ABSTRACT
Recombinant adeno-associated viral (rAAV) vectors have garnered much promise in gene therapy applications. However, widespread clinical use has been limited by transduction efficiency. Previous studies suggested that the majority of rAAV accumulates in the perinuclear region of cells, presumably unable to traffic into the nucleus. rAAV nuclear translocation remains ill-defined; therefore, we performed microscopy, genetic, and biochemical analyses in vitro in order to understand this mechanism. Lectin blockade of the nuclear pore complex (NPC) resulted in inhibition of nuclear rAAV2. Visualization of fluorescently labeled particles revealed that rAAV2 localized to importin-β-dense regions of cells in late trafficking steps. Additionally, small interfering RNA (siRNA) knockdown of importin-β partially inhibited rAAV2 nuclear translocation and inhibited transduction by 50 to 70%. Furthermore, coimmunopreciptation (co-IP) analysis revealed that capsid proteins from rAAV2 could interact with importin-β and that this interaction was sensitive to the small GTPase Ran. More importantly, mutations to key basic regions in the rAAV2 capsid severely inhibited interactions with importin-β. We tested several other serotypes and found that the extent of importin-β interaction varied, suggesting that different serotypes may utilize alternative import proteins for nuclear translocation. Co-IP and siRNA analyses were used to investigate the role of other karyopherins, and the results suggested that rAAV2 may utilize multiple import proteins for nuclear entry. Taken together, our results suggest that rAAV2 interacts with importin-β alone or in complex with other karyopherins and enters the nucleus via the NPC. These results may lend insight into the design of novel AAV vectors that have an enhanced nuclear entry capability and transduction potential.
IMPORTANCE Use of recombinant adeno-associated viral (rAAV) vectors for gene therapy applications is limited by relatively low transduction efficiency, in part due to cellular barriers that hinder successful subcellular trafficking to the nucleus, where uncoating and subsequent gene expression occur. Nuclear translocation of rAAV has been regarded as a limiting step for successful transduction but it remains ill-defined. We explored potential nuclear entry mechanisms for rAAV2 and found that rAAV2 can utilize the classical nuclear import pathway, involving the nuclear pore complex, the small GTPase Ran, and cellular karyopherins. These results could lend insight into the rational design of novel rAAV vectors that can more efficiently translocate to the nucleus, which may lead to more efficient transduction.
INTRODUCTION
Adeno-associated virus (AAV) is a Dependovirus in the family Parvoviridae that cannot replicate on its own (1). For this reason, recombinant adeno-associated viruses (rAAV) have recently garnered much attention in the field of gene therapy (reviewed in reference 2). Several serotypes have been discovered that transduce various tissue types with high efficiency (reviewed in reference 3). Despite its promise, widespread use of AAV vectors has been hindered by their relatively low transduction efficiency. Thus, much interest in the field has been directed toward the rational and combinatorial design of enhanced AAV vectors that can overcome transduction barriers at the level of receptor binding, subcellular trafficking, and transgene expression.
It has become apparent that subcellular trafficking presents multiple barriers to successful rAAV transduction, which involves movement of the vector from the host cell surface into the nucleus, where uncoating and subsequent gene expression occur (4–7). These events are mediated by interactions between host cell proteins and the three capsid proteins, VP1, VP2, and VP3 (8, 9). In the case of rAAV2, the vector binds to primary and coreceptors, such as heparan sulfate proteoglycan and αvβ5-integrins (10–12). The particle is internalized through receptor-mediated endocytosis via clathrin-coated pits or through the clathrin-independent carriers/glycophosphatidylinositol-enriched early endosomal compartments (CLIC/GEEC) pathway (13, 14). rAAV2 then traffics along the endo-lysosomal route, accumulating near the Golgi compartment and the microtubule organizing center (MTOC) (15–20). rAAV2 harbors domains buried within the capsid surface that are critical for further subcellular trafficking and nuclear entry (21–23). At some point prior to escape from the endosome, rAAV2 undergoes a conformational change, leading to the exposure of the unique N-terminal ends of VP1 and VP2; this conformation is termed VP1up (22, 23). VP1up contains a phospholipase A2 (PLA2) domain that is thought to mediate escape from the endosomal compartment (24), as well as 3 putative nuclear localization sequences (NLSs) (25, 26). Upon endosomal escape, rAAV2 enters the nucleus as an intact particle (27), where subsequent uncoating and gene expression occur. Currently, the mechanism of nuclear entry by rAAV vectors is unknown. Studies into the intracellular trafficking of rAAV2 have revealed a common pattern: while the majority of particles traffic to a perinuclear region associated with the MTOC, most of these particles remain distal to the nucleus and never translocate (4, 5, 28). These particles are eventually degraded by host proteasomes and are likely presented as antigens on the cell surface (29). If this barrier to nuclear entry could be overcome, two benefits could be achieved. First, extra vectors in the nucleus could potentially bolster transduction efficiency by contributing to greater transgene expression. Second, movement of particles from the cytosol into the nucleus could reduce the number of capsids susceptible to antigen presentation and host cell immunity. Therefore, an understanding of the mechanism of nuclear entry by AAV would create a foundation to design improved capsids that can overcome this barrier.
The majority of nuclear-targeted cellular proteins utilize the canonical nuclear import pathway (for a review, see references 30 and 31). Proteins with a NLS are bound by karyopherins, most typically a member of the importin-β family or an importin-α adapter/importin-β heterodimer. The importin proteins serve as chaperones to bring the nuclear-bound “cargo” through the nuclear pore complex (NPC), a multiprotein complex that can accommodate import of proteins up to 39 nm (32). This process is dependent on the small GTPase Ran, which provides the energy required for the translocation and mediates the dissociation of the importin-β complex once it is inside the nucleus (33–36).
Many viruses take advantage of all or parts of this pathway (for reviews, see references 37 and 38). For example, the DNA-bound protein VII of adenovirus enters the nucleus through the NPC via interactions with importin-β, importin-α, and importin-7 (39). Herpes simplex virus 1 (HSV-1) mediates nuclear entry of its nucleic acid through directly binding to the cytoplasmic filaments of the NPC in an importin-β-dependent manner (40, 41). The hepadnavirus hepatitis B virus (HBV) has been shown to bind to the NPC in a manner mediated by the phosphorylation of its capsid and the exposure of two NLSs that bind to importin-α and importin-β (42–44). Interestingly, the autonomous parvovirus minute virus of mice (MVM) has been shown to utilize an alternative nuclear entry mechanism involving direct disruption of the nuclear lamina in a caspase-dependent manner (45, 46).
Several studies have yielded insights into how AAV translocates into the nucleus; however, the mechanism of nuclear entry remains ill-defined and could differ between wild-type (wt) virions and AAV vectors. With a capsid diameter of 26 nm, AAV particles are within the size limit to traverse the NPC. The unique N termini of AAV2 VP1 and VP2 contain three basic regions (BRs) that resemble classic NLSs and can support nuclear entry of exogenous proteins (23, 47). Mutations to these basic regions inhibit nuclear entry of rAAV2 and subsequent transduction (25, 26). These results suggest that the mechanism of nuclear entry by rAAV2 involves components of the canonical nuclear import pathway, such as importin proteins and the NPC. In contrast, a study using wt AAV2 showed that inhibition of the nuclear pore by wheat germ agglutinin (WGA) or an antibody against nucleoporin p62 in purified nuclei did not prevent nuclear accumulation of intact virions (7). In a study utilizing wt AAV2 in the presence of adenovirus, inhibition of nuclear entry via the NPC through use of the calcium channel inhibitor thapsigargin did not prevent the appearance of viral genomes in the nucleus but did affect the level of nuclear accumulation and replication (48). Recently, a PDZ-binding domain was discovered on the N terminal of VP1 that was shown to be important for nuclear entry of wt AAV2 (49). While the current study was under review, a recent study revealed that AAV2 that had been acidified and then neutralized could cause nuclear envelope breakdown in permeabilized HeLa cells (50). As in vitro assays become more harmonized with respect to physiological infection, these results, taken together, suggest that AAV may utilize multiple import mechanisms to gain access to the nucleus, or that the mechanism of nuclear entry might vary for wt AAV2 and AAV vectors.
Therefore, we sought to further understand the mechanism of nuclear entry for rAAV in the absence of a helper virus in the context of gene therapy applications. Here we show that rAAV2 utilizes the host cell canonical nuclear entry pathway to enter host cell nuclei. Unlike the autonomous parvovirus MVM, rAAV does not appear to disrupt host cell nuclear lamina and is not dependent on caspase activity for nuclear entry. Rather, inhibition of the nuclear pore through microinjection of WGA partially inhibited the nuclear translocation of viral particles. Moreover, nuclear import of rAAV2 was dependent on importin-β; its import may also be facilitated by several importin-α proteins as well as importin-7. Interactions between rAAV2 capsid proteins and importin-β were mediated by VP1 and VP2, specifically, the BR domains therein. Interestingly, the extent of interaction with importin-β varied among the rAAV serotypes tested. Taken together, our results suggest that entry through the canonical pathway plays a role in rAAV2 nuclear translocation.
MATERIALS AND METHODS
Cell culture.
HeLa and HEK-293 cells were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) that was supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 100 g/ml streptomycin. For imaging experiments, HeLa cells were maintained in phenol red-free DMEM for at least 2 passages prior to plating.
Drug treatment.
HeLa cells (3 × 104 cells/well) were treated with either z-vad-fmk (200 μM; Tocris Bioscience) or dimethyl sulfoxide (DMSO) 2 h prior to infection with rAAV2. To confirm the functionality of z-vad-fmk, HeLa cells were treated with etoposide (200 μM; Tocris) following a 2-h pretreatment with z-vad-fmk or DMSO. Prevention of apoptosis was visually inspected 48 h after etoposide treatment and quantitatively confirmed through use of 7-amino-actinomycin D (Life Technologies) via flow cytometry.
PCR site-directed mutagenesis.
Capsid mutations were generated in the pACG2 backbone as previously described (27), with minor modifications. PCR was performed with the QuikChange Lightning site-directed mutagenesis kit (Stratagene). PCR products were digested with DpnI and transformed into DH10B bacteria, which were selected on ampicillin-containing agar plates. Single colonies were picked and grown in liquid cultures overnight, and plasmids were isolated by column purification (Qiagen). Mutations were verified by sequencing of the plasmids with the Eton Bioscience automated DNA sequencing facility.
Virus production.
Virus was produced in HEK-293 cells as previously described (51). Briefly, polyethylenimine max was used for the triple transfection of the pXR2 cap and rep plasmids, the pXX6-80 helper plasmid, and a TR-luciferase reporter plasmid containing the firefly luciferase (Luc) transgene flanked by inverted terminal repeats. Cells were harvested between 48 and 72 h posttransfection, and virus was purified by iodixanol gradient centrifugation followed by ion-exchange chromatography. After identifying peak fractions by dot blot hybridization, virus was dialyzed into phosphate-buffered saline (PBS) containing 5% sorbitol, MgCl2 (1 mM), and CaCl2 (0.5 mM). Titers were calculated by quantitative PCR (qPCR) according to established procedures (52) using a LightCycler 480 instrument with Sybr green (Roche) and primers designed against the Luc transgene: 5′-AAA AGC ACT CTG ATT GAC AAA TAC-3′ (forward) and 5′-CCT TCG CTT CAA AAA ATG GAA C-3′ (reverse). Conditions used for the reaction were as follows: 1 cycle at 95°C for 10 min; 45 cycles at 95°C for 10 s, 62°C for 10 s, and 72°C for 10 s for acquisition; 1 cycle at 95°C for 30 s, 65°C for 1 min, and 99°C for melting curve analysis. For Cy5-labeled virus, purified rAAV2, VP3-only particles, and BR mutants were labeled with Cy5 dye (GE Amersham) as previously described (28).
Confocal immunofluorescence microscopy.
Similar to what we previously described (26), HeLa cells (3 × 104 cells/well) were plated onto poly-l-lysine-coated 12-mm glass coverslips (no. 1.5) for 16 h before infection. At 30 min before infection, HEPES was added to the medium (10 mM, total concentration). Cy5-labeled recombinant virions were added to cell medium (100,000 vector genome copies [vg]/cell) for 1 h. No virus was added to control wells. Medium was then replaced with prewarmed medium, and cells were placed at 37°C. At the indicated time points, cells were washed three times with PBS and then fixed with 2% paraformaldehyde for 15 min at room temperature. The cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min at room temperature. Following four washes with PBS, the permeabilized cells were blocked with immunofluorescence buffer (IFB; 20 mM Tris [pH 7.5], 137 mM NaCl, 3 mM KCl, 1.5 mM MgCl2, 5 mg/ml bovine serum albumin, 0.05% Tween) for 30 min at room temperature. Where indicated, the cells were incubated with importin-β primary antibody diluted in IFB overnight at 4°C. The cells were then incubated in secondary antibody diluted 1:5,000 in IFB (anti-mouse Dylight 488 or anti-rabbit Dylight 488; Abcam) for 1 h at 37°C. After six washes in PBS, coverslips were mounted cell side down onto glass slides with mounting medium (Prolong Antifade gold with DAPI [4′,6′-diamidino-2-phenylindole]; Molecular Probes). Images were captured on a Zeiss 710 upright laser scanning confocal microscope. Three-dimensional (3D) rendering and image processing of confocal z-stack images were performed using AutoQuant and Imaris software, respectively (UNC Microscopy Services Laboratory).
Microinjections with pulse infection.
HeLa cells (7.5 × 104) were plated on fibronectin-coated 35-mm glass-bottom microwell dishes with a number 1.5 coverglass (MatTek). Cells were microinjected using the FemptoJet system with Femptotips (Eppendorf). For virus studies, 30 min before infection cells were cooled to 4°C. Cy5-labeled AAV2 (100,000 vg/cell) was added to each dish for 1 h. Where indicated, microinjection mixes (10 μl) included fluorescein isothiocyanate (FITC)-dextran (0.8 mg/ml; Life Technologies), wheat germ agglutinin (2 mg/ml; Calbiochem), and phosphate-buffered saline. For rhodamine B-labeled, NLS-conjugated bovine serum albumin (BSA) comicroinjections, mixtures included rhodamine B-BSA (1.33 mg/ml; Sigma). Following microinjection, medium was replaced with fresh, prewarmed medium. At 2 h postmicroinjection, cells were fixed and processed for microscopy as described above.
Transduction assays.
For small interfering RNA (siRNA) studies, HeLa cells were plated in 24-well plates 18 h prior to transfection at densities that approximated 30% confluence (3 × 104 cells/well) in antibiotic-free DMEM. siRNA (total concentration, 25 nM; ON-TargetPlus SmartPool; Dharmacon) to KPNB1 (importin-β), KPNA2 (importin-α1), KPNA4 (importin-α3), KPNA1 (importin-α5), and IPO7 (importin-7) was utilized in combination with 1 μl DharmaFECT 1 (Dharmacon) as per the manufacturer's instructions. At 24 h posttransfection (for importin-β) or 48 h posttransfection (for importin-α1, -α3, -α5, and importin-7), cells were infected with purified rAAV2 at the designated number of vector genomes per cell and typically harvested after 24 h unless otherwise noted. For drug studies, 5 × 104 HeLa cells were plated in 24-well plates 18 h prior to drug treatment. For treatment with z-vad-fmk (Tocris Bioscience; 200 nM in DMSO), cells were treated 2 h prior to infection; for treatment with chloroquine (Sigma; 100 μM in PBS), cells were treated at the same time as infection. Luciferase activity was measured in accordance with the manufacturer's instructions (Promega) with a PerkinElmer 1420 Victor3 automated plate reader. Error bars in the figures represent standard deviations of samples scored in triplicate. Graphs are representative of data sets from at least three independent assays.
Coimmunoprecipitation.
Protein G-Dynabead-antibody complexes were prepared by washing the beads 2 times in PBS and resuspending them in PBS with 0.02% Tween 20 and antibody (30 μl of beads and 3 μg of antibody per reaction mixture). The Dynabeads were rocked at 4°C overnight. For studies of rAAV2 and mutants, HeLa cells (1 × 107 per coimmunoprecipitation [co-IP] mixture) were washed 2 times with ice-cold PBS and harvested in lysis buffer (Tris [pH 7.4], 50 mM; MgCl2, 10 mM; NaCl, 150 mM; Triton X-100, 0.1%; deoxycholate, 0.1%; protease inhibitor cocktail [Pierce], 1×). Lysate was incubated for 30 min at 4°C and clarified by centrifugation. Lysate was precleared by incubating with beads for 30 min at 4°C. rAAV2 or mutants (2.5 × 1011) were boiled in lysis buffer for 10 min. Lysate was then added to virus and incubated for 2 h at 4°C. A 10% aliquot of each co-IP reaction mixture was collected to serve as the input amount. Dynabeads were washed 2 times with lysis buffer and then added to the virus-lysate mix for 2 h at 4°C. Dynabeads were then washed 3 times with lysis buffer and transferred to a new tube. Eluates were collected by resuspending the Dynabeads in 50 μl 2× NuPage lithium dodecyl sulfate sample buffer (Life Technologies), and samples were boiled for 10 min. Samples were then subjected to SDS-PAGE and immunoblot analysis using the B1 antibody (a kind gift from Jürgen A. Kleinschmidt). For RanQ69L studies, HEK-293 cells were used. Cells were plated 24 h prior to transfection (2.5 × 104 cells per 10-cm plate, 2 plates per co-IP). Polyethylenimine Max was used to transfect pmCherry-C1-RanQ69L (a kind gift from Jay Brenman) or pmCherry-C1. At 40 h posttransfection, cells were harvested in lysis buffer and processed as described above. One milligram of total lysate was used for each co-IP reaction.
Statistics.
All statistical analyses were performed using the Student t test. Statistical significance was defined as a P value of <0.05.
RESULTS
Previous studies have shown that the majority of rAAV2 virions traffic to the perinuclear region of cells but never enter the nucleus. These virions likely become degraded by proteases or are subjected to proteasomal degredation and antigen presentation by host cells (29). Current opinion suggests that if the barrier to nuclear entry were to be overcome, more viruses could translocate to the nucleus for subsequent uncoating and transgene expression. To date, the mechanism of nuclear entry by rAAV is largely unknown. However, a few reports have suggested potential mechanisms by which rAAV enters the nucleus, including entry via the NPC through the classic nuclear entry pathway (25, 26), entry through NPC-independent mechanisms (7, 48), and direct lamina disruption, like that of the autonomous parvoviruses (46). Focusing our studies on AAV in the context of gene therapy vectors, we aimed to determine the mechanism for nuclear entry by rAAV2 in the absence of a helper virus.
Analysis of host cell nuclear lamina during rAAV2 infection.
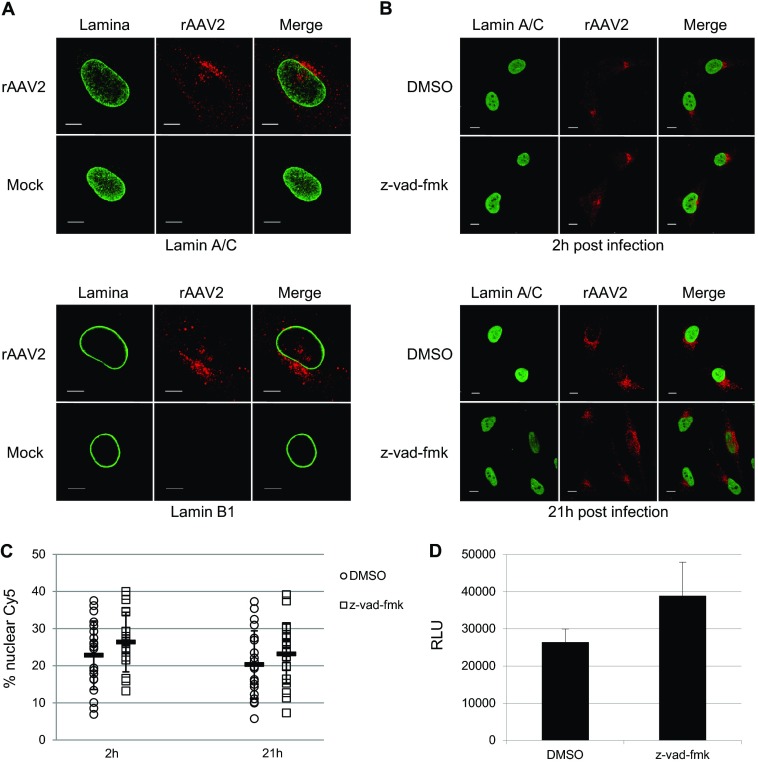
Previous studies have described a unique nuclear entry mechanism for the autonomous parvovirus MVM, whereby MVM enters the nucleus through physical disruption of the nuclear lamina in a caspase-dependent mechanism (46). Since AAV is a parvovirus, we sought to determine whether rAAV2 utilized a similar mechanism. To investigate the integrity of the nuclear lamina upon rAAV2 infection, we utilized confocal microscopy to visualize Lamin A/C and Lamin B1 during infection with Cy5-labeled rAAV2 (Fig. 1A). Immunofluorescence analysis revealed no detectable changes in the appearance of Lamin A/C or Lamin B1 2 h postinfection, a time that has been reported to capture 50% of total viral nuclear entry (28). We next utilized quantitative imaging analysis to measure the percentage of nuclear Cy5 fluorescence within HeLa cells treated with the caspase inhibitor z-vad-fmk. Our lab has established that confocal analysis of Cy5-labeled rAAV2 particles can be used to quantify the percentage of nuclear rAAV2 (28), and our quantification of the percentage of Cy5 fluorescence within nuclei of vehicle-treated cells was consistent with what has been previously reported (53, 54). Treatment with z-vad-fmk, which was shown to inhibit lamina disruption and nuclear entry of MVM, resulted in no change in nuclear Cy5 fluorescence compared with vehicle-treated cells (Fig. 1B and C). In fact, pretreatment of HeLa cells with z-vad-fmk followed by infection with rAAV2-Luc led to a small but nonsignificant increase in transduction compared to the vehicle control (Fig. 1D). We confirmed the functionality of the z-vad-fmk used in these studies through 7-amino-actinomycin D analysis of apoptosis prevention in etoposide-treated cells (data not shown). Taken together, these results suggest that, unlike the autonomous parvovirus MVM, rAAV2 does not overtly disrupt host cell nuclear lamina nor rely on caspase activation for nuclear entry or transduction.
FIG 1.
Nuclear lamina integrity and caspase involvement during rAAV2 infection. (A) Immunofluorescence of Cy5-labeled rAAV2 and intact nuclear lamina 2 h postinfection. Capsids (red) are shown juxtaposed to the nuclear membrane, composed of Lamin A/C (top, green) and Lamin B1 (bottom, green). Bar, 10 μm. (B) Nuclear entry of Cy5-labeled rAAV2 in cells treated with vehicle or z-vad-fmk. HeLa cells were treated with z-vad-fmk or vehicle (DMSO) 1 h prior to and throughout infection. Cells were fixed at 2 h and 21 h postinfection. Red, Cy5-labeled rAAV2; green, Lamin A/C. Bar, 10 μm. (C) Quantification of nuclear Cy5 fluorescence in z-vad-fmk- or vehicle-treated cells. Confocal z-stack images for 20 cells per treatment group were acquired. Images were rendered in 3D, and isosurfaces were created for nuclei and Cy5 fluorescence within each cell. The percent Cy5 fluorescence within nuclei compared to total Cy5 fluorescence within the cell was calculated. (D) Luciferase assay of transduction in HeLa cells. Cells were treated with either z-vad fmk or vehicle 1 h prior to infection and then infected with rAAV2-CBA-Luc (1,000 vg/cell). Luciferase activity was measured 24 h postinfection. Error bars represent standard deviations from a representative experiment performed in triplicate.
rAAV2 can utilize the nuclear pore complex to enter the nucleus.
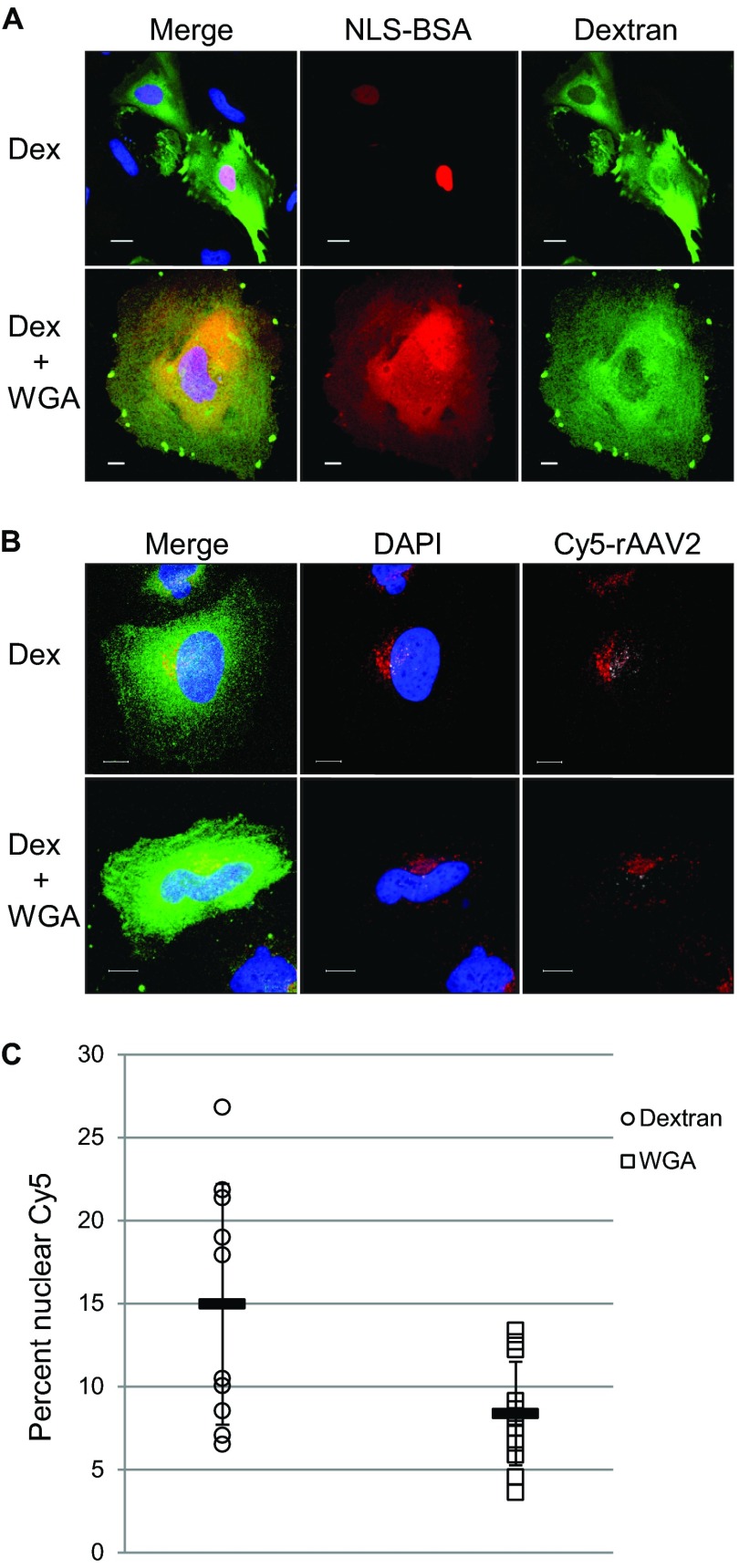
Many viruses utilize the nuclear pore complex to traffic intact particles, viral proteins, or DNA into the nucleus (37, 38). The nuclear pore has been shown to accommodate proteins of sizes up to 39 nm in diameter (32). Because the diameter of the AAV capsid is approximately 26 nm and it has been shown that rAAV2 can enter the nucleus as an intact particle (27), we investigated whether rAAV2 could enter the nucleus by using the NPC. A common method to determine if the NPC is necessary to facilitate nuclear entry is to physically block the outer pore with the lectin WGA (55). WGA binds to O-linked N-acetylglucosamine residues on NPC proteins, thereby blocking any incoming cargo. We confirmed this by microinjecting HeLa cells with rhodamineB-labeled, NLS-conjugated BSA with either FITC-labeled dextran or dextran and WGA (Fig. 2A). In cells that were injected with dextran alone, the NLS-BSA could be visualized inside the nucleus 2 h postmicroinjection. However, in cells that were injected with dextran and WGA, the NLS-BSA was mostly excluded from the nucleus. We then applied this technique to HeLa cells subjected to viral infection with Cy5-labeled rAAV2. We prebound HeLa cells with Cy5-labeled rAAV2 and performed microinjections with either dextran alone or dextran and WGA. At 2 h postmicroinjection, we processed the cells for confocal microscopy to assess nuclear localization through visualization of the Cy5-labeled particles. In cells that were microinjected with dextran, viral particles could be seen inside of the nucleus (Fig. 2B, top). However, in cells that were microinjected with WGA, fewer viral particles could be seen inside the nucleus, suggesting that nuclear entry was limited (Fig. 2B, bottom). We quantified the percentage of nuclear Cy5 fluorescence in microinjected cells and found an average of 15% nuclear Cy5 fluorescence in dextran-injected cells, while cells injected with dextran plus WGA showed an average of 8% nuclear Cy5 fluorescence (Fig. 3C). Taken together, these results suggest that rAAV2 can utilize the NPC to enter the nucleus.
FIG 2.
rAAV2 can enter HeLa cell nuclei through the NPC. (A) Rhodamine-labeled, NLS-conjugated BSA (red) was comicroinjected with dextran (Dex) alone (top, green) or dextran and WGA (bottom, green) into HeLa cells. Cells were fixed and imaged 2 h after microinjection to assess NLS-BSA localization. Bar, 10 μm. (B) Cells were cooled to 4°C and infected with Cy5-labeled rAAV2 (red). One hour later, cells were microinjected with dextran (green, top) or dextran plus WGA (green, bottom). Immunofluorescence microscopy was used to assess Cy5-rAAV2 nuclear entry 2 h postinfection. Shown are maximum intensity projections of 3D-reconstructed cells, where red depicts Cy5 fluorescence within the cytoplasm and white depicts nuclear Cy5 fluorescence. Bar, 10 μm. (C) Quantification of microinjected cells. Confocal z-stack images of microinjected cells were rendered in 3D, and isosurfaces were created for nuclei and Cy5 fluorescence within the cell. The percent Cy5 fluorescence within nuclei compared to total Cy5 fluorescence within the cell was calculated (n = 10 for dextran-injected cells; n = 12 for WGA-injected cells).
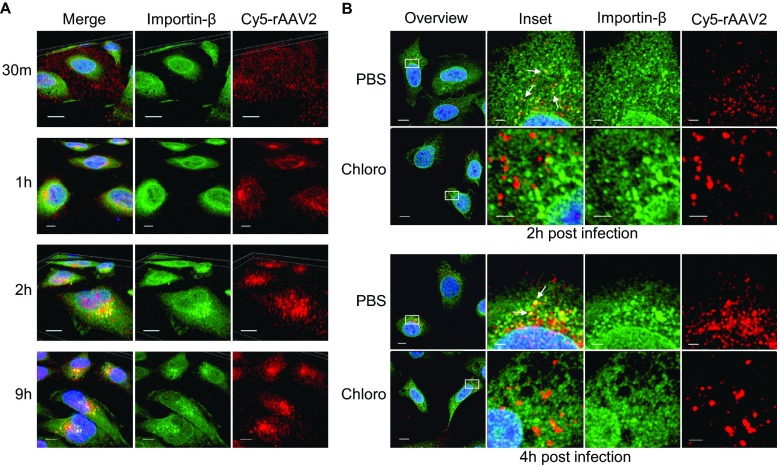
FIG 3.
rAAV2 codistributes with importin-β in later trafficking steps. (A) HeLa cells were pulse-infected with Cy5-labeled rAAV2 for the indicated times. Confocal z-stack images were acquired and rendered in three dimensions to assess the localization of rAAV2 (red) and importin-β (green) in relation to the nucleus (blue). Bar, 10 μm. (B) HeLa cells were treated with chloroquine (100 uM) or vehicle (PBS) 2 h prior to and throughout the duration of infection. Cells were pulse-infected with Cy5-rAAV2 for the indicated times. Confocal images were acquired in the z-plane that harbored the center of the nucleus of the to assess the 2D localization of Cy5-rAAV2 (red) and importin-β (green). Arrows indicate regions of extremely similar spatial distributions. Bar, 10 μm (field) or 2 μm (zoom).
rAAV2 shares spatial distribution with importin-β during later trafficking events.
Importin-β plays a key role in canonical nuclear entry by either directly binding to NLS-harboring cargo or interacting with cargo through an adapter importin-α protein. Importin-β has been shown to be involved in nuclear import for several viral infectious pathways, including facilitating the binding of HSV-1 capsids (41), HBV core particles (42), HIV-1 preintegration complexes (56), and influenza A virus nucleoprotein (57) to host nuclei. Importantly, importin-β has been shown to interact with a DNA-bound protein of the AAV helper virus adenovirus (39). Therefore, we aimed to determine if rAAV2 localized near cellular regions with a high importin-β concentration during the course of infection. We utilized Cy5-labeled rAAV2 and immunofluorescence confocal microscopy to acquire z-stack images of HeLa cells infected with rAAV2 at various time points. These images were rendered in three dimensions by using volume imaging software. During the course of infection, rAAV2 traffics from the cell periphery to the MTOC and nearby cellular compartments before translocating to the nucleus (15, 18, 20). As expected, at 30 min postinfection, when the majority of rAAV2 was detected dispersed throughout the cytoplasm, there appeared to be little to no detectable codistribution between rAAV2 and importin-β (Fig. 3A). However, as early as 1 h postinfection and through 9 h postinfection, Cy5-rAAV2 was localized in cellular regions with a high importin-β concentration. These results suggested that rAAV2 and importin-β interact once AAV2 has accumulated near the MTOC.
Previous reports have suggested that endosomal escape precedes nuclear entry of rAAV2 (16, 23). We therefore investigated whether the codistribution of rAAV2 and importin-β was dependent on later trafficking events that could involve escape from the endosome; we acquired 2D images of cells in the z-plane harboring the center of the nucleus to represent the center of the cell. Endosomal escape has been shown to be inhibited by pharmacological agents that block vesicle acidification (16, 29). We therefore utilized chloroquine, a small molecule that blocks the acidification of endosomes, to inhibit later trafficking events and endosomal escape of rAAV2. Treatment of cells with chloroquine resulted in the loss of codistribution between rAAV2 and importin-β (Fig. 3B). These results suggested that rAAV2 and importin-β interact in later trafficking steps, presumably once rAAV2 has exposed the unique N terminal of VP1 and escaped the endosome.
rAAV2 interacts with importin-β in a Ran-sensitive manner.
Because we observed a codistribution pattern between Cy5-rAAV2 and importin-β in later viral trafficking steps, we sought to determine if rAAV2 and importin-β form a physical interaction. We hypothesized that because rAAV2 contains putative NLSs on VP1 and VP2, an interaction could occur in late trafficking events, perhaps once rAAV2 has undergone conformational changes to expose these regions of the capsid. Previous studies have shown that classical importin-β/cargo complexes are mediated through an NLS on the cargo and can be dissociated by the small GTPase Ran in its GTP-bound form (35, 36). Therefore, we performed co-IP experiments with dissociated capsid proteins (to expose VP1 and VP2) and HEK-293 lysate transfected with a permanently GTP-bound form of Ran (Q69L) or a control plasmid (Fig. 4). rAAV2 capsid proteins were able to co-IP with importin-β; however, this interaction was completely inhibited in the presence of RanQ69L. This result suggested that rAAV2 capsid proteins form a specific interaction with importin-β that can be disrupted by RanGTP.
FIG 4.
rAAV2 capsid proteins form an interaction with importin-β that can be dissociated by RanGTP. rAAV2 particles were dissociated by heat and incubated with HEK-293 cell lysate previously transfected with control plasmid or RanQ69L. Co-IPs were performed using Dynabeads and an anti-importin-β antibody, followed by immunblotting with B1. rAAV2 capsid protein marker, lanes 1 and 4; control, lanes 2 and 5; RanQ69L, lanes 3 and 6. Identities of relevant bands are highlighted in bold.
Knockdown of importin-β inhibits nuclear entry and transduction of rAAV2.
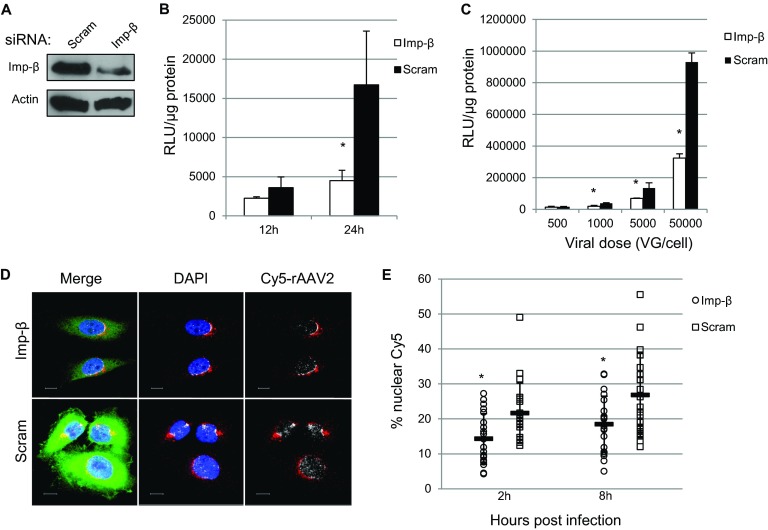
Because we observed similar spatial distributions between rAAV2 and importin-β, we next investigated whether importin-β played a physiological role in rAAV2 trafficking and transduction. We utilized an siRNA approach to transiently silence importin-β expression in HeLa cells. Previous studies have shown that long-term inhibition of importin-β expression can disrupt cellular homeostasis, since importin-β plays a vital role in the nuclear import of essential nuclear proteins. Therefore, we chose to examine the effects of importin-β knockdown on rAAV2 transduction within the first 24 h post-siRNA treatment, and we found approximately 50% knockdown of importin-β at the time of rAAV2 infection (Fig. 4A). While we cannot rule out inhibition of rAAV-specific transcription factor import by importin-β knockdown, we verified that global gene transcription was not altered by utilizing qRT-PCR to measure mRNA levels of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and determined that these levels were similar between results with importin-β knockdown and treatment with a scrambled siRNA (data not shown). To assess effects on transduction, we utilized rAAV2 carrying a CBA-luciferase transgene. rAAV2 transduction was inhibited when luciferase activity was measured at either early (12 h) or late (24 h) time points, with the greatest inhibition (approximately 70%) at 24 h postinfection (Fig. 5B). We next assessed whether this inhibition was maintained at various viral doses. Consistent with our initial results, knockdown of importin-β inhibited rAAV2 transduction to levels between 50 and 70% at all vector doses higher than 500 vg/cell (Fig. 5C). Finally, we knocked down importin-β and investigated changes in the nuclear localization of Cy5-labeled rAAV2. Consistent with the changes in transduction, siRNA knockdown of importin-β inhibited Cy5- rAAV2 nuclear localization (Fig. 5D). To quantify these observations, we determined the percentages of nuclear Cy5 fluorescence in cells treated with siRNA to importin-β or a scramble control. In control cells, nuclear Cy5 fluorescence increased from 21% at 2 h postinfection to 27% at 8 h postinfection. Nuclear Cy5 fluorescence in importin-β knockdown cells remained lower, increasing from 14% to 18% at 8 h postinfection. Complete inhibition of nuclear entry and transduction was not observed, which reflects either the activity of the remaining importin-β in knockdown cells or the utilization of alternative, importin-β-independent nuclear entry pathways by rAAV2. These results do suggest that importin-β plays a role in rAAV2 transduction as a mediator of nuclear entry.
FIG 5.
Importin-β is important for rAAV2 nuclear entry and transduction. (A) HeLa cells were treated with siRNA to importin-β or a scrambled siRNA control. Cell lysate was collected 24 h posttransfection and immunoblotted for importin-β levels. (B) HeLa cells were treated with siRNA to KPNB1 (importin-β) and infected with rAAV2-CBA-Luc (1,000 vg/cell) at the indicated times. Luciferase activity was measured 12 h and 24 h postinfection. (C) HeLa cells were treated with siRNA to KPNB1 and infected with rAAV2-luc at the indicated doses. Luciferase activity was measured 24 h postinfection. (D) HeLa cells were treated with either scrambled siRNA or siRNA to KPNB1 for 24 h. Cells were pulse-infected with Cy5-labeled rAAV2. At 8 h postinfection, cells were fixed, and confocal images were acquired to assess nuclear entry of virions. Red, Cy5 fluorescence within cytoplasm; white, nuclear Cy5 fluorescence; DAPI, nucleus; green, importin-β. Bar, 10 μm. (E) HeLa cells were treated with siRNA to KPNB1 or a scrambled siRNA and pulse-infected with Cy5-rAAV2 for either 2 or 8 h. Confocal z-stack images for at least 20 cells per treatment group were acquired. Images were rendered in 3D, and isosurfaces were created for nuclei and Cy5 fluorescence within each cell. The percent Cy5 fluorescence within nuclei compared to total Cy5 fluorescence within the cell was calculated.
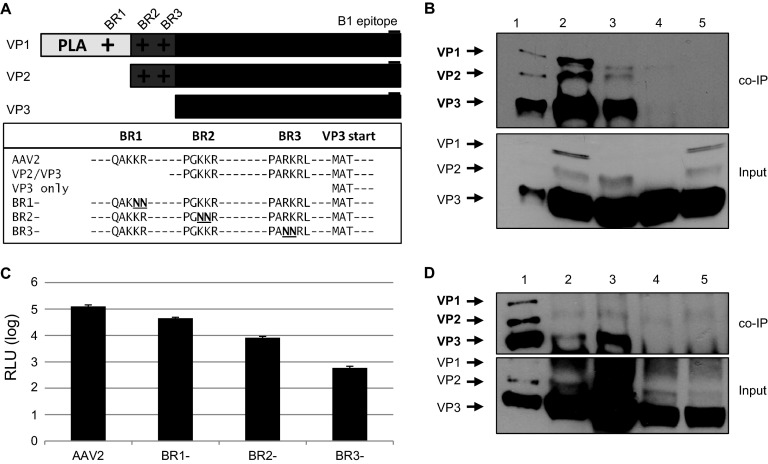
Determination of unique regions of the AAV2 capsid that are important for the interaction with importin-β.
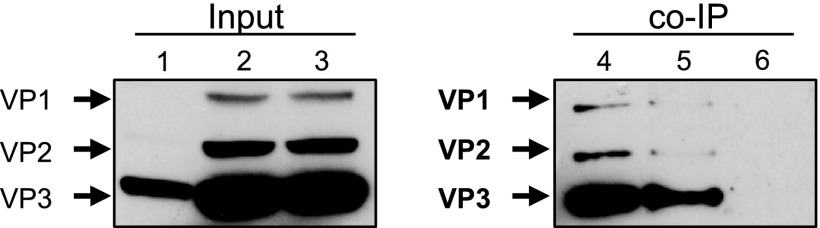
Since we were able to detect a functional role for importin-β in the rAAV2 infectious pathway, we sought to determine which region of the AAV capsid mediates this interaction. Importin-β binds to nucleus-bound cargo via classical NLSs (cNLS) but has also been shown to interact with proteins with nonconventional NLSs. Because the unique N-terminal regions of VP1 and VP2 contain basic stretches of amino acids that resemble cNLSs (Fig. 6A), we hypothesized that one or both of these capsid proteins mediate the interaction with importin-β. To determine which capsid proteins were important for binding to importin-β, we performed co-IPs using dissociated capsids from particles consisting of all three capsid proteins, particles with only VP2/VP3, and VP3-only particles, which were produced as previously described (58). Dissociated rAAV2 particles with all three capsid proteins or VP2/VP3-only particles were able to co-IP with importin-β; however, VP3-only particles resulted in limited to nondetectable co-IP (Fig. 6B). Co-IP utilizing an isotype control antibody also yielded nondetectable product. Interestingly, while co-IP seemed to only require the presence of VP1 and/or VP2, we were able to detect capsid protein VP3 in the IP product. We believe this was due to incomplete capsid dissociation or oligomerization between NLS-containing capsid proteins and VP3, which may occur after capsid dissociation.
FIG 6.
BR domains on VP1/VP2 mediate interactions with importin-beta. (A) Schematic of basic regions in rAAV2 VPs and mutations made to ablate NLS function. The B1 epitope is highlighted. (B) rAAV2 (lane 2), VP2/VP3 (lane 3), and VP3-only (lane 4) particles were dissociated by heat and incubated with HeLa cell lysate. Co-IPs were performed using Dynabeads and an anti-importin-β antibody, followed by immunblotting with B1. Lane 1, capsid protein marker; lane 5, mouse IgG2a isotype control. (C) HeLa cells were infected with rAAV2-CBA-Luc and mutants (1,000 vg/cell). Luciferase activity was measured 24 h postinfection. Error bars represent standard deviations from three independent samples. (D) Co-IPs were performed with rAAV2 (lane 2), BR1− (lane 3), BR2− (lane 4), and BR3− (lane 5), followed by immunoblotting with B1. Lane 1, capsid protein marker. Relevant band identities are highlighted in bold.
AAV2 harbors 4 putative NLSs: BR1, located exclusively in VP1; BR2 and BR3, located in VP1 and VP2; and BR4, located in all 3 capsid proteins (Fig. 6A). The 4 BR domains have been studied, and their implications in AAV infectivity have been characterized in detail (23, 25, 26, 47). Mutations to BR1, BR2, and BR3 have been shown to hinder rAAV2 transduction, with mutations to BR2 and BR3 having the greatest effect on transduction. Since mutations to BR4 result in defects in capsid assembly (25), we only investigated mutations to BR1, BR2, and BR3. Therefore, to assess the contributions of each BR domain in the interaction with importin-β, we produced particles with mutations in each of these BR domains. Consistent with the results presented by Grieger et al. and Johnson et al. (25, 26), mutations to BR1 alone had a modest effect on transduction, while mutations to BR2 or BR3 hindered transduction by over 10-fold in both HeLa cells and HEK-293 cells (Fig. 6C and data not shown).
We next wanted to determine whether the transduction profiles of these mutants are consistent with their capabilities to interact with importin-β. Co-IP analysis revealed that while the capsid proteins of rAAV2 interacted with importin-β, mutations to BR2 and BR3 limited this interaction, while mutations to BR1 had no effect on co-IP (Fig. 6D). The higher-density band seen in the BR1 co-IP was likely due to higher capsid protein input. Taken together, our data support that the BR domains, especially BR2 and BR3, are necessary for transduction as they mediate interactions with importin-β.
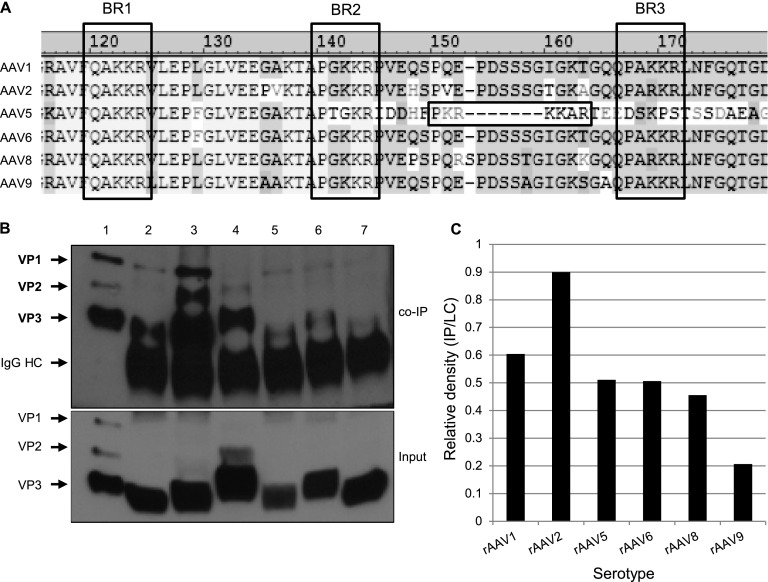
Interaction with importin-β is varied among rAAV serotypes.
The majority of information about the infectious pathway of AAV has been determined for AAV2, with limited understanding of the subcellular trafficking of other serotypes. Comparison of VP1up regions of several common AAV serotypes revealed that most serotypes harbor similar, if not identical, BR domains (Fig. 7A). The only exception is AAV5, which has a bipartite-like NLS between BR2 and BR3 rather than two monopartite-like NLSs. Because these serotypes share similar BR domains, we investigated whether they interact with importin-β. Co-IP analysis between importin-β and dissociated capsid proteins from each serotype revealed that all viral particles tested interacted with importin-β, but to varying extents (Fig. 7B). We measured the co-IP results by quantifying the ratio of capsid protein product in the co-IP and the capsid protein applied to the lysate. We found that rAAV2 had the most robust interaction with importin-β, followed by rAAV1. rAAV5, rAAV6, and rAAV8 were similarly able to interact with importin-β, although to a lesser extent than rAAV2 and rAAV1. Finally, we detected interaction, but to a lesser extent, between rAAV9 and importin-β. Interestingly, while rAAV8 and rAAV9 vectors do not typically transduce HeLa cells, we were still able to detect interactions between these capsid proteins and importin-β. Since our co-IP assay utilizes HeLa cell lysate and denatured capsid proteins, our experimental setup bypasses cellular barriers such as receptor-mediated endocytosis and endosomal escape. Thus, it is possible that the use of import proteins such as importin-β is a shared feature of rAAV vectors but occurs downstream of initial cellular entry and subcellular trafficking. Our results suggest that different AAV serotypes utilize alternative import proteins in addition to importin-β for nuclear entry.
FIG 7.
Interaction with importin-β varies among rAAV serotypes. (A) Alignment of the N-terminal regions of VP1 in AAV serotypes. BR domains are outlined by dashed lines, and a putative bipartite NLS of AAV5 is highlighted with solid lines. (B) rAAV1 (lane 2), -2 (lane 3), -5 (lane 4), -6 (lane 5), -8 (lane 6), and -9 (lane 7) particles were dissociated by heat and incubated with HeLa cell lysate. Co-IP was performed using Dynabeads and an anti-importin-β antibody, followed by immunblotting with B1. Lane 1, capsid protein marker. Relevant band identities are highlighted in bold. (C) Co-IPs were quantified by densitometric calculation of the co-IP product versus initial input.
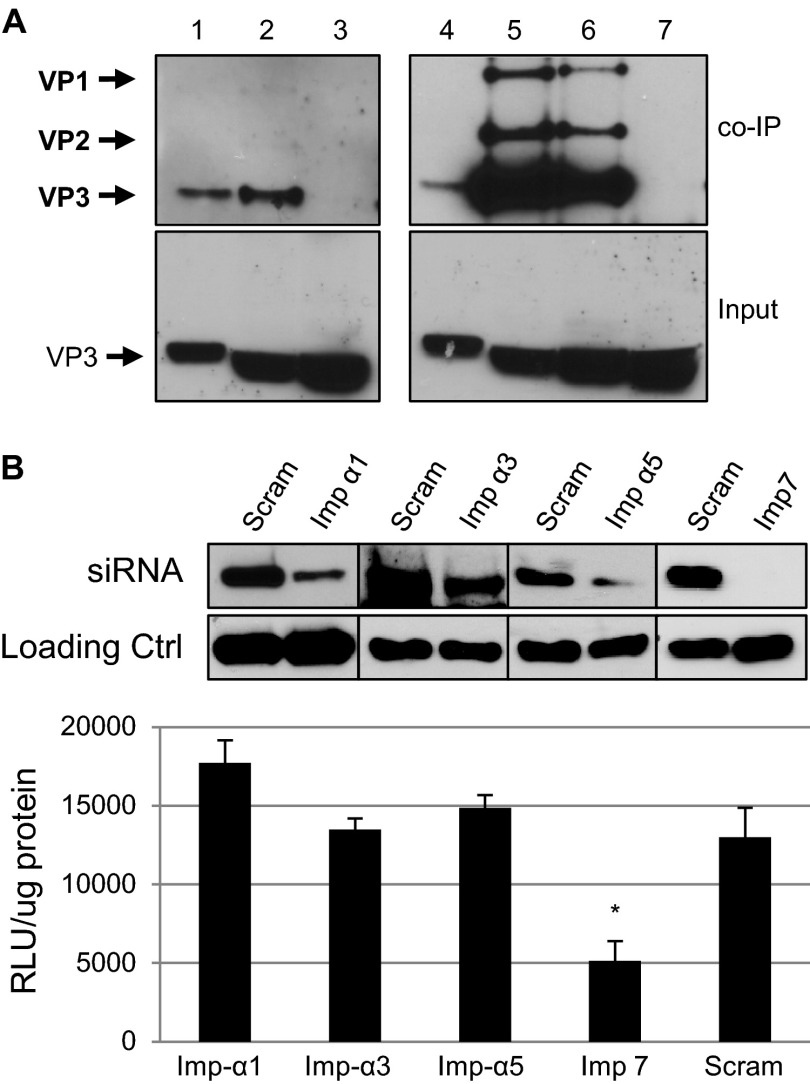
Analysis of the role of other import proteins in rAAV2 nuclear localization.
The import protein superfamily consists of the importin-β proteins, importin-α adapters, transportins, and several other importins, such as importin-7 (59, 60). Importin-α proteins link nuclear-bound cargo to importin-β through an importin-β binding domain. Importin-7 and transportins can mediate nuclear import independently of importin-β (61–63). It has also been shown that importin-7 can form a cooperative dimer with importin-β (62). Studies have shown that importin-α proteins recognize both mono- and bipartite NLSs and can have redundant functions in host cells (64–67). Because we established an interaction between rAAV2 and importin-β, we investigated if this interaction is mediated by the importin-α adapter proteins. Additionally, we wanted to identify any potential interactions with the alternative import proteins importin-7, transportin 1, or transportin 2. Coimmunoprecipitation analysis revealed strong interactions between dissociated rAAV2 capsid proteins and importin-α1, importin-α3, and importin-α5 (Fig. 8A). Under these conditions, we did not detect any coimmunoprecipitation between rAAV2 capsid proteins and importin-7, transportin 1, or transportin 2 (data not shown). Interestingly, siRNA knockdown of importin-α1, importin-α3, or importin-α5 had no effect on rAAV2 transduction 24 h postinfection (Fig. 8B). Similarly, knockdown of all three importin proteins, to eliminate any compensatory function among these proteins, also resulted in no change in transduction of rAAV2 (data not shown). The importin-α superfamily consists of at least 7 known members; therefore, it is possible that other importin-α proteins that were not identified in this study may also mediate nuclear import of rAAV2. In contrast to our other coimmunoprecipitation data, knockdown of importin-7 reduced transduction by approximately 60% (Fig. 8B). Taken together, our results suggest that multiple importin-α proteins play a role in the import of AAV2 and may be able to compensate for the depletion of specific importin-α proteins. Additionally, importin-7 appears to be important for transduction, but it may only weakly interact with rAAV2.
FIG 8.
Analysis of additional importin proteins in rAAV2 nuclear entry. (A) rAAV2 particles were dissociated by heat and incubated with HeLa cell lysate. Co-IPs were performed using antibodies to importin-α1, -3, or -5. Immunoblot assays were performed using the B1 antibody. Lanes 1 and 4, capsid protein marker; lane 2, importin-α1 co-IP; lane 3, mouse IgG1 isotype control; lane 5, importin-α3 co-IP; lane 6, importin-α5 co-IP; lane 7, rabbit polyclonal antibody isotype control. (B) Relevant band intensities are highlighted in bold. HeLa cells were treated with scrambled siRNA (blot, lanes 1, 3, 5, and 7) or to KPNA2 (importin-α1, lane 2), KPNA4 (importin-α3, lane 4), KPNA1 (importin-α5, lane 6), or IPO7 (importin-7, lane 8). Luciferase activity was measured 24 h postinfection. Error bars represent standard deviations from a representative experiment performed in triplicate.
DISCUSSION
Mammalian viruses have been shown to utilize components of the classical nuclear import pathway, including the NPC, nucleoporins, and various import proteins. HSV-1 and adenovirus both directly bind to the cytoplasmic side of the NPC via nucleoporins (40, 41, 68–71). The adenoviral core protein protein VII has been shown to bind to several import proteins, including importin-α, importin-β, importin-7, and transportin, which are thought to mediate entry of the nucleoprotein through the NPC (39). Previous studies have suggested that some viruses, such as HBV and simian virus 40, may enter the nucleus intact. Two NLSs on the core protein of HBV become exposed and mediate movement into the NPC in an importin-α- and importin-β-dependent manner (42–44, 72). Our data suggesting that rAAV2 utilizes components of the classical nuclear import machinery to enter the nucleus are consistent with the mechanism of how other viruses achieve nuclear entry. While the current study is limited to characterization and mechanistic insights utilizing an established cell line, it provides a foundation for further studies in primary cells and in vivo. A growing interest in the AAV vector field has focused on understanding vector trafficking through host cells, with the notion that understanding the viral trafficking pathway can lead to the design of novel vectors that can overcome cellular barriers to transduction. Our study provides insight into a rate-limiting step of AAV vector transduction, nuclear entry, and thus the foundation for novel vector design aimed to expedite this step in subcellular trafficking.
We have shown that blockade of the NPC through microinjection of WGA followed by infection with rAAV2 inhibits the nuclear entry of rAAV2, thus supporting a role for the NPC in rAAV2 nuclear entry. We did not observe complete inhibition of nuclear entry at this time point, suggesting either reversible inhibition of nuclear import by WGA, which has previously been reported (73), or an alternative, NPC-independent pathway that is utilized by rAAV2. The classical import pathway described herein for rAAV2 differs from the mechanism of nuclear entry proposed by Cohen et al. for a related parvovirus, MVM (45), and also recent findings describing nuclear envelope breakdown of permeabilized HeLa cells by AAV2 that had been acidified and then neutralized (50). Future work is required to understand how the acidification and neutralization of rAAV2 mediates nuclear entry under physiological subcellular trafficking conditions. It is possible that wt AAV2 and/or rAAV2 can cause nuclear envelope breakdown during infection, but the effects on the nuclear lamina might be more subtle than what can be detected by fluorescence microscopy. Since the infectious pathway of rAAV2 differs from that of other autonomous parvoviruses, it is not surprising that the mechanisms of nuclear entry may include both shared and disparate features between autonomous parvoviruses and nonautonomous parvoviruses. For instance, while proteosome inhibitors have been shown to inhibit the infectivity of MVM and canine parvovirus (CPV) and have no effect on bovine parvovirus (BPV) (74), it is well established that proteasome inhibition greatly enhances the nuclear entry and transduction of rAAV2 (27, 75). Our results also differ from a previous report that showed that intact AAV2 could enter purified nuclei, despite blocking of the pore with WGA or an antibody to an NPC protein (7), suggesting that the nuclear pore is dispensable for nuclear entry of AAV2. It is important to note, however, that the previous study utilized AAV2 virions that had not gone through physiological endosomal processing, allowing for the conformational change that exposes the basic regions along with the PLA2 domain required for transduction. A recent study by Salganik et al. showed that purified AAV2 virions have external protease activity at physiological pH that is ablated at low pH (76). It is possible that virions that traffic to the nucleus using alternative pathways can exert alternative functions, such as protease-mediated, NPC-independent nuclear entry under certain conditions. Xiao et al. showed that treatment with thapsigargan, an inhibitor of cellular calcium flux and modulator of nuclear pore structure, had no effect on the nuclear entry and subsequent replication of wt AAV2 in the presence of adenovirus, suggesting that the nuclear pore was not necessary for AAV2 nuclear entry in a wild-type context (48). The role of calcium depletion in relation to nuclear entry through the NPC remains controversial (77), and it could vary depending on the protein (or virus) being investigated (78). Furthermore, in addition to inhibiting calcium flux, thapsigargin has many off-target effects, which could have offset any effects on the nuclear entry of AAV2. Finally, it is possible that the mechanism of nuclear entry could differ between wild-type AAV2 in the presence of adenovirus and rAAV2 independent of any helper viruses.
Import proteins, also known as karyopherins, are generally thought of as chaperones that aid in the translocation of nuclear-bound cargo. Karyopherins have been implicated in the import of other viruses, including adenovirus (39), HIV-1 (56, 79–81), influenza A virus (57, 82–84), HBV (42), and HSV-1 (41). We have shown that knockdown of importin-β inhibits the nuclear translocation and transduction of rAAV2, suggesting that importin-β facilitates nuclear import. Furthermore, we have shown that rAAV2 can codistribute with importin-β within cells and form a Ran-sensitive complex, supporting an interaction that may facilitate nuclear entry. Interestingly, while we saw co-IP with importin-α1, -α3, and -α5, knockdown of these proteins did not affect transduction. Currently, seven human importin-α proteins have been identified (85), and it is known that these karyopherins can have both distinct substrate recognition as well as redundant NLS recognition (86). It is likely that despite knockdown, import of rAAV2 is compensated through the presence of other importin-α proteins and importin-β. Furthermore, while we did not observe co-IP between rAAV2 and importin-7 under the conditions tested, transduction was inhibited in cells that had been subjected to importin-7 knockdown. Finally, while a third class of karyopherins, known as the transportins, have been shown to be involved with the import of adenovirus, we saw no coimmunoprecipitation with transportin 1 or transportin 2 under the conditions tested.
Previous reports have shown that AAV2 contains putative NLS domains within the unique N-terminal regions of VP1 and VP2 and that mutations to these domains inhibit nuclear entry and transduction (25, 26). Incorporating one of the BR domains (BR3) into rAAV2 particles lacking VP1up has been shown to partially rescue the infectivity of these mutant virions (87). Additionally, conjugation of BR domains to exogenous green fluorescent protein has been shown sufficient to direct this protein to the nucleus (23, 47). Taken together, these results provide evidence for the role of the BR domains in rAAV2 nuclear entry. Indeed, we have shown that while rAAV2 capsid proteins interact with importin-β in our co-IP assays, this interaction is ablated when VP1 or VP2 is not available or when BR2 or BR3 has been mutated. Intriguingly, even though at least one BR domain was still present in the case of the BR2− and BR3− mutants (i.e., BR3 was available in the BR2− mutant), we were not able to detect an interaction with importin-β in our co-IP analysis. This observation suggests that the interaction between importin-β and rAAV2 capsid proteins is conformation dependent. Indeed, while the other rAAV vectors we tested share fairly conserved BR domains, their abilities to co-IP with importin-β varied. Further understanding of the contributions of serotype-specific domains will be required to understand the differential interactions between importin-β and other AAV serotypes.
Questions remain about the nuclear translocation efficiency of rAAV2. Import mediated by importin-β has been historically regarded as highly efficient and rapid (88). Since our results indicated that the interaction between importin-β and rAAV2 was the most robust compared to the other serotypes tested, one would assume that rAAV2 nuclear entry is the most rapid and efficient as well. However, Keiser et al. showed that in HeLa cells, rAAV1 and rAAV5 nuclear entry was initially more efficient than rAAV2 (89). This same study revealed that rAAV1 and rAAV5 likely utilize different subcellular trafficking pathways in order to gain access to the nucleus. Indeed, recent work has demonstrated that while the microtubule network acts to direct rAAV2 particles to the perinuclear region in cells (17), microtubules within the MTOC may also act as a net, physically trapping virions in this region. This assertion is supported by the fact that application of microtubule-disrupting drugs after rAAV2 has accumulated in the MTOC actually increases transduction (P. J. Xiao and R. J. Samulski, unpublished data). Therefore, it is possible that despite efficient interaction with importin-β, rAAV2 nuclear translocation might be physically impeded by other cellular factors, such as microtubules in the MTOC. Consistent with this notion, we observed similar cellular distributions between AAV2 and importin-β at the nuclear periphery up to 9 h postinfection, suggesting that while interactions may occur between AAV2 and importin-β, translocation might be inhibited by another cellular factor that does not impede translocation of other AAV serotypes. While our data suggest a mechanism for rAAV2 nuclear import through the NPC that is mediated by importin-α and importin-β proteins, our findings may only describe one of several translocation pathways utilized by rAAV vectors. In fact, recent work by Popa-Wagner et al. identified a PDZ-binding motif on VP1 that, when mutated, rendered AAV2 defective in nuclear entry. They concluded that this AAV2 PDZ binding domain might initiate signaling pathways within the cell to facilitate nuclear entry (49). Moreover, previous work has shown that AAV2 can interact with the nucleolar proteins nucleophosmin and nucleolin (27, 90, 91), suggesting that rAAV2 can translocate into the nucleus through interaction with these proteins. In addition to the observations of nuclear envelope breakdown by parvoviruses, including AAV2, Porwal et al. showed that AAV2 can directly interact with nucleoporins that comprise the nuclear pore complex (50). Finally, given that the abundance of karyopherins differs among cell types, it is possible that rAAV serotypes could utilize different karyopherins for nuclear entry, which would further contribute to the differences in cellular tropism historically observed among the rAAV serotypes.
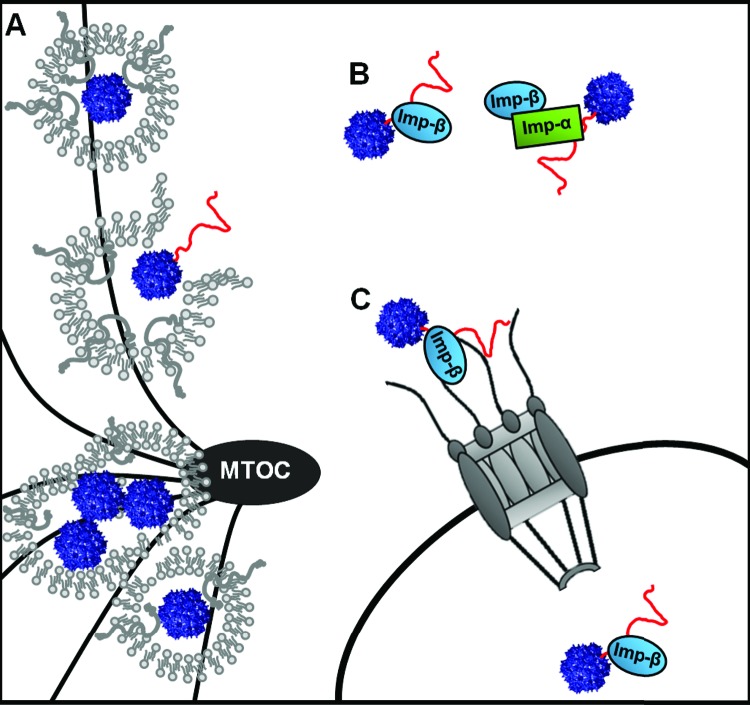
Based on our results, we have constructed a working model for the mechanism of nuclear import by rAAV2 (Fig. 9). Upon acidification of the late endosomal compartment and the conformational change that facilitates the exposure of VP1up, the BR2 and BR3 domains interact with importin-β or an importin-α/β complex. rAAV2 could also interact with importin-7 or an importin-β/importin-7 complex. Once rAAV2 escapes the endosomal compartment or the MTOC, the virus/importin complex translocates to the nucleus via the NPC, where subsequent uncoating and gene expression can occur. Improving the transduction efficiency of AAV vectors has become paramount to successful gene therapy applications. While much work has been devoted to discerning the infectious pathway of AAV vectors, further understanding of specific trafficking events within host cells is necessary to define cellular barriers to transduction. Ultimately, understanding the details of viral trafficking and host cell interactions will facilitate the rational design of AAV vectors that can overcome some of the trafficking inefficiencies currently observed. Vectors that are more efficient at navigating the subcellular space in order to deliver their genetic payload should improve overall gene therapy applications and clinical outcomes.
FIG 9.
Model of nuclear import. rAAV2 traffics to the MTOC in late endosomal or lysosomal compartments. (A) rAAV2 undergoes a conformational change, exposing VP1up (shown in red), which harbors the three NLSs. (B) rAAV2 interacts with importin-β or an importin-α/importin-β heterodimer. rAAV2 could also interact with importin-7 or an importin-7/importin-β heterodimer. (C) rAAV2 translocates to the nucleus through the NPC, with assistance from cellular karyopherins.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01AI080726, R01DK084033, R01AR064369, R01AI072176, and P01HL112761 (to R.J.S.).
We thank the laboratory of James Bear, particularly Elizabeth Haynes, for assistance with microinjection procedures. We thank Ping-Jie Xiao for assistance with the production of ultrapure Cy5-labeled rAAV2. We thank the members of the University of North Carolina at Chapel Hill Gene Therapy Center for productive discussions, particularly Chengwen Li, Matthew Hirsch, Jayme Warischalk, and Angela Mitchell. We greatly appreciate Sophia Shih for calculating virus titers via qPCR. Microscopy equipment and analysis software used in this study were kindly provided by the UNC Microscopy Services Laboratory.
Footnotes
Published ahead of print 29 January 2014
REFERENCES
- 1.Atchison RW, Casto BC, Hammon WM. 1965. Adenovirus-associated defective virus particles. Science 149:754–756 [DOI] [PubMed] [Google Scholar]
- 2.Mitchell AM, Nicolson SC, Warischalk JK, Samulski RJ. 2010. AAV's anatomy: roadmap for optimizing vectors for translational success. Curr. Gene Ther. 10:319–340. 10.2174/156652310793180706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asokan A, Schaffer DV, Samulski RJ. 2012. The AAV vector toolkit: poised at the clinical crossroads. Mol. Ther. 20:699–708. 10.1038/mt.2011.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding W, Yan Z, Zak R, Saavedra M, Rodman DM, Engelhardt JF. 2003. Second-strand genome conversion of adeno-associated virus type 2 (AAV-2) and AAV-5 is not rate limiting following apical infection of polarized human airway epithelia. J. Virol. 77:7361–7366. 10.1128/JVI.77.13.7361-7366.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauck B, Zhao W, High K, Xiao W. 2004. Intracellular viral processing, not single-stranded DNA accumulation, is crucial for recombinant adeno-associated virus transduction. J. Virol. 78:13678–13686. 10.1128/JVI.78.24.13678-13686.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanlioglu S, Benson PK, Yang J, Atkinson EM, Reynolds T, Engelhardt JF. 2000. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 74:9184–9196. 10.1128/JVI.74.19.9184-9196.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen J, Qing K, Srivastava A. 2001. Infection of purified nuclei by adeno-associated virus 2. Mol. Ther. 4:289–296. 10.1006/mthe.2001.0457 [DOI] [PubMed] [Google Scholar]
- 8.Johnson FB, Ozer HL, Hoggan MD. 1971. Structural proteins of adenovirus-associated virus type 3. J. Virol. 8:860–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salo RJ, Mayor HD. 1977. Structural polypeptides of parvoviruses. Virology 78:340–345 [DOI] [PubMed] [Google Scholar]
- 10.Summerford C, Samulski RJ. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Summerford C, Bartlett JS, Samulski RJ. 1999. αVβ5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78–82 [DOI] [PubMed] [Google Scholar]
- 12.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 5:71–77 [DOI] [PubMed] [Google Scholar]
- 13.Duan D, Li Q, Kao AW, Yue Y, Pessin JE, Engelhardt JF. 1999. Dynamin is required for recombinant adeno-associated virus type 2 infection. J. Virol. 73:10371–10376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nonnenmacher M, Weber T. 2011. Adeno-associated virus 2 infection requires endocytosis through the CLIC/GEEC pathway. Cell Host Microbe 10:563–576. 10.1016/j.chom.2011.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bantel-Schaal U, Hub B, Kartenbeck J. 2002. Endocytosis of adeno-associated virus type 5 leads to accumulation of virus particles in the Golgi compartment. J. Virol. 76:2340–2349. 10.1128/JVI.76.5.2340-2349.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartlett JS, Wilcher R, Samulski RJ. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 74:2777–2785. 10.1128/JVI.74.6.2777-2785.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douar AM, Poulard K, Stockholm D, Danos O. 2001. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J. Virol. 75:1824–1833. 10.1128/JVI.75.4.1824-1833.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao PJ, Samulski RJ. 2012. Cytoplasmic trafficking, endosomal escape, and perinuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J. Virol. 86:10462–10473. 10.1128/JVI.00935-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding W, Zhang LN, Yeaman C, Engelhardt JF. 2006. rAAV2 traffics through both the late and the recycling endosomes in a dose-dependent fashion. Mol. Ther. 13:671–682. 10.1016/j.ymthe.2005.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pajusola K, Gruchala M, Joch H, Luscher TF, Yla-Herttuala S, Bueler H. 2002. Cell-type-specific characteristics modulate the transduction efficiency of adeno-associated virus type 2 and restrain infection of endothelial cells. J. Virol. 76:11530–11540. 10.1128/JVI.76.22.11530-11540.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bleker S, Sonntag F, Kleinschmidt JA. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 79:2528–2540. 10.1128/JVI.79.4.2528-2540.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kronenberg S, Bottcher B, von der Lieth CW, Bleker S, Kleinschmidt JA. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 79:5296–5303. 10.1128/JVI.79.9.5296-5303.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonntag F, Bleker S, Leuchs B, Fischer R, Kleinschmidt JA. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 80:11040–11054. 10.1128/JVI.01056-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girod A, Wobus CE, Zadori Z, Ried M, Leike K, Tijssen P, Kleinschmidt JA, Hallek M. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973–978 http://vir.sgmjournals.org/content/83/5/973.long [DOI] [PubMed] [Google Scholar]
- 25.Grieger JC, Snowdy S, Samulski RJ. 2006. Separate basic region motifs within the adeno-associated virus capsid proteins are essential for infectivity and assembly. J. Virol. 80:5199–5210. 10.1128/JVI.02723-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson JS, Li C, DiPrimio N, Weinberg MS, McCown TJ, Samulski RJ. 2010. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. J. Virol. 84:8888–8902. 10.1128/JVI.00687-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson JS, Samulski RJ. 2009. Enhancement of adeno-associated virus infection by mobilizing capsids into and out of the nucleolus. J. Virol. 83:2632–2644. 10.1128/JVI.02309-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao PJ, Li C, Neumann A, Samulski RJ. 2012. Quantitative 3D tracing of gene-delivery viral vectors in human cells and animal tissues. Mol. Ther. 20:317–328. 10.1038/mt.2011.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C, He Y, Nicolson S, Hirsch M, Weinberg MS, Zhang P, Kafri T, Samulski RJ. 2013. Adeno-associated virus capsid antigen presentation is dependent on endosomal escape. J. Clin. Invest. 123:1390–1401. 10.1172/JCI66611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorokin AV, Kim ER, Ovchinnikov LP. 2007. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc.) 72:1439–1457. 10.1134/S0006297907130032 [DOI] [PubMed] [Google Scholar]
- 31.Nigg EA. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779–787 [DOI] [PubMed] [Google Scholar]
- 32.Pante N, Kann M. 2002. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 13:425–434. 10.1091/mbc.01-06-0308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore MS, Blobel G. 1993. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature 365:661–663 [DOI] [PubMed] [Google Scholar]
- 34.Melchior F, Paschal B, Evans J, Gerace L. 1993. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J. Cell Biol. 123:1649–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rexach M, Blobel G. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83:683–692 [DOI] [PubMed] [Google Scholar]
- 36.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. 1996. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15:5584–5594 [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen S, Au S, Pante N. 2011. How viruses access the nucleus. Biochim. Biophys. Acta 1813:1634–1645. 10.1016/j.bbamcr.2010.12.009 [DOI] [PubMed] [Google Scholar]
- 38.Kobiler O, Drayman N, Butin-Israeli V, Oppenheim A. 2012. Virus strategies for passing the nuclear envelope barrier. Nucleus 3:526–539. 10.4161/nucl.21979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wodrich H, Cassany A, D'Angelo MA, Guan T, Nemerow G, Gerace L. 2006. Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. J. Virol. 80:9608–9618. 10.1128/JVI.00850-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sodeik B, Ebersold MW, Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 20:4922–4931. 10.1128/MCB.20.13.4922-4931.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeh CT, Liaw YF, Ou JH. 1990. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J. Virol. 64:6141–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckhardt SG, Milich DR, McLachlan A. 1991. Hepatitis B virus core antigen has two nuclear localization sequences in the arginine-rich carboxyl terminus. J. Virol. 65:575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen S, Behzad AR, Carroll JB, Pante N. 2006. Parvoviral nuclear import: bypassing the host nuclear-transport machinery. J. Gen. Virol. 87:3209–3213. 10.1099/vir.0.82232-0 [DOI] [PubMed] [Google Scholar]
- 46.Cohen S, Marr AK, Garcin P, Pante N. 2011. Nuclear envelope disruption involving host caspases plays a role in the parvovirus replication cycle. J. Virol. 85:4863–4874. 10.1128/JVI.01999-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoque M, Ishizu K, Matsumoto A, Han SI, Arisaka F, Takayama M, Suzuki K, Kato K, Kanda T, Watanabe H, Handa H. 1999. Nuclear transport of the major capsid protein is essential for adeno-associated virus capsid formation. J. Virol. 73:7912–7915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao W, Warrington KH, Jr, Hearing P, Hughes J, Muzyczka N. 2002. Adenovirus-facilitated nuclear translocation of adeno-associated virus type 2. J. Virol. 76:11505–11517. 10.1128/JVI.76.22.11505-11517.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Popa-Wagner R, Porwal M, Kann M, Reuss M, Weimer M, Florin L, Kleinschmidt JA. 2012. Impact of VP1-specific protein sequence motifs on adeno-associated virus type 2 intracellular trafficking and nuclear entry. J. Virol. 86:9163–9174. 10.1128/JVI.00282-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porwal M, Cohen S, Snoussi K, Popa-Wagner R, Anderson F, Dugot-Senant N, Wodrich H, Dinsart C, Kleinschmidt JA, Pante N, Kann M. 2013. Parvoviruses cause nuclear envelope breakdown by activating key enzymes of mitosis. PLoS Pathog. 9(10):e1003671. 10.1371/journal.ppat.1003671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grieger JC, Choi VW, Samulski RJ. 2006. Production and characterization of adeno-associated viral vectors. Nat. Protoc. 1:1412–1428. 10.1038/nprot.2006.207 [DOI] [PubMed] [Google Scholar]
- 52.Gray SJ, Choi VW, Asokan A, Haberman RA, McCown TJ, Samulski RJ. 2011. Production of recombinant adeno-associated viral vectors and use in in vitro and in vivo administration. Curr. Protoc. Neurosci. Chapter 4:Unit 4.17. 10.1002/0471142301.ns0417s57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong L, Li B, Jayandharan G, Mah CS, Govindasamy L, Agbandje-McKenna M, Herzog RW, Weigel-Van Aken KA, Hobbs JA, Zolotukhin S, Muzyczka N, Srivastava A. 2008. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology 381:194–202. 10.1016/j.virol.2008.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong L, Zhao W, Wu J, Li B, Zolotukhin S, Govindasamy L, Agbandje-McKenna M, Srivastava A. 2007. A dual role of EGFR protein tyrosine kinase signaling in ubiquitination of AAV2 capsids and viral second-strand DNA synthesis. Mol. Ther. 15:1323–1330. 10.1038/sj.mt.6300170 [DOI] [PubMed] [Google Scholar]
- 55.Finlay DR, Newmeyer DD, Price TM, Forbes DJ. 1987. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 104:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Popov S, Rexach M, Ratner L, Blobel G, Bukrinsky M. 1998. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J. Biol. Chem. 273:13347–13352 [DOI] [PubMed] [Google Scholar]
- 57.O'Neill RE, Jaskunas R, Blobel G, Palese P, Moroianu J. 1995. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J. Biol. Chem. 270:22701–22704 [DOI] [PubMed] [Google Scholar]
- 58.Warrington KH, Jr, Gorbatyuk OS, Harrison JK, Opie SR, Zolotukhin S, Muzyczka N. 2004. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J. Virol. 78:6595–6609. 10.1128/JVI.78.12.6595-6609.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fried H, Kutay U. 2003. Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60:1659–1688. 10.1007/s00018-003-3070-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macara IG. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570–594. 10.1128/MMBR.65.4.570-594.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gorlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138:65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jakel S, Albig W, Kutay U, Bischoff FR, Schwamborn K, Doenecke D, Gorlich D. 1999. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 18:2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rebane A, Aab A, Steitz JA. 2004. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA 10:590–599. 10.1261/rna.5224304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kohler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. 1997. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 417:104–108 [DOI] [PubMed] [Google Scholar]
- 65.Malik HS, Eickbush TH, Goldfarb DS. 1997. Evolutionary specialization of the nuclear targeting apparatus. Proc. Natl. Acad. Sci. U. S. A. 94:13738–13742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muhlhausser P, Muller EC, Otto A, Kutay U. 2001. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2:690–696. 10.1093/embo-reports/kve168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mosammaparast N, Jackson KR, Guo Y, Brame CJ, Shabanowitz J, Hunt DF, Pemberton LF. 2001. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J. Cell Biol. 153:251–262. 10.1083/jcb.153.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Copeland AM, Newcomb WW, Brown JC. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83:1660–1668. 10.1128/JVI.01139-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wisnivesky JP, Leopold PL, Crystal RG. 1999. Specific binding of the adenovirus capsid to the nuclear envelope. Hum. Gene Ther. 10:2187–2195 [DOI] [PubMed] [Google Scholar]
- 70.Greber UF, Suomalainen M, Stidwill RP, Boucke K, Ebersold MW, Helenius A. 1997. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 16:5998–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trotman LC, Mosberger N, Fornerod M, Stidwill RP, Greber UF. 2001. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 3:1092–1100. 10.1038/ncb1201-1092 [DOI] [PubMed] [Google Scholar]
- 72.Yamada M, Kasamatsu H. 1993. Role of nuclear pore complex in simian virus 40 nuclear targeting. J. Virol. 67:119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoneda Y, Imamoto-Sonobe N, Yamaizumi M, Uchida T. 1987. Reversible inhibition of protein import into the nucleus by wheat germ agglutinin injected into cultured cells. Exp. Cell Res. 173:586–595 [DOI] [PubMed] [Google Scholar]
- 74.Ros C, Kempf C. 2004. The ubiquitin-proteasome machinery is essential for nuclear translocation of incoming minute virus of mice. Virology 324:350–360. 10.1016/j.virol.2004.04.016 [DOI] [PubMed] [Google Scholar]
- 75.Duan D, Yue Y, Yan Z, Yang J, Engelhardt JF. 2000. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Invest. 105:1573–1587. 10.1172/JCI8317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salganik M, Venkatakrishnan B, Bennett A, Lins B, Yarbrough J, Muzyczka N, Agbandje-McKenna M, McKenna R. 2012. Evidence for pH-dependent protease activity in the adeno-associated virus capsid. J. Virol. 86:11877–11885. 10.1128/JVI.01717-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sarma A, Yang W. 2011. Calcium regulation of nucleocytoplasmic transport. Protein Cell 2:291–302. 10.1007/s13238-011-1038-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Strubing C, Clapham DE. 1999. Active nuclear import and export is independent of lumenal Ca2+ stores in intact mammalian cells. J. Gen. Physiol. 113:239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. 1996. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 70:1027–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gallay P, Hope T, Chin D, Trono D. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. U. S. A. 94:9825–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vodicka MA, Koepp DM, Silver PA, Emerman M. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 12:175–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang P, Palese P, O'Neill RE. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 71:1850–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O'Neill RE, Palese P. 1995. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology 206:116–125 [DOI] [PubMed] [Google Scholar]
- 84.Melen K, Fagerlund R, Franke J, Kohler M, Kinnunen L, Julkunen I. 2003. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 278:28193–28200. 10.1074/jbc.M303571200 [DOI] [PubMed] [Google Scholar]
- 85.Kelley JB, Talley AM, Spencer A, Gioeli D, Paschal BM. 2010. Karyopherin alpha7 (KPNA7), a divergent member of the importin alpha family of nuclear import receptors. BMC Cell Biol. 11:63. 10.1186/1471-2121-11-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kohler M, Speck C, Christiansen M, Bischoff FR, Prehn S, Haller H, Gorlich D, Hartmann E. 1999. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 19:7782–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grieger JC, Johnson JS, Gurda-Whitaker B, Agbandje-McKenna M, Samulski RJ. 2007. Surface-exposed adeno-associated virus Vp1-NLS capsid fusion protein rescues infectivity of noninfectious wild-type Vp2/Vp3 and Vp3-only capsids but not that of fivefold pore mutant virions. J. Virol. 81:7833–7843. 10.1128/JVI.00580-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Riddick G, Macara IG. 2007. The adapter importin-alpha provides flexible control of nuclear import at the expense of efficiency. Mol. Syst. Biol. 3:118. 10.1038/msb4100160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keiser NW, Yan Z, Zhang Y, Lei-Butters DC, Engelhardt JF. 2011. Unique characteristics of AAV1, 2, and 5 viral entry, intracellular trafficking, and nuclear import define transduction efficiency in HeLa cells. Hum. Gene Ther. 22:1433–1444. 10.1089/hum.2011.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bevington JM, Needham PG, Verrill KC, Collaco RF, Basrur V, Trempe JP. 2007. Adeno-associated virus interactions with B23/nucleophosmin: identification of sub-nucleolar virion regions. Virology 357:102–113. 10.1016/j.virol.2006.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qiu J, Brown KE. 1999. A 110-kDa nuclear shuttle protein, nucleolin, specifically binds to adeno-associated virus type 2 (AAV-2) capsid. Virology 257:373–382 [DOI] [PubMed] [Google Scholar]