ABSTRACT
Human metapneumovirus (hMPV) is a relatively recently identified paramyxovirus that causes acute upper and lower respiratory tract infection. Entry of hMPV is unusual among the paramyxoviruses, in that fusion is accomplished by the fusion (F) protein without the attachment glycoprotein (G protein). It has been suggested that hMPV F protein utilizes integrin αvβ1 as a cellular receptor. Consistent with this, the F proteins of all known hMPV strains possess an integrin-binding motif (329RGD331). The role of this motif in viral entry, infectivity, and pathogenesis is poorly understood. Here, we show that α5β1 and αv integrins are essential for cell-cell fusion and hMPV infection. Mutational analysis found that residues R329 and G330 in the 329RGD331 motif are essential for cell-cell fusion, whereas mutations at D331 did not significantly impact fusion activity. Furthermore, fusion-defective RGD mutations were either lethal to the virus or resulted in recombinant hMPVs that had defects in viral replication in cell culture. In cotton rats, recombinant hMPV with the R329K mutation in the F protein (rhMPV-R329K) and rhMPV-D331A exhibited significant defects in viral replication in nasal turbinates and lungs. Importantly, inoculation of cotton rats with these mutants triggered a high level of neutralizing antibodies and protected against hMPV challenge. Taken together, our data indicate that (i) α5β1 and αv integrins are essential for cell-cell fusion and viral replication, (ii) the first two residues in the RGD motif are essential for fusion activity, and (iii) inhibition of the interaction of the integrin-RGD motif may serve as a new target to rationally attenuate hMPV for the development of live attenuated vaccines.
IMPORTANCE Human metapneumovirus (hMPV) is one of the major causative agents of acute respiratory disease in humans. Currently, there is no vaccine or antiviral drug for hMPV. hMPV enters host cells via a unique mechanism, in that viral fusion (F) protein mediates both attachment and fusion activity. Recently, it was suggested that hMPV F protein utilizes integrins as receptors for entry via a poorly understood mechanism. Here, we show that α5β1 and αv integrins are essential for hMPV infectivity and F protein-mediated cell-cell fusion and that the integrin-binding motif in the F protein plays a crucial role in these functions. Our results also identify the integrin-binding motif to be a new, attenuating target for the development of a live vaccine for hMPV. These findings not only will facilitate the development of antiviral drugs targeting viral entry steps but also will lead to the development new live attenuated vaccine candidates for hMPV.
INTRODUCTION
Human metapneumovirus (hMPV) is a member of the genus Metapneumovirus in the subfamily Pneumovirinae of the family Paramyxoviridae. hMPV was first identified in infants and children with acute respiratory tract infections in 2001 in the Netherlands (1). Soon after its discovery, hMPV was recognized as a globally prevalent pathogen (2, 3). It is a major causative agent of acute respiratory tract disease in individuals of all ages, especially infants, children, the elderly, and immunocompromised individuals (3–5). Epidemiological studies suggest that 5 to 15% of all respiratory tract infections in infants and young children are caused by hMPV, a proportion second only to that of human respiratory syncytial virus (RSV) (6). The clinical signs and symptoms associated with hMPV are similar to those associated with RSV, ranging from mild respiratory problems to severe cough, bronchiolitis, and pneumonia. Currently, there are no therapeutics or vaccines available for hMPV. The only other member in the genus Metapneumovirus, avian metapneumovirus (aMPV), also known as avian pneumovirus or turkey rhinotracheitis virus, is an economically important pathogen that causes acute respiratory disease in turkeys (7).
All viruses must cross the cell membrane to initiate infection. Paramyxoviruses enter host cells through the fusion of the viral envelope with the cellular membrane. Such fusion is mediated by viral glycoproteins (G proteins) that are located on the surface of the virion. For viruses in the Paramyxovirinae subfamily, membrane fusion requires both the attachment protein (G, H, or HN) and the fusion (F) protein (reviewed in reference 8). The paramyxovirus F protein is a class I fusion protein which is synthesized as a precursor protein, F0, and subsequently cleaved into two disulfide-linked subunits, F1 and F2, by a cellular protease (reviewed in reference 8). This cleavage generates a hydrophobic fusion peptide (FP) at the N terminus of F1. During the fusion process, the FP inserts into an opposing membrane. The paramyxovirus F protein contains two conserved heptad repeat (HR) regions, the N-terminal heptad (HRA) and the C-terminal heptad (HRB), which are located downstream of the fusion peptide and upstream of the transmembrane (TM) domain, respectively (9, 10). Upon triggering, the metastable prefusion F protein undergoes a series of dramatic and irreversible conformational changes (11, 12). HRA and HRB assemble into a highly stable six-helix bundle that brings the two membranes together to initiate fusion (11–13). Currently, the mechanism by which fusion is regulated such that it occurs at the proper time and place remains poorly understood. It is thought that binding of the attachment proteins to the cell surface receptor(s) induces conformational changes in F protein, which in turn trigger membrane fusion (reviewed in references 8 and 12).
Membrane fusion of pneumoviruses is unique among the paramyxoviruses, in that fusion is accomplished by the F protein alone without help from the attachment glycoprotein. This attachment protein-independent fusion activation has been well characterized in human RSV, bovine RSV, and ovine RSV (14–16). Recently, it was found that the F proteins of hMPV and aMPV also induce fusion without their attachment G proteins (17–20), suggesting that the G protein is dispensable for attachment and fusion. Consistent with this observation, recombinant hMPV lacking the G protein was found to replicate efficiently in cell culture (21). Another unique characteristic of hMPV entry is that fusion of some hMPV strains requires low pH, whereas fusion of all other paramyxoviruses occurs at neutral pH (17, 18, 22). In addition, fusion of hMPV in cell culture requires the addition of exogenous protease (17, 18), unlike the F protein of RSV but similar to the F proteins of some of the members of the Paramyxovirinae, such as Sendai virus, parainfluenza virus type 1, and avirulent strains of Newcastle disease virus (16). Together these results suggest that hMPV fusion is characterized by a unique combination of features making it distinct from other paramyxoviruses. A detailed understanding of the mechanisms underlying fusion in hMPV is lacking.
The fact that the F protein of hMPV alone is sufficient for induction of cell-cell fusion and viral entry suggests that the hMPV F protein possesses dual functions, receptor binding and fusion activity. Recently, Cseke et al. (2009) provided evidence that integrin αvβ1 is a functional receptor for hMPV F protein (23). Consistent with this, the F proteins of all known hMPV strains contain a putative integrin-binding motif (329RGD331). Furthermore, the same group of investigators showed that hMPV binds to RGD-binding integrins and that this interaction is necessary for virus attachment, viral RNA transcription, and subsequent productive infection (24). Recently, Chang et al. (2012) proposed that heparan sulfate proteoglycans function as the primary receptor for hMPV and that binding to heparan sulfate proteoglycans is mediated by the F protein (25). They hypothesized that the interaction between integrins and hMPV may occur after the initial binding of hMPV F protein to heparan sulfate. This is consistent with the hypothesis by Cox et al. (2012) that heparan sulfate serves as an adhesion factor for hMPV, with subsequent integrin engagement being critical for entry (24). Clearly, the molecular mechanisms underlying the interaction between hMPV F protein, integrin, and heparan sulfate and their relative roles in cell-cell fusion, the hMPV life cycle, and viral pathogenesis remain to be elucidated.
In this study, we determined the roles of integrin subtypes in hMPV F protein-mediated cell-cell fusion and evaluated the contributions of the 329RGD331 motif in the hMPV F protein to cell-cell fusion, viral infectivity, and pathogenesis. Our results show that, in addition to integrin subtype α5β1, αv is also essential for cell-cell fusion and hMPV infectivity. Moreover, mutations in the first two amino acid residues within the 329RGD331 motif of the hMPV F protein significantly diminished the fusogenic activity of hMPV F protein, whereas mutations at D331 did not have a significant impact on fusion. Finally, the RGD motif with the mutations was introduced into recombinant hMPV (rhMPV). The fusion-defective RGD mutations were either lethal or resulted in recombinant hMPVs that had defects in viral replication in cell culture. Using the cotton rat as an animal model, we found that rhMPV with the R329K mutation in the F protein (rhMPV-R329K) and rhMPV-D331A were attenuated but retained high immunogenicity and are thus good live vaccine candidates for hMPV. Collectively, these results suggest that α5β1 and αv integrins are essential for hMPV entry and that the 329RGD331 motif of hMPV F protein plays a crucial role in cell-cell fusion, viral infectivity, and pathogenesis.
MATERIALS AND METHODS
Cells.
Vero E6 cells (ATCC CRL-1586), GD25 cells, GD1286 cells (a generous gift from Mark Peeples), and BHK-SR19-T7 cells (kindly provided by Apath LLC, Brooklyn, NY) were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Invitrogen). The medium of the BHK-SR19-T7 cells was supplemented with 10 μg/ml neomycin (Invitrogen) every other passage to select for T7 polymerase-expressing cells. LLC-MK2 cells were grown in Opti-modified Eagle's medium (Opti-MEM; Invitrogen) supplemented with 2% (vol/vol) FBS. Cells were grown at 37°C in a 5% CO2 humidified incubator.
Antibodies.
Integrin-specific monoclonal antibodies MAB2021Z (αv, clone AV1), MAB1959 (β1, clone P5D2), MAB1389 (β2, clone WT.3), MAB1976Z (αvβ3, clone LM609), MAB1961 (αvβ5, clone P1F6), MAB2077Z (αvβ6, clone 10D5), MAB1956Z (α5, clone P1D6), and MAB1969 (α5β1, clone JBS5) were purchased from Millipore (Billerica, MA). Monoclonal antibody against hMPV F protein (M3884-4C, clone 9i11) was purchased from U.S. Biological Company. hMPV N protein-specific monoclonal antibody (MAB80138, clone 507) and goat anti-mouse fluorescein isothiocyanate (FITC)-conjugated IgG (H+L) antibody (catalog no. 12-506) were purchased from Millipore (Billerica, MA). Goat anti-mouse IgG (H+L) peroxidase-conjugated antibody was purchased from Thermo Scientific (Rockford, IL).
Plasmids and site-directed mutagenesis.
A plasmid (phMPV) encoding the full-length genomic cDNA of hMPV lineage A strain NL/1/00 was kindly provided by Ron A. M. Fouchier at Erasmus Medical Center, Rotterdam, the Netherlands (26, 27). The F-protein gene was amplified from plasmid phMPV using Supermix polymerase (Qiagen, Valencia, CA), digested by EcoRI and XhoI, and cloned into pCAGGS at the same sites, which resulted in the construction of pCAGGS-F. The F-protein gene mutations were generated by site-directed mutagenesis, using phMPV or pCAGGS-F as the template, by the QuikChange methodology (Stratagene, La Jolla, CA). The presence of the desired mutation was verified by sequencing.
Fusion staining assay.
Monolayers of Vero E6 cells in 6-well plates were transfected with 2 μg of each plasmid using the Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. After transfection, the cells continued to grow in 2 ml of Opti-MEM (Invitrogen) for 24 h and were then washed and incubated at 37°C in Opti-MEM containing 0.2 μg/ml tosylamide-2-phenylethyl chloromethyl ketone (TPCK)–trypsin for another 2 h. Since fusion of hMPV lineage A strain NL/1/00 F protein is low-pH dependent (22), all fusion assays were subjected to low-pH pulses. Briefly, the cells were washed four times with phosphate-buffered saline (PBS; pH 5.0) supplemented with 10 mM HEPES and 5 mM MES (morpholineethanesulfonic acid). Each washing cycle lasted for 4 min and was followed by incubation in 2 ml of Opti-MEM for 2 h. After pH pulses, the cells continued to grow in 2 ml of fresh Opti-MEM until 48 h posttransfection. Finally, the cells were fixed with methanol, and syncytia were visualized by Giemsa staining. Digital photographs of syncytia were taken under a Nikon TS100 inverted phase-contrast microscope mounted with a Nikon Coolpix995 camera.
Fusion inhibition assay.
Confluent monolayers of Vero E6 cells in a 48-well plate were transfected with pCAGGS-F, pCAGGS-F mutants, or pCAGGS (as a control), as described above, with the following modifications. For cells in each well, the transfection mixture contained 0.8 μg of plasmids, 200 μl of Opti-MEM, and 0.2 μl of Lipofectamine. After incubation with the transfection mixture for 6 to 8 h, cells continued to grow in Opti-MEM (with 10 μg/ml of selected integrin antibodies) for 24 h. Subsequently, cells were subjected to four cycles of low-pH pulses. Briefly, the cells were washed four times with PBS (pH 5.0) supplemented with 10 mM HEPES and 5 mM MES. Each washing cycle lasted for 4 min and was followed by incubation in 2 ml of Opti-MEM containing 5 μg/ml of selected integrin antibodies and 0.2 μg/ml of TPCK-trypsin for 2 h. The cells continued to grow in Opti-MEM (with 5 μg/ml of selected integrin antibodies and 0.2 μg/ml of TPCK-trypsin) for another 12 h, followed by cell fixation and syncytium visualization.
Integrin-targeting siRNA transfection.
Twenty picomoles of synthetic small interfering RNAs (siRNAs) targeting either human integrin subtype α5 (sc-29372; Santa Cruz Biotechnology, CA) or αv (sc-29373; Santa Cruz Biotechnology) or control siRNA (sc-37007; Santa Cruz Biotechnology) was transfected into Vero E6 cells (at a confluence of 75%) in 24-well plates using Oligofectamine reagents (Invitrogen) according to the manufacturer's instructions. After treatment with siRNAs for 24 h, Vero E6 cells were transfected with 0.8 μg of pCAGGS-F using Lipofectamine Plus reagents (Invitrogen) for 8 h as described above and then subjected to pH pulses, followed by visualization of syncytia.
Content-mixing assay for fusion.
Confluent Vero E6 cells in 6-well plates were cotransfected with 2 μg each of a plasmid containing the hMPV F-protein gene (pCAGGS-F or F mutant) and a plasmid (pGINT7β-gal) encoding a β-galactosidase gene under the control of the T7 promoter. At 24 h posttransfection, the cells were detached with trypsin, washed twice with DMEM, and resuspended in DMEM plus 10% FBS. The plasmid-transfected cells were mixed with equal numbers of BHK-SR19-T7 cells, which constitutively express T7 RNA polymerase. After incubation at 37°C for another 24 h, the cells were washed 4 times with PBS (pH 5.0) containing 10 mM HEPES and 5 mM MES. Where indicated, the fusion assay was performed in the presence of 10 μg/ml of selected integrin antibodies. The cells were lysed by Nonidet P-40 solution (0.5%). The extent of fusion was quantitated by measuring the β-galactosidase activity using a plate reader (Molecular Devices, Sunnyvale, CA). The fusion activity of the F-protein mutants was determined as a percentage of the β-galactosidase production observed in cells expressing wild-type (wt) F protein.
Cell surface expression.
Surface expression of the wt and mutant F proteins was assayed by fluorescence-activated cell sorting (FACS). Six-well plates were seeded with Vero E6 cells at a density of 1 × 106 cells/well. Transfection was performed using Lipofectamine Plus reagents (Invitrogen) according to the manufacturer's instructions. At 48 h posttransfection, the cells were washed twice with 3% FBS in PBS and incubated in primary monoclonal antibody against hMPV F protein (diluted 1:50 in 3% bovine serum albumin [BSA]–PBS) for 1 h at 37°C. After being washed with FACS medium, the cells were incubated with goat anti-mouse FITC-conjugated secondary antibody (1:100 in 3% BSA–PBS) for 1 h at 37°C. The cells were washed again, detached with EDTA, and washed two more times. Finally, the cells were resuspended in 500 μl of fix solution (2% paraformaldehyde, 1% FBS in PBS) and subjected to FACS analysis using a Becton, Dickinson FACSCalibur analyzer with CellQuest software. By comparison to a negative control, only positive cells were gated to estimate the geometric mean fluorescence intensity to evaluate the cell surface expression level. The results were normalized to the percentage for the wt control, which was set at 100%.
Recovery of recombinant hMPV.
Recovery of recombinant hMPV from the infectious clones was carried out as described previously (27). Briefly, rhMPV was recovered by cotransfection of a plasmid encoding the full-length genomic cDNA of hMPV NL/1/00 (phMPV) and support plasmids encoding viral N (pCITE-N), P (pCITE-P), L (pCITE-L), and M2-1 (pCITE-M2-1) proteins into BHK-SR19-T7 cells (kindly provided by Apath LLC, Brooklyn, NY), which stably express the T7 RNA polymerase. At 6 days posttransfection, the cells were subjected to three freeze-thaw cycles, followed by centrifugation at 3,000 × g for 10 min. The supernatant was subsequently used to infect new LLC-MK2 cells. Since hMPV requires trypsin to grow, TPCK-trypsin was added to the medium to a final concentration of 0.1 μg/ml at day 2 postinfection. Cytopathic effects (CPEs) were observed at 5 days postinfection, and the recovered viruses were amplified further in LLC-MK2 cells. The recovery of recombinant virus was confirmed by immunostaining and direct agarose overlay plaque assays as described previously (28).
Immunostaining of recombinant hMPV.
Immunostaining was used for virus titration as described previously (1, 28). Briefly, LLC-MK2 or Vero E6 cells (at a confluence of 90%) in a 96-well plate were inoculated with 10-fold serial dilutions of recombinant hMPV mutants and incubated at 37°C for 1 h. Infected cells were cultured in fresh Opti-MEM (0.2 ml per well) at 37°C. At day 4 postinfection, the supernatant was removed and cells were fixed in a prechilled acetone-methanol solution (at a ratio of 3:2) at room temperature for 15 min. Cells were permeabilized in PBS containing 0.4% Triton X-100 at room temperature for 10 min and blocked at 37°C for 1 h using 1% BSA in PBS. The cells were then labeled with an anti-hMPV N protein primary monoclonal antibody (Millipore, Billerica, MA) at a dilution of 1:1,000, followed by incubation with horseradish peroxidase (HRP)-labeled rabbit antimouse secondary antibody (Thermo Scientific, Waltham, MA) at a dilution of 1:5,000. After incubation with 3-amino-9-ethylcarbazole (AEC) chromogen substrate (Sigma, St. Louis, MO), positive cells were then visualized under a microscope. Positive plaques were counted to determine the viral titers.
Agarose overlay plaque assay for recombinant hMPV mutants.
The agarose overlay plaque assay was used for determination of the plaque size of recombinant hMPV, as described previously (28). Briefly, Vero E6 cells were seeded into six-well plates (Corning Life Sciences, Wilkes-Barre, PA) at a density of 2 × 106 cells per well. After incubation for 18 h, the medium was removed and cell monolayers were infected with 400 μl of a 10-fold dilution series of each virus. After incubation at 37°C for 1 h with agitation every 10 min, the cells in each well were overlaid with 2.5 ml of Eagle minimum essential medium (MEM) containing 1% agarose, 1% FBS, 0.075% sodium bicarbonate (NaHCO3), 20 mM HEPES (pH 7.7), 2 mM l-glutamine, 12.5 mg/ml of penicillin, 4 mg/ml of streptomycin, and 4 mg/ml of kanamycin. The plates were incubated at 4°C for 30 min to solidify the overlay medium. After incubation at 37°C and 5% CO2 for 6 days, the cells were fixed in 10% (vol/vol) formaldehyde for 2 h, and the plaques were visualized by staining with 0.05% (wt/vol) crystal violet.
Virus-cell infectivity (blocking) assays.
Confluent monolayers of LLC-MK2 cells in 48-well plates were infected with each virus at a multiplicity of infection (MOI) of 100 per well. After incubation on ice for 1 h (with gentle shaking every 15 min), the inoculum was removed and the unbound viruses were removed by washing with Opti-MEM three times. The infected cells were then incubated with fresh Opti-MEM at 37°C in 5% CO2. At 24 h after infection, immunostaining was performed and the positive spots were counted under a microscope. For virus-cell infectivity blocking assays, the cells were pretreated with integrin antibody (20 μg/ml) at 37°C for 1 h. The cells were then infected and analyzed by immunostaining.
Single-step growth of recombinant hMPV.
Viral growth was determined using an immunostaining assay on LLC-MK2 cells. Confluent LLC-MK2 cells in 35-mm dishes were infected with each recombinant hMPV at an MOI of 0.01. After adsorption for 1 h, the inoculum was removed and the cells were washed with 2 ml of Opti-MEM. Subsequently, fresh Opti-MEM (supplemented with 2% FBS) was added and infected cells were incubated at 37°C for various time periods. Aliquots of the cell culture fluid were removed at the intervals indicated below, and the viral titer was determined by an immunostaining assay on LLC-MK2 cells.
Infectivity of recombinant hMPV in a cotton rat model.
Thirty 4-week-old specific-pathogen-free female cotton rats (Harlan Laboratories, Indianapolis, IN) were randomly divided into six groups (5 rats per group). These cotton rats were housed within University Laboratory Animal Resources (ULAR) facilities of The Ohio State University and used under an animal use protocol approved by the Institutional Animal Care and Use Committee (IACUC) of The Ohio State University. Each group was separately housed under biosafety level 2 conditions. Rats in group 1 were inoculated with 2.0 ×105 PFU of the wild-type hMPV and served as positive controls. Rats in groups 2 to 5 were inoculated with 2.0 ×105 PFU of four hMPV mutants (rhMPV-R329K, -D331A, -D331E, and -D331R). Rats in group 6 were inoculated with cell culture medium (DMEM) and served as uninfected controls (healthy controls). Each rat was inoculated intranasally at a dose of 2.0 × 105 PFU in a volume of 100 μl while under isoflurane narcosis. After inoculation, the animals were evaluated on a daily basis for mortality, weight loss, and the presence of any respiratory symptoms of hMPV. At day 4 postinfection, cotton rats were sacrificed, and their lungs and nasal turbinates were collected for both virus isolation and histological analysis, as described below.
Immunogenicity of recombinant hMPV in cotton rats.
Thirty 4-week-old specific-pathogen-free female cotton rats (Harlan Laboratories, Indianapolis, IN) were randomly divided into five groups (5 rats per group). Rats in groups 1 to 3 were immunized with wild-type rhMPV, rhMPV-R329K, and rhMPV-D331A, respectively. Rats in group 4 were inoculated with DMEM and served as the unimmunized but challenged control group. Rats in group 5 were inoculated with DMEM and served as uninfected controls (healthy controls). All rats were immunized intranasally at a dose of 2.0 × 105 PFU per rat. After immunization, the animals were evaluated daily for body weight, mortality, and the presence of any symptoms of hMPV infection. Blood samples were collected from each rat weekly by retro-orbital bleeding while they were under isoflurane narcosis, and serum was separated for antibody detection. At week 4 postimmunization, rats in groups 1 to 5 were challenged intranasally with wild-type hMPV at a dose of 1.0 ×106 PFU per rat. After challenge, the animals were evaluated twice every day for mortality and the presence of any symptoms of hMPV infection. The body weight of each rat was monitored on a daily basis. At day 5 postchallenge, all rats from each group were euthanized. The lungs and nasal turbinates from each rat were collected for virus isolation and histological evaluation.
Determination of viral titer in lungs and nasal turbinates.
Nasal turbinates and the right lung from each cotton rat were removed, weighed, and homogenized in 1 ml of PBS solution. The presence of infectious virus was determined by an immunostaining assay in Vero E6 cells as described above.
RT-PCR and sequencing.
Viral RNA was extracted from each recombinant hMPV mutant or lung or nasal turbinate tissue using an RNeasy minikit (Qiagen) following the manufacturer's recommendation. Reverse transcription-PCR (RT-PCR) was performed using a One Step RT-PCR kit (Qiagen) with two hMPV F-specific primers, 5′-CGGAATTCATGTCTTGGAAAGTGGTGATC-3′ (forward) and 5′-CCGCTCGAGCTAATTATGTGGTATGAAGC-3′ (reverse). The PCR products were purified and sequenced at The Ohio State University Plant Microbe Genetics Facility to confirm the presence of the designed mutations.
Plaque reduction virus neutralization assay.
The hMPV neutralizing antibody was determined using a plaque reduction neutralization assay (29). Briefly, serum samples from cotton rats were heat inactivated at 56°C for 30 min and were subjected to 2-fold serial dilutions on 96-well plates. Each serum dilution was mixed with an equal volume of Opti-MEM containing 100 PFU of wild-type rhMPV, followed by incubation at room temperature for 1 h. The mixture was then transferred to Vero E6 cells in 96-well plates. After 1 h of incubation at 37°C, cells were overlaid with 0.75% methylcellulose in Opti-MEM and incubated at 37°C for 4 days, followed by immunostaining with monoclonal antibody against hMPV N protein as described above. Neutralizing antibody titers were expressed as the reciprocal of the serum dilution that resulted in a 50% reduction in plaque numbers compared to those for the healthy cotton rat serum control. Each serum sample was tested in duplicate.
Histology.
The right lung from each cotton rat was preserved in 4% (vol/vol) phosphate-buffered paraformaldehyde. Fixed tissues were embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin-eosin (H&E) for the examination of histological changes by light microscopy. Histopathological changes were scored, including the extent of inflammation (focal or diffuse), the pattern of inflammation (peribronchiolar, perivascular, interstitial, alveolar), and the nature of the cells in the infiltrate (neutrophils, eosinophils, lymphocytes, macrophages). Deparaffinized sections were also stained with monoclonal antibody against hMPV matrix protein (Virostat, Portland, ME) to determine the distribution of viral antigen. The histological slides were examined independently by three pathologists.
Structure modeling of F protein.
The structure prediction of the hMPV F protein was performed using the Modeler (version 9.0) program on the basis of the prefusion crystal structure of the RSV F protein (PDB accession no. 4JHW) as the template (30, 31).
Statistical analysis.
All experiments were repeated three to five times. Quantitative analysis was performed either by densitometric scanning of autoradiographs or by using a phosphorimager (Typhoon; GE Healthcare, Piscataway, NJ) and ImageQuant TL software (GE Healthcare, Piscataway, NJ). Statistical analysis was performed by one-way multiple comparisons using SPSS (version 8.0) statistical analysis software (SPSS Inc., Chicago, IL). A P value of <0.05 was considered statistically significant.
RESULTS
α5β1 and αv integrin-specific antibodies inhibit hMPV F protein-induced cell-cell fusion.
Previously, it has been shown that hMPV engages αvβ1 integrin to mediate virus infection, suggesting that integrins might serve as functional receptors for hMPV (23). However, it is not known whether integrins are involved in hMPV F protein-mediated cell-cell fusion. To address this question, we first determined whether integrin-specific antibodies can inhibit cell-cell fusion triggered by hMPV F protein. Briefly, Vero E6 cells were transfected with pCAGGS-F, followed by incubation with specific anti-integrin antibodies, and fusion was determined by assaying for syncytium formation. As shown in Fig. 1A, incubation with an antibody specific for α5β1 led to a significant reduction in syncytium formation in comparison with that for the no-antibody control. However, hMPV F protein-transfected cells treated with antibodies against αvβ3, αvβ5, and αvβ6 subtypes displayed syncytia similar to those of the untreated control. Interestingly, incubation with antibodies against either the α5 subunit or β1 subunit alone did not have a significant effect on syncytium formation. Similarly, the combination of anti-β1 and anti-α5 antibodies as well as antibody against the β2 subunit did not exhibit any inhibitory effect. In contrast, incubation of hMPV F protein-transfected cells with an antibody against αv and the combination of antibodies against the αv and β1subunits notably diminished the formation of syncytia. Subsequently, we quantified the extent of cell-cell fusion by the content-mixing fusion assay. As shown in Fig. 1B, the fusion of hMPV F protein-transfected cells was inhibited by approximately 80% in the presence of anti-α5β1 or anti-αv antibody. In contrast, none of the other tested integrin antibodies had a significant effect on fusion in comparison with that for untreated cells (P > 0.05). Taken together, these results demonstrated that anti-α5β1 and anti-αv antibodies specifically inhibited the cell-cell fusion triggered by the hMPV F protein.
FIG 1.
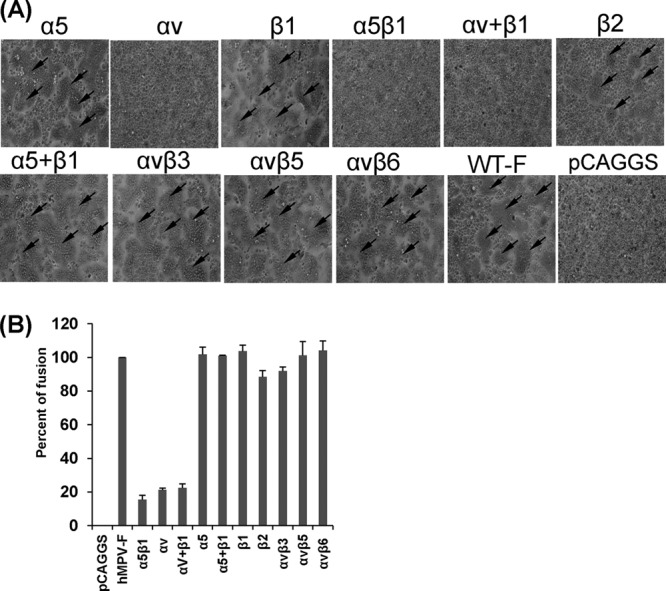
α5β1 and αv integrin-specific antibodies block cell-cell fusion triggered by hMPV F protein. (A) Syncytium formation induced by hMPV F protein expression in the presence of integrin antibodies. Vero E6 cells in 48-well plates were transfected with 0.8 μg of pCAGGS-F or pCAGGS. After incubation with a plasmid-Lipofectamine mixture for 8 h, cells continued to grow in Opti-MEM containing 10 μg/ml of integrin antibody and 0.2 μg/ml of TPCK-trypsin. At 48 h, monolayers were fixed with methanol and stained with Giemsa. Syncytia are indicated by arrows. (B) Content-mixing fusion assay in the presence of integrin antibodies. Vero E6 cells were cotransfected with 2 μg of pCAGGS-F and a reporter gene plasmid (pGINT7). At 24 h posttransfection, the cells were detached with trypsin and mixed with equal numbers of BHK-SR19-T7 cells, which express T7 RNA polymerase. Then, the cells were incubated with 2 ml of Opti-MEM containing 5 μg/ml of selected integrin antibody and 0.2 μg/ml of TPCK-trypsin for 12 h. The cells were lysed and mixed with an equal amount of the β-galactosidase substrate chlorophenol red-β-d-galactopyranoside (16 mM). The extent of fusion was quantitated by use of a microplate spectrophotometer at an absorbance of 570 nm. Percent fusion for each antibody treatment was normalized to the fusion of pCAGGS-F in the absence of integrin antibody. The data shown are averages for three independent experiments.
Downregulating the expression of the α5 or αv subunit diminishes hMPV F protein-triggered cell-cell fusion.
To further determine the role of α5β1 and αv integrins in cell-cell fusion, we knocked down the α5 or αv integrins using siRNA targeting these two subunits. We first transfected Vero E6 cells with α5- or αv-targeting siRNAs, which reduced the expression of the α5 or αv subunit on the cell surface by approximately 85% (Fig. 2A). Subsequently, the siRNA-treated cells were transfected with pCAGGS-F, and syncytium formation was examined. As shown in Fig. 2B, fusion was significantly reduced in cells transfected with siRNAs targeting α5 or αv integrin but not in cells transfected with control siRNA. Further quantitative analysis (Fig. 2C) demonstrated that α5 and αv siRNA reduced fusion activity by approximately 80% and 60%, respectively.
FIG 2.
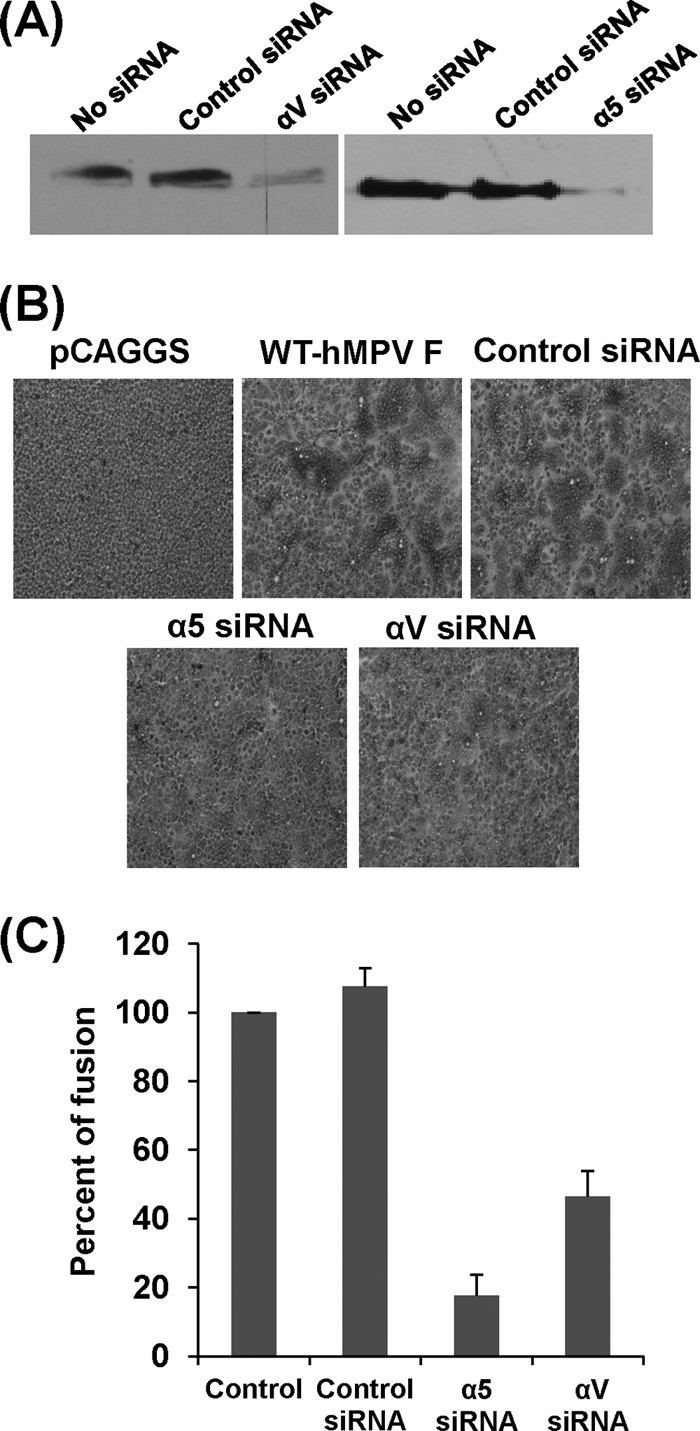
siRNAs targeting α5 and αv back cell-cell fusion triggered by hMPV F protein. (A) Knockdown of integrin α5 and αv expression by siRNA. Twenty picomoles of synthetic siRNA targeting human integrin subtype α5 or αv as well as control siRNA was transfected into Vero E6 cells in 24-well plates using Oligofectamine reagents according to the manufacturer's instructions. After 48 h posttransfection, the expression of α5 or αv was detected by Western blotting. (B) Syncytium formation induced by F protein of hMPV after knockdown of integrins α5 and αv. Vero E6 cells in 24-well plates were transfected with 20 pmol of synthetic siRNA targeting human integrin subtype α5 or αv as well as control siRNA. After treatment with siRNAs for 24 h, Vero E6 cells were transfected with 0.8 μg of pCAGGS-F using Lipofectamine Plus reagents and then subjected to pH pulses (pH 5.0). At 48 h, monolayers were fixed with methanol and stained with Giemsa. (C) Quantitation of syncytium formation after knockdown of integrins α5 and αv. The number of syncytia (≥4 nuclei in each syncytium) was counted under a microscope using six randomly selected fields in each siRNA-treated or untreated well. The mean number of syncytia per field was calculated for each treatment. The percent fusion for each siRNA treatment was normalized by the mean number of syncytia in cells transfected with pCAGGS-F without siRNA treatment. The data shown are averages for three independent experiments.
Specific anti-α5β1 and anti-αv antibodies inhibit hMPV infectivity in Vero E6 cells.
We then investigated the effect of various anti-integrin antibodies on hMPV infection in Vero E6 cells. Briefly, Vero E6 cells were incubated with different anti-integrin antibodies at 37°C for 1 h prior to hMPV infection. Cells were infected with hMPV at an MOI of 100 per well. After incubation on ice for 1 h, the unbound viruses were removed by washing with Opti-MEM three times. At 24 h postinfection, the amount of hMPV bound to Vero E6 cells was determined by immunostaining as described in Materials and Methods. As shown in Fig. 3, antibodies specific for α5β1 and αv integrins inhibited 85% and 75% of the hMPV infectivity in Vero E6 cells, respectively, whereas other integrin antibodies, including antibodies specific for α5, β1, β2, αvβ3, αvβ5, and αvβ6, had no significant effect on hMPV infectivity. These results are consistent with our finding that antibodies specific for α5β1 and αv integrins efficiently inhibited cell-cell fusion triggered by hMPV F protein.
FIG 3.
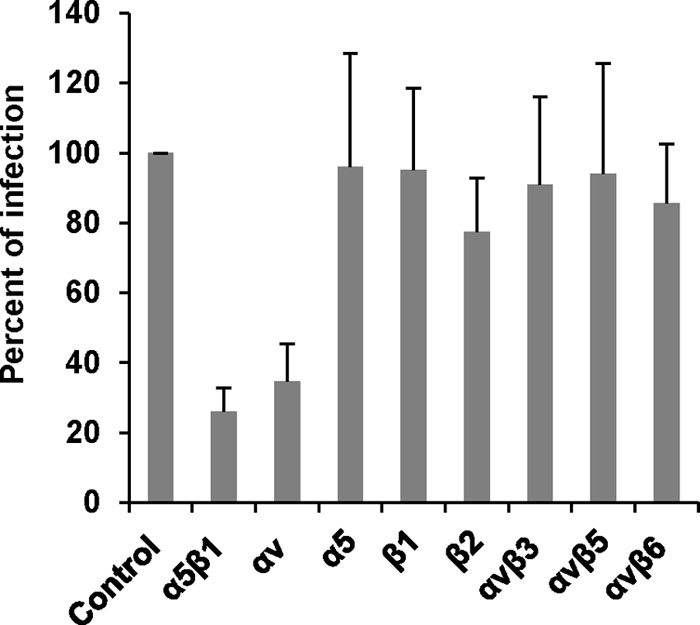
Integrin α5β1and αv antibodies inhibit hMPV infectivity in host cells. Confluent monolayers of LLC-MK2 cells in 96-well plates were pretreated with each integrin antibody (20 μg/ml) at 37°C for 1 h. Then, the cells were shifted to a 4°C incubator for 30 min. The cells were incubated with hMPV at an MOI of 100 PFU/well. After incubation on ice for 1 h (with shaking every 15 min), the inoculum was removed and the cells were washed with cold Opti-MEM 3 times. The infected cells were then incubated with fresh DMEM at 37°C in 5% CO2. After 24 h, the binding and infectivity were determined by counting the number of immunostaining spots. The percent infectivity for each antibody treatment was normalized by the infectivity of hMPV without antibody treatment. The data shown are averages for three independent experiments.
α5β1 integrin-expressing cells enhance viral infectivity.
To further determine the role of α5β1 integrin in viral binding and infectivity, we took advantage of cell lines that were defective in integrin or stably expressing integrin receptors. Specifically, we examined the binding and infectivity of hMPV in cells of the mouse fibroblast cell line GD25, which lack expression of the β1 family of integrin heterodimers, and GD1286 cells, which constitutively express the α5β1 integrin receptor. As shown in Fig. 4A, the infectivity of hMPV in GD1286 cells was approximately 5 times higher than that in GD25 cells, consistent with the idea that the expression of α5β1 integrin significantly increases the binding of hMPV to host cells and the infectivity of hMPV for host cells. Interestingly, the size of the immunospots formed in GD1286 cell monolayers was much larger than that of the immunospots formed in GD25 cell monolayers (Fig. 4B), suggesting that α5β1 integrin may also play a role in cell-to-cell spread.
FIG 4.

Infectivity of hMPV in an integrin-deficient cell line. (A) hMPV has defects in infectivity in an integrin-deficient cell line. Confluent monolayers of GD1286 or GD25 cells in 24-well plates were incubated at 4°C for 30 min. The cells were infected with 100 PFU of hMPV per well. After incubation on ice for 1 h (with shaking every 15 min), the inoculum was removed and the cells were washed with cold DMEM 3 times. The infected cells were then incubated with fresh medium at 37°C in 5% CO2. After 24 h, the binding capacity was determined by counting the number of immunostaining spots. Percent infectivity in GD25 cells was normalized by the infectivity of hMPV in GD1286 cells. The data shown are averages for three independent experiments. (B) hMPV forms much smaller immunospots in an integrin-deficient cell line. Confluent monolayers of GD1286 or GD25 cells were infected with hMPV. After 24 h, immunostaining assay was performed, and immunospots formed by hMPV were visualized.
Mutations in the RGD motif of the hMPV F protein impair fusion activity.
The results presented above demonstrate that α5β1 and αv integrins play essential roles in hMPV F protein-triggered cell-cell fusion, as well as viral binding and infectivity. Consistent with this, a putative integrin-binding motif (329RGD331) is present in the F proteins of all known hMPV strains. Interestingly, the corresponding motif in the F protein of aMPV subtype C strains is 329RSD331. To determine the role of the RGD motif in hMPV cell-cell fusion, we performed an extensive mutagenesis analysis of this motif. As summarized in Table 1, three classes of substitutions were generated: (i) alanine substitutions, including R329A, G330A, and D331A; (ii) mutations that conserve the charge, namely, R329K and D331E; and (iii) mutations that introduce a charge change, including R329D and D331R. In addition, a G330S mutation was introduced into the hMPV F protein to mimic the aMPV subtype C F protein.
TABLE 1.
Summary of phenotypes of the mutations to the RGD motif in hMPV F protein
| F-protein mutation | Rationale | % fusion | Virus recovery | Viral titera | Replication in: |
|
|---|---|---|---|---|---|---|
| LLC-MK2 cellsb | Cotton ratsc | |||||
| WT | 100 | Recovered | 2.5 × 106 | 100% | WT | |
| R329A | Change to neutral | 13 | Lethal | NAd | NA | NA |
| R329K | Maintain charge | 72 | Recovered | 1.2 × 106 | Attenuated | Attenuated |
| R329D | Change to negative charge | 13 | Lethal | NA | NA | NA |
| G330A | Maintain size | 20 | Recovered | 3.0 × 102 | Attenuated | NA |
| G330S | Mimic RSD in aMPV subtype C F protein | 9 | Was unstable | NA | NA | NA |
| D331A | Change to neutral | 58 | Recovered | 1.0 × 106 | Attenuated | Attenuated |
| D331E | Maintain charge | 118 | Recovered | 1.8 × 106 | WT | WT |
| D331R | Change to positive charge | 107 | Recovered | 4.0 × 106 | WT | WT |
Viral titer was determined by immunostaining assay.
Attenuation was judged by plaque size, infectivity, and growth curve; WT, wild-type level of replication in cell culture.
Attenuation was judged by viral replication in nasal turbinates and lungs and histology; WT, wild-type level of replication in cotton rats.
NA, not applicable.
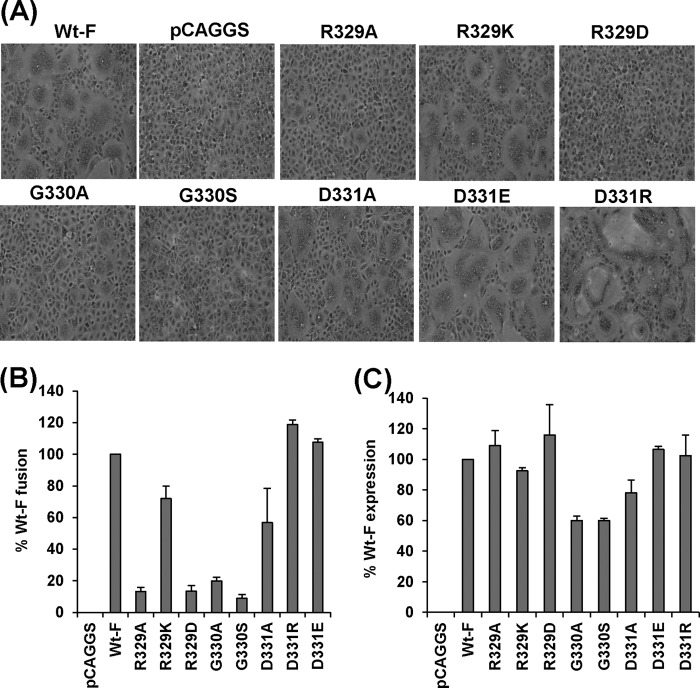
All F-protein mutants were transfected into Vero E6 cells, and their cell surface expression and fusion activities were evaluated. As shown in Fig. 5C, all F-protein mutants were efficiently expressed at the cell surface, as determined by FACS. Interestingly, the R329A and G330A mutations significantly decreased the extent of syncytium formation compared with that for wt hMPV F protein, whereas the R329K and D331A mutations slightly diminished the size of the syncytia (Fig. 5A). However, the R329D and G330S mutations significantly impaired fusion activity; no syncytium formation was observed with these two F-protein mutants until 48 h posttransfection. Interestingly, hMPV with the D331E and D331R mutations, especially the latter, was able to form syncytia that were larger than those of hMPV with wt F protein.
FIG 5.
Effects of mutations to the RGD motif on cell-cell fusion triggered by hMPV F protein. (A) Syncytium formation of hMPV F proteins carrying mutations at the RGD motif. Confluent monolayers of Vero E6 cells were transfected with 2 μg plasmids of pCAGGS-F or F-protein mutants. At 24 h posttransfection, the monolayers were fixed with methanol and stained with Giemsa. (B) Content-mixing fusion assay for hMPV F mutants. The extent of fusion for each F mutant was quantitated with the content-mixing fusion assay at pH 5.0 and normalized by the fusion of wild-type hMPV F protein. The data shown are averages for three independent experiments. (C) Cell surface expression of hMPV F mutants. Cell surface expression was determined by FACS using monoclonal antibody against hMPV F protein and normalized by the expression level of wild-type F protein at the cell surface.
The extent of fusion was also quantified in a content-mixing fusion assay. As shown in Fig. 5B, the R329A, R329D, G330A, and G330S mutations decreased the level of fusion to less than 20% of that for hMPV with wt F protein, while the R329K and D331A mutations decreased fusion activity to 50 to 80% of that of the wt. Consistent with the results from staining, the mutants with the D331R and D331E mutations exhibited even higher fusion activity than hMPV with wt F protein. Taken together, these data demonstrate that (i) alanine substitution for the first two amino acid residues (R329 and G330) in the RGD motif dramatically reduces fusion activity, whereas an alanine substitution for the third amino acid residue (D331) results in a less dramatic decrease in fusion; (ii) maintenance of the positive charge at amino acid position 329 is essential for fusion; (iii) replacement of the RGD motif found in hMPV F protein with the RSD motif found in aMPV F protein significantly impairs the fusion activity; and (iv) replacement of D331 with a positively or negatively charged residue does not affect fusion. These results suggest that amino acid residues R329 and G330 within the RGD motif are essential for fusion activity, whereas the identity of the amino acid residue at position 331 appears to be less important.
Recovery of rhMPV carrying mutations in the RGD motif of F protein.
To evaluate the role of the RGD motif in viral infectivity and pathogenesis, we constructed and recovered recombinant hMPV (rhMPV) carrying mutations in the RGD motif of the F protein. Each of the above-described mutations was introduced into an infectious cDNA clone of hMPV NL/1/00, and recombinant hMPV isolates carrying individual RGD mutations were recovered. The successful recovery of recombinant viruses was initially identified by an immunostaining assay using a monoclonal antibody against hMPV N protein and further confirmed by a direct agarose overlay plaque assay. Using these approaches, we successfully recovered rhMPV carrying R329K, G330A, G330S, D331A, D331R, and D331E mutations in the RGD motif (Fig. 6). However, we failed to recover recombinant viruses with the R329A and R329D mutations after multiple attempts, suggesting that these mutations are lethal to hMPV. As shown above, these F-protein mutants retained less than 20% of the fusion activity of hMPV with wild-type F protein (Fig. 5). As shown in Fig. 6 (top), rhMPV-G330A, -D331A, and -R329K formed much smaller immunospots at day 7 postinfection, whereas rhMPV-D331R and -D331E formed immunospots that were comparable to those formed by rhMPV. The ability of these recombinant viruses to form plaques in Vero E6 cells was determined by an agarose overlay plaque assay (Fig. 6, bottom). After 7 days of incubation, the average plaque sizes for rhMPV-R329K and -D331A were 0.91 mm and 0.75 mm in diameter, respectively, which were significantly smaller than the plaque size for rhMPV (1.10 mm). In contrast, rhMPV-D331E and -D331R formed plaques which were significantly larger than the rhMPV plaque. Interestingly, rhMPV-G330A was not able to form plaques, even though it could form immunospots by an immunostaining assay (Fig. 6). Finally, the entire F-protein gene of each recombinant virus was amplified by RT-PCR and sequenced. All recombinant viruses except for rhMPV-G330S contained the desired mutation in the RGD motif. Subsequently, we sequenced rhMPV-G330S from all passages and found that it had reverted back to wild type after the third passage in Vero E6 cells. Since rhMPV-G330S is not genetically stable, we did not include this mutant in subsequent studies.
FIG 6.
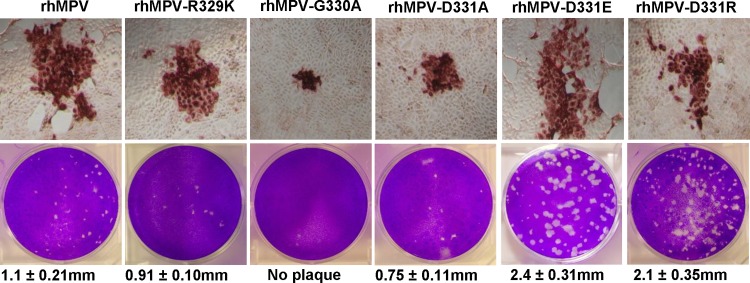
Recovery of recombinant hMPVs carrying mutations in the RGD motif. (Top) Immunostaining spots formed by recombinant hMPVs. LLC-MK2 cells were infected with recombinant hMPV mutants and incubated at 37°C for 1 h. At day 4 postinfection, the supernatant was removed and cells were fixed. The cells were then labeled with an anti-hMPV N protein primary monoclonal antibody, followed by incubation with HRP-labeled rabbit antimouse secondary antibody. After incubation with AEC chromogen substrate, positive cells with immunostaining spots were visualized under a microscope. (Bottom) Plaque morphology of recombinant hMPVs. An agarose overlay plaque assay was performed in monolayer Vero E6 cells. Viral plaques were developed at day 7 postinfection.
Single mutations in the RGD motif in recombinant hMPV result in defects in viral replication.
The replication kinetics of recombinant hMPV mutants with single mutations in the RGD motif were determined in LLC-MK2 cells. Briefly, LLC-MK2 cells were infected with each recombinant virus at an MOI of 0.01. At the indicated time points, the amount of virus in the supernatant was determined by immunostaining. As shown in Fig. 7, a significant release of infectious virus particles was detected by day 2 postinfection for rhMPV, rhMPV-D331E, and rhMPV-D331R, while the first release of rhMPV-R329K and rhMPV-D331A was observed only at day 3 postinfection. Interestingly, the release of rhMPV-G330A was not detected during the entire 8-day period. Overall, the growth curves of rhMPV-D331R and -D331E were not significantly different from the growth curve of rhMPV (P > 0.05). In contrast, rhMPV-R329K and -D331A had significant defects in viral replication compared to the replication of rhMPV (P > 0.05). Since rhMPV-G330A was not detectable in the supernatant, we harvested this virus at day 14 postinfection by three freeze-thaw cycles followed by low-speed centrifugation. A low viral titer (102 PFU/ml) was detected by immunostaining (Table 1). Figure 8 shows the cytopathic effects (CPEs) caused by each recombinant virus. rhMPV-R329K and -D331A exhibited delayed CPEs compared to rhMPV. However, the CPE caused by rhMPV-D331E was earlier than that caused by rhMPV. Collectively, these results are consistent with rhMPV-R329K, -G330A, and -D331A exhibiting defects in their viral replication kinetics, whereas rhMPV-D331R and -D331E demonstrate wild-type replication.
FIG 7.
Single-step growth curve of recombinant hMPVs carrying mutations in the RGD motif. LLC-MK2 cells in 35-mm dishes were infected with each recombinant hMPV at an MOI of 0.01. After adsorption for 1 h, the inocula were removed and the infected cells were washed 3 times with Opti-MEM. Then, fresh Opti-MEM containing 2% FBS was added and cells were incubated at 37°C for various time periods. Aliquots of the cell culture fluid were removed at the indicated intervals. Viral titer was determined by an immunostaining assay in LLC-MK2 cells.
FIG 8.
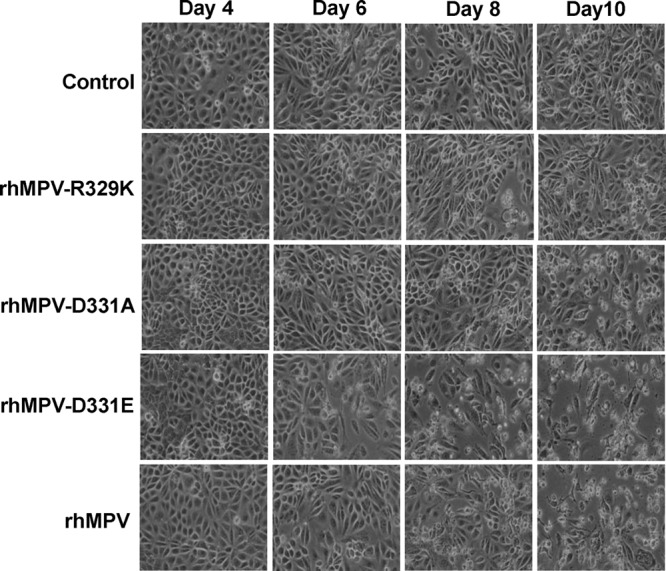
CPEs caused by hMPVs carrying mutations in the RGD motif. LLC-MK2 cells were infected with each recombinant hMPV at an MOI of 0.01. The CPE was monitored on a daily basis. Pictures were taken at days 4, 6, 8, and 10 postinfection.
Infectivity of rhMPV mutants in a cotton rat model.
To determine whether rhMPV carrying mutations in the RGD motif was attenuated in vivo, all recombinant viruses were inoculated into cotton rats and viral replication and pathogenesis were examined. Neither wt nor mutant rhMPV caused any detectable clinical symptoms of respiratory tract infection in cotton rats. At day 4 postinfection, cotton rats from each group were sacrificed, viral replication in nasal turbinates and lungs was determined, and pulmonary histology was examined. wt rhMPV replicated efficiently in the nasal turbinates and lungs of all five cotton rats (Table 2). Average viral titers of 105.19 and 104.08 were found in the nasal turbinates and lungs, respectively. rhMPV-D331E and -D331R replicated as efficiently as rhMPV in cotton rats, producing similar titers in both the nasal turbinates and the lungs. For the rhMPV-R329K group, only one out of five cotton rats had detectable infectious virus in the lung with a titer of 104.16, and two out of five rats had infectious virus in the nasal turbinates with an average titer of 102.87. For the rhMPV-D331A group, only two out of five cotton rats had infectious virus in the lungs with an average titer of 102.1, although all five rats had infectious virus in the nasal turbinates with an average titer comparable to that for wild-type rhMPV.
TABLE 2.
Replication of rhMPV carrying mutations in RGD motif in cotton ratsf
| Virusa | No. of cotton rats/group | Viral replication in: |
Lung histologyd | Lung IHCe | |||
|---|---|---|---|---|---|---|---|
| Nasal turbinateb |
Lungc |
||||||
| % infected animals | Mean log titer (PFU/g) | % infected animals | Mean log titer (PFU/g) | ||||
| rhMPV | 5 | 100 | 5.19 ± 0.48a | 100 | 4.08 ± 0.26a | 1.5a | 2.5a |
| rhMPV-R329K | 5 | 40 | 2.87b | 20 | 4.16b | 0.5b | 0.5b |
| rhMPV-D331A | 5 | 100 | 4.74 ± 0.43a | 40 | 2.1b | 0.6b | 0.5b |
| rhMPV-D331E | 5 | 100 | 5.12 ± 0.22a | 100 | 4.33 ± 0.51a | 1.2a | 2.5a |
| rhMPV-D331R | 5 | 100 | 5.20 ± 0.24a | 100 | 4.64 ± 0.27a | 1.0a | 3.0a |
Cotton rats were inoculated intranasally with DMEM or 2 × 105 PFU wild-type rhMPV or rhMPV mutants. At day 4 postimmunization, animals were euthanized for pathology study.
For rhMPV-R329K, two out of five cotton rats had detectable virus with an average titer of 2.87 log units.
For rhMPV-R329K, one out of five cotton rats had detectable virus with a titer of 4.16 log units. For rhMPV-D331A, two out of five cotton rats had detectable virus with an average titer of 2.1 log units.
The severity of lung histology was scored for each lung tissue specimen. The average score for each group is shown. 0, no change; 1, mild change; 2, moderate change; 3, severe change.
IHC, immunohistobiochemistry. The amount of hMPV antigen expression in lung tissue was scored. The average score for each group is shown. 0, no antigen; 1, small amount of antigen; 2, moderate amount of antigen; 3, large amount of antigen.
Values within a column followed by different lowercase letters (a and b) are significantly different (P < 0.05).
Pulmonary histology showed that rhMPV caused moderate histological changes, including interstitial pneumonia, peribronchial lymphoplasmacytic infiltrates, mononuclear cell infiltrate, and edematous thickening of the bronchial submucosa (Fig. 9). rhMPV-D331E and -D331R caused less interstitial pneumonia, but cotton rats infected with these recombinants had significantly increased peribronchial and perivascular inflammation. In contrast, rhMPV-R329K and -D331A caused no to mild pulmonary histological changes.
FIG 9.
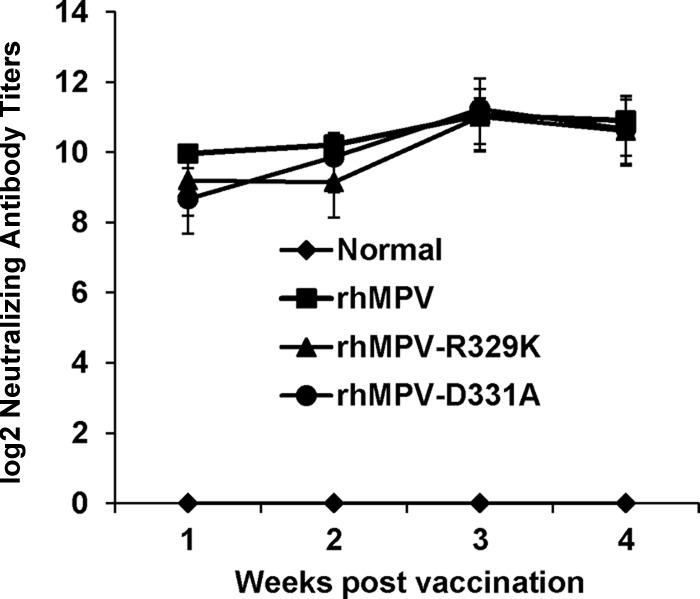
Recombinant hMPVs triggered a high neutralizing antibody titer in cotton rats. Cotton rats were immunized with each recombinant hMPV intranasally at a dose of 2.0 × 105 PFU per rat. Blood samples were collected from each rat weekly by retro-orbital bleeding. The hMPV neutralizing antibody was determined using a plaque reduction neutralization assay, as described in Materials and Methods.
Immunohistobiochemistry analysis found that rhMPV deposited a large amount of viral antigen in epithelial cells in lung tissues, whereas significantly less viral antigen was found in the lungs of rhMPV-D331A- and rhMPV-R329K-infected animals (Fig. 10). The viral antigen expression of rhMPV-D331E and -D331R in lung epithelial cells was similar to that of wild-type hMPV (data not shown). Taken together, these results confirm that rhMPV-D331E and -D331R replicated as efficiently as rhMPV but that rhMPV-R329K and -D331A had significant defects in viral replication in cotton rats. These results also suggest that rhMPV-R329K and -D331A are attenuated both in vitro and in vivo.
FIG 10.
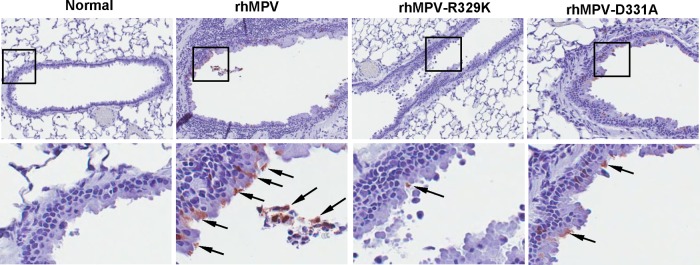
Lung histology of recombinant hMPVs. The right lung from each cotton rat was preserved in 4% (vol/vol) phosphate-buffered paraformaldehyde. Fixed tissues were embedded in paraffin, sectioned at 5 μm, and stained with H&E for the examination of histological changes by light microscopy.
Immunogenicity of rhMPV-R329K and -D331A in cotton rats.
Since rhMPV-R329K and -D331A were attenuated in cotton rats, we next examined the immunogenicity of these recombinant viruses. Cotton rats were inoculated intranasally with wild-type rhMPV or rhMPV mutants. Serum samples were collected weekly for the detection of a humoral immune response. At week 4 postinoculation, animals were challenged with 106 PFU of rhMPV. At day 4 postchallenge, all the animals were sacrificed and nasal turbinate and lung samples were collected for virus detection and pathological examination. As shown in Fig. 11, rhMPV-R329K and -D331A elicited high levels of neutralizing antibody that were comparable to those elicited by rhMPV. No hMPV-specific antibody was detected in unvaccinated control animals. As shown in Table 3, cotton rats vaccinated with rhMPV-D331A did not have any detectable infectious virus particles in either nasal turbinates or lungs after challenge with rhMPV on day 5. Only one out of five animals in the rhMPV-R329K-vaccinated group had detectable virus in nasal turbinate but not in the lung. In contrast, unvaccinated challenge control animals had average titers of 104.99 and 104.74 in nasal turbinate and lung, respectively. Pulmonary histology showed that the unvaccinated challenged control animals had moderate pathological changes characterized by interstitial pneumonia, mononuclear cell infiltrate, and edematous thickening of the bronchial submucosa. In contrast, no or mild histological changes were found in the lungs of cotton rats vaccinated with rhMPV-R329K and -D331A. Collectively, these results demonstrate that rhMPV-R329K and -D331A provide complete protection from virulent challenge and are potential vaccine candidates for hMPV.
FIG 11.
Lung histobiochemistry of recombinant hMPVs. Lung tissues were fixed in 4% (vol/vol) phosphate-buffered paraformaldehyde. Deparaffinized sections were stained with monoclonal antibody against hMPV matrix protein (Virostat, Portland, ME) to determine the distribution of viral antigen.
TABLE 3.
Immunogenicity of rhMPV carrying mutations in RGD motif in cotton rats
| Virusa | No. of cotton rats/group | Nasal turbinateb |
Lung |
Lung histologyc | ||
|---|---|---|---|---|---|---|
| % infected animals | Mean titer log (PFU/g) | % infected animals | Mean titer log (PFU/g) | |||
| DMEM | 5 | 100 | 4.99 ± 0.30 | 100 | 4.74 ± 0.50 | 1.5a |
| rhMPV | 5 | 0 | NDd | 0 | ND | 0.5b |
| rhMPV-R329K | 5 | 20 | 4.27 | 0 | ND | 0.4b |
| rhMPV-D331A | 5 | 0 | ND | 0 | ND | 0.5b |
Animals were immunized intranasally with DMEM or 2 × 105 PFU wild-type rhMPV or rhMPV mutants. At day 28 postimmunization, animals were challenged with 106 PFU wild-type rhMPV.
One out of five cotton rats challenged with rhMPV-R329K had detectable virus with a titer of 4.27 log units.
The severity of lung histology was scored for each lung tissue specimen. The average score for each group is shown. 0, no change; 1, mild change; 2, moderate change; 3, severe change. Values followed by different lowercase letters (a and b) are significantly different (P < 0.05).
ND, nondetectable.
DISCUSSION
In this study, we have demonstrated that α5β1 and αv integrins are essential for cell-cell fusion triggered by hMPV F protein. Moreover, we have shown that amino acid residues R329 and G330 within the putative integrin-binding motif (329RGD331) in the F protein play crucial roles in cell-cell fusion and viral infectivity. This conclusion is supported by multiple lines of evidence. First, hMPV F protein-mediated cell-cell fusion was inhibited by antibodies against integrin α5β1 or αv, as well as by siRNA-mediated knockdown of α5 or αv expression. Second, integrin α5β1- or αv-specific antibodies reduced hMPV infection, and expression of integrin α5β1 on the cell surface enhanced hMPV infectivity in cell culture. Third, amino acid substitutions in R329 and G330 within the RGD motif of the F protein significantly impaired fusion activity. Fourth, mutations to the RGD motif that reduced fusion were either lethal to the virus or resulted in recombinant viruses (rhMPV-R329K and -D331A) that were attenuated in viral replication in cell culture. Finally, rhMPV-R329K and -D331A were attenuated in viral replication in a cotton rat model in vivo. Collectively, our data support the idea that α5β1 and αv integrins serve as receptors for hMPV.
Integrins are a family of cell surface heterodimeric glycoproteins composed of 18 α and 8 β subunits that are expressed in nearly all cell types and facilitate cellular adhesion to and migration on the extracellular matrix proteins found in intercellular spaces and basement membranes (reviewed in reference 32). Previous studies have demonstrated that several integrin subtypes function as receptors for viral attachment and entry in a number of nonenveloped viruses (such as rotavirus, foot-and-mouth disease virus, and human adenovirus) and enveloped viruses (Epstein-Barr virus, Kaposi's sarcoma-associated herpesvirus, Sindbis virus, and yellow fever virus) (33–38). Integrin binding is mediated by a specific interaction between integrin receptors and the RGD tripeptide motif in viral glycoproteins or capsid proteins.
Cseke et al. (2009) first showed that αvβ1 integrin is able to promote hMPV infection, suggesting that αvβ1 integrin may function as a receptor for hMPV (23). Subsequently, the same group showed that multiple integrins can mediate hMPV binding and entry through interaction with the RGD domain in hMPV F protein (24). Since viral infectivity was determined by an immunostaining assay which measures viral entry and gene expression, it is not clear which step(s) in virus entry is promoted by αvβ1 integrin. For instance, these integrins could serve as either an initial or a secondary receptor for hMPV. Alternatively, it is possible that integrin expression increases expression of a viral receptor or increases overall viral gene expression rather than directly mediates attachment and/or virus entry.
We hypothesized that disruption of the interaction between integrin receptors and the RGD motif of hMPV F protein would inhibit fusion activity, which in turn would inhibit viral entry if integrin receptors indeed play direct roles in viral attachment and entry. Since paramyxoviruses enter cells by fusing their viral envelope with a host cell membrane, the role of integrin receptors and their RGD motif-binding activity can be directly measured by a cell-cell fusion assay. Using this assay, we first demonstrated that both αv and α5β1 integrin receptors are essential for cell-cell fusion mediated by hMPV F protein. Consistent with this, mutation of the integrin-binding motif (329RGD331), especially residues R329 and G330, significantly inhibited cell-cell fusion without significantly altering cell surface expression of F protein. Replacing the positively charged side chain of R329 with an uncharged side chain (R329A) or a negatively charged side chain (R329D) abolished the ability of hMPV F protein to promote cell-cell fusion. We failed to recover viable recombinant virus with these mutations, suggesting that they are lethal to the virus. Similarly, introducing mutations at residue 330 (G330A or G330S) significantly impaired the fusion activity of hMPV F protein. While we successfully recovered rhMPV-G330A and -G330S, rhMPV-G330A replicated extremely poorly in cell culture and rhMPV-G330S was genetically unstable. Compared to residues R329 and G330, residue D331 appeared to play a minor role in fusion. Replacement of the negatively charged side chain of D331 with positively or negatively charged side chains (D331R and D331E, respectively) led to enhanced cell-cell fusion. Consistent with this, recombinant hMPVs carrying these mutations replicated as efficiently as wild-type hMPV in cell culture and in cotton rats. Previously, it was shown that virus with the D331A mutation retains approximately 80% of fusion activity (25), which was consistent with our observation. In addition, we found that rhMPV-D331A was defective in viral replication in cell culture and cotton rats. Overall, the extent of fusion activity for each F-protein mutant was consistent with its replication ability in vitro and in vivo. For example, F-protein mutations that promoted less than 20% of wt fusion activity were lethal to the virus, whereas F-protein mutants (R329K and D331A) that had 60 to 70% of wild-type fusion activity were attenuated in viral replication in cell culture and cotton rats. Our results support the idea that residues R329 and G330 within the RGD motif are essential for cell-cell fusion and viability of the virus and that the RGD motif in the F protein is required for hMPV infectivity in vitro and in vivo.
Although the RGD motif is conserved in a number of viral proteins, the contributions of these three residues to virus entry and infectivity differ from virus to virus. For example, a change of RGD to RGE or RGK in the envelope protein of Murray Valley encephalitis virus (MVEV) resulted in a plaque size similar to that of the wt, whereas mutation of RGD to RGA reduced the plaque size (39). Mutating the RGD motif to RGE resulted in a larger plaque morphology for MVEV (39). In yellow fever virus, mutating the RGD motif to RGE had no effect on viral titers, whereas changing RGD to TGD, TGE, TAD, TAE, or RGS led to reduced titers (40). In hepatitis C virus (HCV), the two amino acid substitutions generated at the first position, AGE and KGE, completely abrogated HCV infectivity (41). Changing the glycine to an alanine at the second position also reduced infectivity to 18%. Interestingly, replacement of the negatively charged aspartic acid (D) or glutamic acid (E) with a neutral alanine (A) did not impair the infectivity of HCV; however, a positive lysine (K) at this position reduced the infectivity of HCV to 14% of that of the wt (41). Interestingly, mutating the RGD motif to RAD or RGA was lethal to foot-and-mouth disease virus (42). Many bacterial pathogens target cell adhesion molecules via integrin receptors to establish an intimate contact with host cells and tissues (reviewed in references 43 and 44). Mutagenesis and biochemical studies found that integrin receptors exhibited variable degrees of affinity to organisms with the analogs or mutants of the RGD motif, depending on the property of the integrin-binding protein (44). Individual residues within the RGD motif in surface proteins of many bacteria (such as Shigella, Leptospira, and Streptococcus species) played a dominant role in binding to integrin receptors (45–47). However, in the type IV secretion system (T4SS) protein CagL of Helicobacter pylori, all three residues in the RGD motif are important for integrin binding, as evidenced that the finding that replacement of each of these residues with alanine abolished the adhesion function (48).
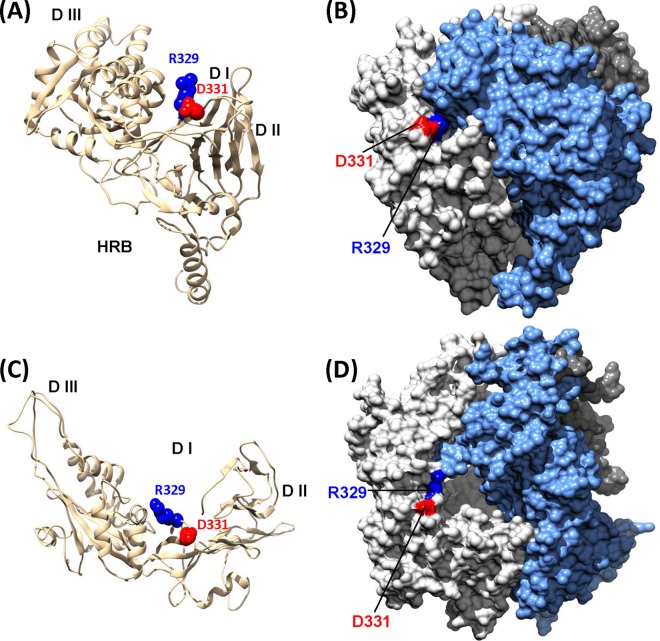
To gain possible structural insight into the role of the RGD motif in viral binding, we constructed a model structure of hMPV F protein using the atomic coordinates of the crystal structure of the prefusion form of RSV F protein as the template (PDB accession no. 4JHW) (30, 31). As in RSV F protein, each F-protein monomer is composed of five well-structured domains, namely, DI to DIII, HRA, and HRB, and one relatively loose loop connecting the C terminus of DII and HRB, which has been named the “HRB linker.” The RGD motif is located in the loop between strands β14 and β15 in the HRA region in the predicted structure of the hMPV F monomer (Fig. 12A). In the model structure of the hMPV F-protein trimer (Fig. 12B), the RGD motif is located in the contact region of each subunit of the F-protein trimer. In addition, residues R329 and G330 are exposed on the surface of the F-protein trimer, suggesting that the RGD motif may make contact with integrin receptors on the cell surface. An electrostatic interaction between R329 of the F protein and specific integrin subtypes and the small steric hindrance at D331 may favor integrin binding and the subsequent promotion of cell-cell fusion, whereas D331 may be not directly involved in binding to integrin. Most recently, a partial structure of hMPV F protein composed of three domains (DI to DIII) has been solved (49). In this structure, R329 and D331 in the RGD motif are exposed on the surface of the F-protein trimer (Fig. 12C and D). It is not clear if this structure represents the prefusion or postfusion form of the F protein (49). However, the location of the RGD motif suggests that it could directly contact a cell surface receptor.
FIG 12.
Location of the RGD motif in the predicted structure of hMPV F protein. (A) Predicted hMPV F-protein monomer. The structure was predicted using the Modeler (version 9.0) program on the basis of the prefusion crystal structure of RSV F protein (PDB accession no. 4JHW) as the template. The putative integrin-binding sites (R329 and D331) are highlighted. (B) Predicted hMPV F-protein trimer. The surface of each monomer in the F-protein trimer is highlighted in a different color. The RGD motif is located on the contact region of each subunit of the F-protein trimer. (C) Location of the RGD motif in hMPV F-protein monomer. The partial structure of hMPV F protein containing DI, DII, and DIII was solved (PDB accession no. 4DAG) (49). The location of the RGD motif is highlighted. (D) The location of the RGD motif in hMPV F-protein trimer. The surface of each monomer in the F-protein trimer is highlighted in a different color.
Recently, Chang et al. (2012) suggested that heparan sulfate proteoglycans may function as the primary receptor for hMPV F protein (25). Mutant CHO cell lines lacking the ability to synthesize glycosaminoglycans (GAGs), particularly heparan sulfate proteoglycans, were resistant to hMPV binding and infection, although wild-type CHO-K1 cells were permissive. In addition, hMPV infection was abolished when CHO-K1 cells were treated with heparinases to remove heparan sulfate proteoglycans from the cell surface. Thus, it was proposed that the interaction between integrins and hMPV occurs after the initial binding of hMPV F to heparan sulfate proteoglycans (25). Perhaps the simplest explanation is that hMPV F protein uses integrins and heparan sulfate proteoglycans as coreceptors. Previously, Cseke et al. (2009) showed that soluble hMPV F protein carrying D331E (F-D331E) abolished the binding of F protein to integrin receptors in vitro (23). However, our results clearly show that F-D331E triggers a level of fusion activity higher than that of wild-type F protein and that rhMPV-D331E replicates more efficiently in vitro and in vivo than rhMPV. These results strongly suggest that F-D331E may utilize alternative receptors to gain entry. It has been shown that viral glycoprotein and heparan sulfate binding is reversible (50, 51). It is possible that heparan sulfate proteoglycans first localize virus on the cell surface, enhancing opportunities for direct interactions between virion glycoproteins (such as hMPV F protein) and specific cellular entry receptors (such as integrins). It is also possible that hMPV, or any virus, may utilize a different receptor(s) in vitro and in vivo. This hypothesis is supported by the observation that the glycosaminoglycan moiety cannot be detected on the apical surface of primary well-differentiated human airway epithelial (HAE) cultures (52, 53). These cultures are derived directly from human lung airway tissue and closely resemble the differentiated airway epithelium in vivo with a columnar morphology, including goblet cells that produce mucus and ciliated cells whose cilia move the mucus around the apical surface of the culture. It is of interest to determine the binding and entry of hMPV in HAE culture in the future.
The development of a vaccine to protect against hMPV infection and disease is a major goal. The two most common vaccine strategies against infectious diseases are inactivated and live attenuated viruses. For safety, inactivated vaccines are generally preferred. However, development of an inactivated vaccine for human paramyxoviruses has been a problem. A formalin-inactivated RSV vaccine developed and tested in the 1960s not only failed to induce a protective immune response in a human trial but also led to enhanced respiratory disease upon natural infection with RSV (54). A recent study showed that cotton rats immunized with formalin-inactivated hMPV vaccine were protected against infection but developed increased lung damage (55). These observations suggest that, similar to RSV, an inactivated vaccine may not be the best choice for hMPV.
Like RSV, a live attenuated vaccine for hMPV would appear to be the best vaccine approach because enhanced lung damage has not been observed with virus infection of naive animals following challenge with the same virus (21, 56, 57). However, it has been technically difficult to isolate a virus that has an optimal balance between attenuation and immunogenicity. Remarkably in the present study, recombinants rhMPV-R329K and -D331A were significantly defective in viral replication in cell culture as well as in cotton rats. Importantly, immunization of cotton rats with rhMPV-R329K and -D331A not only triggered a high level of neutralizing antibody but also provided complete protection against hMPV infection. These results demonstrate that rhMPV-R329K and -D331A are attenuated and retain high immunogenicity. Therefore, inhibition of the integrin-F protein interaction might serve as an approach to attenuate hMPV for the development of live attenuated vaccines.
In summary, our study highlights a major role for integrins in hMPV F protein-mediated membrane fusion, infection, and pathogenesis. Specifically, we show that α5β1 and αv integrins are essential for hMPV infectivity and F protein-mediated cell-cell fusion and that the integrin-binding motif in the F protein plays a crucial role in these functions. Our results also identify the integrin-binding motif as a new, attenuating target for the development of a live vaccine for hMPV.
ACKNOWLEDGMENTS
This study was supported in part by grants from the NIH (R01AI090060) and the USDA Agriculture and Food Research Initiative (2010-65119-20602) and by a pilot grant from the OSU Center for Clinical and Translational Science (CCTS) to J.L.
We thank Ron A. M. Fouchier for the infectious cDNA clone of hMPV. We are grateful to members of the J. Li laboratory for critical readings of the manuscript. We thank Steven Krakowka for his help on histology.
Footnotes
Published ahead of print 29 January 2014
REFERENCES
- 1.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719–724. 10.1038/89098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van den Hoogen BG, van Doornum GJ, Fockens JC, Cornelissen JJ, Beyer WE, de Groot R, Osterhaus AD, Fouchier RA. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J. Infect. Dis. 188:1571–1577. 10.1086/379200 [DOI] [PubMed] [Google Scholar]
- 3.Crowe JE., Jr 2004. Human metapneumovirus as a major cause of human respiratory tract disease. Pediatr. Infect. Dis. J. 23:S215–S221. 10.1097/01.inf.0000144668.81573.6d [DOI] [PubMed] [Google Scholar]
- 4.Biacchesi S, Skiadopoulos MH, Boivin G, Hanson CT, Murphy BR, Collins PL, Buchholz UJ. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315:1–9. 10.1016/S0042-6822(03)00528-2 [DOI] [PubMed] [Google Scholar]
- 5.Boivin G, Abed Y, Pelletier G, Ruel L, Moisan D, Cote S, Peret TC, Erdman DD, Anderson LJ. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330–1334. 10.1086/344319 [DOI] [PubMed] [Google Scholar]
- 6.Falsey AR, Erdman D, Anderson LJ, Walsh EE. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785–790. 10.1086/367901 [DOI] [PubMed] [Google Scholar]
- 7.Govindarajan D, Buchholz UJ, Samal SK. 2006. Recovery of avian metapneumovirus subgroup C from cDNA: cross-recognition of avian and human metapneumovirus support proteins. J. Virol. 80:5790–5797. 10.1128/JVI.00138-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb RA. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1–11. 10.1006/viro.1993.1561 [DOI] [PubMed] [Google Scholar]
- 9.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U. S. A. 102:9288–9293. 10.1073/pnas.0503989102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, Lamb RA. 2012. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc. Natl. Acad. Sci. U. S. A. 109:16672–16677. 10.1073/pnas.1213802109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. 2006. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 103:17903–17908. 10.1073/pnas.0608678103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb RA, Jardetzky TS. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17:427–436. 10.1016/j.sbi.2007.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YH, Donald JE, Grigoryan G, Leser GP, Fadeev AY, Lamb RA, DeGrado WF. 2011. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc. Natl. Acad. Sci. U. S. A. 108:20992–20997. 10.1073/pnas.1116034108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallipeddi SK, Samal SK. 1993. Structural difference in the fusion protein among strains of bovine respiratory syncytial virus. Vet. Microbiol. 36:359–367. 10.1016/0378-1135(93)90102-D [DOI] [PubMed] [Google Scholar]
- 15.Pastey MK, Samal SK. 1997. Analysis of bovine respiratory syncytial virus envelope glycoproteins in cell fusion. J. Gen. Virol. 78(Pt 8):1885–1889 [DOI] [PubMed] [Google Scholar]
- 16.Chaiwatpongsakorn S, Epand RF, Collins PL, Epand RM, Peeples ME. 2011. Soluble respiratory syncytial virus fusion protein in the fully cleaved, pretriggered state is triggered by exposure to low-molarity buffer. J. Virol. 85:3968–3977. 10.1128/JVI.01813-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schowalter RM, Smith SE, Dutch RE. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J. Virol. 80:10931–10941. 10.1128/JVI.01287-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. 2009. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J. Virol. 83:1511–1522. 10.1128/JVI.01381-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mas V, Herfst S, Osterhaus AD, Fouchier RA, Melero JA. 2011. Residues of the human metapneumovirus fusion (F) protein critical for its strain-related fusion phenotype: implications for the virus replication cycle. J. Virol. 85:12650–12661. 10.1128/JVI.05485-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Y, Feng K, Yao X, Cai H, Li J, Mirza AM, Iorio RM, Li J. 2012. Localization of a region in the fusion protein of avian metapneumovirus that modulates cell-cell fusion. J. Virol. 86:11800–11814. 10.1128/JVI.00232-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biacchesi S, Skiadopoulos MH, Yang L, Lamirande EW, Tran KC, Murphy BR, Collins PL, Buchholz UJ. 2004. Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J. Virol. 78:12877–12887. 10.1128/JVI.78.23.12877-12887.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herfst S, Mas V, Ver LS, Wierda RJ, Osterhaus AD, Fouchier RA, Melero JA. 2008. Low-pH-induced membrane fusion mediated by human metapneumovirus F protein is a rare, strain-dependent phenomenon. J. Virol. 82:8891–8895. 10.1128/JVI.00472-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cseke G, Maginnis MS, Cox RG, Tollefson SJ, Podsiad AB, Wright DW, Dermody TS, Williams JV. 2009. Integrin alphavbeta1 promotes infection by human metapneumovirus. Proc. Natl. Acad. Sci. U. S. A. 106:1566–1571. 10.1073/pnas.0801433106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox RG, Livesay SB, Johnson M, Ohi MD, Williams JV. 2012. The human metapneumovirus fusion protein mediates entry via an interaction with RGD-binding integrins. J. Virol. 86:12148–12160. 10.1128/JVI.01133-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang A, Masante C, Buchholz UJ, Dutch RE. 2012. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. J. Virol. 86:3230–3243. 10.1128/JVI.06706-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Hoogen BG, Bestebroer TM, Osterhaus AD, Fouchier RA. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119–132. 10.1006/viro.2001.1355 [DOI] [PubMed] [Google Scholar]
- 27.Herfst S, de Graaf M, Schickli JH, Tang RS, Kaur J, Yang CF, Spaete RR, Haller AA, van den Hoogen BG, Osterhaus AD, Fouchier RA. 2004. Recovery of human metapneumovirus genetic lineages A and B from cloned cDNA. J. Virol. 78:8264–8270. 10.1128/JVI.78.15.8264-8270.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Wei Y, Li J, Li J. 2012. Development and optimization of a direct plaque assay for human and avian metapneumoviruses. J. Virol. Methods 185:61–68. 10.1016/j.jviromet.2012.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Graaf M, Herfst S, Schrauwen EJ, van den Hoogen BG, Osterhaus AD, Fouchier RA. 2007. An improved plaque reduction virus neutralization assay for human metapneumovirus. J. Virol. Methods 143:169–174. 10.1016/j.jviromet.2007.03.005 [DOI] [PubMed] [Google Scholar]
- 30.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. 2013. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342:592–598. 10.1126/science.1243283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. 10.1126/science.1234914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Triantafilou K, Takada Y, Triantafilou M. 2001. Mechanisms of integrin-mediated virus attachment and internalization process. Crit. Rev. Immunol. 21:311–322. 10.1615/CritRevImmunol.v21.i4.10 [DOI] [PubMed] [Google Scholar]
- 33.Asokan A, Hamra JB, Govindasamy L, Agbandje-McKenna M, Samulski RJ. 2006. Adeno-associated virus type 2 contains an integrin alpha5beta1 binding domain essential for viral cell entry. J. Virol. 80:8961–8969. 10.1128/JVI.00843-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berryman S, Clark S, Monaghan P, Jackson T. 2005. Early events in integrin alphavbeta6-mediated cell entry of foot-and-mouth disease virus. J. Virol. 79:8519–8534. 10.1128/JVI.79.13.8519-8534.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chesnokova LS, Hutt-Fletcher LM. 2011. Fusion of Epstein-Barr virus with epithelial cells can be triggered by alphavbeta5 in addition to alphavbeta6 and alphavbeta8, and integrin binding triggers a conformational change in glycoproteins gHgL. J. Virol. 85:13214–13223. 10.1128/JVI.05580-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dicara D, Burman A, Clark S, Berryman S, Howard MJ, Hart IR, Marshall JF, Jackson T. 2008. Foot-and-mouth disease virus forms a highly stable, EDTA-resistant complex with its principal receptor, integrin alphavbeta6: implications for infectiousness. J. Virol. 82:1537–1546. 10.1128/JVI.01480-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gianni T, Campadelli-Fiume G. 2012. alphaVbeta3-integrin relocalizes nectin1 and routes herpes simplex virus to lipid rafts. J. Virol. 86:2850–2855. 10.1128/JVI.06689-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Summerford C, Bartlett JS, Samulski RJ. 1999. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78–82. 10.1038/4768 [DOI] [PubMed] [Google Scholar]
- 39.Lee E, Lobigs M. 2000. Substitutions at the putative receptor-binding site of an encephalitic flavivirus alter virulence and host cell tropism and reveal a role for glycosaminoglycans in entry. J. Virol. 74:8867–8875. 10.1128/JVI.74.19.8867-8875.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Most RG, Corver J, Strauss JH. 1999. Mutagenesis of the RGD motif in the yellow fever virus 17D envelope protein. Virology 265:83–95. 10.1006/viro.1999.0026 [DOI] [PubMed] [Google Scholar]
- 41.Rothwangl KB, Rong L. 2009. Analysis of a conserved RGE/RGD motif in HCV E2 in mediating entry. Virol. J. 6:12. 10.1186/1743-422X-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leippert M, Beck E, Weiland F, Pfaff E. 1997. Point mutations within the betaG-betaH loop of foot-and-mouth disease virus O1K affect virus attachment to target cells. J. Virol. 71:1046–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnaout MA, Goodman SL, Xiong JP. 2002. Coming to grips with integrin binding to ligands. Curr. Opin. Cell Biol. 14:641–651. 10.1016/S0955-0674(02)00371-X [DOI] [PubMed] [Google Scholar]
- 44.Hauck CR, Agerer F, Muenzner P, Schmitter T. 2006. Cellular adhesion molecules as targets for bacterial infection. Eur. J. Cell Biol. 85:235–242. 10.1016/j.ejcb.2005.08.002 [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Zhang C, Ojcius DM, Sun D, Zhao J, Lin X, Li L, Li L, Yan J. 2012. The mammalian cell entry (Mce) protein of pathogenic Leptospira species is responsible for RGD motif-dependent infection of cells and animals. Mol. Microbiol. 83:1006–1023. 10.1111/j.1365-2958.2012.07985.x [DOI] [PubMed] [Google Scholar]
- 46.Allaoui A, Sansonetti PJ, Menard R, Barzu S, Mounier J, Phalipon A, Parsot C. 1995. MxiG, a membrane protein required for secretion of Shigella spp. Ipa invasins: involvement in entry into epithelial cells and in intercellular dissemination. Mol. Microbiol. 17:461–470 [DOI] [PubMed] [Google Scholar]
- 47.Stockbauer KE, Magoun L, Liu M, Burns EH, Jr, Gubba S, Renish S, Pan X, Bodary SC, Baker E, Coburn J, Leong JM, Musser JM. 1999. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins alphavbeta3 and alphaIIbbeta3. Proc. Natl. Acad. Sci. U. S. A. 96:242–247. 10.1073/pnas.96.1.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barden S, Lange S, Tegtmeyer N, Conradi J, Sewald N, Backert S, Niemann HH. 2013. A helical RGD motif promoting cell adhesion: crystal structures of the Helicobacter pylori type IV secretion system pilus protein CagL. Structure 21:1931–1941. 10.1016/j.str.2013.08.018 [DOI] [PubMed] [Google Scholar]
- 49.Wen X, Krause JC, Leser GP, Cox RG, Lamb RA, Williams JV, Crowe JE, Jr, Jardetzky TS. 2012. Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nat. Struct. Mol. Biol. 19:461–463. 10.1038/nsmb.2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akula SM, Pramod NP, Wang FZ, Chandran B. 2001. Human herpesvirus 8 envelope-associated glycoprotein B interacts with heparan sulfate-like moieties. Virology 284:235–249. 10.1006/viro.2001.0921 [DOI] [PubMed] [Google Scholar]
- 51.Spear PG, Longnecker R. 2003. Herpesvirus entry: an update. J. Virol. 77:10179–10185. 10.1128/JVI.77.19.10179-10185.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L, Bukreyev A, Thompson CI, Watson B, Peeples ME, Collins PL, Pickles RJ. 2005. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 79:1113–1124. 10.1128/JVI.79.2.1113-1124.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76:5654–5666. 10.1128/JVI.76.11.5654-5666.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 89:422–434 [DOI] [PubMed] [Google Scholar]
- 55.Yim KC, Cragin RP, Boukhvalova MS, Blanco JC, Hamlin ME, Boivin G, Porter DD, Prince GA. 2007. Human metapneumovirus: enhanced pulmonary disease in cotton rats immunized with formalin-inactivated virus vaccine and challenged. Vaccine 25:5034–5040. 10.1016/j.vaccine.2007.04.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchholz UJ, Nagashima K, Murphy BR, Collins PL. 2006. Live vaccines for human metapneumovirus designed by reverse genetics. Expert Rev. Vaccines 5:695–706. 10.1586/14760584.5.5.695 [DOI] [PubMed] [Google Scholar]
- 57.Biacchesi S, Pham QN, Skiadopoulos MH, Murphy BR, Collins PL, Buchholz UJ. 2005. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J. Virol. 79:12608–12613. 10.1128/JVI.79.19.12608-12613.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]