ABSTRACT
Recognition of viral pathogens by the retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) family results in the activation of type I interferon (IFN) responses. To avoid this response, most viruses have evolved strategies that target different essential steps in the activation of host innate immunity. In this study, we report that the nonstructural protein NSs of the newly described severe fever with thrombocytopenia syndrome virus (SFTSV) is a potent inhibitor of IFN responses. The SFTSV NSs protein was found to inhibit the activation of the beta interferon (IFN-β) promoter induced by viral infection and by a RIG-I ligand. Astonishingly, we found that SFTSV NSs interacts with and relocalizes RIG-I, the E3 ubiquitin ligase TRIM25, and TANK-binding kinase 1 (TBK1) into SFTSV NSs-induced cytoplasmic structures. Interestingly, formation of these SFTSV NSs-induced structures occurred in the absence of the Atg7 gene, a gene essential for autophagy. Furthermore, confocal microscopy studies revealed that these SFTSV NSs-induced structures colocalize with Rab5 but not with Golgi apparatus or endoplasmic reticulum markers. Altogether, the data suggest that sequestration of RIG-I signaling molecules into endosome-like structures may be the mechanism used by SFTSV to inhibit IFN responses and point toward a novel mechanism for the suppression of IFN responses.
IMPORTANCE The mechanism by which the newly described SFTSV inhibits host antiviral responses has not yet been fully characterized. In this study, we describe the redistribution of RIG-I signaling components into virus-induced cytoplasmic structures in cells infected with SFTSV. This redistribution correlates with the inhibition of host antiviral responses. Further characterization of the interplay between the viral protein and components of the IFN responses could potentially provide targets for the rational development of therapeutic interventions.
INTRODUCTION
An important component of the innate immune response to viral infection is the production of type I interferons (IFNs). Activation of this response occurs when cytosolic receptors sense the presence of viral nucleic acids or other virus-specific components (1–5). The retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) family constitutes one of these groups of cytosolic receptors that are important in sensing the viral RNA of invading pathogens. To date, three members of the RLR family have been described: RIG-I, melanoma differentiation-associated gene 5 (MDA-5), and laboratory of genetics and physiology 2 (LGP2) (1, 6–8). Both RIG-I and MDA-5 have been shown to play a pivotal role in the recognition of RNA viruses, and they may differentially contribute to this function in a cell type-specific manner (9–11). How these viral sensors distinguish self RNA from viral RNA has been an area of extensive study. It is now well accepted that 5′-triphosphate groups within viral genomic RNAs and short, double-stranded RNA (dsRNA) are recognized by RIG-I, whereas the long dsRNA that is thought to be produced during replication of many positive-strand RNA viruses serves as a ligand for MDA-5 (12–16). RIG-I has been shown to be critical for the recognition of most single-stranded RNA (ssRNA) viruses studied to date, including all negative-strand RNA viruses and some positive-strand RNA viruses, such as hepatitis C virus and Japanese encephalitis virus (17, 18).
In regard to RIG-I activation, studies have shown that upon RNA binding, the E3 ubiquitin ligase Riplet mediates K63-linked polyubiquitination of the RIG-I repressor domain, leading to the interaction with the E3 ubiquitin ligase, tripartite motif 25 (TRIM25), and TANK-binding kinase 1 (TBK1). The N-terminal caspase recruitment domains (CARDs) of RIG-I undergo Lys-63-linked ubiquitination by TRIM25, inducing RIG-I oligomerization, interaction with the mitochondrial antiviral signaling protein (MAVS), and downstream signaling events that lead to the transcription of IFNs and other innate response genes (8, 19, 20). Recently, it has also been shown that TRIM25-synthesized K63-polyubiquitin binds directly to the exposed CARDs of RIG-I, mediating downstream binding to MAVS without the need of ubiquitin conjugation to RIG-I (21–23).
Due to the critical role of IFN responses in controlling virus infection, many pathogens have developed strategies to evade or counteract these responses (18, 24, 25). Among bunyaviruses, the nonstructural protein NSs has been found to be a major virulence factor acting as a global inhibitor of host cell transcription and antagonist of the IFN system (26–28). The Rift Valley fever virus (RVFV) NSs protein is probably one of the best-characterized examples and has been shown to be a multifunctional protein with the capacity to evade host antiviral responses through different mechanisms, including inhibition of beta interferon (IFN-β) gene transcription through a Sin3A-associated protein 30 (SAP30)-dependent mechanism, inhibition of host cell transcription by sequestering the p44 subunit, and promotion of the proteasomal degradation of the p62 subunit of the basal transcription factor TFIIH. Furthermore, RVFV NSs inhibits the phosphorylation of eukaryotic initiation factor 2 alpha (eIF2α) by posttranslational downregulation of the dsRNA-dependent protein kinase R (PKR) through the proteasomal pathway (29–34).
Recently, the isolation of a novel bunyavirus closely related to Uukuniemi virus and RVFV was documented (35, 36). The severe fever with thrombocytopenia syndrome virus (SFTSV) was isolated from patients presenting with hemorrhagic manifestations (35, 37). The disease was first recognized in China in 2009, and an initial case fatality rate of 12 to 30% was reported (37). Due to the recent emergence of this pathogen, limited knowledge about the mechanisms involved in disease pathogenesis is currently available. A recent report suggests that key innate immune molecules are downregulated in human monocytes upon infection with SFTSV (38, 39); however, the molecular mechanism(s) by which SFTSV suppresses innate immune responses has not been well defined. SFTSV is known to encode an NSs protein which differs significantly from RVFV NSs, having only 11% similarity at the amino acid sequence level (35). It has previously been reported that this protein inhibits the activation of the IFN-β promoter induced by viral infection and dsRNA and that it interacts with TBK1, a protein critical for the activation of IFN responses (39). However, it is not yet clear whether this interaction affects TBK1 functions accounting for the inhibition of IFN-β responses. Furthermore, it is unknown if other RLR proteins may be targeted by SFTSV NSs to inhibit IFN responses.
In this study, we describe the SFTSV NSs protein to be a potent inhibitor of IFN responses. Surprisingly, the inhibition of IFN responses by SFTSV NSs correlates with the sequestration of not only TBK1 but also that of RIG-I and TRIM25 within NSs-induced LC3-positive (LC3+), Rab5-positive (Rab5+) cytoplasmic structures. Hence, SFTSV has adopted an elegant, previously undescribed mechanism to circumvent IFN responses.
MATERIALS AND METHODS
Cells, plasmids, and viruses.
Human embryonic kidney (HEK293T) cells were obtained from ATCC and maintained with Dulbecco's minimal essential medium (Lonza) supplemented with l-glutamine, sodium pyruvate, and 10% fetal bovine serum (Atlanta Biologicals). HeLa and Vero76 cells were obtained from ATCC and maintained with Eagle minimal essential medium (Lonza) supplemented with l-glutamine, penicillin-streptomycin, and 10% fetal bovine serum. The Atg7+/+ and Atg7−/− mouse embryonic fibroblast (MEF) cells have been previously described and were kindly provided by Masaaki Komatsu (40). The plasmids carrying SFTSV NSs and nucleoprotein (NP) were constructed by PCR using overlapping deoxyoligonucleotides corresponding to a published GenBank sequence (accession no. NC_018137.1). An expression plasmid carrying green fluorescent protein (GFP)-tagged LC3 (GFP-LC3) (plasmid 21073) was obtained from Addgene (41), and plasmids carrying RIG-I, TRIM25, MAVS, TBK1, interferon regulatory factor 3 (IRF3), constitutively activated IRF3 (IRF3-5D), dominant negative Rab5 (S34N), and the corresponding fusion proteins were previously described (42–44) or constructed following standard cloning procedures. Plasmids carrying red fluorescent protein (RFP)-tagged B4GalT1 (B4GalT1-RFP), TGOLN-RFP, CALR-C–RFP, Rab5-RFP, and Rab4-RFP were obtained from OriGene. The Sendai virus (SeV) Cantell strain was obtained from ATCC and grown in embryonated chicken eggs to generate viral stocks. SFTSV strain HB29 was provided by the Chinese Center for Disease Control and Prevention and passaged twice in Vero E6 cells to generate viral stocks for this study. The SFTSV titers were determined by plaque assays. Briefly, virus stock was diluted 10-fold, added to 1 ml of medium containing 1.5 × 105 BHK21 cells, and then incubated at 37°C with 5% CO2 for 3 h. A 1-ml volume of 4% methylcellulose was placed in each well, and the plates were incubated for 6 days at 37°C. The plates were then fixed with methanol-acetone (1:1), and foci were stained immunologically using anti-SFTSV mouse hyperimmune ascitic fluid and counted to determine virus titers.
Reporter assays and detection of IFN-β.
Reporter assays and assays for detection of IFN-β were performed as previously described (45, 46). Chloramphenicol acetyltransferase (CAT) activity was assessed by using a CAT enzyme-linked immunosorbent assay (ELISA) kit following the manufacturer's recommendations (Roche). To confirm that the presence of SFTSV NSs inhibits production of IFN-β, HeLa cells expressing SFTSV NSs-mCherry or mCherry alone were infected with SeV, and at 24 h after infection, cell supernatants were cleared by centrifugation and production of IFN-β was measured using a human IFN-β ELISA kit (PBL Assay Science, Piscataway, NJ). Statistical analyses were carried out using one-way analysis of variance followed by Dunnett's multiple-comparison test implemented in GraphPad Prism (version 6) software for Windows (GraphPad Software, La Jolla, CA). P values of <0.05 were considered statistically significant.
Transfection, immunoblotting, and immunoprecipitation.
Unless otherwise stated, all transfections were carried out by using 500 ng of plasmid DNA and the Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's recommendations. At 16 to 24 h posttransfection, the cells were harvested and lysed with NP-40 lysis buffer containing 1% Halt protease inhibitor cocktail (Thermo Scientific). For immunoblotting, proteins were separated by SDS-PAGE and subsequently transferred into a 0.2-μm-pore-size polyvinylidene difluoride (PVDF) membrane (Millipore). The PVDF membranes were blocked for 45 min with 5% nonfat dry milk in Tris-buffered saline with 1% Tween 20 (TBST). Membranes were incubated with primary antibodies for 16 to 18 h at 4°C. The membranes were washed in TBST and incubated with secondary horseradish peroxidase (HRP)-conjugated antibodies for 1 h. Blots were developed by using Western Lightning ECL substrate (PerkinElmer) according to the manufacturer's recommendations. For protein immunoprecipitation, lysates were incubated in the presence of anti-FLAG M2 affinity gel (Sigma) or anti-hemagglutinin (anti-HA) agarose (Pierce). Eluted proteins and the corresponding lysates were separated by SDS-PAGE, followed by immunoblotting as described above. The following primary antibodies were utilized for immunoblotting procedures: rabbit anti-FLAG (1:1,000; Sigma), rabbit anti-HA (1:1,000; Sigma), mouse anti-V5 (1:5,000; Invitrogen), rabbit antiactin (1:1,000; Cell Signaling), mouse anti-β-tubulin (1:1,000; Abcam), rabbit anti-pIRF3 (1:10,000; Epitomics), and rabbit anti-IRF3 (1:1,000; Cell Signaling). The secondary antibodies utilized were donkey anti-rabbit IgG HRP-conjugated antibody (1:5,000) and sheep anti-mouse IgG HRP-conjugated antibody (1:5,000) (GE Healthcare).
Immunofluorescence.
HeLa, Vero76, and Atg7+/+ and Atg7−/− MEF cells were seeded in poly-l-lysine (Sigma)-treated coverslips and transfected following standard procedures. Cells were incubated overnight at 37°C in 5% CO2. Following this incubation, cells were either fixed with 4% paraformaldehyde or infected with SFTSV (multiplicity of infection [MOI] = 0.5) for 48 h and then fixed and permeabilized with a solution of phosphate-buffered saline (PBS) containing 0.1% Triton X-100 (Sigma). Following permeabilization, the cells were washed and blocked with 10% goat serum and 3% bovine serum albumin (BSA; Thermo Scientific) in PBS (Lonza). If required by the experiment, cells were stained for 1 h with primary antibodies. Cell nuclei were stained with Hoechst 33342 (1:1,000; Invitrogen), and the coverslips were mounted onto glass slides by using ProLong Gold antifade reagent (Invitrogen). The following primary antibodies were used in immunofluorescence experiments: polyclonal rabbit anti-SFTSV NSs generated by GenScript (Piscataway, NJ) (1:500), rabbit anti-FLAG (1:100; Sigma), and mouse anti-HA (1:1,000; Covance). The following antibodies were used as secondary antibodies for immunofluorescence experiments: Alexa Fluor 488-conjugated goat antirabbit (1:1,000; Molecular Probes) and Alexa Fluor 594-conjugated goat antirabbit (1:2,000; Molecular Probes) antibodies. All samples were visualized in a Zeiss LSM510META laser scanning confocal microscope or an Olympus spinning disc confocal microscope.
Immunogold electron microscopy studies.
For visualization of SFTSV NSs cytoplasmic vesicles, we modified the protocol of Cao et al. (47). Briefly, HeLa cells expressing SFTSV NSs fused to mCherry were rinsed three times in PBS, followed by one rinse in solution D (78 mM KCl, 4 mM MgCl2, 8 mM CaCl2, 10 mM EDTA, 50 mM HEPES-KOH, pH 7.0). Cells were then scraped off in 10 ml of solution D, centrifuged at 400 relative centrifugal force (rcf) for 5 min, and resuspended in 25 ml of solution D. Cells were lysed with an equal volume of glass beads and vortexed at top speed. Supernatant was transferred to a Beckman tube and subjected to sequential centrifugation and OptiPrep discontinuous gradient centrifugation. After centrifugation, fractions were collected, resolved by SDS-PAGE, and blotted for SFTSV NSs. The 35% top fraction was used for visualization of vesicles by electron microscopy. Negative staining was carried out on both the pellet and the purified fraction. Briefly, 10 μl of pellet and fraction was allowed to adsorb onto nickel grids for 10 min, followed by incubation in primary antibody (rabbit anti-NSs, 1:10) for 30 min at room temperature in a dark moist chamber. After incubation, grids were washed 4 times with 1% BSA-PBS and incubated with secondary antibody (goat antirabbit, 15-nm gold particles, electron microscopy grade, 1:20 dilution; Electron Microscopy Sciences, Hatfield, PA) for 30 min at room temperature. Grids were washed with ultrapure water 3 times for 1 min each time and fixed with 2% aqueous glutaraldehyde for 10 min. Following fixation, the grids were then washed 3 times with ultrapure water. Lastly, the grids were stained with 2% aqueous uranyl acetate for 30 s and blotted dry.
RESULTS
SFTSV NSs inhibits type I IFN responses.
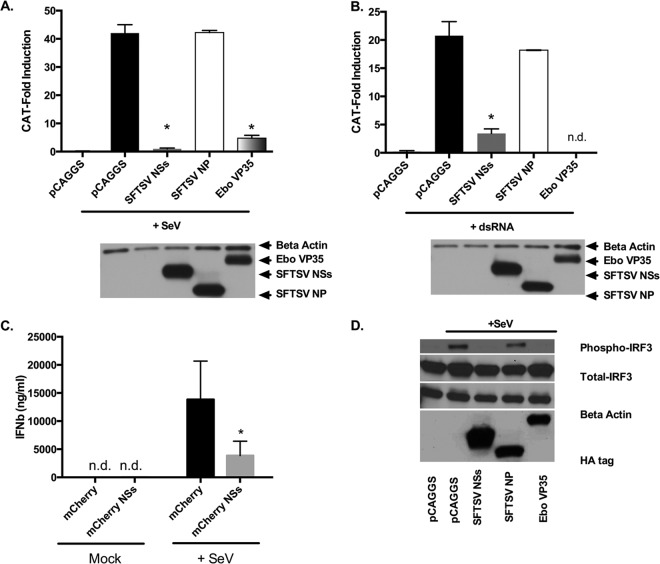
It was previously reported that both SFTSV NSs and NP inhibit the activation of the IFN-β promoter induced by virus infection and dsRNA treatment (39). To confirm those findings, we conducted reporter gene assays with the goal of determining whether SFTSV NSs and NP were able to prevent in cells the establishment of an IFN-α/β-induced antiviral state by blocking the activation of the IFN-α/β promoter. Thus, HEK293T cells were cotransfected with plasmids expressing Ebola virus VP35 (VP35), SFTSV NSs, or SFTSV NP, alongside an IFN-β promoter CAT reporter plasmid. VP35 has previously been described to inhibit the activation of the IFN-β promoter by binding dsRNA and by suppressing the kinase activity of the IkappaB kinase epsilon (IKKepsilon) (46, 48–50). Consistent with previous observations (39, 46), VP35 and SFTSV NSs inhibited the activation of the IFN-β promoter induced following Sendai virus (SeV) infection or transfection with the RIG-I ligand short dsRNA (Fig. 1A and B). The magnitude of this inhibition, resulting in ∼5- to 20-fold differences in reporter activation between the empty vector pCAGGS and SFTSV NSs, was comparable to that observed for VP35. To confirm that the expression of SFTSV NSs results in inhibition of IFN-β production in the context of viral infection, HeLa cells expressing either mCherry or the SFTSV NSs fused to mCherry were infected with SeV for 24 h, and supernatants were collected to measure the production of IFN-β. Interestingly, HeLa cells expressing the SFTSV NSs secreted approximately 3.5-fold less IFN-β than mCherry-expressing cells following SeV infection (Fig. 1C). It should be noted that these experiments, which rely upon transient transfection, leave some percentage of cells untransfected and therefore likely underestimate the inhibitory effect of SFTSV NSs on IFN-β production. These results further suggest that SFTSV NSs interferes with the induction of type I IFN responses. Contrary to a previous report (39), we did not observe inhibition of the activation of the IFN-β promoter induced by virus infection or dsRNA in the presence of SFTSV NP (Fig. 1A and B).
FIG 1.
SFTSV NSs inhibits type I IFN. (A and B) HEK293T cells were transfected with an IFN-β-CAT plasmid, along with the indicated expression plasmids. At 24 h posttransfection, cells were infected with SeV (A) or transfected with dsRNA (B), and at 24 h after infection or dsRNA treatment, cell lysates were collected and assessed for CAT activity. Error bars represent the standard deviation of the mean percentage in three independent experiments. Asterisks specify statistically significant differences (P < 0.05) between the indicated group and the pCAGGS-treated group. (C) HeLa cells were transfected with a plasmid expressing mCherry or SFTSV NSs fused to mCherry and infected with SeV. At 24 h postinfection, supernatants were collected and assessed for IFN-β by ELISA. Error bars represent the standard deviation of the mean percentage in three independent experiments. Asterisks indicate statistically significant differences (P < 0.05) in the production of IFN-β between the mCherry- and SFTSV NSs-mCherry-expressing cells. (D) HEK293T cells were transfected with plasmids carrying IRF3 and the indicated HA-tagged expression plasmids. At 24 h after transfection, cells were infected with SeV, and 10 h later cell lysates were collected and assayed for IRF3 phosphorylation by using phosphor and total IRF3 antibodies. Expression of the viral proteins was confirmed by using antibodies against the HA tag, and loading was verified with antibodies against β-actin. n.d., not detected.
Importantly, we also attempted to measure the production of IFN-β in SFTSV-infected HEK293T and HeLa cells; however, despite high levels of virus replication (>105 focus-forming units [FFU]/ml), IFN-β levels were undetectable.
SFTSV NSs inhibits phosphorylation of IRF3.
It is well-known that induction of the IFN-β promoter following SeV infection is the culmination of a signaling cascade that begins with viral RNA recognition by RIG-I and results in the activation of IRF3 (5, 12). IRF3 is a transcription factor that is present in an inactivated state in the cytoplasm and whose activation occurs through hyperphosphorylation of its autoinhibitory, carboxy-terminal domain, resulting in IRF3 dimerization and nuclear accumulation (51, 52). In light of our and previous observations (39) concerning the inhibition of IFN-α/β promoter activation by SFTSV NSs, we employed previously described methodologies to assess if SFTSV NSs inhibits SeV infection-induced activation of IRF3 (48). Results from cotransfection assays that used expression plasmids carrying VP35 (a known inhibitor of IRF3 phosphorylation [48]), SFTSV NP, and SFTSV NSs together with an IRF3 expression plasmid showed a marked inhibition of SeV-mediated phosphorylation of IRF3 by SFTSV NSs. Notably, the SeV-mediated activation of IRF3 was similarly inhibited in cells expressing either VP35 or SFTSV NSs, and this inhibition was not due to reduced IRF3 expression levels (Fig. 1D). Consistent with the results from the IFN reporter gene assays, we did not observe inhibition of IRF3 phosphorylation by SFTSV NP. This finding suggests that inhibition of type I IFN by SFTSV NSs occurs upstream of IRF3 phosphorylation.
SFTSV NSs inhibits activation of the IFN-β promoter induced by signaling components upstream of IRF3.
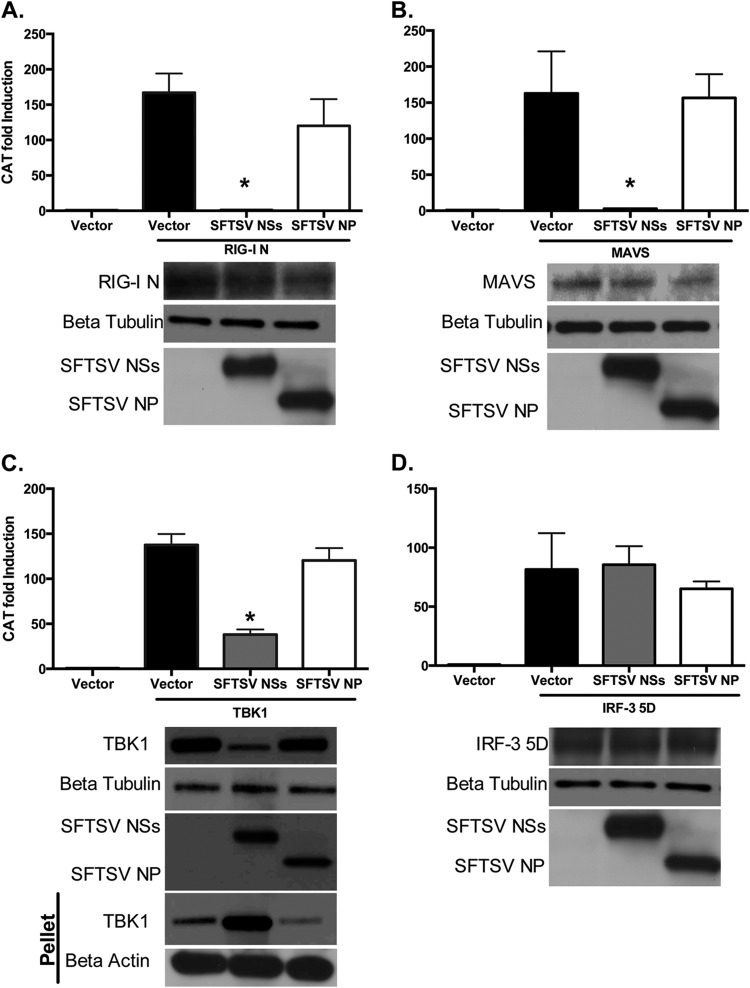
RIG-I activation results in IRF3 phosphorylation due to a cascade of signaling events involving binding of activated RIG-I to MAVS followed by activation of IRF3 kinases, such as TBK1. It has also been previously reported that SFTSV NSs interacts with TBK1, but how this interaction affects TBK1 functions has not been elucidated (39). To gain insights into the mechanism by which SFTSV NSs inhibits IRF3 phosphorylation, we conducted reporter gene-based assays to determine if the interaction of SFTSV NSs with TBK1 affects TBK1 signaling. Furthermore, we examined another relevant innate signaling component(s) as a potential target of the SFTSV NSs. HEK293T cells were cotransfected with an IFN-β promoter fused to CAT, an empty vector, or plasmids carrying genes expressing SFTSV NSs and SFTSV NP, together with a plasmid carrying a gene expressing constitutively activated RIG-I–FLAG (RIG-I–CARDs), MAVS-HA, TBK1-FLAG, or constitutively activated IRF3 (IRF3-5D), whose overexpression results in activation of the IFN-β promoter. Notably, SFTSV NSs, but not SFTSV NP, inhibited the activation of the IFN-β promoter induced by RIG-I–CARDs, MAVS, and TBK1 (Fig. 2). These results indicate that SFTSV NSs specifically inhibits the RIG-I signaling pathway upstream of IRF3, resulting in the suppression of IFN responses. Interestingly, a substantial portion of the expressed TBK1 protein, but not RIG-I or TRIM25, was retained in the cellular pellet (Fig. 2C and data not shown). Since it has previously been shown that SFTSV NSs interacts with TBK1 (39), these results suggest that the SFTSV NSs/TBK1 interaction negatively impacts TBK1 signaling functions by relocalizing the signaling protein into a different compartment. However, our data suggest that it may be possible for SFTSV NSs to target other RLR signaling components besides TBK1.
FIG 2.
SFTSV NSs inhibits activation of IFN-β induced by different signaling factors. HEK293T cells were transfected with the indicated viral protein expression plasmids and an IFN-β–CAT plasmid, along with the RIG-I N terminus (RIG-I N) (A), MAVS (B), TBK1 (C), or IRF3-5D (D), and at 16 h after transfection, cell lysates were collected and assessed for CAT activity. Error bars represent the standard deviation of the mean percentage in three independent experiments. Asterisks specify statistically significant differences (P < 0.05) between the indicated group and the pCAGGS-treated group. Expression of the proteins was confirmed by Western blotting using anti-FLAG, anti-HA, anti-IRF3, and anti-β-tubulin antibodies.
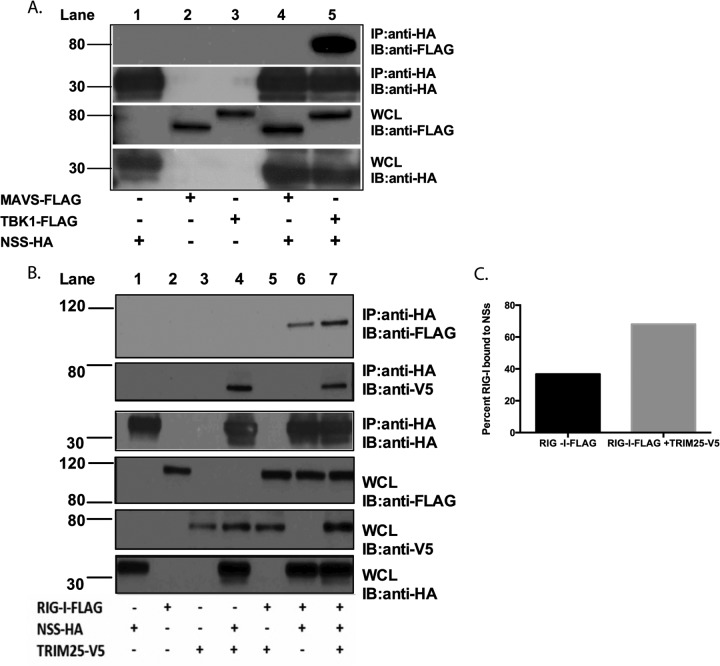
SFTSV NSs interacts with TBK1, RIG-I, and TRIM25.
In light of the findings described above, we sought to determine the possible physical interactions between SFTSV NSs, RIG-I, MAVS, TBK1, and TRIM25. In these experiments, SFTSV NSs-HA, RIG-I–FLAG, MAVS-FLAG, TBK1-FLAG, and TRIM25-V5 were cotransfected in HEK293T cells. SFTSV NSs was precipitated from cellular lysates by using anti-HA-tagged beads. Protein complexes resulting from the protein-protein interaction with the SFTSV NSs were resolved by SDS-PAGE and Western blotting. As previously reported (39), we observed that TBK1 coprecipitated with SFTSV NSs (Fig. 3A, lane 5). Interestingly, we also observed that TRIM25, but not MAVS, coprecipitated with SFTSV NSs (Fig. 3B, lane 4, and A, lane 4, respectively). Furthermore, RIG-I also coprecipitated with SFTSV NSs (Fig. 3B, lane 6); however, the amount of coprecipitated RIG-I (relative to its total expression levels) was smaller than the amount of TBK1 and TRIM25 that coprecipitated with SFTSV NSs. Interestingly, when TRIM25 and RIG-I were expressed together with SFTSV NSs-HA, the amount of coprecipitated RIG-I, determined using anti-HA antibodies, increased by approximately 2-fold (Fig. 3B [compare lanes 6 and 7] and C). Taken together, these data suggest that SFTSV NSs interacts primarily with TBK1 and TRIM25 and that the interaction with RIG-I may be indirect and promoted via TRIM25.
FIG 3.
SFTSV NSs interacts with TBK1, RIG-I, and TRIM25. (A) HEK293T cells were transfected with the indicated expression plasmids, and at 24 h after transfection, cell lysates were collected and subjected to immunoprecipitation (IP) by using anti-HA antibodies. Immunoprecipitates were analyzed by immunoblotting (IB) using anti-FLAG and anti-HA antibodies. Whole-cell lysates (WCL) were immunoblotted by using anti-FLAG and anti-HA antibodies. (B) HEK293T cells were transfected with the indicated expression plasmids, and at 24 h after transfection, cell lysates were collected and subjected to immunoprecipitation by using anti-HA antibodies. Immunoprecipitates were analyzed by immunoblotting using anti-FLAG, anti-V5, and anti-HA antibodies. Whole-cell lysates were immunoblotted by using anti-FLAG, anti-V5, and anti-HA antibodies. Numbers to the left of the gels in panels A and B are molecular masses (in kilodaltons). (C) Densitometry analysis of immunoprecipitated RIG-I for panel B. The band signal intensity of the immunoprecipitated RIG-I protein was normalized to the signal intensity of the RIG-I protein expressed in the whole-cell lysate. Signal intensities were obtained by using Image Studio Lite software.
SFTSV NSs sequesters RIG-I signaling molecules into NSs-induced LC3+ and Rab5+ cytoplasmic structures.
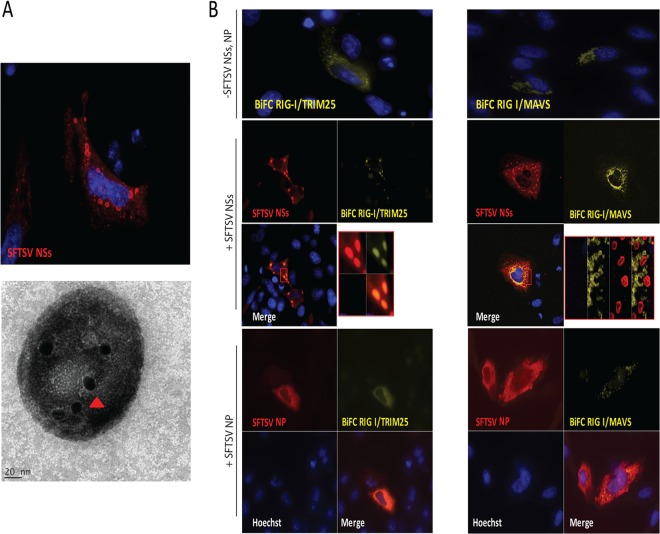
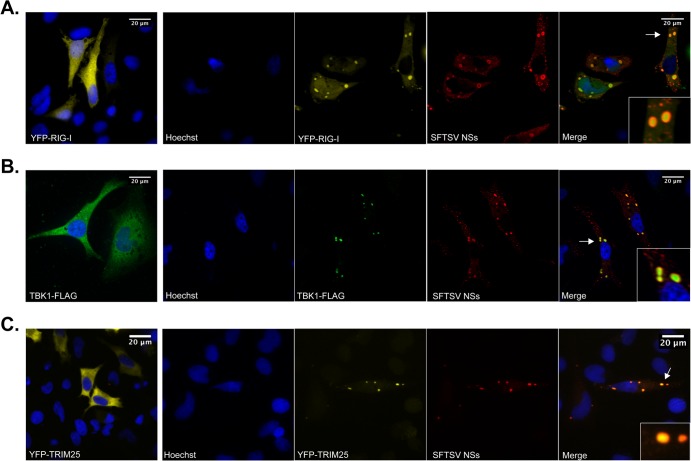
Upon viral RNA recognition, RIG-I switches from a passive to an active conformation, exposing the CARDs for interaction with downstream signaling proteins, including the adaptor protein MAVS. In addition, interaction of the exposed CARDs of RIG-I with the E3 ubiquitin ligase TRIM25 results in RIG-I conjugation and/or binding to K63 polyubiquitin, and this is critical for optimal RIG-I interaction with MAVS and activation of downstream signaling events (21–23). In light of the results described above and to gain more insight into the mechanism by which SFTSV NSs inhibits IFN responses, we conducted a bimolecular fluorescence complementation (BiFC) assay (53) to investigate whether the inhibition of IFN responses by SFTSV NSs has an inhibitory effect on RIG-I/TRIM25 or RIG-I/MAVS interaction. In this cell-based assay, detection of yellow fluorescent protein (YFP) is achieved upon RIG-I/TRIM25 (Fig. 4B, upper left) or RIG-I/MAVS (Fig. 4B, upper right) interaction, which occurs as a result of activation of RLR signaling due to overexpression of these proteins. Thus, it would be predicted that if SFTSV NSs inhibits the RIG-I interaction with TRIM25 or MAVS, YFP expression would not be detected. Notably, transfection assays using an expression plasmid carrying SFTSV NSs alone resulted in the formation of cytoplasmic structures with a vesicle-like morphology, as visualized by confocal and immunogold electron microscopy studies (Fig. 4A). In the presence of SFTSV NSs, the formation of the BiFC RIG-I/TRIM25 and BiFC RIG-I/MAVS complexes was not inhibited. However, we observed that the RIG-I/TRIM25 complex, but not the RIG-I/MAVS complex, relocalized within the SFTSV NSs-induced structures (Fig. 4B, left and right, respectively). These results differ from those observed in SFTSV NP-expressing cells, where the localization and formation of the BiFC RIG-I/TRIM25 and BiFC RIG-I/MAVS complexes were not affected (Fig. 4B, bottom). Furthermore, results from cotransfection assays using expression plasmids carrying an enhanced yellow fluorescent protein-tagged RIG-I (YFP–RIG-I), TRIM25 (TRIM25-V5), or TBK1 (TBK1-FLAG) together with SFTSV NSs resulted in the colocalization of RIG-I, TRIM25, and TBK1 within these SFTSV NSs-induced structures (Fig. 5A to C). The SFTSV NSs was observed lining the membrane of these cytoplasmic structures, suggesting that there might be a spatial organization between the viral protein and the signaling components. Taken together, these data suggest that the expression of SFTSV NSs promotes redistribution of the cytoplasmic proteins RIG-I, TRIM25, and TBK1, but not the mitochondrial signaling protein MAVS, into SFTSV NSs-induced cytoplasmic structures.
FIG 4.
SFTSV NSs targets the RIG-I/TRIM25 complex to cytoplasmic structures. (A) HeLa cells were transfected with a plasmid carrying SFTSV NSs fused to mCherry, and at 24 h after transfection, cells were fixed and analyzed by confocal microscopy (top). In another set of experiments, the SFTSV NSs-induced structures were purified from cells stably expressing SFTSV NSs-mCherry using serial centrifugation and OptiPrep density gradient centrifugation and subjected to immunogold labeling and electron microscopy analyses (bottom). Red arrowhead, SFTSV NSs protein staining. (B) HeLa cells were transfected with BiFC RIG-I/TRIM25 (left) or BiFC RIG-I/MAVS (right) construct pairs along with an empty plasmid or a plasmid carrying SFTSV NSs-HA. Transfected cells were incubated for 24 h at 37°C and then incubated at 30°C for 2 h to promote fluorophore maturation. Cells were fixed and analyzed by confocal microscopy. Rabbit anti-HA and Alexa Fluor 633 were used for detection of SFTSV NSs. Cell nuclei were visualized by using Hoechst dye.
FIG 5.
SFTSV NSs targets TBK1, RIG-I, and TRIM25 to cytoplasmic structures. (A) HeLa cells were transfected with a plasmid carrying YFP–RIG-I (A), TBK1-FLAG (B), or YFP-TRIM25 (C) along with an empty vector or a plasmid carrying SFTSV NSs-HA. Then, at 24 h after transfection, cells were fixed, stained with anti-FLAG and anti-HA antibodies, and analyzed by confocal microscopy. Cell nuclei were visualized by using Hoechst dye. Arrows, sections selected for the insets.
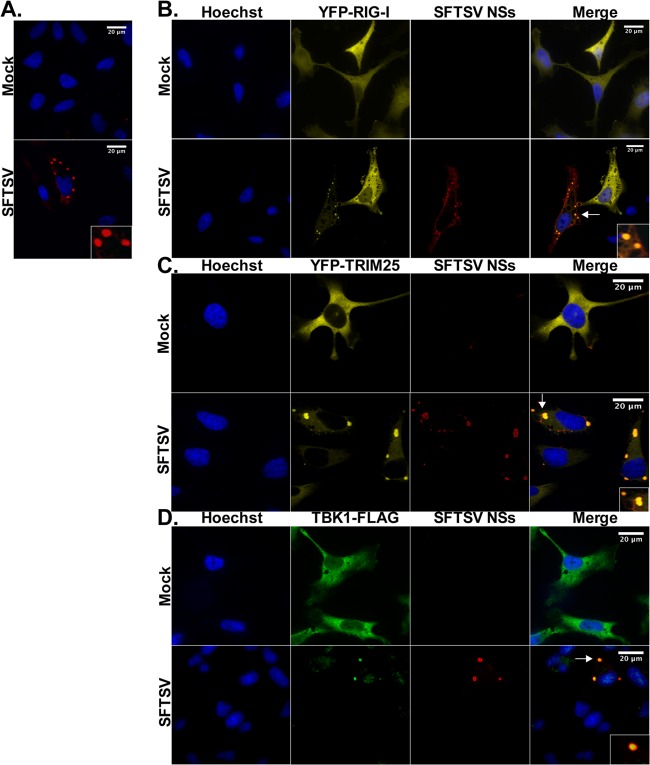
Due to the observed colocalization of the signaling components and their interaction with the SFTSV NSs-containing structures, we sought to further characterize the source of these puncta. Formation of NSs containing cytoplasmic vesicles in the context of SFTSV infection was first confirmed by indirect immunofluorescence assays (IFA) using antibodies raised against the viral protein NSs (Fig. 6A). Colocalization of RIG-I, TRIM25, and TBK1 within these SFTSV NSs-induced structures was also confirmed in SFTSV-infected cells (Fig. 6B to D). We observed that over 90% of SFTSV-infected cells expressing these IFN signaling proteins displayed 3 or more colocalization events. Furthermore, the spatial organization between the SFTSV NSs and the signaling components was confirmed in the context of SFTSV infection.
FIG 6.
RIG-I and TRIM25 colocalize in SFTSV NSs-induced cytoplasmic structures following SFTSV infection. (A) HeLa and Vero cells were infected with SFTSV (MOI = 0.5), and at 24 h postinfection, cells were fixed and immunofluorescence staining was performed with antibodies against SFTSV NSs. Cell nuclei were visualized by using Hoechst dye. Results from HeLa cells are shown. (B and D) HeLa cells were transfected with YFP-RIG-I (B), YFP-TRIM25 (C), or TBK1-FLAG (D) expression plasmids and then mock infected or infected with SFTSV (MOI = 0.5). Then, at 24 h postinfection, cells were fixed and immunofluorescence staining was performed by using antibodies against SFTSV NSs and FLAG. Cell nuclei were visualized by using Hoechst dye. Arrows, sections selected for the insets.
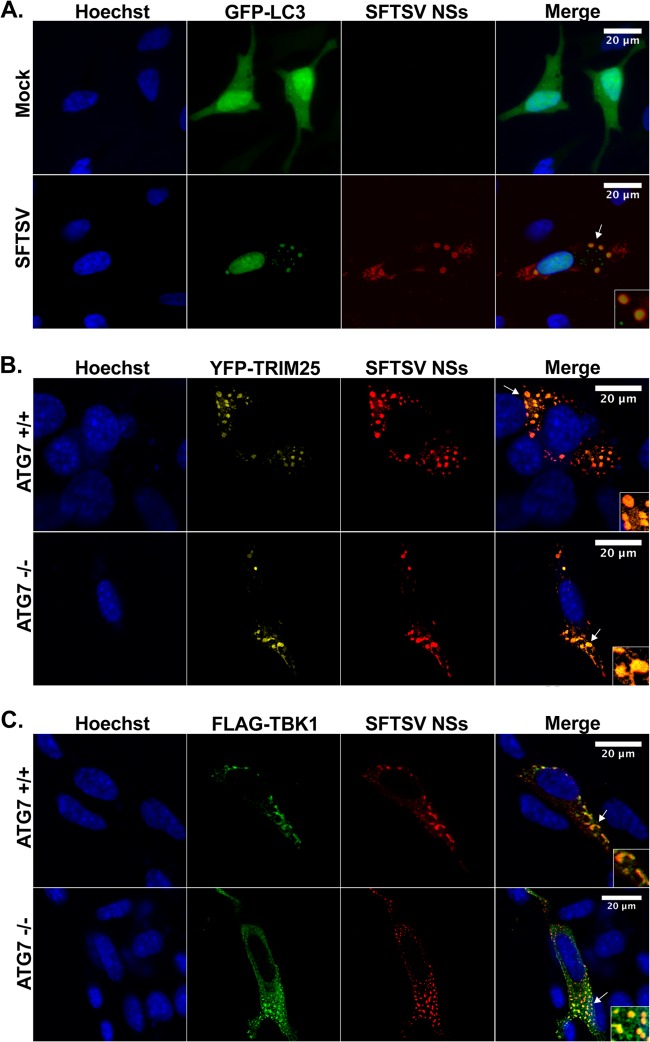
Since some RNA viruses induce the formation of cytoplasmic vesicles that display hallmarks of cellular autophagosomes, including positive staining for LC3 (54), we next examined colocalization of SFTSV NSs with GFP-LC3 following infection with SFTSV. It is known that LC3 has a cytosolic distribution in its inactive state, which is reflected as a diffuse low-level GFP-LC3 expression pattern; however, upon activation during upregulation of autophagy, GFP-LC3 accumulates in autophagosomal membranes, resulting in the presence of GFP-LC3 puncta. Accordingly, cells expressing GFP-LC3 showed low levels of diffuse fluorescence in their cytoplasm, and only a minor percentage of the cells displayed GFP-LC3 puncta. However, when these cells were infected with SFTSV, we observed a substantial increase in the number of cells with cytoplasmic structures containing GFP-LC3, consistent with the formation of autophagosomes. More importantly, colocalization of GFP-LC3 with SFTSV NSs-induced structures was evident (Fig. 7A), suggesting that the SFTSV NSs-induced cytoplasmic structures may be autophagosomes. To test this possibility, we next investigated whether the knockdown of Atg7, a gene essential for conventional autophagy (40), disrupted the formation of the SFTSV NSs-induced cytoplasmic structures. Thus, Atg7−/− and Atg7+/+ MEF cells were transfected with an expression plasmid carrying SFTSV NSs alone or in the presence of plasmids carrying GFP-LC3, YFP-TRIM25, or TBK1-FLAG, and the formation of cytoplasmic structures and relocalization of TRIM25 and TBK1 within these structures were evaluated by confocal microscopy. Surprisingly, the formation of SFTSV NSs-induced cytoplasmic structures and the redistribution of RLR signaling proteins within these structures were evident in both Atg7+/+ and Atg7−/− cells (Fig. 7B and C). Furthermore, colocalization of GFP-LC3 structures with SFTSV NSs was still observed in Atg7−/− cells (data not shown). It is worth noting that the size and morphology of the SFTSV NSs-induced cytoplasmic structures differed from those observed in human cell lines and the structures appeared to be more heterogeneous in shape (compare Fig. 7 to Fig. 4 to 6). Taken together, the results suggest that SFTSV NSs-induced structures are not conventional autophagosomes.
FIG 7.
SFTSV NSs-induced cytoplasmic structures colocalize with LC3 but are not autophagosomes. (A) HeLa cells were transfected with a plasmid carrying GFP-LC3, and at 24 h after transfection, cells were mock infected or infected with SFTSV (MOI = 0.5). Cells were fixed, and immunofluorescence staining was performed by using antibodies against SFTSV NSs. Cell nuclei were visualized by using Hoechst dye. (B and C) Atg7+/+ and Atg7+/+ MEF cells were transfected with plasmids carrying YFP-TRIM25 (B) or TBK1-FLAG (C), together with a plasmid carrying SFTSV NSs-HA. At 24 h posttransfection, cells were fixed and immunofluorescence staining was performed with antibodies against SFTSV NSs and FLAG. Cell nuclei were visualized by using Hoechst dye. Arrows, sections selected for the insets.
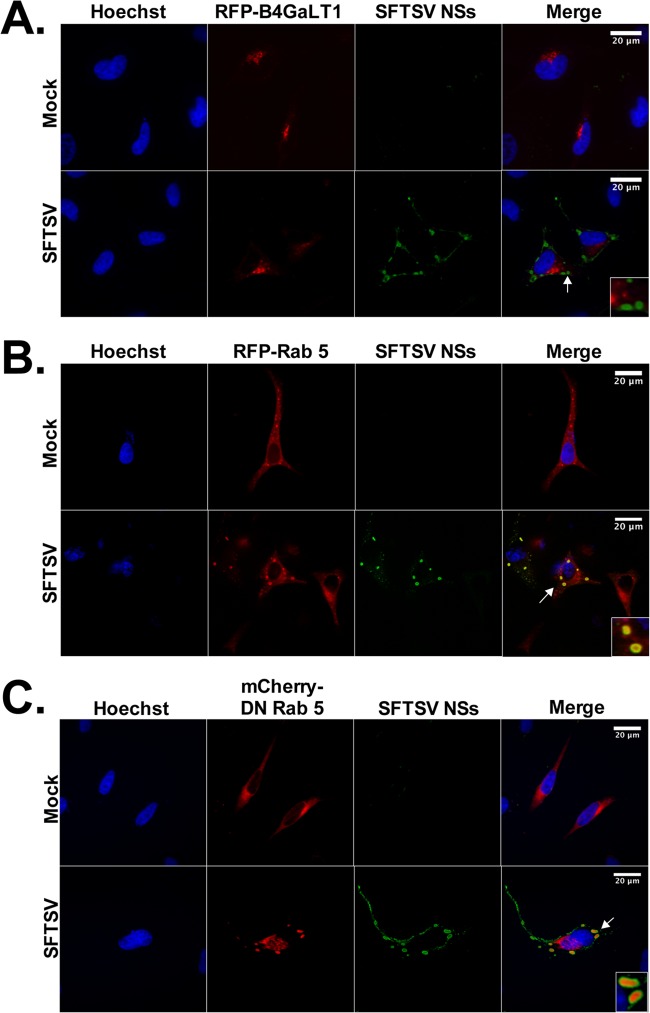
Since it is known that bunyaviruses assemble in association with Golgi membranes and components of the endoplasmic reticulum (ER) (55–57), we next investigated if the SFTSV NSs-induced cytoplasmic structures colocalized with Golgi apparatus and ER markers. HeLa cells were transfected with the Golgi apparatus markers B4GalT1-RFP and TGOLN-RFP or the ER marker CALR-C–RFP, along with a plasmid carrying SFTSV NSs. Interestingly, confocal microscopy studies did not reveal an association of the SFTSV NSs-induced structures with the Golgi apparatus or ER membranes (data not shown). The lack of colocalization between SFTSV NSs and the corresponding markers was confirmed in the context of SFTSV infection (Fig. 8A). Next, we evaluated whether other cellular markers (including mitochondria, peroxisomes, lysosomes, EDEMosomes, and early and late endosome markers) colocalized with the SFTSV NSs-induced cytoplasmic structures, and we observed that the early endosome marker Rab5, but not Rab4, colocalized with the SFTSV NSs-induced cytoplasmic structures (Fig. 8B), suggesting that SFTSV NSs uses the endosomal system to inhibit IFN responses.
FIG 8.
The early endosomal marker Rab5 is associated with the SFTSV NSs-induced cytoplasmic vesicles. HeLa cells were transfected with plasmid carrying the Golgi apparatus marker B4GalT1-RFP (A), the endosomal marker Rab5-RFP (B), or the a dominant negative (DN) variant of Rab5 fused to mCherry (C). At 24 h posttransfection, cells were infected with SFTSV (MOI = 0.5). At 24 h postinfection, the cells were fixed, stained with anti-SFTSV NSs antibody, and analyzed by confocal microscopy. Cell nuclei were visualized by using Hoechst dye. Arrows, sections selected for the insets.
Rab5 plays a central role in endosome biogenesis (58, 59). In addition, the endosomal pathway has been shown to promote internalization and/or virus replication (60–63). Indeed, entry into Rab5+ early endosomes has been previously described for Uukuniemi virus, another member of the genus Phlebovirus (64). Thus, we sought to determine if Rab5 was required for the formation of the SFTSV NSs-induced endosome-like structures. Interestingly, we observed that Rab5 gene knockdown by transfection of exogenous Rab5 small interfering RNA did not affect the formation of cytoplasmic structures in SFTSV NSs-transfected cells (data not shown). Furthermore, transient transfection with a dominant negative (S34N) Rab5 mutant, which has been shown to preferentially bind GDP over GTP, keeping Rab5 in an inactive state (65), did not prevent the formation of the SFTSV NSs-induced cytoplasmic structures in SFTSV NSs-transfected or SFTSV-infected cells (data not shown and Fig. 8C). Taken together, these data suggest that activation of Rab5 and/or downstream endocytic trafficking is not necessary for the formation of the SFTSV NSs-induced cytoplasmic structures.
DISCUSSION
Recognition of viral pathogens by RLRs results in the activation of type I IFN responses. To avoid this response, most viruses have evolved strategies that target different essential steps in the activation of host innate immunity. Our understanding of how phleboviruses counteract host IFN responses has come principally from studies involving RVFV. RVFV activates the cytoplasmic sensor RIG-I through RIG-I recognition of its viral RNA (66). However, it is known that the NSs protein of RVFV is a major virulence factor with the capacity to block IFN-β gene expression early (3 to 4 h) after infection (30). Mechanistically, RVFV NSs was found to interact with Sin3A-associated protein 30 (SAP30), a subunit of the Sin3A/nuclear receptor corepressor (NCoR)/histone deacetylase repressor complex and a partner of the transcription factor YY1 (67). The Sin3A-associated corepressor factors SAP30 and NSs are then recruited to the IFN-β promoter site through YY1, thereby inhibiting the recruitment of the coactivator CREB-binding protein, histone acetylation, and transcriptional activation (30). RVFV NSs was also shown to inhibit host cell gene transcription by sequestering the p44 subunit of the TFIIH basal transcription factor into NSs filamentous structures that are characteristic of RVFV infection (31). This process occurs relatively late (8 to 9 h) after viral infection. Recently, RVFV NSs was also shown to promote the posttranslational downregulation of PKR through the proteasomal pathway, which prevents the phosphorylation of eIF2α and supports viral translation in infected cells (29, 32, 33). In addition, RVFV NSs-mediated posttranslational downregulation of the p62 subunit of TFIIH has recently been described (34). Interestingly, this mechanism was not shared by sand fly fever Sicilian phlebovirus, another member of the genus Phlebovirus (68).
In this study, we sought to identify whether the novel phlebovirus SFTSV encodes proteins with the ability to inhibit IFN responses and to determine the mechanism by which the viral protein exerts its function. Not surprisingly, we found that the SFTSV NSs serves as a potent inhibitor of host IFN responses, in agreement with the role of other bunyavirus NSs proteins in suppressing host cellular responses (28). We observed that cells expressing SFTSV NSs produced less IFN-β in response to virus infection than cells expressing an irrelevant protein. This result is also consistent with that of a previous study reporting the ability of SFTSV NSs to suppress activation of the IFN-β promoter induced by viral infection and dsRNA treatment (39). Interestingly, we also attempted to measure the production of IFN-β in cells infected with SFTSV, but IFN-β was undetected, despite high levels of virus replication. This result provides further support for the ability of SFTSV to block IFN-β responses. It was also previously reported that the SFTSV NP had the capacity to inhibit activation of the IFN-β promoter upon viral infection and poly(I·C) treatment (39). However, the findings from our study contradicted those observations, because in our assays the SFTSV NP was not able to inhibit the phosphorylation of IRF3 induced by SeV infection or the activation of the IFN-β promoter induced by dsRNA, RIG-I, MAVS, TBK1, IRF3, or viral infection.
Consistent with previous observations, we confirmed the capacity of SFTSV NSs to interact with TBK1 (39). Astonishingly, we found that SFTSV NSs induces the formation of cytoplasmic structures and that it targets TBK1 into these structures. In an attempt to determine whether SFTSV NSs may affect the function of other RLR signaling components, we observed that SFTSV NSs also interacts with and relocalizes the viral recognition receptor RIG-I and the E3 ubiquitin ligase TRIM25 into these SFTSV NSs-induced structures. The interaction of SFTSV NSs with RIG-I appears to be indirect on the basis of the fact that (i) some protein still remains in the cytoplasm, (ii) RIG-I is still able to interact with MAVS, as evidenced by the BiFC data, and (iii) TRIM25 enhances this interaction. Since the association of SFTSV NSs with TBK-1 and other IFN signaling components was assessed only upon overexpression, it is also possible that these associations are not direct and may be mediated through another still unidentified cellular factor(s). Future studies involving recombinant proteins are needed to fully characterize these associations.
In our study, we also observed that SFTSV NSs colocalizes with GFP-LC3 structures, prompting us to investigate if SFTSV NSs might target TBK1, RIG-I, and TRIM25 to autophagosomes. This possibility was explored in cells lacking Atg7, a gene essential for conventional autophagy, and the results demonstrated that the SFTSV NSs-induced structures were still formed in Atg7−/− cells. Furthermore, redistribution of TBK1, RIG-I, and TRIM25 into these SFTSV NSs-induced structures still occurred in cells lacking Atg7, suggesting that the cytoplasmic structures induced by SFTSV NSs are not conventional autophagosomes. An alternative Atg7-independent autophagy pathway has recently been recognized. In this pathway, autophagosomes are generated in a Rab9-dependent manner and do not involve the lipidation of LC3-I to LC3-II and, thus, lack the classical formation of LC3 puncta (69, 70). The SFTSV NSs-induced structures do not appear to be unconventional autophagosomes, since formation of GFP-LC3 structures was still observed in Atg7−/− cells in the presence of SFTSV NSs. Notably, it has recently been recognized that LC3 can also play an autophagy-independent role in vesicle-trafficking events (71, 72). Specifically, it has been shown that LC3 is associated with EDEMosomes, which are double-membrane vesicles released from the ER and contain certain ER-associated degradation (ERAD) factors, such as EDEM1 and OS-9. In our studies, we did not find colocalization of EDEMosome markers with SFTSV NSs-induced LC3+ cytoplasmic structures.
In an attempt to identify the source of the SFTSV NSs-induced structures, we investigated the involvement of the endosomal pathway and observed the colocalization of SFTSV NSs with the early endosome marker Rab5 but not Rab4. Some of the SFTSV NSs-induced cytoplasmic structures are reminiscent of vesicles produced upon overexpression of a constitutive active form of Rab5 (73). However, the formation of SFTSV NSs-induced structures was still observed even in the presence of dominant negative (S34N) Rab5, suggesting that activation of Rab5 and downstream endocytic trafficking are not necessary for the formation of these cytoplasmic structures. Thus, it is possible that SFTSV NSs could be functioning in a Rab5-like manner. This could potentially open interesting avenues to explore in future studies.
It was earlier reported that cytomegalovirus redirects the NF-κB essential modulator (NEMO), a regulatory subunit of the IKK complex, to autophagosomes to avoid activation of the inflammatory cascade (74). The cytomegalovirus protein M45 was found to bind NEMO and to target the protein to autophagosomes for degradation, blocking the host inflammatory response. However, redirection of critical components of the IFN signaling pathways to LC3+ and Rab5+ structures as a potential mechanism to inhibit host innate immune responses has not yet been described. These results show a correlation between the sequestration of RIG-I signaling proteins and the inhibition of IFN responses. This points toward a novel mechanism for the suppression of IFN responses by a viral pathogen. Future studies are needed to conclusively demonstrate that the sequestration of these signaling proteins is conducive to the inhibition of IFN responses.
ACKNOWLEDGMENTS
We thank Thorsten Wolff for providing bimolecular fluorescence complementation plasmids, Richard Cadagan and Osman Lizardo for excellent technical assistance, and Adriana Paulucci and Leoncio Vergara from the Optical Microscopy Core Facility at The University of Texas Medical Branch for assistance with the confocal microscopy studies. We also thank Juan Ayllon for providing the dominant negative Rab5 plasmid. We thank Shinji Makino for critical review of the manuscript.
This research was supported by grants from the Institute for Human Infections and Immunity, National Institute of Allergy and Infectious Diseases (NIAID), through the Western Regional Center for Excellence for Biodefense and Emerging Infectious Disease Research, National Institutes of Health (NIH) grant number U54AI057156 (to P.V.A.), U19 AI083025 (to A.G.-S.), NIH contract HHSN27220100 0040I/HHSN27200004/D04, and startup funds from The University of Texas Medical Branch to P.V.A. F.W.S. was supported by an NIH T32 training grant (AI007536) in emerging and reemerging infectious diseases. J.R.P. was supported by an NIH T32 training grant (AI007647) in virus-host interactions. This project was also supported by the Cytometry and Cell Sorting Core at the Baylor College of Medicine with funding from the NIH (AI036211, CA125123, and RR024574) and the expert assistance of Joel M. Sederstrom.
Footnotes
Published ahead of print 29 January 2014
REFERENCES
- 1.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- 3.Meylan E, Tschopp J, Karin M. 2006. Intracellular pattern recognition receptors in the host response. Nature 442:39–44. 10.1038/nature04946 [DOI] [PubMed] [Google Scholar]
- 4.Yoneyama M, Fujita T. 2007. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev. 18:545–551. 10.1016/j.cytogfr.2007.06.023 [DOI] [PubMed] [Google Scholar]
- 5.Baum A, García-Sastre A. 2010. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids 38:1283–1299. 10.1007/s00726-009-0374-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi O, Akira S. 2009. Innate immunity to virus infection. Immunol. Rev. 227:75–86. 10.1111/j.1600-065X.2008.00737.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoneyama M, Fujita T. 2008. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity 29:178–181. 10.1016/j.immuni.2008.07.009 [DOI] [PubMed] [Google Scholar]
- 8.Yoneyama M, Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 227:54–65. 10.1111/j.1600-065X.2008.00727.x [DOI] [PubMed] [Google Scholar]
- 9.Yount JS, Gitlin L, Moran TM, López CB. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai virus defective interfering particles. J. Immunol. 180:4910–4918 [DOI] [PubMed] [Google Scholar]
- 10.Melchjorsen J, Jensen SB, Malmgaard L, Rasmussen SB, Weber F, Bowie AG, Matikainen S, Paludan SR. 2005. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J. Virol. 79:12944–12951. 10.1128/JVI.79.20.12944-12951.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19–28. 10.1016/j.immuni.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 12.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397–408. 10.1016/j.cell.2010.01.020 [DOI] [PubMed] [Google Scholar]
- 13.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- 14.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- 15.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- 16.Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJ. 2012. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2:1187–1196. 10.1016/j.celrep.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato H, Takahasi K, Fujita T. 2011. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 243:91–98. 10.1111/j.1600-065X.2011.01052.x [DOI] [PubMed] [Google Scholar]
- 18.Habjan M, Andersson I, Klingström J, Schümann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Mühlberger E, Mirazimi A, Weber F. 2008. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One 3:e2032. 10.1371/journal.pone.0002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131–137. 10.1038/ni1303 [DOI] [PubMed] [Google Scholar]
- 20.Oshiumi H, Miyashita M, Matsumoto M, Seya T. 2013. A distinct role of Riplet-mediated K63-linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 9:e1003533. 10.1371/journal.ppat.1003533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. 10.1038/nature05732 [DOI] [PubMed] [Google Scholar]
- 22.Gack MU, Kirchhofer A, Shin YC, Inn KS, Liang C, Cui S, Myong S, Ha T, Hopfner KP, Jung JU. 2008. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. U. S. A. 105:16743–16748. 10.1073/pnas.0804947105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141:315–330. 10.1016/j.cell.2010.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García-Sastre A, Biron CA. 2006. Type 1 interferons and the virus-host relationship: a lesson in détente. Science 312:879–882. 10.1126/science.1125676 [DOI] [PubMed] [Google Scholar]
- 25.Haller O, Kochs G, Weber F. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119–130. 10.1016/j.virol.2005.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 98:664–669. 10.1073/pnas.98.2.664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blakqori G, Delhaye S, Habjan M, Blair CD, Sánchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, Weiss S, Michiels T, Staeheli P, Weber F. 2007. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J. Virol. 81:4991–4999. 10.1128/JVI.01933-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elliott RM, Weber F. 2009. Bunyaviruses and the type I interferon system. Viruses 1:1003–1021. 10.3390/v1031003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. 2009. Dual functions of Rift Valley fever virus NSs protein: inhibition of host mRNA transcription and post-transcriptional downregulation of protein kinase PKR. Ann. N. Y. Acad. Sci. 1171(Suppl. 1):E75–E85. 10.1111/j.1749-6632.2009.05054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le May N, Mansuroglu Z, Léger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 4:e13. 10.1371/journal.ppat.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, Egly JM. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550. 10.1016/S0092-8674(04)00132-1 [DOI] [PubMed] [Google Scholar]
- 32.Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365–4375. 10.1128/JVI.02148-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. 2009. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 5:e1000287. 10.1371/journal.ppat.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalveram B, Lihoradova O, Ikegami T. 2011. NSs protein of Rift Valley fever virus promotes posttranslational downregulation of the TFIIH subunit p62. J. Virol. 85:6234–6243. 10.1128/JVI.02255-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, Zhang L, Zhang QF, Popov VL, Li C, Qu J, Li Q, Zhang YP, Hai R, Wu W, Wang Q, Zhan FX, Wang XJ, Kan B, Wang SW, Wan KL, Jing HQ, Lu JX, Yin WW, Zhou H, Guan XH, Liu JF, Bi ZQ, Liu GH, Ren J, Wang H, Zhao Z, Song JD, He JR, Wan T, Zhang JS, Fu XP, Sun LN, Dong XP, Feng ZJ, Yang WZ, Hong T, Zhang Y, Walker DH, Wang Y, Li DX. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 364:1523–1532. 10.1056/NEJMoa1010095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang YZ, He YW, Dai YA, Xiong Y, Zheng H, Zhou DJ, Li J, Sun Q, Luo XL, Cheng YL, Qin XC, Tian JH, Chen XP, Yu B, Jin D, Guo WP, Li W, Wang W, Peng JS, Zhang GB, Zhang S, Chen XM, Wang Y, Li MH, Li Z, Lu S, Ye C, de Jong MD, Xu J. 2012. Hemorrhagic fever caused by a novel bunyavirus in China: pathogenesis and correlates of fatal outcome. Clin. Infect. Dis. 54:527–533. 10.1093/cid/cir804 [DOI] [PubMed] [Google Scholar]
- 37.Zhang YZ, Zhou DJ, Xiong Y, Chen XP, He YW, Sun Q, Yu B, Li J, Dai YA, Tian JH, Qin XC, Jin D, Cui Z, Luo XL, Li W, Lu S, Wang W, Peng JS, Guo WP, Li MH, Li ZJ, Zhang S, Chen C, Wang Y, de Jong MD, Xu J. 2011. Hemorrhagic fever caused by a novel tick-borne bunyavirus in Huaiyangshan, China. Zhonghua Liu Xing Bing Xue Za Zhi 32:209–220 (In Chinese.) [PubMed] [Google Scholar]
- 38.Jin C, Liang M, Ning J, Gu W, Jiang H, Wu W, Zhang F, Li C, Zhang Q, Zhu H, Chen T, Han Y, Zhang W, Zhang S, Wang Q, Sun L, Liu Q, Li J, Wang T, Wei Q, Wang S, Deng Y, Qin C, Li D. 2012. Pathogenesis of emerging severe fever with thrombocytopenia syndrome virus in C57/BL6 mouse model. Proc. Natl. Acad. Sci. U. S. A. 109:10053–10058. 10.1073/pnas.1120246109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qu B, Qi X, Wu X, Liang M, Li C, Cardona CJ, Xu W, Tang F, Li Z, Wu B, Powell K, Wegner M, Li D, Xing Z. 2012. Suppression of the interferon and NF-κB responses by severe fever with thrombocytopenia syndrome virus. J. Virol. 86:8388–8401. 10.1128/JVI.00612-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434. 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, García-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. 10.1016/j.chom.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pythoud C, Rodrigo WW, Pasqual G, Rothenberger S, Martínez-Sobrido L, de la Torre JC, Kunz S. 2012. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKε. J. Virol. 86:7728–7738. 10.1128/JVI.00187-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel JR, Jain A, Chou YY, Baum A, Ha T, García-Sastre A. 2013. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 14:780–787. 10.1038/embor.2013.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aguilar PV, Weaver SC, Basler CF. 2007. Capsid protein of eastern equine encephalitis virus inhibits host cell gene expression. J. Virol. 81:3866–3876. 10.1128/JVI.02075-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Basler CF, Wang X, Mühlberger E, Volchkov V, Paragas J, Klenk HD, García-Sastre A, Palese P. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. U. S. A. 97:12289–12294. 10.1073/pnas.220398297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Z, Li C, Higginbotham JN, Franklin JL, Tabb DL, Graves-Deal R, Hill S, Cheek K, Jerome WG, Lapierre LA, Goldenring JR, Ham AJ, Coffey RJ. 2008. Use of fluorescence-activated vesicle sorting for isolation of Naked2-associated, basolaterally targeted exocytic vesicles for proteomics analysis. Mol. Cell. Proteomics 7:1651–1667. 10.1074/mcp.M700155-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Mühlberger E, Bray M, Klenk HD, Palese P, García-Sastre A. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945–7956. 10.1128/JVI.77.14.7945-7956.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cárdenas WB, Loo YM, Gale M, Hartman AL, Kimberlin CR, Martínez-Sobrido L, Saphire EO, Basler CF. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80:5168–5178. 10.1128/JVI.02199-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prins KC, Cárdenas WB, Basler CF. 2009. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J. Virol. 83:3069–3077. 10.1128/JVI.01875-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Servant MJ, ten Oever B, LePage C, Conti L, Gessani S, Julkunen I, Lin R, Hiscott J. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355–363. 10.1074/jbc.M007790200 [DOI] [PubMed] [Google Scholar]
- 52.Weaver BK, Kumar KP, Reich NC. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerppola TK. 2006. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7:449–456. 10.1038/nrm1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374. 10.1038/nrmicro1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cash P. 1982. Inhibition of La Crosse virus replication by monensin, monovalent ionophore. J. Gen. Virol. 59:193–196. 10.1099/0022-1317-59-1-193 [DOI] [PubMed] [Google Scholar]
- 56.Nakitare GW, Elliott RM. 1993. Expression of the Bunyamwera virus M genome segment and intracellular localization of NSm. Virology 195:511–520. 10.1006/viro.1993.1402 [DOI] [PubMed] [Google Scholar]
- 57.Lappin DF, Nakitare GW, Palfreyman JW, Elliott RM. 1994. Localization of Bunyamwera bunyavirus G1 glycoprotein to the Golgi requires association with G2 but not with NSm. J. Gen. Virol. 75(Pt 12):3441–3451. 10.1099/0022-1317-75-12-3441 [DOI] [PubMed] [Google Scholar]
- 58.Zeigerer A, Gilleron J, Bogorad RL, Marsico G, Nonaka H, Seifert S, Epstein-Barash H, Kuchimanchi S, Peng CG, Ruda VM, Del Conte-Zerial P, Hengstler JG, Kalaidzidis Y, Koteliansky V, Zerial M. 2012. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature 485:465–470. 10.1038/nature11133 [DOI] [PubMed] [Google Scholar]
- 59.Woodman PG. 2000. Biogenesis of the sorting endosome: the role of Rab5. Traffic 1:695–701. 10.1034/j.1600-0854.2000.010902.x [DOI] [PubMed] [Google Scholar]
- 60.Stone M, Jia S, Heo WD, Meyer T, Konan KV. 2007. Participation of Rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J. Virol. 81:4551–4563. 10.1128/JVI.01366-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng CY, Shih WL, Huang WR, Chi PI, Wu MH, Liu HJ. 2012. Bovine ephemeral fever virus uses a clathrin-mediated and dynamin 2-dependent endocytosis pathway that requires Rab5 and Rab7 as well as microtubules. J. Virol. 86:13653–13661. 10.1128/JVI.01073-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang WR, Wang YC, Chi PI, Wang L, Wang CY, Lin CH, Liu HJ. 2011. Cell entry of avian reovirus follows a caveolin-1-mediated and dynamin-2-dependent endocytic pathway that requires activation of p38 mitogen-activated protein kinase (MAPK) and Src signaling pathways as well as microtubules and small GTPase Rab5 protein. J. Biol. Chem. 286:30780–30794. 10.1074/jbc.M111.257154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garrison AR, Radoshitzky SR, Kota KP, Pegoraro G, Ruthel G, Kuhn JH, Altamura LA, Kwilas SA, Bavari S, Haucke V, Schmaljohn CS. 2013. Crimean-Congo hemorrhagic fever virus utilizes a clathrin- and early endosome-dependent entry pathway. Virology 444:45–54. 10.1016/j.virol.2013.05.030 [DOI] [PubMed] [Google Scholar]
- 64.Lozach PY, Mancini R, Bitto D, Meier R, Oestereich L, Overby AK, Pettersson RF, Helenius A. 2010. Entry of bunyaviruses into mammalian cells. Cell Host Microbe 7:488–499. 10.1016/j.chom.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stenmark H, Olkkonen VM. 2001. The Rab GTPase family. Genome Biol. 2:REVIEWS3007. 10.1186/gb-2001-2-5-reviews3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, García-Sastre A, Weber F. 2013. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe 13:336–346. 10.1016/j.chom.2013.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang NE, Lin CH, Lin YS, Yu WC. 2003. Modulation of YY1 activity by SAP30. Biochem. Biophys. Res. Commun. 306:267–275. 10.1016/S0006-291X(03)00966-5 [DOI] [PubMed] [Google Scholar]
- 68.Lihoradova OA, Indran SV, Kalveram B, Lokugamage N, Head JA, Gong B, Tigabu B, Juelich TL, Freiberg AN, Ikegami T. 2013. Characterization of Rift Valley fever virus MP-12 strain encoding NSs of Punta Toro virus or sandfly fever Sicilian virus. PLoS Negl. Trop. Dis. 7:e2181. 10.1371/journal.pntd.0002181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. 2009. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461:654–658. 10.1038/nature08455 [DOI] [PubMed] [Google Scholar]
- 70.Shimizu S, Arakawa S, Nishida Y. 2010. Autophagy takes an alternative pathway. Autophagy 6:290–291. 10.4161/auto.6.2.11127 [DOI] [PubMed] [Google Scholar]
- 71.de Haan CA, Molinari M, Reggiori F. 2010. Autophagy-independent LC3 function in vesicular traffic. Autophagy 6:994–996. 10.4161/auto.6.7.13309 [DOI] [PubMed] [Google Scholar]
- 72.Reggiori F, de Haan CA, Molinari M. 2011. Unconventional use of LC3 by coronaviruses through the alleged subversion of the ERAD tuning pathway. Viruses 3:1610–1623. 10.3390/v3091610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blümer J, Rey J, Dehmelt L, Mazel T, Wu YW, Bastiaens P, Goody RS, Itzen A. 2013. RabGEFs are a major determinant for specific Rab membrane targeting. J. Cell Biol. 200:287–300. 10.1083/jcb.201209113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fliss PM, Jowers TP, Brinkmann MM, Holstermann B, Mack C, Dickinson P, Hohenberg H, Ghazal P, Brune W. 2012. Viral mediated redirection of NEMO/IKKγ to autophagosomes curtails the inflammatory cascade. PLoS Pathog. 8:e1002517. 10.1371/journal.ppat.1002517 [DOI] [PMC free article] [PubMed] [Google Scholar]