systemic leaves of three plants inoculated per recombinant clone at 15 d.p.i. (Fig. 4). The data were analyzed with a generalized linear model with full-factorial design, using a log-link function and gamma distributed error structure. Overall, the recombinant clone (χ2 = 2,383.451, 10 df, P < 0.001) and the plant genotype (χ2 = 35.356, 1 df, P < 0.001) had significant effects on anthocyanin accumulation, and there was a significant interaction between these two factors as well (χ2 = 419.276, 10 df, P < 0.001). A Tukey post hoc test highlights the existence of three nonoverlapping groups of recombinant clones (Fig. 4A): TEVΔNIb-Ros1-NIb1 and TEVΔNIb-Ros1-NIb2 showed the same expression level as the wild-type TEV-Ros1, TEVΔNIb-Ros1-NIb3 showed an intermediate expression level that depended on the plant genotype (explaining the significant interaction term), and the other six recombinant clones were not significantly different from the mock inoculated plants in either plant genotype.
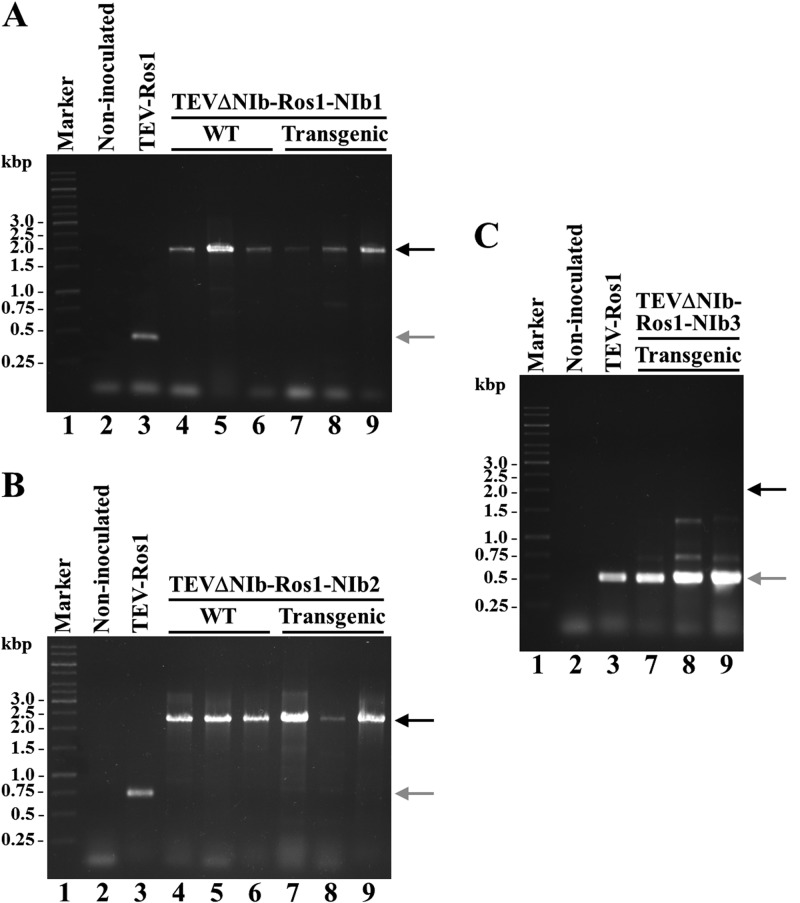
FIG 3.
Stability of the relocated NIb gene in the viral progeny at 15 d.p.i. RNA from systemic leaves of three representative inoculated plants was purified. cDNAs amplified by RT-PCR using primers flanking the new NIb location for TEVΔNIb-Ros1-NIb1 (A), TEVΔNIb-Ros1-NIb2 (B), and TEVΔNIb-Ros1-NIb3 (C) were separated by electrophoresis in 1% agarose gels, and stained with ethidium bromide. Lanes 1, DNA marker with the sizes of some of the components indicated on the left; lanes 2, noninoculated negative control; lanes 3, parental virus positive control (TEV-Ros1); lanes 4 to 6, three infected wild-type plants; lanes 7 to 9, three infected transgenic plants. Note that in panel C, lanes 4 to 6 (wild-type plants) are missing because none of these plants was infected. The black and gray arrows indicate the positions of the expected RT-PCR products if the relocated NIb is present or absent, respectively. Primers used to amplify the different cDNAs were, for RT reactions, 5′-CTTTACATAGTTTTTTTCCAACATTTCATG-3′, and for PCRs, 5′-AAAATAACAAATCTCAACACAACATATAC-3′ and 5′-CTCTTGCCATGGGTGAGCGCGCGAC-3′ (A), 5′-CTCAACTCCAAGAATTTC-3′ and 5′-GTCGCGCGCTCACCCATGGCAAGAG-3′ (B), and 5′-GTGGCCAACAATGCAAGATGTTGC-3′ and 5′-GTTAACTTTTGCGCCAAGGCTGAC-3′ (C).
FIG 4.
Anthocyanin accumulation at 15 d.p.i. in systemic leaves of tobacco plants inoculated with TEV recombinant clones in which the NIb gene was relocated to different intercistronic positions. (A) The absorbance at 530 nm of extracts from systemic leaves from three inoculated wild-type and transgenic plants constitutively expressing TEV NIb was plotted. Error bars represent ±1 standard deviation among experimental replicates. Letters a, b, and c over the bars represent groups of inoculations with homogeneous accumulation of anthocyanins according to a Tukey honestly significant difference test. (B) Pictures of systemic leaves harvested at 15 d.p.i. from three wild-type and three transgenic tobacco plants that were noninoculated and inoculated with TEV-Ros1 and TEVΔNIb-Ros1-NIb1 to -3 as indicated.
These results suggest the existence of many restrictions to the organization of the potyviral genome. Even though NIb can be provided in trans (8, 9), it can only be relocated to the amino terminus of the polyprotein or in between P1 and HC-Pro without affecting virus viability. The relocation of NIb to the other seven positions renders nonviable viruses, even in a transgenic plant constitutively expressing TEV NIb that can be infected by a TEV mutant with a complete NIb deletion (8). The relatively late infection of approximately half of the transgenic plants inoculated with TEVΔNIb-Ros1-NIb3 probably resulted from a sporadic recombination event in which the relocated NIb was deleted. The resulting virus (TEVΔNIb-Ros1) was then able to infect the plant, but only when NIb was provided from a transgene. We amplified by RT-PCR a cDNA fragment corresponding to the HC-Pro/P3 intercistronic region from the TEVΔNIb-Ros1-NIb3 progeny arising in the NIb-expressing transgenic plants and cloned it. Sequencing of three independent clones showed that recombination cleanly restored the wild-type HC-Pro/P3 junction in all cases. Interestingly, no such recombination events were observed in the case of any of the other recombinant clones. In fact, the inoculated leaves of both wild-type and transgenic plants inoculated with TEVΔNIb-Ros1-NIb3 showed slight anthocyanin accumulation, suggesting some replication capacity of this chimera.
The relocation of NIb to seven intercistronic positions probably results in nonviable viruses because of the induction of lethal defects in polyprotein processing, disruption of partially processed gene products with distinctive roles during infection, or disruption of regulatory RNA elements in the potyviral genome. A related potyvirus (Turnip mosaic virus) was found to support the expression of heterologous proteins between HC-Pro and P3 and between 6K1 and CI in Nicotiana benthamiana and Chenopodium quinoa (13). These differences may be due to the use of different virus species and host plants in both studies. On the other hand, in the previous work, heterologous genes were inserted into the potyviral genome (13), while here, a potyviral gene was relocated along the genome. Processing of the potyviral polyprotein seems to be a finely regulated process that produces the right amounts of the different gene products in time and space (14). Regulation is mainly based on the specific amino acid sequence recognized by the viral NIaPro, with some processing sites being cleaved faster than others (15–17). The insertion of NIb in some of the intercistronic positions of the polyprotein may have fatal effects on this regulation. Partially processed products from the potyviral polyprotein may have distinctive roles in the infectious cycle, different from those of the final processing products. This seems to be the case for the 6K2/VPg/NIaPro polyprotein that has been suggested to anchor TEV replication complexes to cellular endomembranes while recruiting NIb for replication (18). Insertion of NIb may therefore be lethal because it interrupts functional polyproteins. Finally, the potyviral genome contains regulatory RNA elements overlapping the ORF, including a series of RNA hairpins at the end of the CP cistron and 3′ UTR that are involved in TEV replication (19). Disruption of these elements may also have fatal consequences for the virus. Our results highlight the complexity of the potyviral genome organization, suggesting the existence of many more regulatory elements and functional entities than those currently recognized. Our results seem also to reflect the genomic organization of the different Potyviridae genera, apparently consisting of two genome blocks, a more conserved block from P3 to the end and the block including P1 and HC-Pro (in the genus Potyvirus) that is more variable between the different genera and even species.
Potyviruses have been used as expression vectors in plants (20–22). Their expression strategy, mainly based on the production of a large polyprotein, makes them particularly attractive to simultaneously produce equimolar amounts of several heterologous proteins (9, 23–25). Our results show the potentials and limitations inherent in expressing heterologous proteins from potyviral vectors. According to our results, the only positions where sequences coding heterologous proteins can be inserted without completely compromising viral viability are the amino terminal end of the polyprotein, between P1 and HC-Pro, and between NIb and CP. The P1/HC-Pro and NIb/CP intercistronic positions have been used with great frequency to express heterologous proteins in many potyviruses (21, 24, 26–28). However, the outermost amino terminal end of the polyprotein has never been used yet, although green fluorescent protein has been successfully expressed close to the amino terminus of potato potyvirus A polyprotein (29).
Supplementary Material
ACKNOWLEDGMENTS
We thank Verónica Aragonés for excellent technical assistance.
This research was supported by grant BIO2011-26741 from the Spanish Ministerio de Economía y Competitividad (MINECO) to J.-A.D., grant PROMETEO/2010/019 from Generalitat Valenciana to S.F.E. and J.-A.D., and grants BFU2012-30805 and 22371 from MINECO and the John Templeton Foundation, respectively, to S.F.E. E.M. was supported by a predoctoral fellowship (AP2012-3751) from the Spanish Ministerio de Educación, Cultura y Deporte. M.P.Z. was supported by a Juan de la Cierva postdoctoral contract (JCI-2011-10379) from MINECO and a Rubicon grant from the Netherlands Organization for Scientific Research (www.nwo.nl).
The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the John Templeton Foundation.
Footnotes
Published ahead of print 22 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03336-13.
REFERENCES
- 1.Roossinck MJ. 2011. The big unknown: plant virus biodiversity. Curr. Opin. Virol. 1:63–67. 10.1016/j.coviro.2011.05.022 [DOI] [PubMed] [Google Scholar]
- 2.Chirico N, Vianelli A, Belshaw R. 2010. Why genes overlap in viruses. Proc. R. Soc. B 277:3809–3817. 10.1098/rspb.2010.1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belshaw R, Pybus OG, Rambaut A. 2007. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 17:1496–1504. 10.1101/gr.6305707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen M, Haenni AL. 2003. Expression strategies of ambisense viruses. Virus Res. 93:141–150. 10.1016/S0168-1702(03)00094-7 [DOI] [PubMed] [Google Scholar]
- 5.Riechmann JL, Laín S, García JA. 1992. Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 73:1–16. 10.1099/0022-1317-73-1-1 [DOI] [PubMed] [Google Scholar]
- 6.Urcuqui-Inchima S, Haenni AL, Bernardi F. 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74:157–175. 10.1016/S0168-1702(01)00220-9 [DOI] [PubMed] [Google Scholar]
- 7.Chung BYW, Miller WA, Atkins JF, Firth AE. 2008. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U. S. A. 105:5897–5902. 10.1073/pnas.0800468105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li XH, Carrington JC. 1995. Complementation of tobacco etch potyvirus mutants by active RNA polymerase expressed in transgenic cells. Proc. Natl. Acad. Sci. U. S. A. 92:457–461. 10.1073/pnas.92.2.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedoya L, Martínez F, Rubio L, Daròs JA. 2010. Simultaneous equimolar expression of multiple proteins in plants from a disarmed potyvirus vector. J. Biotechnol. 150:268–275. 10.1016/j.jbiotec.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 10.Bedoya LC, Martínez F, Orzáez D, Daròs JA. 2012. Visual tracking of plant virus infection and movement using a reporter MYB transcription factor that activates anthocyanin biosynthesis. Plant Physiol. 158:1130–1138. 10.1104/pp.111.192922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majer E, Daròs JA, Zwart MP. 2013. Stability and fitness impact of the visually discernible Rosea1 marker in the Tobacco etch virus genome. Viruses 5:2153–2168. 10.3390/v5092153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedoya LC, Daròs JA. 2010. Stability of Tobacco etch virus infectious clones in plasmid vectors. Virus Res. 149:234–240. 10.1016/j.virusres.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 13.Chen CC, Chen TC, Raja JA, Chang CA, Chen LW, Lin SS, Yeh SD. 2007. Effectiveness and stability of heterologous proteins expressed in plants by Turnip mosaic virus vector at five different insertion sites. Virus Res. 130:210–227. 10.1016/j.virusres.2007.06.014 [DOI] [PubMed] [Google Scholar]
- 14.Merits A, Rajamäki ML, Lindholm P, Runeberg-Roos P, Kekarainen T, Puustinen P, Mäkeläinen K, Valkonen JP, Saarma M. 2002. Proteolytic processing of potyviral proteins and polyprotein processing intermediates in insect and plant cells. J. Gen. Virol. 83:1211–1221 [DOI] [PubMed] [Google Scholar]
- 15.Parks TD, Howard ED, Wolpert TJ, Arp DJ, Dougherty WG. 1995. Expression and purification of a recombinant tobacco etch virus NIa proteinase: biochemical analyses of the full-length and a naturally occurring truncated proteinase form. Virology 210:194–201. 10.1006/viro.1995.1331 [DOI] [PubMed] [Google Scholar]
- 16.Kim DH, Park YS, Kim SS, Lew J, Nam HG, Choi KY. 1995. Expression, purification, and identification of a novel self-cleavage site of the Nla C-terminal 27-kDa protease of turnip mosaic potyvirus C5. Virology 213:517–525. 10.1006/viro.1995.0024 [DOI] [PubMed] [Google Scholar]
- 17.Carrington JC, Haldeman R, Dolja VV, Restrepo-Hartwig MA. 1993. Internal cleavage and trans-proteolytic activities of the VPg-proteinase (NIa) of tobacco etch potyvirus in vivo. J. Virol. 67:6995–7000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daròs JA, Schaad MC, Carrington JC. 1999. Functional analysis of the interaction between VPg-proteinase (NIa) and RNA polymerase (NIb) of tobacco etch potyvirus, using conditional and suppressor mutants. J. Virol. 73:8732–8740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haldeman-Cahill R, Daròs JA, Carrington JC. 1998. Secondary structures in the capsid protein coding sequence and 3′ nontranslated region involved in amplification of the tobacco etch virus genome. J. Virol. 72:4072–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolja VV, McBride HJ, Carrington JC. 1992. Tagging of plant potyvirus replication and movement by insertion of β-glucuronidase into the viral polyprotein. Proc. Natl. Acad. Sci. U. S. A. 89:10208–10212. 10.1073/pnas.89.21.10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández-Fernández MR, Mouriño M, Rivera J, Rodriguéz F, Plana-Durán J, García JA. 2001. Protection of rabbits against rabbit hemorrhagic disease virus by immunization with the VP60 protein expressed in plants with a potyvirus-based vector. Virology 280:283–291. 10.1006/viro.2000.0762 [DOI] [PubMed] [Google Scholar]
- 22.Sánchez F, Sáez M, Lunello P, Ponz F. 2013. Plant viral elongated nanoparticles modified for log-increases of foreign peptide immunogenicity and specific antibody detection. J. Biotechnol. 168:409–415. 10.1016/j.jbiotec.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 23.Masuta C, Yamana T, Tacahashi Y, Uyeda I, Sato M, Ueda S, Matsumura T. 2000. Development of clover yellow vein virus as an efficient, stable gene-expression system for legume species. Plant J. 23:539–546. 10.1046/j.1365-313x.2000.00795.x [DOI] [PubMed] [Google Scholar]
- 24.Beauchemin C, Bougie V, Laliberté JF. 2005. Simultaneous production of two foreign proteins from a polyvirus-based vector. Virus Res. 112:1–8. 10.1016/j.virusres.2005.03.001 [DOI] [PubMed] [Google Scholar]
- 25.Kelloniemi J, Mäkinen K, Valkonen JP. 2008. Three heterologous proteins simultaneously expressed from a chimeric potyvirus: infectivity, stability and the correlation of genome and virion lengths. Virus Res. 135:282–291. 10.1016/j.virusres.2008.04.006 [DOI] [PubMed] [Google Scholar]
- 26.Whitham SA, Yamamoto ML, Carrington JC. 1999. Selectable viruses and altered susceptibility mutants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 96:772–777. 10.1073/pnas.96.2.772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolja VV, Hong J, Keller KE, Martin RR, Peremyslov VV. 1997. Suppression of potyvirus infection by coexpressed closterovirus protein. Virology 234:243–252. 10.1006/viro.1997.8660 [DOI] [PubMed] [Google Scholar]
- 28.Dietrich C, Maiss E. 2003. Fluorescent labelling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. J. Gen. Virol. 84:2871–2876. 10.1099/vir.0.19245-0 [DOI] [PubMed] [Google Scholar]
- 29.Rajamäki ML, Kelloniemi J, Alminaite A, Kekarainen T, Rabenstein F, Valkonen JP. 2005. A novel insertion site inside the potyvirus P1 cistron allows expression of heterologous proteins and suggests some P1 functions. Virology 342:88–101. 10.1016/j.virol.2005.07.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.