ABSTRACT
A key step of retroviral replication is packaging of the viral RNA genome during virus assembly. Specific packaging is mediated by interactions between the viral protein Gag and elements in the viral RNA genome. In HIV-1, similar to most retroviruses, the packaging signal is located within the 5′ untranslated region and extends into the gag-coding region. A recent study reported that a region including the Gag-Pol ribosomal frameshift signal plays an important role in HIV-1 RNA packaging; deletions or mutations that affect the RNA structure of this signal lead to drastic decreases (10- to 50-fold) in viral RNA packaging and virus titer. We examined here the role of the ribosomal frameshift signal in HIV-1 RNA packaging by studying the RNA packaging and virus titer in the context of proviruses. Three mutants with altered ribosomal frameshift signal, either through direct deletion of the signal, mutation of the 6U slippery sequence, or alterations of the secondary structure were examined. We found that RNAs from all three mutants were packaged efficiently, and they generate titers similar to that of a virus containing the wild-type ribosomal frameshift signal. We conclude that although the ribosomal frameshift signal plays an important role in regulating the replication cycle, this RNA element is not directly involved in regulating RNA encapsidation.
IMPORTANCE To generate infectious viruses, HIV-1 must package viral RNA genome during virus assembly. The specific HIV-1 genome packaging is mediated by interactions between the structural protein Gag and elements near the 5′ end of the viral RNA known as packaging signal. In this study, we examined whether the Gag-Pol ribosomal frameshift signal is important for HIV-1 RNA packaging as recently reported. Our results demonstrated that when Gag/Gag-Pol is supplied in trans, none of the tested ribosomal frameshift signal mutants has defects in RNA packaging or virus titer. These studies provide important information on how HIV-1 regulates its genome packaging and generate infectious viruses necessary for transmission to new hosts.
INTRODUCTION
An essential step in generating infectious HIV-1 particles is the packaging of the viral RNA genome. The genome packaging of HIV-1 is mediated by interactions between the structural protein Gag and elements in the viral RNA (1–6). Of the various domains in HIV-1 Gag, nucleocapsid (NC) has been shown to play a critical role in genome encapsidation (7–9). Mutations in NC can drastically reduce the RNA genome packaged into virions, and replacing NC with a leucine zipper motif results in particles that contain little RNA (10–12). Similar to many other retroviruses, the major HIV-1 packaging signal is located at the 5′ untranslated region (UTR) and extends into a portion of the gag gene (13–19). Viral RNA is well-structured near the 5′ end of the HIV-1 genome, and maintaining the proper structure is important for RNA packaging (13, 17, 20–22). Because the full-length RNA also serves as the template for protein translation, the relationship between packaging and translation has been explored. Using a competition assay that coexpresses two proviruses, one encoding a functional Gag and the other containing a mutation to disrupt Gag translation, it was observed that HIV-1 RNA with an AUG mutation at the Gag translation start site or a premature stop codon in gag can be packaged with an efficiency similar to that of RNA with the wild-type gag gene (23). Hence, translation is not required for RNA packaging (23, 24).
With the exception of spumaviruses, retroviral pol genes are expressed as Gag-Pol polyproteins. In some retroviruses, such as murine leukemia virus (MLV), gag and pol are in the same reading frame; a programmed translational readthrough occurs to generate Gag-Pol polyproteins (25, 26). In the case of HIV-1, the pol gene is in the −1 reading frame of gag; ribosomal frameshift occasionally occurs at a UUUUUU (6 U) slippery sequence to generate Gag-Pol polyproteins (27). Both the translational readthrough and the ribosomal frameshift events are highly regulated processes, which are governed by RNA secondary structures. Hence, altering the RNA structures near the translational readthrough or ribosomal frameshift signal can affect Gag-Pol expression and consequently viral replication.
A recent study by Chamanian et al. reported that in addition to regulating the expression of pol, the HIV-1 gag-pol ribosomal frameshift signal plays an important role in viral RNA packaging (28). Using a system that coexpressed a truncated mutant genome along with a near full-length genome, it was found that deletion of the ribosomal frameshift signal led to severe RNA packaging defects, up to a 50-fold decrease compared to RNAs that retained the signal. Furthermore, substitution mutations showed that changing as few as 3 nucleotides (nt) could destroy the RNA structure of the ribosomal frameshift signal and cause severe (28-fold) defects in viral RNA packaging.
The observed drastic effects of the gag-pol ribosomal frameshift signal mutants were puzzling to us for the following reasons. First, it has been previously shown that deleting the central portion of the HIV-1 genome, including the gag-pol ribosomal frameshift signal, has little effect on RNA packaging efficiency (29); our unpublished results also support this conclusion. In addition, we have recently developed a single virion analyses system that contains a 3-nt synonymous mutation to abolish the 6 U ribosomal frameshift signal and showed that most (>94%) HIV-1 particles contain viral genomes (30).
We postulated that perhaps the effects of the ribosomal frameshift signal are dependent upon the specific mutations introduced into the system. It was also possible that the proposed packaging defects could only be observed using a competition system where HIV-1 constructs containing the mutation were coexpressed along with a construct containing the wild-type signal as the one used in the recent report. To determine whether the Gag-Pol ribosomal frameshift signal is important for RNA packaging, we coexpressed two HIV-1 proviruses, one containing the wild-type and the other containing a mutated ribosomal frameshift signal, and then compared the RNA packaging and virus titers of these viruses. Included in our study are two of the HIV-1 constructs containing mutations reported to have severe packaging defects (28). Our results showed that when Gag/Gag-Pol is supplied in trans, none of the tested ribosomal frameshift signal mutants had defects in RNA packaging or virus titer.
MATERIALS AND METHODS
Plasmid construction.
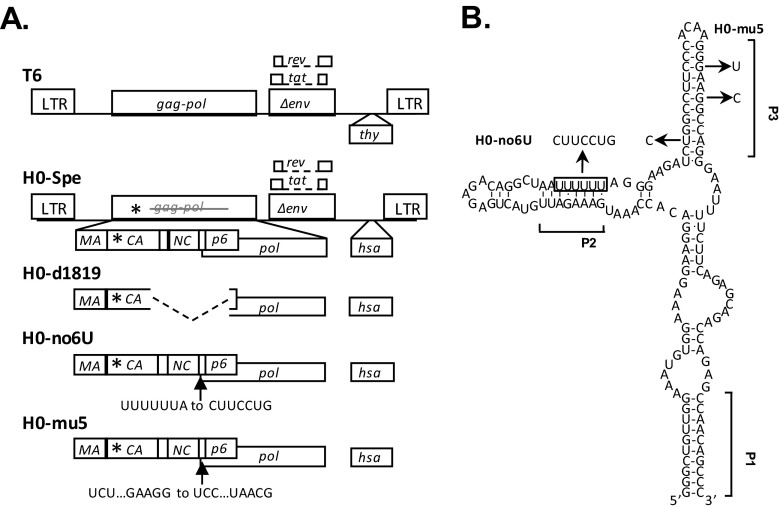
The modified HIV-1 constructs used in the present study were derived from previously described NL4-3-based T6 and H0 vectors (31), which express Gag/Gag-Pol, Tat, and Rev, and contain all cis-acting elements essential for virus replication. T6 carries in the nef reading frame a marker cassette consisting of a mouse CD90.2 gene, referred to as thy in this report, followed by an internal ribosome entry site (IRES) and an inactivated green fluorescent protein (gfp) gene. H0 has a structure similar to T6 except the thy gene is replaced with a mouse heat-stable antigen (hsa) gene and the inactivating mutation in gfp is located at a different position. Because the inactivated gfp marker genes are not used as a readout in the present study, for simplicity, the IRES-gfp is not shown in Fig. 1A. The previously described H0-Spe contains a 4-bp insertion in the capsid coding region of gag, which generates a frameshift and leads to a premature stop codon in the Gag reading frame (23).
FIG 1.
General structures of HIV-1 constructs (A) and predicted RNA structure of the HIV-1 Gag-Pol ribosomal frameshift signal (B). Asterisks denote locations of the stop codon insertion in the CA-coding region. Dashed lines indicate deletions, and arrows indicate the locations of point mutations. The predicted secondary structure of NL4-3 ribosomal frameshift signal is modified from an earlier study (42), with square brackets indicating positions of proposed RNA helices P1, P2, and P3. The boxed sequence denotes the stretch of six U nucleotides that constitute the ribosome slippery sequence. The locations of point mutations and the names of the mutants are shown. Three nucleotide changes were introduced into the 6 U stretch to generate H0-no6U, whereas three nucleotide mutations were introduced into the P3 helix to generate H0-mu5.
Mutations in the ribosome frameshift signals were introduced by overlapping PCR using the H0-Spe plasmid as a template. PCR products were digested with SphI and SbfI restriction enzymes and cloned into a SphI-SbfI-digested H0 plasmid. Plasmids were constructed using standard molecular cloning techniques (32). All constructs were characterized by restriction mapping, and the PCR-amplified regions were verified by DNA sequencing.
Cell culture, DNA transfections, infections, and flow cytometry analysis.
All cultured cells were maintained in humidified 37°C incubators with 5% CO2. The modified human embryonic kidney cell line 293T (33) was maintained in Dulbecco modified Eagle medium (CellGro) supplemented with 5% fetal calf serum (HyClone), 5% calf serum (HyClone), penicillin (50 U/ml; Gibco), and streptomycin (50 U/ml; Gibco). The human T-cell line HUT/R5, derived from HUT78 to express chemokine receptor CCR5 (34), was maintained in RPMI medium (CellGro) supplemented with 10% fetal calf serum, penicillin (50 U/ml), streptomycin (50 U/ml), 1 μg of puromycin/ml, and 500 μg of G418/ml.
DNA transfection experiments were performed using TransIT LT1 reagent (Mirus) according to the manufacturer's instructions. Viral supernatants were harvested 48 h posttransfection, clarified through a 0.45-μm-pore-size filter to remove cellular debris, and used immediately or stored at −80°C prior to infection.
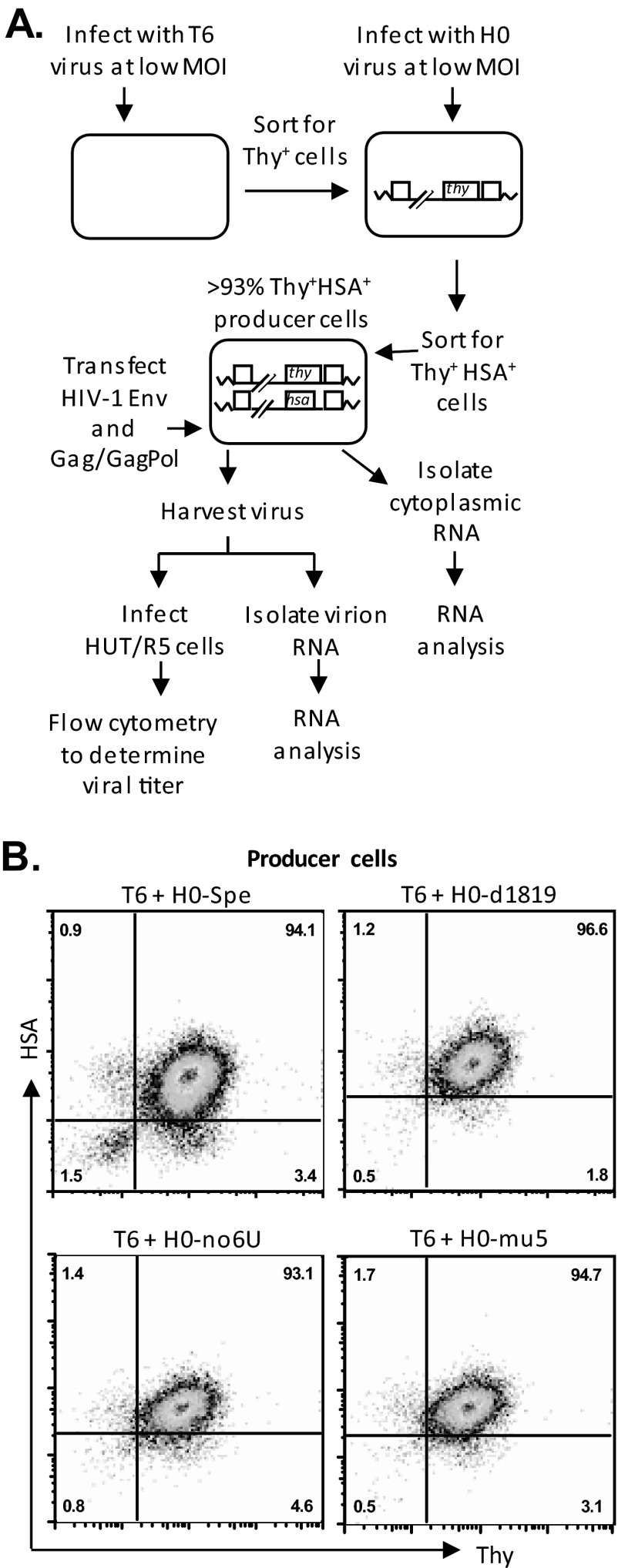
The producer cell lines were generated by sequential infection of 293T cells with two viruses at a low multiplicity of infection (MOI) of <0.15 (31). Viral stocks used to generate producer cell lines were produced from 293T cells transfected with the corresponding HIV-1 plasmid, along with pVSV-G (35) and pSYNGP (36), which express G protein from vesicular stomatitis virus and codon-optimized HIV-1 Gag/Gag-Pol, respectively. Viruses were harvested 48 h posttransfection, serially diluted, and used to infect fresh 293T cells; infection levels were determined 72 h postinfection using flow cytometry to detect Thy and/or HSA expression. Dually infected cells were enriched by several rounds of cell sorting so that >93% of the cells expressed markers encoded by both vectors. Cell line T6/H0-Spe containing proviruses T6 and H0-Spe has been described previously (23). To determine virus titers, producer cells were transfected with pSYNGP and pIIINL(AD8)env (37) that express HIV-1 Gag/Gag-Pol and Env, respectively. Viruses were harvested 48 h posttransfection, clarified through 0.45-μm-pore-size filter and used to infect target HUT/R5 cells by spinoculation at 1,200 × g for 1 h at 25°C. Infected cells were analyzed 72 h postinfection by using flow cytometry.
To detect marker gene expression, cells were stained with phycoerythrin-conjugated α-HSA antibody (Becton Dickinson Biosciences) and allophycocyanin-conjugated α-Thy1.2 antibody (eBioscience) at 0.4 and 2.0 μg/ml, respectively. Flow cytometry analyses were performed on a FACSCalibur system (BD Biosciences), whereas cell sorting was performed on an ARIA II system (BD Biosciences). Flow cytometry data were analyzed using FlowJo software (Tree Star).
Viral RNA isolation and analysis.
RNA was isolated from virions using a QIAmp viral RNA minikit (Qiagen) according to the manufacturer's instructions. Cytoplasmic RNA was isolated from producer cells according to a standard protocol (38). Briefly, 1 × 106 to 2 × 106 cells were washed twice with cold phosphate-buffered saline, collected by centrifugation, and resuspended in lysis buffer; nuclei and cellular debris were removed by centrifugation. The supernatant was treated with proteinase K; RNA was extracted with a phenol-chloroform-isoamyl alcohol mixture (25:24:1) and precipitated with ethanol. All RNA samples were treated with the Turbo DNA-free kit (Ambion) to remove any contaminating DNA.
Reverse transcription-PCR (RT-PCR) sequencing was performed to determine ratios of wild-type to mutant viral RNAs as described previously (23, 39). RNA was converted to cDNA using random hexamer primers and a Transcriptor kit (Roche). The region encompassing the 4-nt insertion in CA was amplified using the primers NL1075S (5′-GACACCAAGGAAGCCTTA-3′) and NL1564A (5′-CTACTGGGATAGGTG-3′); PCR product was gel purified and sequenced using the primer NL1143S (5′-AGCAGCTGACACAGGAAACAAC-3′). The PCR amplified both the wild-type and mutant templates, and the ratio of polymorphisms was detected as the relative heights of peaks on the sequencing chromatogram.
The following control experiment was performed to determine the accuracy and the dynamic range of the RT-PCR sequencing method. Cytoplasmic RNAs were isolated from cell lines containing either T6 provirus or H0-Spe provirus. RNAs were first converted into cDNAs as described above, and the amount of HIV-1 full-length RNA was quantified by real-time PCR using a LightCycler Probes master kit (Roche) with the previously described gag primers HIV-gag-F1 and HIV-gag-R1 and the gag probe P-HUS-103 (40, 41). RNAs from the two samples were mixed at various ratios and analyzed using the RT-PCR sequencing method.
RESULTS
Experimental system used to examine the role of the ribosomal frameshift signal in HIV-1 RNA packaging.
To examine the effects of the gag-pol frameshift signal on the efficiency of HIV-1 RNA packaging, we generated variants containing mutations in this region based on a previously described near full-length HIV-1 construct H0-Spe (23). H0-Spe contains HIV-1 sequences from the 5′ long terminal repeat (5′LTR) to the end of pol, as well as tat, rev, a partially deleted env, and 3′LTR (Fig. 1A). This construct also contains a heat stable antigen (hsa) gene in the nef reading frame, the expression of which can be detected using antibody staining and flow cytometry. In addition, a 4-nt insertion in gag generates a frameshift and a premature stop codon in the capsid-coding region. Thus, H0-Spe does not express functional Gag/Gag-Pol; however, when Gag/Gag-Pol is provided in trans, the RNA generated from H0-Spe is packaged efficiently, at a level similar to that of a structurally similar construct that expresses functional Gag (23). We chose to use H0-Spe as a backbone to eliminate complications associated with expressing mutant proteins so that the effects observed will be associated with viral RNA rather than its encoded proteins.
The ribosomal frameshift signal in HIV-1 is predicted to form a three-helix junction (Fig. 1B) (42). The base-pairing of the 5′ and 3′ ends of the sequence forms the anchoring helix P1, the slippery sequence UUUUUU is located in the helix P2, whereas sequences 3′ to the slippery sequence form helix P3. It is thought that this RNA structure slows down translation, allowing the ribosome to pause, form alternative tRNA pairing, and shift the reading frame. To examine the role of the ribosomal frameshift signal in HIV-1 RNA packaging, we generated three mutants which disrupted the ribosomal frameshift signal either through deletion, mutation of the slippery 6 U sequence, or alteration of the secondary structure. Mutant H0-d1819 contains a large 432-bp deletion, including the entire ribosomal frameshift signal from nt 1819 to nt 2251 (NL4-3 numbering); H0-no6U contains a 3-nt substitution that changed the slippery sequence from UUUUUUA to CUUCCUG and likely alters the P2 helix of the predicted ribosomal frameshift signal structure (Fig. 1B); H0-mu5 contains a 3-nt mutation that distorts the P3 helix of the frameshift RNA structure (28). The d1819 and mu5 mutations are identical to those previously reported by Chamanian et al. as RTΔ1819-2251 and mu5, which caused 10- and 28-fold defects in RNA packaging compared to their counterparts with wild-type sequences, respectively (28).
To test the packaging efficiencies of RNAs containing the aforementioned ribosomal frameshift signal mutations, we compared their encapsidation in competition with a structurally similar HIV RNA derived from T6 (Fig. 1A). T6 is identical to H0-Spe except that it retains the wild-type gag-pol gene, including the ribosomal frameshift signal and encodes a mouse thy gene marker in the nef gene; Thy expression can be detected by antibody staining and flow cytometry.
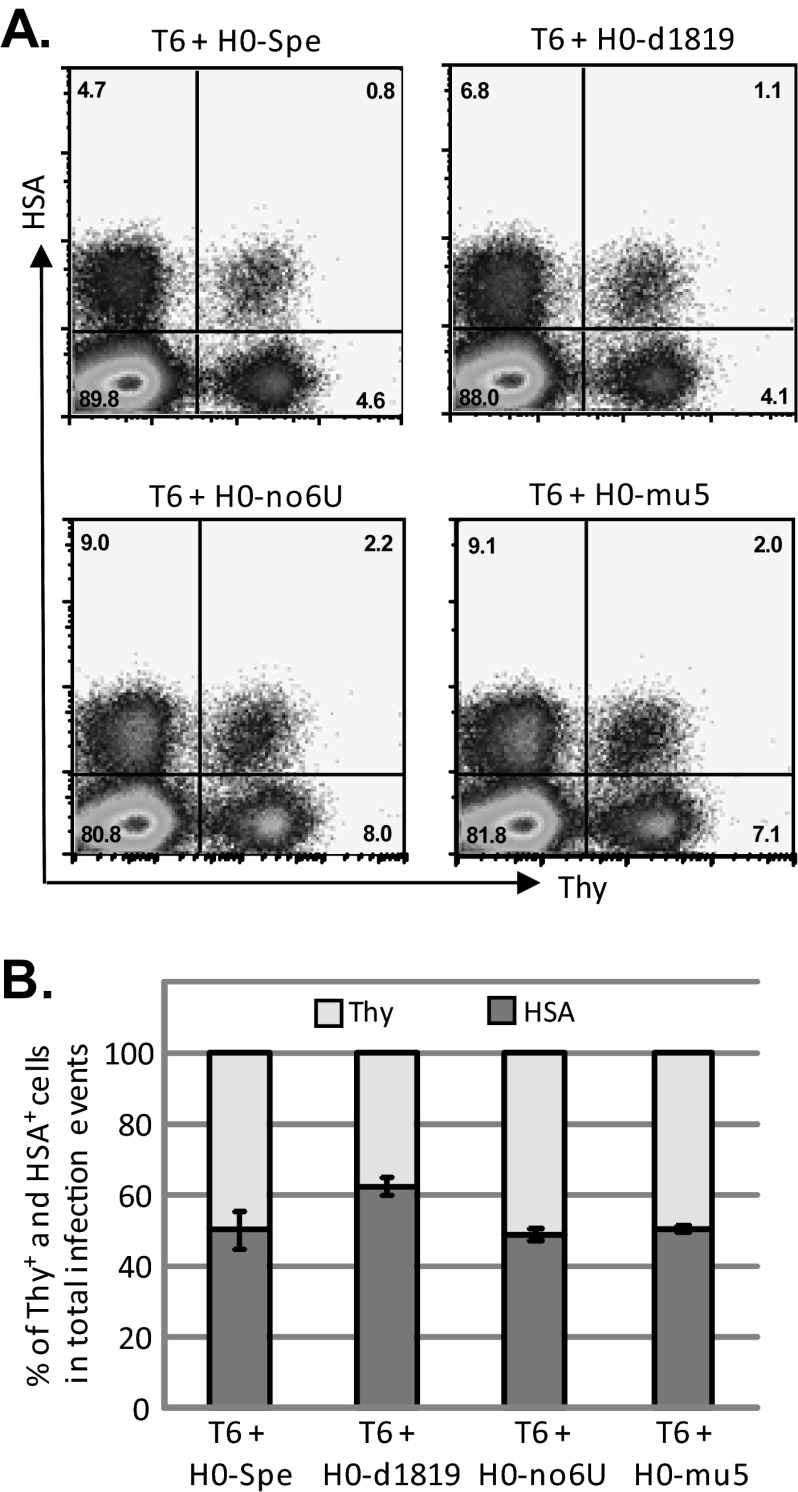
To mimic the conditions during HIV replication, we examined the RNA packaging efficiencies in the context of proviruses introduced into the cells by infection. For this purpose, we generated dually infected producer cells by first infecting the 293T cells with T6 viruses at low MOI; the infected cells were enriched by sorting based on Thy expression (Fig. 2A). These cells were then infected at a low MOI with H0-Spe-derived viruses containing mutant ribosomal frameshift signals; dually infected cells were enriched by cell sorting by selecting for cells expressing both Thy and HSA markers until >93% of the producer cells were positive with both markers (Thy+/HSA+) (Fig. 2B). To generate infectious viruses, we transfected these dually infected producer cells with two plasmids, pSYNGP and pIIINL(AD8)env, that express HIV-1 Gag/Gag-Pol and CCR5-tropic HIV-1 Env, respectively. Viruses from these cells were harvested and used to infect HUT/R5 cells; the titers of T6 and H0-Spe-derived viruses were determined by the number of cells that express Thy and HSA, respectively (Fig. 3A). As 293T cells do not express CD4, HIV-1 Env containing viruses cannot reinfect the producer cells; hence, the titer measurements reflect one round of viral replication.
FIG 2.
Experimental protocol used to generate producer cell lines (A) and flow cytometry analyses of producer cell lines (B). Thy and HSA expression are shown on the x and y axes, respectively; all producer cell lines contain >93% Thy+/HSA+ cells.
FIG 3.
Effects of mutations in the ribosomal frameshift signals on HIV-1 titers. Viruses were generated by transfecting producer cell lines with helper plasmids expressing HIV-1 Env and Gag/Gag-Pol and were used to infect HUT/R5 cells. Virus titers were determined using flow cytometry by detecting cells that express HSA and Thy markers. (A) Representative flow cytometry analyses of HUT/R5 cells infected with viruses harvested from producer cells. (B) Relative contribution of Thy+ and HSA+ cells in all infection events. Total infection events are set as 100%. Error bars represent standard deviations of three infection experiments.
Mutations of the Gag-Pol ribosomal frameshift signal do not affect virus titers.
We compared the virus titers from producer cells containing T6 and various H0-Spe-derived constructs; a summary of the findings from three sets of infection experiments is shown in Fig. 3B. When the producer cells coexpress T6 and H0-Spe proviruses, both containing wild-type ribosomal frameshift sequences, the Thy and the HSA titers were very similar. Of the total infection events (Thy titer plus HSA titer), 52% was from HSA titer. When the producer cells express T6 and H0-d1819 that contains a 432-bp deletion, including the ribosomal frameshift signal, 62% of the total infection events were from HSA titers. Mutating 3 nt to abolish the 6 Us used for frameshift (H0-noU6) or to disrupt the RNA structure of the ribosomal frameshift signal (H0-mu5) did not affect titers of these viruses. When coexpressed with T6, H0-no6U and H0-mu5 generated 54 and 56% of the infectious events, respectively. Therefore, none of the ribosomal frameshift signal mutants has a decreased virus titer compared to that from H0-Spe, which contains a wild-type ribosomal frameshift signal.
Examining the packaging efficiencies of RNAs containing mutations in the ribosomal frameshift signal.
To directly compare the packaging efficiency of HIV-1 RNA with or without mutations in the ribosomal frameshift signal, we isolated RNA from the cytoplasm of the producer cells and from cell-free virions and analyzed the ratios of T6 and H0-Spe RNA by the RT-PCR sequencing method (23, 39). Briefly, RNA samples were reverse transcribed, amplified by PCR using primers in gag, and the PCR products were gel purified and sequenced. The sequence variation between T6 and H0-Spe RNAs was used to determine the ratios of the two RNAs by measuring the heights of individual nucleotide signals on the sequencing chromatograms. Because the gag sequences were analyzed, this method examines the full-length unspliced HIV-1 RNAs.
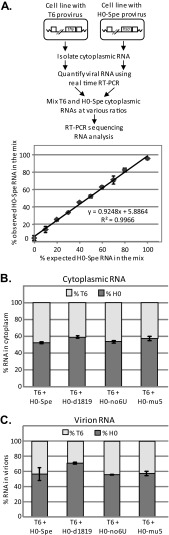
To verify the accuracy and the dynamic range of the RT-PCR sequencing method, we performed the following experiments (Fig. 4A). Cytoplasmic RNAs were isolated from a producer cell line expressing the T6 provirus and another cell line expressing the H0-Spe provirus. The amounts of full-length HIV-1 RNA were quantified by real-time RT-PCR using gag-specific primers and probe (40, 41). These two RNA samples were then mixed at various ratios and analyzed by the RT-PCR sequencing method. We found that the RT-PCR sequencing method accurately measures the proportion of different viral RNA species in the mixture when one of the RNA species is present within the 10 to 90% range (Fig. 4A). These results are similar to the previously reported sensitivity of the RT-PCR sequencing method (23).
FIG 4.
Effects of mutations in the ribosomal frameshift signal on viral RNA packaging. (A) Control experiment examining the accuracy and dynamic range of the RNA detection method. Cytoplasmic RNAs were extracted from cell lines containing either T6 provirus or H0-Spe provirus. The amount of full-length HIV-1 RNA in each sample was quantified by real-time RT-PCR using primers and probe annealing to gag. RNAs from the two samples were mixed at various ratios and analyzed using the RT-PCR sequencing method. The expected and observed percentages of H0-Spe RNA are shown in the x and y axes, respectively. (B and C) Relative percentages of T6 and H0-Spe full-length RNAs in the cytoplasm of producer cells (B) and in cell-free virions (C). Total full-length HIV-1 RNA was set as 100%. Error bars represent standard deviations of two independent experiments.
We isolated cytoplasmic RNAs from the producer cell lines and RNAs from the cell-free virions and analyzed these samples by the RT-PCR sequencing method. When both proviruses, T6 and H0-Spe, contain the wild-type ribosomal frameshift signal, these two RNAs were expressed at similar levels; 49% of the total HIV-1 full-length RNAs were from H0-Spe (Fig. 4B). Virion RNA analyses indicated that T6 and H0-Spe RNAs were also packaged at similar ratios; 55% of the packaged HIV-1 RNAs were from H0-Spe. These results are similar to previous findings (23). We then analyzed RNA from cell lines coexpressing T6 and H0-Spe-derived ribosomal frameshift signal mutants. The H0-d1819 RNA constituted 60% of full-length HIV-1 RNA in the cytoplasm of producer cells and 73% of RNA in cell-free virions (Fig. 4B and C, respectively), indicating that the deletion did not result in a loss of RNA packaging efficiency. The cytoplasm of producer cells expressing T6 and H0-no6U proviruses contained 57% of the H0-no6U RNAs, whereas cell-free virions contained 61% of the H0-no6U RNA. Similarly, when coexpressed with T6, the H0-mu5 provirus generated 61% of the HIV-1 full-length RNA in the cytoplasm and 62% of the RNA in the released virions. In all of our experiments, the proportion of full-length H0-Spe-derived viral RNAs in cytoplasm ranged between 49 and 60% regardless of whether they contain ribosomal frameshift signals, indicating that these mutations did not affect the cytoplasmic RNA levels. Furthermore, virions released from these producer cells contain ratios of T6 and H0-Spe RNAs similar to those in the cytoplasm, indicating that H0-Spe RNAs do not have packaging defects regardless of whether the wild-type ribosomal frameshift signal is present in the RNA. Furthermore, the RNA measurements are consistent with our virus titer results. Together, these findings demonstrate that, in our system, the absence of the wild-type ribosomal frameshift signal does not impede the packaging of the HIV-1 full-length RNA.
DISCUSSION
The cis-acting elements of HIV-1 RNA regulate multiple aspects of viral replication, from the efficient generation of viral RNA transcripts to conversion of the viral RNA into a DNA copy. Some of the elements in viral RNA interact with Gag to ensure the packaging of viral genome into virions. Although the HIV-1 major packaging signal was identified 2 decades ago, it is unclear whether other signals exist that also regulate the encapsidation process. Several groups have reported that the Rev protein and the rev response element (RRE) are important for viral RNA packaging (43, 44). However, the export function of RRE and Rev can be replaced by the constitutive transport element (CTE) from Mason-Pfizer monkey virus, which bears little sequence homology to RRE (45). Furthermore, HIV-1 RNAs containing the CTE instead of the RRE are efficiently packaged into virions (46, 47). These studies clarified that the essential function of RRE is transport of the viral RNA and its role in RNA packaging is indirect.
Chamanian et al. recently reported that the HIV-1 ribosomal frameshift signal is an important element for RNA genome packaging (28). This effect was identified by systematically deleting the HIV-1 genome starting from the 3′ end. Compared to near full-length RNA that contained sequences from the 5′ end to the nef gene, deletion of the entire pol and env including the RRE had no effect in RNA expression and packaging, whereas further deletion into the NC-coding region had a 27-fold decrease in viral RNA packaging. These experiments showed that deletion of an RNA region including the ribosomal frameshift signal caused severe packaging defects, whereas deletion of the RRE, without supplementing with another RNA export element, does not affect RNA expression/packaging.
In the current report, we examined the importance of the gag-pol ribosomal frameshift signal on RNA packaging. We generated a mutant containing the previously described 432-bp deletion, including the ribosomal frameshift signal, and found that this mutant does not have a decreased virus titer or a defect in RNA packaging, in contrast to the previously reported 10-fold decrease in RNA packaging. Similarly, substitution mutations that destroyed the predicted RNA secondary structures of the ribosomal frameshift signal, which was reported to have a 28-fold defect in RNA packaging, also did not display any defects in our experiments. At this time, the cause of the discrepancies between the two studies is unclear. There are differences in the experimental systems used by the two studies. Chamanian et al. used transient transfections in all experiments and in many, but not all, experiments used a copackaging system to measure viral RNA and titers. We studied HIV-1 in the context of proviruses that are stably integrated into the host chromosomes and compared the cellular RNA expression with the viral RNA packaged into the virions. It is unclear whether these are the reasons for the observed discrepancies; further experiments are needed to determine the cause of these differences.
RNA packaging in HIV-1 is a highly regulated event. Recent studies demonstrated that most HIV-1 particles contain RNA genomes (30); furthermore, two copies of viral RNA forming a dimer are packaged even when the RNA size is one-third of the wild-type genome (48). These results support the hypothesis that recognition of one dimeric RNA by Gag is a key point of viral RNA encapsidation. Recent advances shed light on how Gag recognizes dimeric RNA; for example, studies of MLVs have revealed that dimerization of the viral RNA exposes high-affinity Gag binding sites to allow the initiation of RNA packaging (49–51). Studies in HIV-1 have also identified structural elements important in dimerization, Gag binding, and packaging (17, 20, 21). Nevertheless, it is still unclear how HIV-1 achieves the regulation of packaging just one dimer; future studies are needed to define the mechanism of this regulation.
ACKNOWLEDGMENTS
We thank Vinay K. Pathak for discussions and suggestions throughout the project and Krista Delviks-Frankenberry and Vinay Pathak for critical reading of the manuscript.
This study was funded in part by the Intramural AIDS Targeted Antiviral Program, National Institutes of Health, and the Intramural Research Program of the Center for Cancer Research, Nation Cancer Institute.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 22 January 2014
REFERENCES
- 1.Berkowitz R, Fisher J, Goff SP. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177–218 [DOI] [PubMed] [Google Scholar]
- 2.D'Souza V, Summers MF. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3:643–655. 10.1038/nrmicro1210 [DOI] [PubMed] [Google Scholar]
- 3.Johnson SF, Telesnitsky A. 2010. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog. 6:e1001007. 10.1371/journal.ppat.1001007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lever AM. 2007. HIV-1 RNA packaging. Adv. Pharmacol. 55:1–32. 10.1016/S1054-3589(07)55001-5 [DOI] [PubMed] [Google Scholar]
- 5.Moore MD, Hu WS. 2009. HIV-1 RNA dimerization: it takes two to tango. AIDS Rev. 11:91–102 [PMC free article] [PubMed] [Google Scholar]
- 6.Rein A. 1994. Retroviral RNA packaging: a review. Arch. Virol. 1994(Suppl 9):513–522 [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz RD, Ohagen A, Hoglund S, Goff SP. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69:6445–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorelick RJ, Nigida SM, Jr, Bess JW, Jr, Arthur LO, Henderson LE, Rein A. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Barklis E. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71:6765–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crist RM, Datta SA, Stephen AG, Soheilian F, Mirro J, Fisher RJ, Nagashima K, Rein A. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 83:2216–2225. 10.1128/JVI.02031-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorelick RJ, Gagliardi TD, Bosche WJ, Wiltrout TA, Coren LV, Chabot DJ, Lifson JD, Henderson LE, Arthur LO. 1999. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology 256:92–104. 10.1006/viro.1999.9629 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Qian H, Love Z, Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. 2005. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry 44:9058–9066. 10.1021/bi0502588 [DOI] [PubMed] [Google Scholar]
- 14.Clever JL, Miranda D, Jr, Parslow TG. 2002. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 76:12381–12387. 10.1128/JVI.76.23.12381-12387.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clever JL, Taplitz RA, Lochrie MA, Polisky B, Parslow TG. 2000. A heterologous, high-affinity RNA ligand for human immunodeficiency virus Gag protein has RNA packaging activity. J. Virol. 74:541–546. 10.1128/JVI.74.1.541-546.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lever A, Gottlinger H, Haseltine W, Sodroski J. 1989. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 63:4085–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu K, Heng X, Summers MF. 2011. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 410:609–633. 10.1016/j.jmb.2011.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luban J, Goff SP. 1994. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 68:3784–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride MS, Panganiban AT. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 70:2963–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, Edme K, Telesnitsky A, Summers MF. 2012. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 417:224–239. 10.1016/j.jmb.2012.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6:e96. 10.1371/journal.pbio.0060096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ooms M, Huthoff H, Russell R, Liang C, Berkhout B. 2004. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 78:10814–10819. 10.1128/JVI.78.19.10814-10819.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolaitchik O, Rhodes TD, Ott D, Hu WS. 2006. Effects of mutations in the human immunodeficiency virus type 1 gag gene on RNA packaging and recombination. J. Virol. 80:4691–4697. 10.1128/JVI.80.10.4691-4697.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butsch M, Boris-Lawrie K. 2000. Translation is not required to generate virion precursor RNA in human immunodeficiency virus type 1-infected T cells. J. Virol. 74:11531–11537. 10.1128/JVI.74.24.11531-11537.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshinaka Y, Katoh I, Copeland TD, Oroszlan S. 1985. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl. Acad. Sci. U. S. A. 82:1618–1622. 10.1073/pnas.82.6.1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houck-Loomis B, Durney MA, Salguero C, Shankar N, Nagle JM, Goff SP, D'Souza VM. 2011. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature 480:561–564. 10.1038/nature10657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280–283. 10.1038/331280a0 [DOI] [PubMed] [Google Scholar]
- 28.Chamanian M, Purzycka KJ, Wille PT, Ha JS, McDonald D, Gao Y, Le Grice SF, Arts EJ. 2013. A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation. Cell Host Microbe 13:181–192. 10.1016/j.chom.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride MS, Schwartz MD, Panganiban AT. 1997. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 71:4544–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen JB, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. 2009. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U. S. A. 106:13535–13540. 10.1073/pnas.0906822106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhodes TD, Nikolaitchik O, Chen JB, Powell D, Hu WS. 2005. Genetic recombination of human immunodeficiency virus type 1 in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 79:1666–1677. 10.1128/JVI.79.3.1666-1677.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu L, Martin TD, Vazeux R, Unutmaz D, KewalRamani VN. 2002. Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. J. Virol. 76:5905–5914. 10.1128/JVI.76.12.5905-5914.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yee JK, Miyanohara A, LaPorte P, Bouic K, Burns JC, Friedmann T. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 91:9564–9568. 10.1073/pnas.91.20.9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotsopoulou E, Kim VN, Kingsman AJ, Kingsman SM, Mitrophanous KA. 2000. A Rev.-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 74:4839–4852. 10.1128/JVI.74.10.4839-4852.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang M, Orenstein JM, Martin MA, Freed EO. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilman M. 2000. Preparation of RNA from eukaryotic and prokaryotic cells, p 4.1.2–4.1.6 In Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. (ed), Current protocols in molecular biology. John Wiley & Sons, Inc, Hoboken, NJ [Google Scholar]
- 39.Leitner T, Halapi E, Scarlatti G, Rossi P, Albert J, Fenyo EM, Uhlen M. 1993. Analysis of heterogeneous viral populations by direct DNA sequencing. Biotechniques 15:120–127 [PubMed] [Google Scholar]
- 40.Nikolaitchik OA, Gorelick RJ, Leavitt MG, Pathak VK, Hu WS. 2008. Functional complementation of nucleocapsid and late domain PTAP mutants of human immunodeficiency virus type 1 during replication. Virology 375:539–549. 10.1016/j.virol.2008.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buckman JS, Bosche WJ, Gorelick RJ. 2003. Human immunodeficiency virus type 1 nucleocapsid Zn2+ fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 77:1469–1480. 10.1128/JVI.77.2.1469-1480.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716. 10.1038/nature08237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cockrell AS, van Praag H, Santistevan N, Ma H, Kafri T. 2011. The HIV-1 Rev./RRE system is required for HIV-1 5′ UTR cis elements to augment encapsidation of heterologous RNA into HIV-1 viral particles. Retrovirology 8:51. 10.1186/1742-4690-8-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandt S, Blissenbach M, Grewe B, Konietzny R, Grunwald T, Uberla K. 2007. Rev. proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 3:e54. 10.1371/journal.ppat.0030054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, Hammarskjold ML. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. U. S. A. 91:1256–1260. 10.1073/pnas.91.4.1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore MD, Nikolaitchik OA, Chen J, Hammarskjold ML, Rekosh D, Hu WS. 2009. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 5:e1000627. 10.1371/journal.ppat.1000627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blissenbach M, Grewe B, Hoffmann B, Brandt S, Uberla K. 2010. Nuclear RNA export and packaging functions of HIV-1 Rev. revisited. J. Virol. 84:6598–6604. 10.1128/JVI.02264-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolaitchik OA, Dilley KA, Fu W, Gorelick RJ, Tai SHS, Soheilian F, Ptak RG, Nagashima K, Pathak VK, Hu WS. 2013. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 9:e1003249. 10.1371/journal.ppat.1003249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JW, Jr, Gorelick RJ, Rein A, Weeks KM. 2010. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc. Natl. Acad. Sci. U. S. A. 107:19248–19253. 10.1073/pnas.1006897107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gherghe C, Leonard CW, Gorelick RJ, Weeks KM. 2010. Secondary structure of the mature ex virio Moloney murine leukemia virus genomic RNA dimerization domain. J. Virol. 84:898–906. 10.1128/JVI.01602-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D'Souza V, Summers MF. 2004. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431:586–590. 10.1038/nature02944 [DOI] [PubMed] [Google Scholar]