ABSTRACT
We previously showed that prototype macaque-tropic human immunodeficiency virus type 1 (HIV-1) acquired nonsynonymous growth-enhancing mutations within a narrow genomic region during the adaptation process in macaque cells. These adaptive mutations were clustered in the 3′ region of the pol gene, encoding a small portion of the C-terminal domain of integrase (IN). Mutations in HIV-1 IN have been reported to have pleiotropic effects on both the early and late phases in viral replication. cis-acting functions in the IN-coding sequence for viral gene expression have also been reported. We here demonstrated that the adaptive mutations promoted viral growth by increasing virion production with no positive effects on the early replication phase. Synonymous codon alterations in one of the adaptive mutations influenced virion production levels, which suggested nucleotide-dependent regulation. Indeed, when the single-nucleotide natural polymorphisms observed in the 3′ regions of 196 HIV-1/simian immunodeficiency virus (SIVcpz) pol genes (nucleotides [nt] 4895 to 4929 for HIV-1 NL4-3) were introduced into macaque- and human-tropic HIV-1 clones, more than half exhibited altered replication potentials. Moreover, single-nucleotide mutations caused parallel increases or decreases in the expression levels of viral late proteins and viral replication potentials. We also showed that the overall expression profiles of viral mRNAs were markedly changed by single-nucleotide mutations. These results demonstrate that the 3′ region of the HIV-1 pol gene (nt 4895 to 4929) can alter viral replication potential by modulating the expression pattern of viral mRNAs in a nucleotide-dependent manner.
IMPORTANCE Viruses have the plasticity to adapt themselves under various constraints. HIV-1 can mutate and evolve in growth-restrictive cells by acquiring adaptive changes in its genome. We have previously identified some growth-enhancing mutations in a narrow region of the IN-coding sequence, in which a number of cis-acting elements are located. We now focus on the virological significance of this pol gene region and the mechanistic basis underlying its effects on viral replication. We have found several naturally occurring synonymous mutations within this region that alter viral replication potentials. The effects caused by these natural single-nucleotide polymorphisms are linked to the definite expression patterns of viral mRNAs. We show here that the nucleotide sequence of the pol gene (nucleotides 4895 to 4929 for HIV-1 NL4-3) plays an important role in HIV-1 replication by modulating viral gene expression.
INTRODUCTION
The gene expression process of human immunodeficiency virus type 1 (HIV-1) (transcription, capping, polyadenylation, splicing, nuclear export, and translation) is highly coordinated and regulated by interactions between host/viral proteins and cis-acting elements located within the viral genome (1, 2). During this process, more than 40 mRNA species with nine viral genes are generated by alternative splicing (3, 4). These mRNA species are divided into three major groups: ∼9-kb mRNAs (unspliced form) encoding Gag and Gag-Pol proteins, ∼4-kb mRNAs (singly spliced form) encoding Vif, Vpr, Vpu, and Env proteins, and ∼1.8-kb mRNAs (completely spliced form) encoding Tat, Rev, and Nef proteins. In the early phase of HIV-1 gene expression, ∼1.8-kb mRNA species are transported to the cytoplasm and translated to synthesize Tat, Rev, and Nef proteins. Tat, along with some host factors, trans-activates HIV-1 transcription (5, 6). Rev facilitates the nuclear export of ∼4-kb and ∼9-kb HIV-1 mRNAs, and their encoded proteins are subsequently produced (7, 8). Alterations in the tightly regulated process of HIV-1 gene expression can affect viral replication (3, 4, 9–11).
HIV-1 integrase (IN) is generated from a Gag-Pol precursor and mediates integration, a hallmark of retroviruses. HIV-1 IN is involved not only in integration but also in reverse transcription, viral DNA nuclear import, and virion assembly/production (12–19). The deletion or C-terminal truncation of HIV-1 IN has been shown to reduce virion production in producer cells (20, 21). Although mutations in IN have negative effects on virion production, they also affect the early phase of viral replication (15, 22). Different amino acid substitutions at the same sites often have diverse effects on viral replication potential (13, 15, 23, 24). Furthermore, a number of splicing sites and cis-acting elements have been identified in the IN-coding sequence (3, 4, 25–30). Therefore, mutations in the IN-coding sequence can change the nucleotide sequence important for viral replication as well as the protein-coding sequence associated with IN activity. The replication-defective mutant IN E246K represents a good example. It showed a processing defect in Gag, and its virion production level was markedly reduced (15). The E246K mutation, which is located within splicing site D2 and affects viral RNA splicing, was shown to result in the loss of viral infectivity (31). Thus, cis-acting functions in the IN-coding sequence must be considered to delineate its possible roles in viral replication.
In a previous virus adaptation study, we demonstrated that growth-enhanced viruses, which emerged following a long-term culture of cells infected with macaque-tropic HIV-1 NL-DT5R (5R) or NL-DT562 (562), frequently and reproducibly acquired mutations in the 3′ region of the pol gene (nucleotides [nt] 4889 to 4923 for 5R [Fig. 1] and nt 4895 to 4929 for the standard HIV-1 NL4-3), which encodes a small portion of the IN C-terminal domain (CTD) (32). Four adaptive mutations (N222K, F223Y, D229E, and V234I) in the region were identified in our repeated virus adaptation experiments and were responsible for viral growth enhancements (32). In this study, we aimed to elucidate how mutations in the 3′ region of the pol gene promoted viral replication and what the virological significance of this region was. The four mutations mentioned above were found to augment virus replication potential by increasing infectious virion production without any effects on the early replication phase. The CTD has been reported to be the least conserved sequence of the three domains in IN (15, 17, 19), with the 234th amino acid in IN being polymorphic (33). Codon alterations in V234I from ATT to ATC and ATA influenced virion production, which indicated regulation by a single-nucleotide change. An investigation of the sequences in the 3′ region of the pol gene in 196 HIV-1/simian immunodeficiency virus (SIVcpz) genomes (HIV sequence compendium 2011, Los Alamos National Laboratory, NM, USA) revealed that natural variants carried ATT or ATC at amino acid position 234. Based on these findings, single-nucleotide natural variations in the 3′ region of the pol gene (nt 4895 to 4929 for NL4-3) were introduced into macaque- and human-tropic HIV-1 clones. We identified several natural variations that alter virion production/replication efficiency. The observed effects of single-nucleotide variations were attributed to an increase or decrease in the expression levels of viral late proteins (Gag, Gag-Pol, Vpu, and Env), whereas early proteins (Nef and Rev) were invariably expressed. Moreover, single-nucleotide variations caused changes in the viral mRNA expression pattern. Taken together, our results showed that the nucleotide sequence of the 3′ region of the HIV-1 pol gene (nt 4895 to 4929 for NL4-3) play an important role in viral replication by modulating viral gene expression.
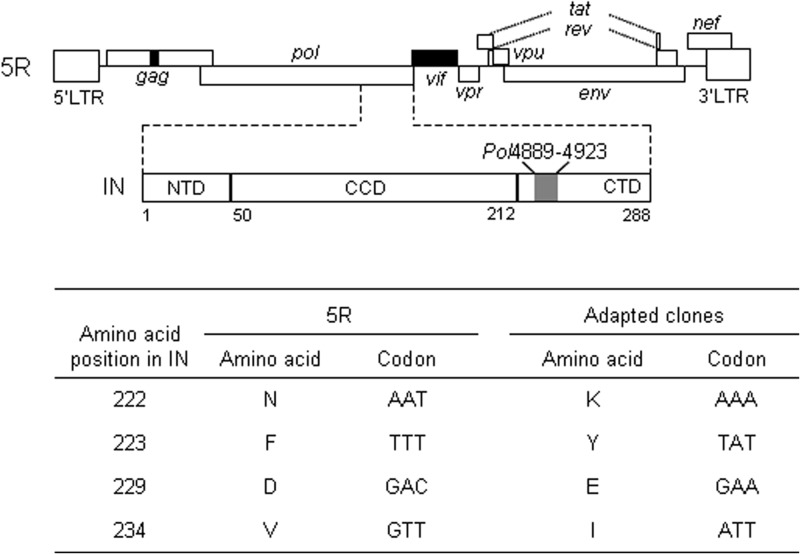
FIG 1.
Growth-enhancing mutations identified in the 3′ region of the pol gene. The genome structure of 5R (34) (GenBank accession no. AB266485) is schematically depicted. Black areas show the sequence derived from SIVmac239 (56) (GenBank accession no. M33262). The pol region encoding IN and the IN domain structure are indicated. The gray area represents a region in the pol gene (nt 4889 to 4923) designated Pol4889-4923, in which adaptive mutations are located. NTD, N-terminal domain; CCD, catalytic core domain; CTD, C-terminal domain. Details of the adaptive mutations found in the Pol4889-4923 region are shown at the bottom.
MATERIALS AND METHODS
Plasmid DNAs.
The proviral clones pNL-DT5R (34) and pNL4-3 (35) were used as parental clones in the present study. Proviral clones carrying each mutation in the 3′ region of the pol gene were constructed with the QuikChange site-directed mutagenesis kit (Agilent Technologies Inc., Santa Clara, CA). Proviral env-deficient clones were constructed by deleting the NdeΙ (nt 6584)-NheΙ (nt 7435) fragment of pNL-DT5R. Reporter clones were constructed by introducing the luciferase gene into the nef gene of env-deficient proviral clones as described previously (36). A long terminal repeat (LTR)-driven luciferase reporter clone (5RLTR-Luc) was constructed by replacing the AatII (5′end of the proviral genome)-NcoI (5′end of the luciferase gene in the nef gene) fragment of the pNL-DT5R luciferase reporter clone with the LTR region (nt 1 to 789) of pNL-DT5R. A proviral gag/gag-pol frameshift clone (gtg-Spe) was constructed by cutting the V234gtg clone with SpeI (nt 1501) within the gag capsid and inserting four nucleotides with T4 DNA polymerase.
Cells.
Human kidney 293T cells were cultured in minimal essential medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum (hiFBS). Human lymphocyte M8166 cells were cultured in RPMI 1640 supplemented with 10% hiFBS. Human lymphocyte MT4/CCR5 cells (MT4 cells stably expressing CCR5) were maintained in RPMI 1640 containing 10% hiFBS and 200 μg/ml of hygromycin B (Sigma-Aldrich Co., St. Louis, MO).
Analysis of virus growth kinetics.
Virus stocks were prepared from 293T cells transfected with proviral clones as previously described (34, 35). Virion-associated reverse transcriptase (RT) activity was measured as previously described (37). M8166 and MT4/CCR5 cells (105) were infected with equal amounts of NL-DT5R and its derivative viruses (35 RT units and 5 × 105 RT units for M8166 and MT4/CCR5 cells, respectively), as previously described (38, 39). Equal amounts (105 RT units) of HIV-1 NL4-3 and its derivative viruses were inoculated into MT4/CCR5 cells (105). Virus replication was monitored by RT activity released into the culture supernatants.
Analysis of single-cycle viral infectivity.
Vesicular stomatitis virus G protein (VSV-G)-pseudotyped viruses were prepared from 293T cells cotransfected with an env-deficient luciferase reporter clone and pCMV-G (40) at a molar ratio of 1:1. Virus amounts on day 2 posttransfection were measured with an HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) kit (ZeptoMetrix Corporation, Buffalo, NY). M8166 cells (105) were infected with equal amounts of pseudotyped viruses (30 pg of p24), and cells were lysed on day 1 postinfection for luciferase assays (Promega Corporation, Madison, WI).
Analysis of viral cDNA synthesis.
DNase I-treated pseudoviruses (150 pg of p24) were inoculated into M8166 cells (5 × 105), and total DNA was extracted on day 1 postinfection using the DNeasy blood and tissue kit (Qiagen GmbH, Hilden, Germany). Quantitative analyses of viral cDNA products using real-time quantitative PCR (ABI7500; Life Technologies Corporation, Carlsbad, CA) were performed as previously described (14).
Analysis of virion production.
M8166 cells (106) were cotransfected with equal amounts of env-deficient proviral clones (1 μg) and the pGL3 luciferase reporter vector (Promega Corporation) (1 μg) using the Amaxa human T cell Nucleofector kit (Lonza Ltd., Basel, Switzerland) with Nucleofector II (Lonza Ltd.). Culture supernatants were collected on day 2 posttransfection, and virion production was measured using the HIV-1 p24 antigen ELISA kit (ZeptoMetrix Corporation). Cell lysates were prepared with 1× CCLR buffer (Promega Corporation) and subjected to luciferase assays (Promega Corporation). The luciferase activity in cell lysates was used to normalize the transfection efficiency.
Analysis of viral protein expression.
293T cells for Western blot analysis were transfected with equal amounts of proviral clones by using Lipofectamine 2000 (Life Technologies Corporation) in the absence or presence of 2 μM saquinavir (SQV) (Sigma-Aldrich Co.). On day 1 posttransfection, cells were lysed in 1× TNE buffer (10 mM Tris-HCl [pH 8.0], 1% Nonidet P-40, 150 mM NaCl, 1 mM EDTA [pH 8.0], and 1% protease inhibitor cocktail [Sigma-Aldrich Co.]). The total protein amounts in the cell lysates were measured with the DC protein assay (Bio-Rad Laboratories Inc., Hercules, CA), and equal amounts were loaded onto Mini-Protean TGX gels (Bio-Rad Laboratories Inc.) for electrophoresis (0.5 μg for the anti-gp160 antibody, 1 μg for the anti-Vpu antibody, 2 μg for the anti-p24, anti-Nef, or anti-β-actin antibody, 5 μg for the anti-Rev antibody, and 20 μg for the anti-RT antibody). Following blotting onto Immobilon-P transfer membranes (Merck KGaA, Darmstadt, Germany), the membranes were treated with the anti-β-actin clone AC-15 (Sigma-Aldrich Co.), anti-HIV-1 p24 (183-H12-5C) (catalog number 3537; NIH Research and References Reagent Program), anti-HIV-1 RT (MP Biomedicals, Santa Ana, CA), HIV-1 NL4-3 Vpu antiserum (catalog number 969; NIH Research and References Reagent Program), anti-HIV-1 gp160 (ADP409; Immuno Ltd./the MRC AIDS Directed Programme Reagent Project), anti-Rev (ab25871; Abcam PLC, Cambridge, England), or anti-HIV-1 Nef (Advanced Biotechnologies Inc., Columbia, MD) antibody and visualized with the Amersham ECL Plus Western blotting detection system (GE Healthcare UK Ltd., Buckinghamshire, England). A GS-800 calibrated densitometer and Quantity One software (Bio-Rad Laboratories Inc.) were used to quantify signal intensities. To monitor the expression levels of viral proteins, 293T cells were transfected with proviral clones by using Lipofectamine 2000 (Life Technologies Corporation), and on day 2 posttransfection, samples were prepared as described above. The amounts of cell-associated Gag-p24 and Pol-RT were measured using the HIV-1 p24 antigen ELISA kit (ZeptoMetrix Corporation) and RT capture ELISA kit (ImmunoDX, LLC, Woburn, MA), respectively. To monitor Tat activity, 293T cells were cotransfected with proviral clones (0.2 μg) and the 5RLTR-Luc clone (0.05 μg) by using Lipofectamine 2000 (Life Technologies Corporation). Cells were lysed with 1× CCLR buffer (Promega Corporation) on day 1 posttransfection for luciferase assays (Promega Corporation). 293T cells for the interference experiments were transfected with an appropriate amount of proviral clones (5R and its derivatives) by using Lipofectamine 2000 (Life Technologies Corporation). Cells were lysed with 1× TNE buffer on day 1 posttransfection, and the amount of Gag-p24 in cell lysates was measured using the HIV-1 p24 antigen ELISA kit (ZeptoMetrix Corporation).
Northern blot analysis.
293T cells were transfected with equal amounts of proviral clones by using Lipofectamine 2000 (Life Technologies Corporation), and total RNA was extracted at 10 to 20 h posttransfection using the RNeasy Plus Minikit (Qiagen GmbH). Poly(A)+ RNA was isolated with the Oligotex-dT30 Super mRNA purification kit (TaKaRa Bio Inc., Otsu, Japan) and then treated with DNase I (TaKaRa Bio Inc.). Equal amounts of RNA samples were loaded on a glyoxal denatured 1% agarose gel prepared with NorthernMax-Gly 10× Gel Prep/running buffer (Life Technologies Corporation), electrophoresed, and blotted onto a positively charged nylon membrane (Roche Diagnostics GmbH, Mannheim, Germany). The digoxigenin (DIG)-labeled universal probe (U probe) to detect all HIV-1 mRNA species was prepared by using a PCR DIG probe synthesis kit (Roche Diagnostics GmbH.) with pNL4-3 as a template and primers 5′-GAGGATTGTGGAACTTCTGG-3′ and 5′-CTTTGGGAGTGAATTAGCCC-3′. The DIG-labeled Rev-responsive element (RRE) probe, vif probe, and vpr probe were synthesized using templates and primer pairs as follows: RRE probe, pNL4-3, forward primer 5′-CCATTAGGAGTAGCACCCAC-3′, and reverse primer 5′-GTTCCAGAGATTTATTACTCC-3′; vif probe, pNL-DT5R, forward primer 5′-ATGGAGGAGGAAAAGAGGTGG-3′, and reverse primer 5′-CTGCATAAGTACTGAGCCAC-3′; and vpr probe, pNL4-3, forward primer 5′-ATGGAACAAGCCCCAGAAG-3′, and reverse primer 5′-GCAGAATTCTTATTATGGCTTCC-3′. The membrane was hybridized with the DIG-labeled probe in DIG Easy Hyb (Roche Diagnostics GmbH), and visualized with the DIG High Prime DNA labeling and detection starter kit II (Roche Diagnostics GmbH). To monitor and normalize loading amounts of RNAs, membranes were hybridized with the random-prime DIG-labeled GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe prepared with the DIG High Prime DNA labeling and detection starter kit II (Roche Diagnostics GmbH) and visualized as described above.
RESULTS
Growth-enhancing adaptive mutations increase virion production but have no effects on the early phase of viral replication.
We previously obtained adapted viruses with enhanced growth potential from long-term cultures of macaque cells infected with macaque-tropic HIV-1 5R (34) or its R5-tropic version 562 (41). While proviral clones generated from adapted viruses contained a number of mutations in scattered regions of the viral genomes, only mutations in pol-IN or env-Gp120 were found to contribute to viral growth enhancement (32). In this study, we investigated the mechanistic basis for acceleration of viral replication by the adaptive mutations in the IN-coding sequence. As indicated in Fig. 1, four mutations (N222K, F223Y, D229E, and V234I in the 3′ region of the pol gene designated Pol4889-4923) were previously shown to enhance viral growth in macaque cells (32). The introduction of both N222K and V234I into human-tropic HIV-1 NL4-3 had a growth-enhancing effect in human cells similar to that observed for 5R (32). We first studied the effect of the adaptive mutations in the Pol4889-4923 region on viral replication in M8166 cells. As shown in Fig. 2A, all mutants grew more efficiently than the parental 5R virus. To examine the early replication phase, single-cycle viral infectivity was determined by infection with VSV-G-pseudotyped viruses containing a luciferase gene in the nef gene. All mutant clones exhibited infectivity similar to that of 5R (Fig. 2B). Furthermore, the four mutations did not have positive effects on viral cDNA synthesis, as measured by real-time quantitative PCR (Fig. 2C). All four growth-enhancing mutations resulted in an increase in virion production in transfected M8166 cells (Fig. 2D). An enhancement in virion production was consistently observed for pseudotyped and proviral clones in transfected 293T cells (data not shown). These results showed that the acceleration of viral replication by the adaptive mutations in the Pol4889-4923 region could be attributed to the increase in infectious virion production in producer cells.
FIG 2.
Effect of the adaptive mutations in the Pol4889-4923 region on different stages of virus replication in M8166 cells. (A) Replication kinetics. Input viruses were prepared from 293T cells transfected with the indicated proviral clones, and equal amounts were inoculated into M8166 cells. Virus replication was monitored by RT activity released into the culture supernatants. Representative data from at least three independent experiments are shown. (B) Single-cycle viral infectivity. VSV-G-pseudotyped viruses were prepared from 293T cells transfected with the indicated clones, and equal amounts were inoculated into M8166 cells. Cell lysates were prepared on day 1 postinfection and subjected to luciferase assays. Infectivity is presented as luciferase activity relative to that exhibited by 5R. Mean values from three independent experiments are shown with the standard deviations (SD). (C) Viral cDNA synthesis. VSV-G-pseudotyped viruses were prepared from 293T cells transfected with the indicated clones, DNase Ι treated, and inoculated into M8166 cells. Total DNA was extracted from infected cells on day 1 postinfection and subjected to real-time quantitative PCR analyses with a primer pair specific for the late form (R/gag) of viral cDNA. The DNA copy number relative to that of 5R is presented. Mean values ± SD from three independent experiments are shown. (D) Virion production. M8166 cells were cotransfected with the indicated env-deficient proviral clones and a luciferase reporter vector (pGL3) by using a Nucleofector. Virion production on day 2 posttransfection was measured by the amount of Gag-p24 in the culture supernatants. The amount of p24 was normalized by luciferase activity in cell lysates. The amount of p24 relative to that produced by 5R is presented. Mean values ± SD from four independent experiments are shown. Significance relative to 5R as calculated by the Student t test is shown (*, P < 0.01; **, P < 0.02).
Amino acid substitutions caused by the adaptive mutations may not be responsible for the enhancement in virion production/replication ability.
All growth-enhancing mutations in the Pol4889-4923 region were nonsynonymous changes (Fig. 1). To determine whether altered amino acids were critical for the enhancement in virion production/replication efficiency, each amino acid at the four sites (N222, F223, D229, and V234) was replaced by various amino acids with different sizes and chemical properties (detailed in Table 1). The effects of amino acid substitutions at these positions on virion production and viral replication (Fig. 3) were determined as described above. As shown in Fig. 3A, for N222, the virion production level relative to that of 5R was enhanced by the substitution with Ala (A) and Gly (G) as well as the growth-enhancing adaptive mutation Lys (K) but was decreased by the substitution with Phe (F) and Tyr (Y). The increase and decrease in virion production caused by these amino acid substitutions at N222 positively correlated with viral growth potential (Fig. 3A). For F223 (Fig. 3B), all mutant clones carrying Ala (A), Glu (E), Gly (G), His (H), Lys (K), and Ser (S) in addition to the adaptive mutation Tyr (Y) produced more virions than 5R, whereas only F223Y augmented viral replication efficiency. To study the cause of this apparent discrepancy, we compared the effects of F223Y and F223A/E/H/S on the early replication phase (Fig. 4). While F223Y showed levels of single-cycle viral infectivity and viral cDNA synthesis (late and integrated forms) similar to those for 5R, marked differences were observed in the behaviors of the other mutants. The virus infectivity of F223H was approximately 30% to that of 5R, and the infectivity of F223A/E/S was undetectable (Fig. 4A). A reduction in viral cDNA levels was also observed for F223A/E/H/S (Fig. 4B and C). These results indicate that the F223 residue is essential for the function of IN in the early replication phase. For D229 (Fig. 3C), virion production/replication ability was enhanced only by the adaptive mutation Glu (E), and the other mutants carrying D229A/G/T/W had reduced virion production/replication potential. For V234 (Fig. 3D), the Gly (G) substitution as well as the adaptive mutation Ile (I) augmented virion production/replication capability, but V234A/W decreased these. Because mutants D229K and V234E produced virions at the 5R level upon transfection (Fig. 3C and D), they are most likely to be defective for the early replication phase like the F223 mutants (Fig. 3B and 4). The results in Fig. 3 clarify the clear correlation between virion production and viral growth potential, except for the F223 mutants and the D229K/V234E mutants. However, increases or decreases in virion production were not dependent on altered amino acid sizes or chemical properties (Table 1). For example, virion production levels were enhanced by any altered amino acids tested at F223, and V234G and V234A increased and decreased virion production levels, respectively, despite their similar amino acid properties. Therefore, these results indicate that amino acid residues may not be a determinant for virion production.
TABLE 1.
Characteristics of viral clones carrying various mutations in the 3′ region of the pol gene
| Amino acid position in IN | Viral clone name | Amino acid | Codon | Size of amino acid | Chemical property(ies) of amino acid | Early replicationa | Virion productiona | Growthb |
|---|---|---|---|---|---|---|---|---|
| 222 | 5R | N | AAT | Medium-small | Neutral, hydrophilic | ++ | ++ | ++ |
| N222Kc | K | AAA | Medium-large | Basic | ++ | +++ | +++ | |
| N222K-2 | K | AAG | Medium-large | Basic | ++ | +++ | +++ | |
| N222A | A | GCT | Small | Aliphatic, hydrophobic | NDd | +++ | +++ | |
| N222G | G | GGT | Small | Aliphatic, hydrophobic | ND | +++ | +++ | |
| N222F | F | TTC | Large | Aromatic, hydrophobic | ND | + | − | |
| N222Y | Y | TAT | Large | Aromatic | ND | ++ | + | |
| 223 | 5R | F | TTT | Large | Aromatic, hydrophobic | ++ | ++ | ++ |
| F223Yc | Y | TAT | Large | Aromatic | ++ | +++ | +++ | |
| F223Y-2 | Y | TAC | Large | Aromatic | ++ | +++ | +++ | |
| F223H | H | CAT | Large | Basic | + | +++ | + | |
| F223G | G | GGT | Small | Aliphatic, hydrophobic | ND | +++ | − | |
| F223A | A | GCT | Small | Aliphatic, hydrophobic | − | +++ | − | |
| F223S | S | TCT | Small | Neutral, hydrophilic | − | +++ | − | |
| F223E | E | GAA | Medium-large | Acidic | − | +++ | − | |
| F223K | K | AAA | Medium-large | Basic | ND | +++ | − | |
| 229 | 5R | D | GAC | Medium-small | Acidic | ++ | ++ | ++ |
| D229Ec | E | GAA | Medium-large | Acidic | ++ | +++ | +++ | |
| D229E-2 | E | GAG | Medium-large | Acidic | ++ | +++ | +++ | |
| D229K | K | AAA | Medium-large | Basic | ND | ++ | + | |
| D229A | A | GCC | Small | Aliphatic, hydrophobic | ND | + | − | |
| D229G | G | GGC | Small | Aliphatic, hydrophobic | ND | + | + | |
| D229T | T | ACC | Medium-small | Neutral, hydrophilic | ND | − | − | |
| D229W | W | TGG | Large | Aromatic, hydrophobic | ND | − | − | |
| 234 | 5R | V | GTT | Medium-small | Aliphatic, hydrophobic | ++ | ++ | ++ |
| V234Ic | I | ATT | Medium-small | Aliphatic, hydrophobic | ++ | +++ | +++ | |
| V234I-2 | I | ATC | Medium-small | Aliphatic, hydrophobic | ++ | ++ | + | |
| V234I-3 | I | ATA | Medium-small | Aliphatic, hydrophobic | ++ | +++ | +++ | |
| V234G | G | GGT | Small | Aliphatic, hydrophobic | ND | +++ | +++ | |
| V234A | A | GCT | Small | Aliphatic, hydrophobic | ND | + | + | |
| V234E | E | GAA | Medium-large | Acidic | ND | ++ | + | |
| V234W | W | TGG | Large | Aromatic, hydrophobic | ND | + | + |
+++, >150% of 5R activity; ++, >70 to 150% of 5R activity; +, 10 to 70% of 5R activity; −, <10% of 5R activity. Data were obtained in M8166 cells.
+++, replication peaked earlier or virus production levels on the peak day were higher than those of 5R; ++, replication kinetics were similar to those of 5R; +, replication peaked later or virus production levels on the peak day were lower than those of 5R; −, replication was not detected during the observation period. Data were obtained in M8166 cells.
Adaptive (growth-enhancing) mutation.
ND, not done.
FIG 3.
Effect of various amino acid substitutions at the N222 (A), F223 (B), D229 (C), and V234 (D) sites in the Pol4889-4923 region on virion production and replication kinetics. Details of the substitution for each mutant are shown in Table 1. Left panels, virion production. M8166 cells were cotransfected with the indicated env-deficient proviral clones and a luciferase reporter vector (pGL3) by using a Nucleofector. Virion amounts in the culture supernatants on day 2 posttransfection were measured. The amount of p24 was normalized by luciferase activity in cell lysates. The amount of p24 relative to that produced by 5R is presented. Mean values ± SD from at least two independent experiments are shown. Results for N222K, F223Y, D229E, and V234I shown in Fig. 2D (underlined) are incorporated into each panel for an easy comparison. *, under the detection limit. Right panels, viral replication kinetics. Viruses were prepared from transfected 293T cells, and equal amounts were inoculated into M8166 cells. Virus replication was monitored by RT activity released into the culture supernatants. Data in panels A and D were obtained from the same experiment, and the same result for 5R is shown separately in panels A and D as a control. Representative data from three independent experiments are shown.
FIG 4.
Effect of various amino acid substitutions at the F223 site on the early viral replication phases. (A) Single-cycle viral infectivity. VSV-G-pseudotyped viruses were prepared from transfected 293T cells, and equal amounts were inoculated into M8166 cells. Cells were collected and lysed on day 1 postinfection for luciferase assays. Infectivity is presented as luciferase activity relative to that exhibited by 5R. Mean values ± SD from at least four independent experiments are shown. *, mean values are <0.1%. (B and C) Monitoring viral cDNA synthesis. VSV-G-pseudotyped viruses from transfected 293T cells were treated with DNase Ι, and equal amounts were inoculated into M8166 cells. Total DNA was extracted from infected cells on day 1 postinfection and subjected to real-time quantitative PCR analyses with primer pairs specific for the late (B) and integrated (C) forms of viral cDNA. The DNA copy number relative to that of 5R is presented. Mean values ± SD from at least four independent experiments are shown.
Synonymous codon changes in V234I alter virion production/replication ability.
Although no clear relationship was observed between the amino acids in the Pol4889-4923 region and increase in virion production (Table 1), virion production levels were markedly affected by the substitutions with different amino acids (Fig. 3). Since nucleotide mutations in the IN-coding sequence can induce viral phenotypic changes as described above, we speculated about the possible involvement of codon/nucleotide sequences in virion production enhancements. Thus, proviral clones carrying different codons for growth-enhancing mutations were constructed (N222K-2, F223Y-2, D229E-2, V234I-2, and V234I-3), and their growth potentials were compared to those of 5R and parental clones with each adaptive mutation. As shown in Fig. 5A, the growth of codon-altered viral clones (N222K-2, F223Y-2, and D229E-2) was more efficient than that of 5R, as observed for N222K, F223Y, and D229E. A fluctuation in viral replication potential among the V234I codon variants was noted: the growth abilities of V234I and V234I-3 were higher than that of 5R, but that of for V234I-2 was slightly impeded (Fig. 5A). While the single-cycle early infectivities of 5R and these codon variants were similar (Fig. 5B), their virion production levels varied in parallel with their replication potentials (Fig. 5A and C). Only V234I-2 exhibited a virion production level similar to that of 5R, and the other codon-altered mutants showed an enhanced level of virion production. The results for the V234I codon variants demonstrate the alteration in virion production levels in a nucleotide sequence-dependent manner, resulting in the modulation of viral replication ability.
FIG 5.
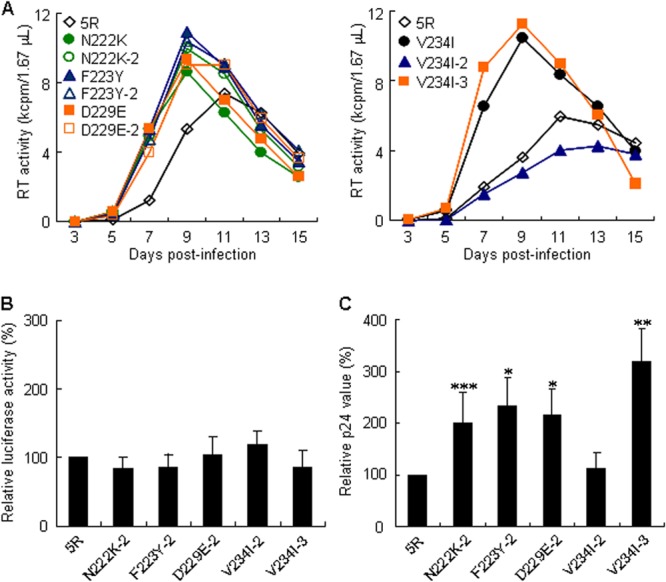
Effect of codon alterations at N222K, F223Y, D229E, and V234I sites on different stages of viral replication. N222K-2, F223Y-2, D229E-2, V234I-2, and V234I-3 indicate mutant clones containing an altered codon. The codon used for each mutant is shown in Table 1. (A) Viral replication kinetics. Input viruses were prepared from transfected 293T cells, and equal amounts were inoculated into M8166 cells. Virus replication was monitored by RT activity released into the culture supernatants. Representative data from at least two independent experiments are shown. (B) Single-cycle viral infectivity. VSV-G-pseudotyped viruses were prepared from transfected 293T cells, and equal amounts were inoculated into M8166 cells. Cell lysates were prepared on day 1 postinfection and subjected to luciferase assays. Infectivity is presented as luciferase activity relative to that exhibited by 5R. Mean values ± SD from at least three independent experiments are shown. (C) Virion production. M8166 cells were cotransfected with the indicated env-deficient proviral clones and a luciferase reporter vector (pGL3) by using a Nucleofector. Virion production on day 2 posttransfection was measured by the amount of p24 in the culture supernatants. The amount of p24 was normalized by luciferase activity in cell lysates. The amount of p24 relative to that produced by 5R is presented. Mean values ± SD from at least three independent experiments are shown. Significance relative to 5R as calculated by the Student t test is shown (*, P < 0.01; **, P < 0.05; ***, P < 0.1).
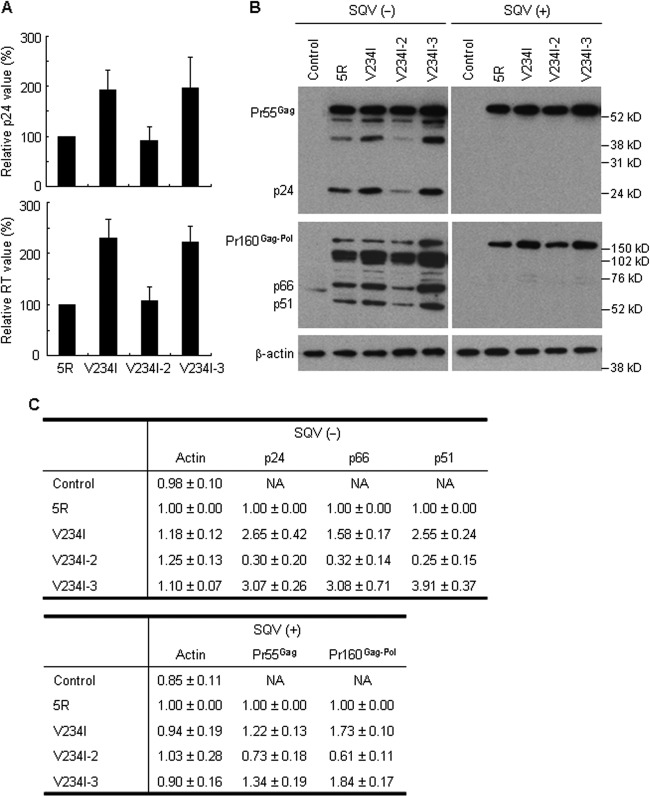
Virion production levels of V234I codon variants correlate with Gag and Gag-Pol expression levels in producer cells.
Synonymous codon changes in Ile (I) at amino acid position 234 in pol-IN caused an alteration in infectious virion production levels (Fig. 5C). We assumed that at least Gag and Gag-Pol expression levels, prerequisites for virion formation, correlatively varied with the amount of progeny virions. It has also been shown that cell-associated Gag was decreased for pol-IN deletion mutant viruses defective in virion production (20). To determine intracellular Gag and Gag-Pol expression levels, the proviral clones of 5R and V234I codon variants were transfected into 293T cells in the presence or absence of the HIV-1 protease inhibitor SQV. First, intracellular Gag-p24 and Pol-RT in the absence of SQV were measured by ELISA. While V234I and V234I-3, with improved virion production potential, expressed higher levels of Gag-p24 and Pol-RT than 5R, V234I-2 and 5R generated similar amounts (Fig. 6A). We then examined the intracellular expression patterns of Gag and Gag-Pol by Western blotting analyses. The expression profiles of Pr55Gag/p24 and Pr160Gag-Pol/p66/p51 for 5R and V234I codon variants were similar in the absence of SQV, which strongly suggested the absence of an effect of V234I mutations on viral protein processing [Fig. 6B SQV(−)]. The expression levels of Gag/Gag-Pol-related proteins were higher for V234I and V234I-3 than for 5R [Fig. 6B and C, SQV(−)]. In contrast, V234I-2 appeared to express slightly lower levels of these viral proteins than 5R [Fig. 6B and C, SQV(−)]. This small decrease in Gag/Gag-Pol expression levels may be consistent with the similar difference observed for the viral replication kinetics of 5R and V234I-2 (Fig. 5A). The variations in Pr55Gag and Pr160Gag-Pol expression levels in the presence of SQV appeared to be smaller than those obtained by Western blotting analyses [Fig. 6B and C, SQV(+)]. This may have been due to the weak recognition of Pr55Gag and Pr160Gag-Pol by the antibodies used. The results described above show that the alteration in the virion production/replication potential of V234I codon variants is in parallel with the increase or decrease in Gag/Gag-Pol expression levels.
FIG 6.
Effect of codon alterations at the V234I site on intracellular viral protein expression. (A) Expression levels of Gag-p24 and Pol-RT as determined by ELISA. 293T cells were transfected with the indicated proviral clones, and on day 2 posttransfection, lysates were prepared from transfected cells for the analysis of Gag-p24 and Pol-RT. The amounts of Gag-p24 and Pol-RT relative to those expressed by 5R are presented. Mean values ± SD from three independent experiments are shown. (B) Expression pattern of viral proteins as analyzed by Western blotting. 293T cells were transfected with the indicated proviral clones in the absence (−) or presence (+) of 2 μM SQV. Cell lysates were prepared from transfected cells on day 1 posttransfection, and analyzed by Western blotting using anti-p24 (upper panels), anti-RT (middle panels), and anti-β-actin (lower panels) antibodies. The migration positions of mass standards are indicated on the right. Representative data from two independent transfection experiments are shown. Control, pUC19. (C) Quantitative analysis of the Western blot. Signal intensities of viral proteins were quantitated, and the intensities relative to those with 5R proteins are shown. Mean values ± SD obtained from the two independent transfection experiments in panel B are indicated. NA, not applicable.
Viral replication capability can be altered by natural variations in the sequences of the 3′ region of the HIV-1 pol gene.
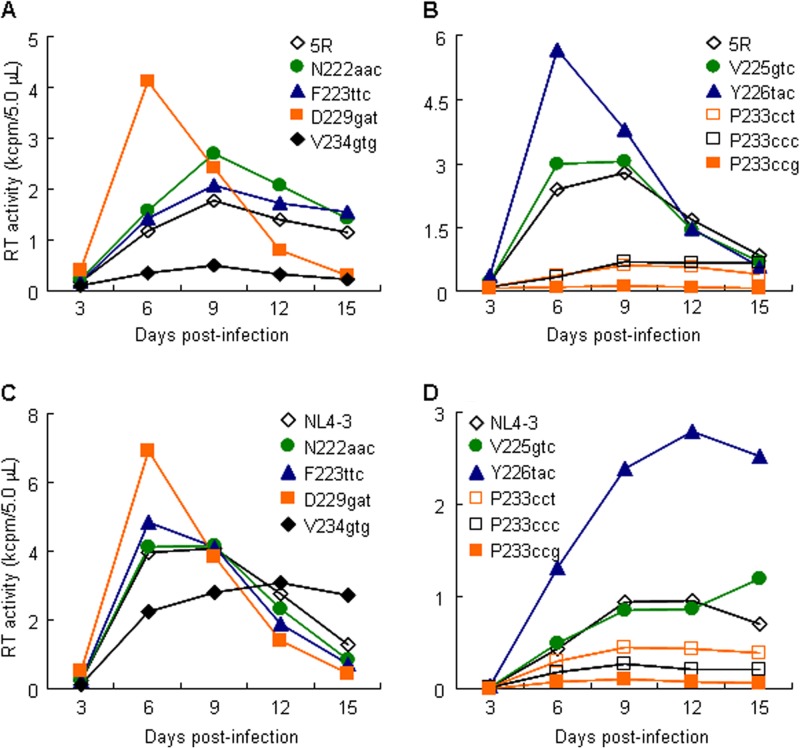
As observed for V234I codon variants, single-nucleotide changes resulted in alterations in virion production/replication potential through the modulation of Gag/Gag-Pol expression levels, which indicated the importance of the nucleotide sequence in the Pol4889-4923 region (Fig. 5 and 6). Recent studies revealed that a naturally occurring synonymous polymorphism in some genes can influence the expression levels, structures, and functions of their encoded proteins (42–44). In addition, the IN CTD sequence is more heterogenous than those of the other IN domains (15, 17, 19). To clarify the significance of the nucleotide sequence in the Pol4889-4923 region for viral replication, we examined HIV-1 sequences within the region obtained from the HIV Sequence Compendium (Los Alamos National Laboratory, NM, USA). As shown in Table 2, while viruses carrying F223Y or D229E, which we identified as growth-enhancing mutations, were not found, those with N222K or V234I were present. We noted that codon variants with distinct growth abilities, V234I (ATT) and V234I-2 (ATC), coexisted in a viral population with different frequencies (Table 2). This suggested that there may be natural variants of HIV-1 with distinct replication potentials. Moreover, a sequence comparison in the Pol4889-4923 region revealed the presence of natural synonymous variations for parental clone 5R-encoded amino acid residues, even though the frequency was lower than that of 5R (Table 2). Thus, we examined whether viral replication can be affected by natural synonymous changes at the sites of adaptive mutations (N222, F223, D229, and V234) as well as other sites (V225, Y226, and P233) within the Pol4889-4923 region. The viral growth kinetics of 5R and its natural variants at adaptive mutation sites (N222aac, F223ttc, D229gat, and V234gtg) were determined in human MT4/CCR5 cells (Fig. 7A and Table 2). While N222aac and F223ttc exhibited growth kinetics similar or slightly better than those of 5R, the viral replication potentials of D229gat and V234gtg was markedly higher and lower, respectively, than that of 5R (Fig. 7A and Table 2). We next determined the viral growth kinetics of additional natural synonymous variants (V225gtc, Y226tac, and P233cct/ccc/ccg). Alterations in the viral replication potentials of these clones were evident: the viral replication kinetics of V225gtc were similar to those of 5R, whereas growth ability was higher for Y226tac and lower for P233cct/ccc/ccg than for 5R (Fig. 7B and Table 2). On the other hand, the genome structure of 5R is different from that of natural human-tropic HIV-1 due to the change of the cyclophilin A-binding loop-coding region in gag and of an entire vif to the corresponding regions of SIVmac239 (34). Thus, we determined whether these natural synonymous variations also affect the viral replication of human-tropic HIV-1 (NL4-3 clone). The viral growth potential of NL4-3 was altered similarly as that of 5R by natural synonymous changes: the replication abilities of D229gat and Y226tac were higher than that of NL4-3, and those of V234gtg and P233cct/ccc/ccg were lower than that of NL4-3 (Fig. 7C and D). We conclude from these results that natural variations (single-nucleotide synonymous mutations) in the 3′ region of the HIV-1 pol gene (nt 4889 to 4923 for 5R and nt 4895 to 4929 for NL4-3) can alter viral replication potential.
TABLE 2.
Amino acid/codon frequency in the 3′ region of the pol gene (nt 4889 to 4923 for 5R and nt 4895 to 4929 for NL4-3) of HIV-1/SIVcpza
| Amino acid position in IN | Amino acid frequencyb |
Codon frequencyb |
Growthc | ||||
|---|---|---|---|---|---|---|---|
| Amino acid | No. | % | Codon | No. | % | ||
| 222 | Nd | 179 | 91.3 | AATd | 175 | 89.3 | ++ |
| AAC | 4 | 2.0 | ++/+++ | ||||
| Ke | 14 | 7.1 | AAAe | 14 | 7.1 | +++ | |
| Q | 2 | 1.0 | CAA | 2 | 1.0 | NDf | |
| D | 1 | 0.5 | GAT | 1 | 0.5 | ND | |
| 223 | Fd | 196 | 100.0 | TTTd | 183 | 93.4 | ++ |
| TTC | 13 | 6.6 | ++/+++ | ||||
| Ye | 0 | 0.0 | TATe | 0 | 0.0 | +++ | |
| 225 | Vd | 195 | 99.5 | GTTd | 175 | 89.3 | ++ |
| GTC | 20 | 10.2 | ++ | ||||
| L | 1 | 0.5 | CTT | 1 | 0.5 | ND | |
| 226 | Yd | 194 | 99.0 | TATd | 193 | 98.5 | ++ |
| TAC | 1 | 0.5 | +++ | ||||
| F | 1 | 0.5 | TTT | 1 | 0.5 | ND | |
| H | 1 | 0.5 | CAT | 1 | 0.5 | ND | |
| 229 | Dd | 196 | 100.0 | GACd | 194 | 99.0 | ++ |
| GAT | 2 | 1.0 | +++ | ||||
| Ee | 0 | 0.0 | GAAe | 0 | 0.0 | +++ | |
| 233 | Pd | 194 | 99.0 | CCAd | 135 | 68.9 | ++ |
| CCT | 35 | 17.9 | + | ||||
| CCC | 23 | 11.7 | + | ||||
| CCG | 1 | 0.5 | + | ||||
| S | 1 | 0.5 | TCA | 1 | 0.5 | ND | |
| T | 1 | 0.5 | ACC | 1 | 0.5 | ND | |
| 234 | Vd | 26 | 13.3 | GTTd | 25 | 12.8 | ++ |
| GTG | 1 | 0.5 | + | ||||
| Ie | 128 | 65.3 | ATTe | 127 | 64.8 | +++ | |
| ATC | 1 | 0.5 | + | ||||
| L | 40 | 20.4 | CTT | 39 | 19.9 | ND | |
| CTG | 1 | 0.5 | ND | ||||
| S | 1 | 0.5 | AGC | 1 | 0.5 | ND | |
| T | 1 | 0.5 | ACT | 1 | 0.5 | ND | |
For 196 sequences of HIV-1/SIVcpz complete genomes from the HIV Sequence Compendium, 2011 (Los Alamos National Laboratory; http://www.hiv.lanl.gov).
Relative to 196 sequences of HIV-1/SIVcpz strains.
+++, replication peaked earlier or virus production levels on the peak day were higher than those of the parental clones; ++, replication kinetics were similar to those of the parental clones; +, replication peaked later or virus production levels on the peak day were lower than those of the parental clones. Data were obtained in M8166 or MT4/CCR5 cells.
Amino acid encoded or codon usage in parental clones.
Amino acid encoded or codon usage in adapted (growth-enhanced) viruses.
ND, not done.
FIG 7.
Effect of single-nucleotide synonymous changes in the Pol4889-4923 region on viral replication potential. Viruses were prepared from transfected 293T cells, and equal amounts were inoculated into MT4/CCR5 cells. Virus replication was monitored by RT activity released into the culture supernatants. Representative data from two independent experiments are shown.
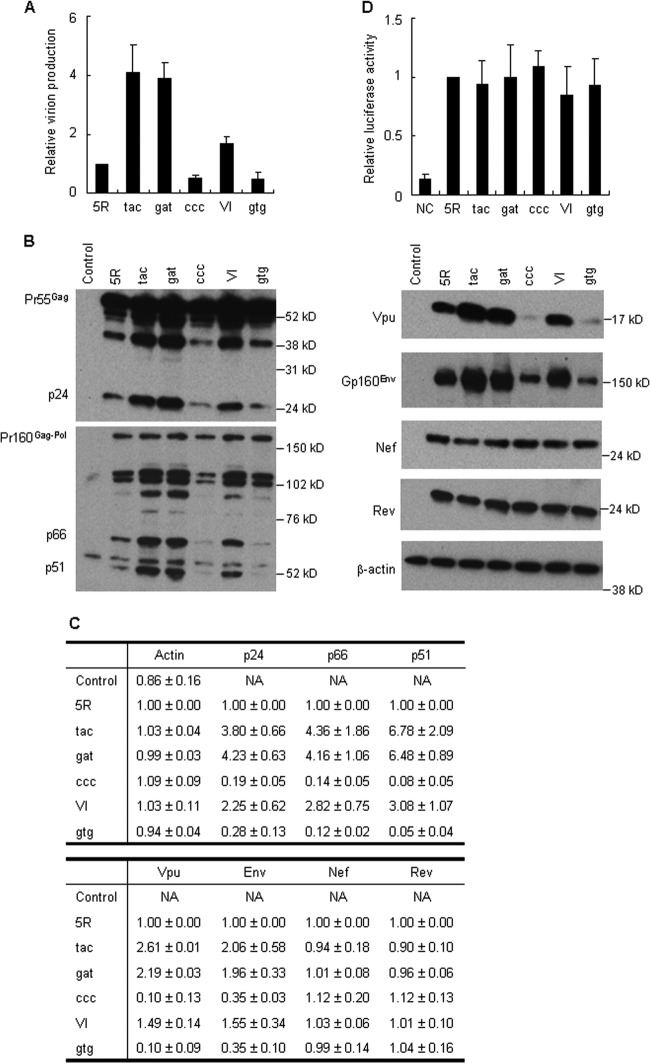
Natural synonymous changes influence the expression levels of intracellular viral proteins (Gag, Gag-Pol, Vpu, and Env) but not those of Nef and Rev.
Gag/Gag-Pol expression levels in V234I codon variants varied correlatively with their virion production/replication abilities (Fig. 5 and 6). Env, in addition to Gag/Gag-Pol, is of course required for infectious virion formation. HIV-1-encoded proteins are translated from distinct viral mRNA species, and the regulation of gene expression is different for early transcripts (completely spliced form) and late transcripts (singly spliced and unspliced forms). To investigate further the virological effect of single-nucleotide natural variations, we determined the expression levels of products from various mRNA species: Gag and Gag-Pol from the unspliced form, Vpu and Env from the singly spliced form, and Nef and Rev from the completely spliced form. Proviral clones (5R, Y226tac, D229gat, P233ccc, V234I, and V234gtg) were transfected into 293T cells, and cell lysates prepared on day 1 posttransfection were analyzed by Western blotting. Although V234I was not a synonymous change in the Pol4889-4923 region, this clone was included in this analysis because V234I is a single-nucleotide natural variant of V234 in 5R, and its virion production/replication ability was increased or decreased by single-nucleotide substitutions at this position (Table 2). As shown in Fig. 8A, we confirmed that virion production levels in transfected 293T cells correlated with viral replication potential (Fig. 2, 7, and 8A) (upregulated, Y226tac, D229gat, and V234I; downregulated, P233ccc and V234gtg). We then examined the intracellular expression level of each viral protein (Fig. 8B). The intracellular expression levels of Gag, Gag-Pol, Vpu, and Env varied among the clones tested and correlated with viral replication potential: clones with enhanced growth efficiencies expressed higher levels of these proteins, and vice versa (Fig. 2, 7, and 8B and C). In contrast, the expression levels of Nef and Rev by all proviral clones tested were similar to that of 5R, and they were constant for all variants with different virion production/replication potentials (Fig. 2, 7, and 8). Furthermore, Tat activity was determined by cotransfection assays with 5R, its single-nucleotide variants, and the 5RLTR-Luc reporter construct. No significant difference in the abilities of these proviral clones to trans-activate luciferase gene expression was observed, which indicated that 5R and its variants had similar Tat activity (Fig. 8D). These results suggest that single-nucleotide changes in the Pol4889-4923 region can alter virion production/replication potential by modulating the expression levels of late (but not early) viral proteins.
FIG 8.
Effect of single-nucleotide changes on virion production and intracellular expression levels of viral proteins. (A) Virion production in transfected cells. 293T cells were transfected with the indicated proviral clones, and on day 1 posttransfection, virion production was monitored by the amount of Gag-p24 in the culture supernatants. The amount of p24 relative to that produced by 5R is presented. Mean values ± SD from three independent experiments are shown. (B) Expression of various viral proteins in transfected cells. 293T cells were transfected with the indicated proviral clones, and on day 1 posttransfection, cell lysates were prepared for Western blotting using anti-Gag-p24, anti-RT, anti-Vpu, anti-Env-gp160, anti-Nef, anti-Rev, and anti-β-actin antibodies. The migration positions of mass standards are indicated on the right. Representative data from two independent transfection experiments are shown. Control, pUC19; tac, Y226tac; gat, D229gat; ccc, P233ccc; VI, V234I; gtg, V234gtg. (C) Quantitative analysis of the Western blot. Signal intensities of viral proteins were quantitated, and the intensities relative to those for 5R proteins are shown. Mean values ± SD obtained from the two independent transfection experiments in panel B are indicated. NA, not applicable. (D) Analysis of Tat activity. 293T cells were cotransfected with the indicated proviral clones and an LTR-driven luciferase reporter clone, and on day 1 posttransfection, cell lysates were prepared for luciferase assays. Luciferase activity relative to that exhibited by 5R is presented. Mean values ± SD from three independent experiments are shown. NC, negative control (basal luciferase activity of the LTR-driven luciferase reporter clone in the absence of proviral clones).
Naturally occurring single-nucleotide variations change the expression patterns of HIV-1 mRNA species.
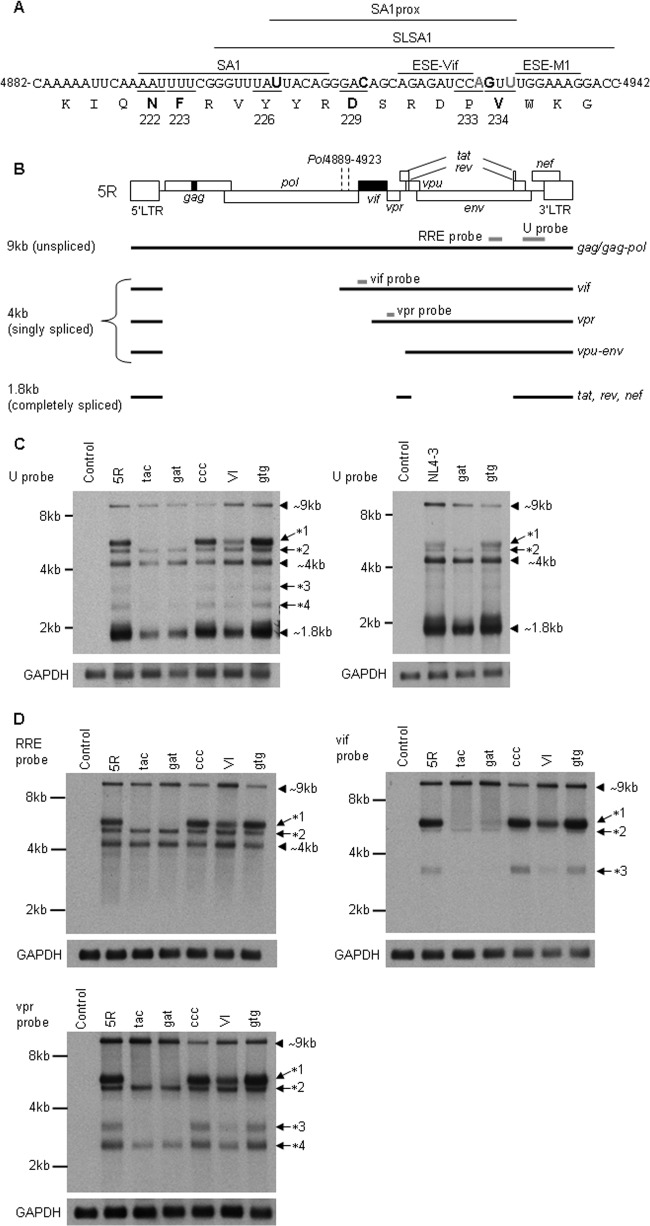
Naturally occurring single-nucleotide mutations that alter the expression levels of viral late proteins were Y226tac, D229gat, V234I, P233ccc, and V234gtg (Fig. 8). We noted that these mutations clustered in the region proximal to the splice acceptor A1 (SA1) site (designated SA1prox in Fig. 9A). To determine the mechanistic basis for the altered phenotype, we analyzed the effect of single-nucleotide changes on the profiles of viral mRNA expression. 293T cells were transfected with parental clones (5R and NL4-3) or their mutants, and DNase I-treated poly(A)+ RNAs were prepared from total cell RNAs at 10 to 20 h posttransfection. Northern blot analysis of these samples using a probe (U probe) to detect all HIV-1 mRNA species (Fig. 9B) was then performed (Fig. 9C). Because no significant difference in the RNA expression patterns for the samples of each clone prepared at 10 and 20 h posttransfection was noted (data not shown), we comparatively analyzed viral mRNAs in cells at the latter time point. As shown in Fig. 9C, the expression levels of viral late proteins did not directly correlate with the steady-state levels of their respective transcripts (9 kb and 4 kb) (Fig. 8B and 9C, left panel). The total amounts of viral mRNAs, especially the 1.8-kb mRNA species, were lower for the clones with increased viral late protein expression (Y226tac, D229gat, and V234I) than for 5R and the other mutants (P233ccc and V234gtg). However, single-nucleotide mutations always gave unique viral mRNA expression profiles. Several bands (*1 to *4 in Fig. 9C, left panel) other than the three major viral RNA species (1.8 kb, 4 kb, and 9 kb) were more or equally intense for P233ccc and V234gtg, with the poor expression of late proteins, and less intense for Y226tac, D229gat, and V234I, with the high expression of late proteins, than for 5R. Similar results, albeit to a lesser extent, were obtained for NL4-3 and its mutants with higher or lower abilities to replicate in cells (Fig. 9C, right panel). As such, viruses with a relatively high replication ability in each group (5R and NL4-3) appeared to be tuned up to have three major mRNA species. Northern blot analysis using HeLa cells was performed to exclude the possibility that the viral mRNA expression pattern described above was 293T cell specific. Results similar to those in 293T cells were obtained for 5R, NL4-3, and their mutants (D229gat and V234gtg) (data not shown). Taken together, our results here show that single-nucleotide changes in the SA1prox affect the expression patterns of viral mRNA species and suggest that this transition may lead to the enhancement or reduction in viral late protein expression/replication efficiency.
FIG 9.
Effect of single-nucleotide changes on viral mRNA expression. (A) Nucleotide and amino acid sequences in the 3′ region of the pol gene. Nucleotides 4882 to 4942 correspond to the NL4-3 sequence (35) (GenBank accession no. AF324493). Black and gray bold letters in the nucleotide sequence show the sites at which the single-nucleotide substitution promoted or decreased viral replication efficiency, respectively. Bold letters in the amino acid sequence represent adaptive mutation sites. The SA1prox (this study), SA1 site (3, 4), ESE-Vif (27), ESE-M1 (28), and SLSA1 region (45) are as indicated. Numbers and underlines show the positions of amino acids in IN and their codons, respectively. (B) Schematic presentation of the 5R genome organization and various HIV-1 mRNA species. The genome structure of 5R is shown as in Fig. 1. Gray bars represent the regions used as probes for Northern blot analyses. The Pol4889-4923 region is also indicated. RRE, Rev-responsive element; U, universal. (C and D) Steady-state expression levels of HIV-1 mRNA species. Total RNA was prepared from 293T cells transfected with the indicated proviral clones, and poly(A)+ RNA was selected. After the DNase I treatment, samples were subjected to Northern blot analysis using the indicated probe. GAPDH was used as an internal standard. Three major species of viral mRNA (∼9 kb, ∼4 kb, and ∼1.8 kb) are shown by arrowheads. The other extra bands *1 to *4 are indicated by arrows. RNA size markers (8 kb, 4 kb, and 2 kb) are on the left. Representative data from four independent experiments (C, left panel [5R and its mutants]) and from two independent experiments (C, right panel [NL4-3 and its mutants] and D) are shown. Control, pUC19; tac, Y226tac; gat, D229gat; ccc, P233ccc; VI, V234I; gtg, V234gtg.
To identify the nature of transcripts detected as extra bands (*1 to *4) (Fig. 9C), we performed Northern blot analysis using RRE, vif, and vpr probes (Fig. 9B). As expected, 9-kb transcripts but not 1.8-kb transcripts were detected by the RRE, vif, and vpr probes (Fig. 9D). While all transcripts longer than 4 kb were detected by the RRE probe, the vif and vpr probes recognized the species *1 to *3 and *1 to *4, respectively. The *2 band was more intense with the vpr probe than with the vif probe. Thus, the *1 species contained the vif transcript (Fig. 9B), and the *2 band consisted mainly of the vpr transcript (Fig. 9B). Transcripts *3 and *4 contained the Vif/Vpr-coding region and Vpr-coding region, respectively, without the RRE region.
Northern blot analysis revealed that the lower expression of late proteins by 5R, P233ccc, and V234gtg was linked to the abundance of transcripts, especially the *1 and *3 species. We hypothesized that these abundant transcripts may disturb the expression of late proteins. To test this possibility, we performed interference assays by the cotransfection of D229gat and 5R/V234gtg (Fig. 10). If the extra transcripts *1 and *3 interfere with translation from transcripts corresponding to late proteins, the Gag-p24 expression levels of D229gat would be proportionally decreased upon cotransfection with increasing amounts of 5R or V234gtg. As shown in Fig. 10 (top and middle panels), p24 expression levels were increased in cells upon single transfection of D229gat, 5R, or V234gtg with an increasing DNA amount, and marked differences were observed between each clone (D229gat > 5R > V234gtg). When a constant amount of D229gat and an increasing amount of 5R or V234gtg were cotransfected, p24 expression levels reflected just the addition of the amount produced by each clone. Virion production from cells correlated well with the intracellular p24 expression levels (data not shown). Cotransfection assays using D229gat and the gag/gag-pol frameshift mutant of V234gtg (gtg-Spe), which is incapable of producing p24, were performed to confirm this result. As is clearly observed in Fig. 10 (bottom panel), the increase in the amount of the gtg-Spe clone did not affect p24 expression levels produced from D229gat. Taken together, these results suggest that a large amount of transcripts (especially the *1 and *3 species) in 5R and V234gtg clones does not interfere with Gag-p24 expression and also that the single-nucleotide changes in the SA1prox act on the expression of late proteins in cis.
FIG 10.
Expression level of Gag-p24 in singly or doubly transfected cells. 293T cells were transfected with proviral clones as indicated, and on day 1 posttransfection, Gag-p24 expression level in cell lysates was determined. The amount of p24 relative to that produced upon transfection with 0.2 μg of the D229gat (gat) clone is presented. Mean values ± SD from three independent experiments are shown. gat, D229gat; gtg, V234gtg; gtg-Spe, a frameshift mutant of V234gtg.
DISCUSSION
In this study, we demonstrated that four adaptive mutations in the 3′ region of the pol gene encoding IN upregulated the viral replication potential by increasing virion production levels without any effects on the early replication phase (Fig. 2). Moreover, the identification of V234I codon variants that have different abilities to produce virions and replicate in cells suggested regulation by single-nucleotide changes (Fig. 5). A comparative investigation of nucleotide sequences in the 3′ region of the pol gene (nt 4889 to 4923 for 5R and nt 4895 to 4929 for NL4-3) has revealed that these variants naturally coexist in a viral population (Table 2). We show here that naturally occurring synonymous changes (Y226tac, D229gat, P233cct/ccc/ccg, and V234gtg) can alter the viral replication potentials of HIV-1 5R and NL4-3 (Fig. 7 and Table 2).
The naturally occurring single-nucleotide variations that alter viral replication potential clustered in the SA1prox (Fig. 9A). These mutations were identified by both viral adaptation experiments (32) and comparative analysis of numerous HIV-1/SIVcpz genomes (Table 2). These findings suggest that the nucleotide sequence in the SA1prox may be involved in the viral adaptation/evolution process. Recent RNA structure analysis has shown that the nucleotide sequence proximal to SA1 within the HIV-1 genome forms a stem-loop structure (45, 46). Moreover, Pollom et al. reported that this stem-loop structure, designated “SLSA1,” was conserved between HIV-1 NL4-3 and SIVmac239, suggesting the virological importance of the SLSA1 structure (45). Interestingly, all single-nucleotide mutations analyzed in Fig. 8 and 9 were mapped onto the SLSA1 sequence (at positions 4901 to 4942 in Fig. 9A). The emergence of novel mutations within this region may be limited due to its effect on IN functions, SA1 functions, and the SLSA1 structure. This may explain the low frequency of single-nucleotide mutations that alter viral replication efficiencies among HIV-1 genomes (Table 2). Nevertheless, the presence of such single-nucleotide variations in SLSA1 represents the plasticity of viruses with the ability to adapt themselves under various constraints. Our results on the replication-altering mutations within SLSA1 may also be useful for analyzing changes in the RNA sequence/structure and their effects on viral replication. Because the SLSA1 structure is conserved between HIV-1 NL4-3 and SIVmac239, it is of interest to determine whether naturally occurring single-nucleotide synonymous mutations in this region affect the replication efficiency of SIVmac239 and its closely related primate lentiviruses.
The change in virion production/replication ability was reflected in the expression levels of viral late proteins (Gag, Gag-Pol, Vpu, and Env) but not in those of the early proteins (Nef and Rev) (Fig. 7 and 8). However, a direct positive correlation between the steady-state levels of viral mRNAs and their corresponding late proteins was not observed (Fig. 8 and 9). More viral RNAs and a large number of viral RNA species were synthesized in cells producing low expressors of viral late proteins (5R, P233ccc, and V234gtg) than in those producing high expressors (Y226tac, D229gat, and V234I). This may imply that the expression level of mRNAs directed by high expressors is necessary and sufficient for the efficient expression of viral proteins and optimal viral replication. We initially assumed that various kinds of viral transcripts by low expressors may hinder the efficient translation of viral late proteins. However, interference assays between variants (Fig. 10) showed that this prediction may not be accurate. Moreover, no significant difference in the Tat activity of 5R and its variants was observed (Fig. 8D). Therefore, it is reasonable to assume that single-nucleotide changes in the SA1prox act in cis and may influence splicing, mRNA stability, mRNA transport, and/or translation efficiency from mRNAs.
The abundance of the vif transcript (species *1 in Fig. 9C and D) observed for 5R and low expressors (P233ccc and V234gtg) suggests enhanced splicing at the SA1 site. On one hand, a combination of two synonymous mutations in SLSA1 that changes its RNA structure was reported to affect splicing at the SA3 site downstream of SA1 but not that at the SA1 site, showing long-range RNA interactions and cross talk between splicing sites (45). This may explain variations in the expression levels of transcripts (the *3, *4, and 1.8-kb species) among 5R/low expressors and high expressors. Since these transcripts do not contain the RRE region, it is clear that they are generated via splicing downstream of SA1. The abundance of these transcripts (*3, *4, and 1.8-kb species) for 5R and low expressors may also imply that the overall splicing efficiency of these clones is higher than that of high expressors. Efficient splicing at SA sites may compete with Rev function, and equilibrium between the strength of splicing acceptors and Rev function for the nuclear export of Rev-dependent mRNAs is important for virus replication (28). Thus, increased splicing for 5R and low expressors may obstruct the function of Rev, which results in a decrease in the Rev-dependent expression of late proteins from RRE-containing transcripts. On the other hand, while the amounts of 1.8-kb mRNAs of 5R and low expressors were larger than those of high expressors, the expression levels of viral early proteins were similar among 5R and its variants. A high concentration of Rev was previously shown to inhibit the translation from various RNAs (47). It is possible that the expression of viral early proteins may be regulated at an optimal level for viral replication. Alternatively, the translation efficiency of ∼40 mRNA isoforms synthesized by alternative splicing events may vary due to differences in their noncoding sequences and/or structures. Viral mRNA species within 1.8-kb and 4-kb RNAs were shown to be altered by mutations that change splicing efficiency at SA1 or the structure of SLSA1 (27, 45, 48). Viral mRNA isoforms with a low translation efficiency, even if present in abundance, may not express a high level of their corresponding proteins.
vif mRNA expression is strongly influenced by splicing efficiency at the SA1 site. The regulation of splicing at SA1 is complicated and is determined by various elements, including three different exonic splicing enhancers (ESE-Vif and ESE-M1 [Fig. 9A] and ESE-M2 [nt 4956 to 4962 in NL4-3]), a suboptimal D2 splicing site (nt 4960 to 4970 in NL4-3), a GGGG silencer (nt 4968 to 4971 in NL4-3), and a G run (GI2-1, nt 5034 to 5038 in NL4-3), which are located within the region from SA1 to just upstream of the vif start codon (nt 5041 in NL4-3) (27, 28, 48). The proviral clone 5R was constructed by introducing SIVmac239 vif into the downstream region of the pol open reading frame in the NL4-3 genome (Fig. 1 and 9B) (34). As a result, while the pol and vif genes of NL4-3 partially overlap, those of 5R do not. Since splicing efficiency is dependent on the sequence around the splice sites and their distance from the regulatory elements, the insertion of SIVmac239 vif into NL4-3 may have changed the splicing event at SA1. Indeed, 5R produced abundant amounts of the vif transcript (the *1 species in Fig. 9). The increase in vif mRNA was previously shown to decrease virion production, and the proportion between unspliced and spliced mRNAs has been suggested to be important for virion production (27). In agreement with this finding, we found that the virion production level from 293T cells transfected with 5R was lower than that from cells transfected with NL4-3 (data not shown). The decrease in vif transcript (*1 species) expression for high expressors may have caused the increase in virion production.
The splicing balance of viral mRNAs has been suggested to have biologically significant effects on viral replication (4, 9–11). Accumulating evidence has shown that HIV-1 gene expression processes, composed of transcription, poly(A) tailing, splicing, mRNA export, and subsequent translation, are mutually affected and coupled, even though these processes are biochemically distinguished (1, 2, 49). In addition, various elements within the HIV-1 genome and a number of virus/host factors have been shown to be involved in HIV-1 gene expression (3, 4, 9–11, 25–30, 50–55). The virological importance of the nucleotide sequence in the SA1prox is evident from the increase or decrease in viral replication caused by naturally occurring single-nucleotide changes. Further studies are needed to elucidate the molecular mechanism underlying the modulation of overall HIV-1 gene expression generated by single-nucleotide changes in the SA1prox.
ACKNOWLEDGMENTS
This study was supported in part by a grant from the Ministry of Health, Labor and Welfare of Japan (Research on HIV/AIDS project no. H24-005).
We are indebted to the NIH AIDS Research and Reference Reagent Program and Immuno Ltd./the MRC AIDS Directed Programme Reagent Project for antibodies. We thank Kazuko Yoshida for her editorial assistance.
We declare that no competing interests exist.
Footnotes
Published ahead of print 29 January 2014
REFERENCES
- 1.Caputi M. 2011. The regulation of HIV-1 mRNA biogenesis, p 79–100 In Grabowski P. (ed), RNA processing. InTech, Rijeka, Croatia: http://www.intechopen.com/books/rna-processing/the-regulation-of-hiv-1-mrna-biogenesis [Google Scholar]
- 2.Karn J, Stoltzfus CM. 2012. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb. Perspect. Med. 4:a006916. 10.1101/cshperspect.a006916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amendt BA, Si ZH, Stoltzfus CM. 1995. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 15:4606–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purcell DFJ, Martin MA. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication and infectivity. J. Virol. 67:6365–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dayton AI, Sodroski JG, Rosen CA, Goh WC, Haseltine WA. 1986. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell 44:941–947. 10.1016/0092-8674(86)90017-6 [DOI] [PubMed] [Google Scholar]
- 6.Fisher AG, Feinberg MB, Josephs SF, Harper ME, Marselle LM, Reyes G, Gonda MA, Aldovini A, Debouk C, Gallo RC, Wong-Staal F. 1986. The trans-activator gene of HTLV-III is essential for virus replication. Nature 320:367–371. 10.1038/320367a0 [DOI] [PubMed] [Google Scholar]
- 7.Felber BK, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis GN. 1989. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. U. S. A. 86:1495–1499. 10.1073/pnas.86.5.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sodroski J, Goh WC, Rosen C, Dayton A, Terwilliger E, Haseltine W. 1986. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature 321:412–417. 10.1038/321412a0 [DOI] [PubMed] [Google Scholar]
- 9.Jablonski JA, Caputi M. 2009. Role of cellular RNA processing factors in human immunodeficiency virus type 1 mRNA metabolism, replication, and infectivity. J. Virol. 83:981–992. 10.1128/JVI.01801-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquenet S, Decimo D, Muriaux D, Darlix JL. 2005. Dual effect of the SR proteins ASF/SF2, SC35 and 9G8 on HIV-1 RNA splicing and virion production. Retrovirology 2:33. 10.1186/1742-4690-2-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wentz PM, Moore BE, Cloyd MV, Berget SM, Donehower LA. 1997. A naturally arising mutation of a potential silencer of exon splicing in human immunodeficiency virus type 1 induces domain aberrant splicing and arrests virus production. J. Virol. 71:8542–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ao Z, Fowke KR, Cohen EA, Yao X. 2005. Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology 2:62. 10.1186/1742-4690-2-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda T, Nishitsuji H, Zhou X, Nara N, Ohashi T, Kannagi M, Masuda T. 2004. Evaluation of the functional involvement of human immunodeficiency virus type 1 integrase in nuclear import of viral cDNA during acute infection. J. Virol. 78:11563–11573. 10.1128/JVI.78.21.11563-11573.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R, Ghory HZ, Engelman A. 2005. Genetic analyses of conserved residues in the carboxyl-terminal domain of human immunodeficiency virus type 1 integrase. J. Virol. 79:10356–10368. 10.1128/JVI.79.16.10356-10368.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuda T, Planelles V, Krogstad P, Chen IS. 1995. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 69:6687–6696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammed KD, Topper MB, Muesing MA. 2011. Sequential deletion of the integrase (Gag-Pol) carboxyl terminus reveals distinct phenotypic classes of defective HIV-1. J. Virol. 85:4654–4666. 10.1128/JVI.02374-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin CG, Taddeo B, Haseltine WA, Farnet CM. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol. 68:1633–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu K, Dobard C, Chow SA. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 78:5045–5055. 10.1128/JVI.78.10.5045-5055.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bukovsky A, Göttlinger H. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820–6825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao WH, Wang CT. 2004. Characterization of human immunodeficiency virus type 1 Pr160gag-pol mutants with truncations downstream of the protease domain. Virology 329:180–188. 10.1016/j.virol.2004.08.010 [DOI] [PubMed] [Google Scholar]
- 22.Lu R, Limón A, Devroe E, Silver PA, Cherepanov P, Engelman A. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 78:12735–12746. 10.1128/JVI.78.23.12735-12746.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannon PM, Byles ED, Kingsman SM, Kingsman AJ. 1996. Conserved sequences in the carboxyl terminus of integrase that are essential for human immunodeficiency virus type 1 replication. J. Virol. 70:651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannon PM, Wilson W, Byles E, Kingsman SM, Kingsman AJ. 1994. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J. Virol. 68:4768–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charneau P, Alizon M, Clavel F. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66:2814–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cochrane AW, Jones KS, Beidas S, Dillon PJ, Skalka AM, Rosen CA. 1991. Identification and characterization of intragenic sequences which repress human immunodeficiency virus structural gene expression. J. Virol. 65:5305–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Exline CM, Feng Z, Stoltzfus CM. 2008. Negative and positive mRNA splicing elements act competitively to regulate human immunodeficiency virus type 1 vif gene expression. J. Virol. 82:3921–3931. 10.1128/JVI.01558-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kammler S, Otte M, Hauber I, Kjems J, Hauber J, Schaal H. 2006. The strength of the HIV-1 3′ splice sites affects Rev function. Retrovirology 3:89. 10.1186/1742-4690-3-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maldarelli F, Martin MA, Strebel K. 1991. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: novel level of gene regulation. J. Virol. 65:5732–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider R, Campbell M, Nasioulas G, Felber BK, Pavlakis GN. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71:4892–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandal D, Feng Z, Stoltzfus CM. 2008. Gag-processing defect of human immunodeficiency virus type 1 integrase E246 and G247 mutants is caused by activation of an overlapping 5′ splice site. J. Virol. 82:1600–1604. 10.1128/JVI.02295-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nomaguchi M, Doi N, Fujiwara S, Saito A, Akari H, Nakayama EE, Shioda T, Yokoyama M, Sato H, Adachi A. 2013. Systemic biological analysis of the mutations in two distinct HIV-1mt genomes occurred during replication in macaque cells. Microbes Infect. 15:319-328. 10.1016/j.micinf.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 33.Rhee SY, Liu TF, Kiuchi M, Zioni R, Gifford RJ, Holmes SP, Shafer RW. 2008. Natural variation of HIV-1 group M integrase: implications for a new class of antiretroviral inhibitors. Retrovirology 5:74. 10.1186/1742-4690-5-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamada K, Igarashi T, Martin MA, Khamsri B, Hatcho K, Yamashita T, Fujita M, Uchiyama T, Adachi A. 2006. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. U. S. A. 103:16959–16964. 10.1073/pnas.0608289103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashita M, Emerman M. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78:5670–5678. 10.1128/JVI.78.11.5670-5678.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doi N, Fujiwara S, Adachi A, Nomaguchi M. 2010. Growth ability in various macaque cell lines of HIV-1 with simian cell-tropism. J. Med. Invest. 57:284-292. 10.2152/jmi.57.284 [DOI] [PubMed] [Google Scholar]
- 39.Kawamura M, Ishizaki T, Ishimoto A, Shioda T, Kitamura T, Adachi A. 1994. Growth ability of human immunodeficiency virus type 1 auxiliary gene mutants in primary blood macrophage cultures. J. Gen. Virol. 75:2427–2431. 10.1099/0022-1317-75-9-2427 [DOI] [PubMed] [Google Scholar]
- 40.Yee JK, Miyanohara A, LaPorte P, Bouic K, Burns JC, Friedmann T. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 91:9564–9568. 10.1073/pnas.91.20.9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamashita T, Doi N, Adachi A, Nomaguchi M. 2008. Growth ability in simian cells of monkey cell-tropic HIV-1 is greatly affected by downstream region of the vif gene. J. Med. Invest. 55:236-240. 10.2152/jmi.55.236 [DOI] [PubMed] [Google Scholar]
- 42.Chamary JV, Parmley JL, Hurst LD. 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 7:98–108. 10.1038/nrg1770 [DOI] [PubMed] [Google Scholar]
- 43.Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 12:32–42. 10.1038/nrg2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauna ZE, Kimchi-Sarfaty C. 2011. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 12:683–691. 10.1038/nrg3051 [DOI] [PubMed] [Google Scholar]
- 45.Pollom E, Dang KK, Potter EL, Gorelick RJ, Burch CL, Weeks KM, Swanstrom R. 2013. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog. 9:e1003294. 10.1371/journal.ppat.1003294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716. 10.1038/nature08237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groom HC, Anderson EC, Dangerfield JA, Lever AM. 2009. Rev. regulates translation of human immunodeficiency virus type 1 RNAs. J. Gen. Virol. 90:1141–1147. 10.1099/vir.0.007963-0 [DOI] [PubMed] [Google Scholar]
- 48.Widera M, Erkelenz S, Hillebrand F, Krikoni A, Widera D, Kaisers W, Deenen R, Gombert M, Dellen R, Pfeiffer T, Kaltschmidt B, Münk C, Bosch V, Köhrer K, Schaal H. 2013. An intronic G run within HIV-1 intron 2 is critical for splicing regulation of vif mRNA. J. Virol. 87:2707–2720. 10.1128/JVI.02755-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cochrane AW, McNally MT, Mouland AJ. 2006. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology 3:18. 10.1186/1742-4690-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, Mouland AJ. 2008. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 14:914–927. 10.1261/rna.829208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Tang H, Mullen TM, Westberg C, Reddy TR, Rose DW, Wong-Staal F. 1999. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc. Natl. Acad. Sci. U. S. A. 96:709–714. 10.1073/pnas.96.2.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swanson CM, Sherer NM, Malim MH. 2010. SRp40 and SRp55 promote the translation of unspliced human immunodeficiency virus type 1 RNA. J. Virol. 84:6748–6759. 10.1128/JVI.02526-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tazi J, Bakkour N, Marchand V, Ayadi L, Aboufirassi A, Branlant C. 2010. Alternative splicing: regulation of HIV-1 multiplication as a target for therapeutic action. FEBS J. 277:867–876. 10.1111/j.1742-4658.2009.07522.x [DOI] [PubMed] [Google Scholar]
- 54.Yedavalli VS, Jeang KT. 2011. Matrin 3 is a co-factor for HIV-1 Rev in regulating post-transcriptional viral gene expression. Retrovirology 8:61. 10.1186/1742-4690-8-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119:381–392. 10.1016/j.cell.2004.09.029 [DOI] [PubMed] [Google Scholar]
- 56.Shibata R, Kawamura M, Sakai H, Hayami M, Ishimoto A, Adachi A. 1991. Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol. 65:3514–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]