ABSTRACT
The herpes simplex virus 1 (HSV-1) UL51 gene encodes a 244-amino-acid (aa) palmitoylated protein that is conserved in all herpesviruses. The alphaherpesvirus UL51 (pUL51) protein has been reported to function in nuclear egress and cytoplasmic envelopment. No complete deletion has been generated because of the overlap of the UL51 coding sequence 5′ end with the UL52 promoter sequences, but partial deletions generated in HSV and pseudorabies virus (PrV) suggest an additional function in epithelial cell-to-cell spread. Here we show partial uncoupling of the replication, release, and cell-to-cell spread functions of HSV-1 pUL51 in two ways. Viruses in which aa 73 to 244 were deleted from pUL51 or in which a conserved YXXΦ motif near the N terminus was altered showed cell-specific defects in spread that cannot be accounted for by defects in replication and virus release. Also, a cell line that expresses C-terminally enhanced green fluorescent protein (EGFP)-tagged pUL51 supported normal virus replication and release into the medium but the formation of only small plaques. This cell line also failed to support normal localization of gE to cell junctions. gE and pUL51 partially colocalized in infected cells, and these two proteins could be coimmunoprecipitated from infected cells, suggesting that they can form a complex during infection. The cell-to-cell spread defect associated with the pUL51 mutation was more severe than that associated with gE-null virus, suggesting that pUL51 has gE-independent functions in epithelial cell spread.
IMPORTANCE Herpesviruses establish and reactivate from lifelong latency in their hosts. When they reactivate, they are able to spread within their hosts despite the presence of a potent immune response that includes neutralizing antibody. This ability is derived in part from a specialized mechanism for virus spread between cells. Cell-to-cell spread is a conserved property of herpesviruses that likely relies on conserved viral genes. An understanding of their function may aid in the design of vaccines and therapeutics. Here we show that one of the conserved viral genes, UL51, has an important role in cell-to-cell spread in addition to its previously demonstrated role in virus assembly. We find that its function depends on the type of cell that is infected, and we show that it interacts with and modulates the function of another viral spread factor, gE.
INTRODUCTION
All of the manifestations of herpes simplex virus (HSV) disease result from the ability of the virus to spread from the initially infected cell to other cells at mucosal surfaces and to and from sensory neurons that enervate the site of primary replication. Similarly, recurrence of symptoms and consequent spread of the virus to new hosts require the ability to spread from neurons in the sensory ganglion to cells at the periphery and among the cells on the mucosal surface. Spread and shedding of the virus in recurrent infection occur in the face of an adaptive immune response including an antibody response, which should neutralize virus released from the cell. Therefore, the disease-causing properties and transmission of HSVs depend on the mechanisms used for the spread of the virus from cell to cell that protect the virus from exposure to effectors of the adaptive immune response. The passage of virus between adjacent cells is the result of a specialized process called cell-to-cell spread (CCS), in which virus is specifically trafficked to and released at junctional surfaces of cells.
While CCS has been most thoroughly explored in the alphaherpesviruses, the problem of viral spread in the presence of immune effectors is common to most, and perhaps all, of the human herpesviruses. The signature characteristic of herpesvirus infections is their ability to establish and then reactivate from latency. Upon reactivation, these viruses may cause symptoms and can be shed throughout the life of the host. Human cytomegalovirus (HCMV) and Epstein-Barr virus (EBV) transmission is thought to result from virus shedding from productively infected epithelial cells in a number of different tissues (1, 2). It is likely that this productive epithelial infection also requires CCS and that there may be a common herpesviral mechanism for accomplishing this.
Epithelial CCS in alphaherpesvirus replication has been shown to depend on the function of glycoprotein E (gE) and gI, which form a heterodimeric complex (3–7). The gE/gI complex is found on most of the cytoplasmic membranes of infected cells, but it concentrates at adherens junctions, where it colocalizes with beta-catenin, and trafficking to junctions has been shown to be essential for gE's role in CCS (5, 8–10). Exactly how gE functions in epithelial spread is unclear, but it apparently facilitates trafficking of virions to cell junctions and may also interact with factors on the surface of an adjacent cell.
While gE and gI play an important role in epithelial CCS, the encoding genes are present only in the alphaherpesviruses and so cannot be at the root of any conserved CCS pathway. This raises the question of whether there are conserved gene products involved in CCS and, if so, which genes these are. We have reported evidence that the product of the conserved UL34 gene is specifically required for CCS (11). This gene was the first of the so-called “core” herpesvirus genes to have an unambiguously demonstrated role in CCS. Identification of CCS functions for core genes represents one avenue for identifying conserved herpesviral CCS mechanisms.
Our studies on UL34 function in CCS highlighted two important points. First, in studying multifunctional gene products, a gene deletion will reveal the earliest important function and could mask later functions. Second, we observed that reductions in replication as high as 50-fold compared to the replication of wild-type (WT) virus did not affect CCS within epithelial cells, as measured by plaque size. This led us to further explore the literature on HSV assembly and egress proteins and identify other conserved genes whose deletion results in a replication defect of <100-fold but that nonetheless cause the formation of small plaques. The proteins encoded by these genes include UL51, UL11, UL49, and possibly others (12–15). These gene products are candidates for important mediators of CCS. A specific function in CCS was recently demonstrated for pUL11 (16), but UL51 function has not been well characterized.
Recombinant viruses containing deletions or stop mutations in the UL51 gene orthologs of HSV, pseudorabies virus (PrV), and human cytomegalovirus (in which the homologous gene is UL71) have been characterized (14, 15, 17, 18). In each case, deletion results in a more or less severe replication defect that is apparently due to a defect in secondary envelopment in the cytoplasm. In each case, the replication defect is accompanied by the formation of small plaques, suggesting the possibility of a CCS defect. We tested the hypothesis that partial deletion or point mutation of the UL51 gene might reveal a specific defect in CCS. We find that pUL51 does indeed have a specific function in CCS and that different mutations affect spread differently in different cell types.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 and Vero cells were maintained as previously described (19). The properties of HSV-1 strain F [HSV-1(F)] were described previously (19, 20).
Generation of anti-pUL51 antiserum.
A PCR amplicon was generated from purified HSV-1(F) viral DNA by using primers ATATCTCGAGTGCGGTTGGGGAGGCTGTAGC and ATATGAATTCAGGAGGCCCTGGCGGTCGTT. The product, which contained codons 36 to 244 of UL51, was digested with XhoI and EcoRI (sites in the primers are underlined) and cloned into the same restriction sites within the multiple-cloning region of pGEX 4T-2 such that the UL51 coding sequences were placed in frame with the gene encoding glutathione S-transferase (GST). The GST fusion protein was expressed in the BL21 strain of Escherichia coli and purified on glutathione-Sepharose beads. Two New Zealand White rabbits were inoculated with the fusion protein emulsified in complete Freund's adjuvant, followed by 3 injections 2 weeks apart of the protein emulsified in incomplete Freund's adjuvant. Two weeks later, rabbit antisera were collected and tested for reactions with UL51 by immunoblotting.
Construction of a UL51 complementing cell line.
Plasmid pRR1117 was constructed by ligation of the 11.44-kb BclI fragment of HSV-1(F) into the BamHI site of pGEM-3Z(F+). pRR1382, containing the UL51 gene, was constructed by digesting pRR1117 with HindIII and StuI, blunting the fragments by treatment with Klenow enzyme in the presence of deoxynucleoside triphosphates (dNTPs), and then ligating the 1.42-kb fragment between the NruI and EcoRV sites of pcDNA3. The resulting plasmid lacks the CMV promoter and has the complete UL51 coding sequence driven by its own promoter/regulatory sequences. Clonal cell line UL51#39 was constructed by transfection of pRR1382 into Vero cells, followed by selection with G418 and isolation of clones by limiting dilution. Clones were initially screened for their ability to complement plaque formation by a UL51 deletion virus.
Construction of recombinant mutant viruses.
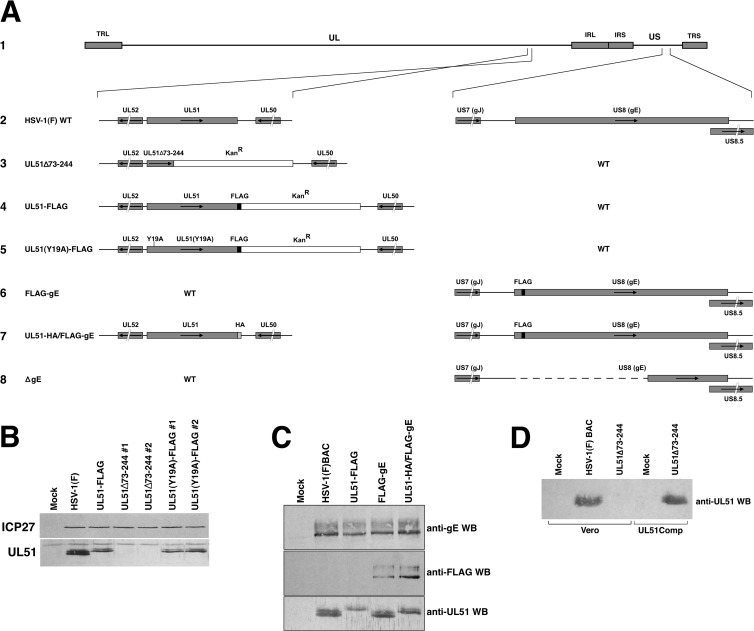
Viruses that carried various alterations to the UL51 and gE coding sequences were constructed. Viruses encoding C-terminally truncated UL51 (UL51Δ73–244), C-terminally FLAG-tagged WT UL51, a deletion of sequences encoding amino acids (aa) 1 to 335 of gE, or FLAG-tagged gE were constructed by using an HSV-1(F) bacterial artificial chromosome (BAC) and methods reported previously by Tischer et al. (21), as previously described (11). The virus encoding FLAG-tagged UL51 with a substitution of alanine for tyrosine 19 (UL51Y19A) was constructed by sequentially introducing the C-terminal FLAG tag into UL51 and then mutating the codon encoding tyrosine 19. The virus encoding FLAG-tagged gE and HA-tagged UL51 was constructed by sequentially introducing the FLAG tag sequence into US8 and then introducing the hemagglutinin (HA) tag sequence into UL51. The sequences of primers used for virus construction are available upon request. Proper structure of the recombinant BACs was determined by sequencing of the UL51 and/or gE gene region. The structures of the altered UL51 and gE genes are indicated in Fig. 1.
FIG 1.
Construction of recombinant viruses. (A) Schematic diagram of the HSV-1(F) genome (line 1) and of the recombinant viruses constructed for this study. The positions of the terminal and internal repeats that flank the long genome component (TRL and IRL, respectively) and the short genome component (IRS and TRS, respectively) are indicated with gray bars. (Line 2) The structures of the wild-type sequences in the regions of UL51 and US8 are shown. (Line 3) The UL51Δ73–244 virus carries a stop codon and a kanamycin resistance cassette in place of the sequences coding for amino acids 73 to 244 of pUL51. (Line 4) The UL51-FLAG virus carries a FLAG tag at the C terminus of UL51 followed by a kanamycin resistance cassette. (Line 5) The UL51(Y19A)-FLAG virus was constructed by mutating the Y19 codon in the context of the UL51-FLAG virus shown in line 4. (Line 6) The FLAG-gE virus was constructed by the insertion of a FLAG-coding sequence between the codons for amino acids 20 and 21 of gE. This was predicted to yield an N-terminally FLAG-tagged gE protein after signal peptide cleavage. (Line 7) The UL51-HA/FLAG-gE virus was constructed by introducing an HA epitope-coding sequence at the C terminus of the UL51 protein-coding sequence in the context of the FLAG-gE virus shown in line 6. (Line 8) the ΔgE virus was constructed by scarless removal of the sequences encoding amino acids 1 to 335 of gE. (B) Expression of UL51 by mutant recombinant viruses. Lysates from Vero cells infected for 16 h with the indicated viruses were probed for either ICP27 to control for the extent of infection and loading (top) or UL51 using anti-UL51 polyclonal antiserum (bottom). (C) Expression of epitope-tagged proteins by recombinant viruses. Lysates from Vero cells infected for 16 h with the indicated viruses were probed for gE (top), the FLAG epitope (middle), or UL51 (bottom). (D) Expression of UL51 by a complementing cell line. Lysates of either Vero or UL51-complementing cells that had been infected with the indicated viruses were probed with anti-UL51 polyclonal antiserum. WB, Western blot.
Recombinant viruses were reconstituted by transfection of BAC DNA into Vero cells. Viruses containing alterations of the UL51 gene sequence were amplified on UL51-complementing cells to minimize selection for phenotypic revertants. Maintenance of mutations in the amplified recombinant viruses was confirmed by PCR amplification and sequencing of the UL51 region.
Construction of a pUL51-EGFP-expressing cell line.
To construct an infection-inducible UL51-enhanced green fluorescent protein (EGFP)-expressing cell line, we built plasmid pRR1381. A PCR product was amplified from the HSV-1(F) genome containing UL51 gene sequences from position −400 (with respect to the UL51 start codon) down to, but not including, the stop codon and flanked by AseI and AgeI restriction sites. This product was cloned between the AseI and AgeI sites of pEGFP-C1 (Clontech). The resulting plasmid expresses UL51 using its own promoter/regulatory sequences following HSV infection. Clonal cell line UL51-EGFP#9 was constructed by transfection of pRR1381 into Vero cells, followed by selection with G418 and isolation of clones by limiting dilution. Expressing cell clones were screened by assays for EGFP expression 20 h after infection with HSV-1(F).
Single-step growth measurements.
Measurement of replication of HSV-1(F), UL51Δ73–244, and UL51Y19A viruses on Vero and HEp-2 cells after infection at a high multiplicity of infection (5 PFU/cell) was performed as previously described (19). Virus release efficiency was calculated as PFU in the culture medium at 24 h (Vero) or 48 h (HEp-2) postinfection (p.i.)/peak PFU produced in the total culture. The statistical significance of single-step growth data was determined by using a Student t test.
Immunostaining of plaques.
Cell monolayers containing the wild type and syncytial variants of HSV-1(F) were fixed by incubation for 15 min in 3.7% formaldehyde in phosphate-buffered saline (PBS). After fixation, monolayers were washed three times with 2 ml PBS. Plaques were stained by indirect immunofluorescence using a 1:5,000 dilution of mouse monoclonal antibody DL6 directed against HSV gD (kind gift of G. Cohen and R. Eisenberg) as a primary antibody and a 1:1,000 dilution of alkaline phosphatase-conjugated goat anti-mouse IgG (Invitrogen) as a secondary antibody.
Quantitative plaque size assays.
Six-well tissue culture plates were seeded with 1.8 × 106 Vero or 2.5 × 106 HEp-2 cells the day before infection. Infection was initiated by removal of the growth medium and addition of 1 ml of virus diluted in V medium (Dulbecco's modified Eagle medium [DMEM] containing 1% heat-inactivated calf serum). The virus inoculum was removed after 90 min and replaced with 2.5 ml V medium containing a 1:250 dilution of pooled human immunoglobulin as a source of HSV-neutralizing antibody (GammaSTAN S/D; Talecris Biotherapeutics). At 2 days after infection, monolayers were washed twice with PBS and then fixed by incubation for 15 min in 3.7% formaldehyde in PBS. After fixation, monolayers were stained as described above, except using 1:2,500 dilution of mouse monoclonal anti-HSV 45-kDa protein (scaffolding protein) antibody (Serotec) as a primary antibody and a 1:1,000 dilution of Alexa Fluor 488 goat anti-mouse IgG (Invitrogen) as a secondary antibody. Plaques were photographed by using an inverted fluorescence microscope. Plaque images were opened in ImageJ and outlined by using the freehand tool. The number of pixels contained within the outline was used as the plaque area. Since plaque areas were not always normally distributed, the nonparametric, distribution-free Kolmogorov-Smirnov test, rather than a t test, was used to determine statistical significance, using a Web-implemented version (http://www.physics.csbsju.edu/stats/KS-test.html).
Selection of syncytial variants of HSV-1(F).
HSV-1(F) was plated onto Vero cells, and several thousand plaques were screened to find 12 well-isolated plaques that showed syncytial phenotypes of various severities. Plaques were picked and then reisolated twice more to obtain virus populations that each had a uniform syncytial phenotype.
Indirect immunofluorescence.
Immunofluorescence for colocalization was performed as previously described, using either a 1:2,000 dilution of mouse monoclonal anti-gE ascites (Goodwin Cancer Institute) or a 1:1,000 dilution of mouse monoclonal anti-FLAG M2 antibody (Sigma) (22, 23).
Immunopurification.
FLAG-gE and pUL51-FLAG were purified from Vero or HEp-2 cells that had been infected with 5 PFU/cell of wild-type or recombinant HSV-1 encoding tagged protein for 16 h. Infected cell monolayers from 100-mm cultures were washed with 5 ml of PBS and then scraped into 3 ml of PBS and pelleted at 1,200 rpm for 10 min. The cell pellets were resuspended in 1.5 ml coimmunoprecipitation (co-IP) buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1× Sigma protease inhibitor cocktail), transferred into microcentrifuge tubes, and incubated on ice for 3 min. Nuclei and other cellular debris were pelleted by centrifugation at 10,000 rpm in a microcentrifuge for 10 min, and the supernatant was transferred into a fresh tube. After removal of a fraction of the sample as a lysate control, 15 μl of an anti-FLAG magnetic bead suspension (Sigma) was added to the remainder of each sample, and the tubes were placed in an end-over-end rotator at 4°C overnight. Magnetic beads were separated from the lysate by using a magnetic separator, and the supernatant containing unbound proteins was discarded. Magnetic beads were washed three times each with 1.5 ml of co-IP buffer, and bound proteins were then eluted with three washes of co-IP buffer containing 100 μg/ml competitor 3×FLAG peptide (Sigma). Lysate and purified protein samples were separated on SDS-PAGE gels, followed by immunoblotting.
Immunoblotting.
Nitrocellulose sheets bearing proteins of interest were blocked in 5% nonfat milk plus 0.2% Tween 20 for at least 2 h. The membranes were probed with either a rabbit polyclonal antiserum raised against a UL51-GST fusion protein (1:1,000 dilution), a rabbit polyclonal antiserum raised against gE (kind gift of H. Friedman) (1:500), mouse anti-FLAG M2 monoclonal antibody (1:1,000; Sigma-Aldrich), or goat polyclonal anti-HA antiserum (1:1,000), followed by reaction with an alkaline phosphatase-conjugated secondary antibody.
RESULTS
Deletion of most of the UL51 protein-coding sequence causes cell-specific defects in virus replication, release, and cell-to-cell spread.
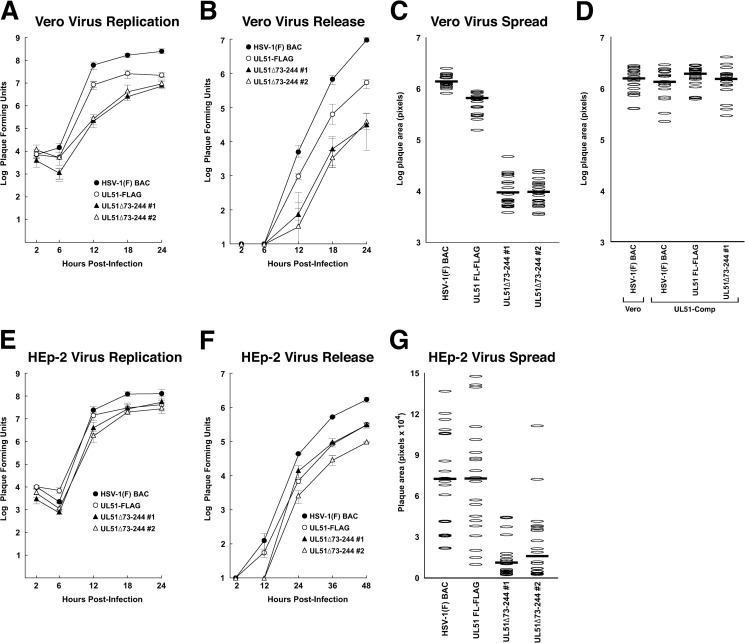
Nozawa et al. reported that the deletion of all but the N-terminal 42 amino acids of HSV-1 UL51 resulted in a roughly 100-fold single-step growth defect and the formation of very small plaques (15). Klupp et al. reported that deletion of all but the first 62 amino acids of pseudorabies virus (PrV) UL51 resulted in only a 6-fold growth defect (14). While those results were obtained by using different viruses in different cell types, they suggested the hypothesis that growth and spread functions of pUL51 might be partially or completely uncoupled by a partial deletion of the UL51 protein-coding sequence. To determine whether the two functions could be uncoupled, we made two independently constructed viruses in which the sequences coding for amino acids 73 to 244 were deleted and replaced by a kanamycin resistance cassette (Fig. 1A). These viruses did not express UL51 protein that could be detected by Western blotting (Fig. 1B). We measured virus single-step growth and CCS compared to those of wild-type HSV-1(F) and a recombinant virus in which the full-length pUL51 protein was FLAG tagged at the C terminus. The C-terminally FLAG-tagged UL51 virus showed a significant defect in single-step growth on Vero cells (Fig. 2A), achieving a peak titer roughly 10-fold lower than that of the WT control. This defect may be due to a somewhat lower expression level of FLAG-tagged UL51 than of the untagged protein (Fig. 1B and C), or it may be that the presence of the FLAG tag interferes with pUL51 function. The deletion viruses also showed a significant growth defect on Vero cells (Fig. 2A). The deletion viruses took a few hours longer to reach their peak titer but achieved nearly the same peak titer as the UL51-FLAG virus.
FIG 2.
Growth and spread of UL51 deletions on Vero and HEp-2 cells. (A) Single-step growth of BAC-derived HSV-1(F), UL51-FLAG, and two independently isolated UL51 deletion viruses was measured on Vero cells. Stocks were prepared from the total infected culture (cells and medium). (B) Virus released into the medium during the single-step growth experiment shown in panel A. (C) Sizes of plaques formed by wild-type and mutant viruses on Vero cells. Plaque areas were measured 2 days following low-multiplicity infection as described in Materials and Methods. Each oval represents the area of a single plaque. Twenty plaques were measured for each virus. Note that the y axis has a logarithmic scale. (D) Same as panel C except that plaques were measured on Vero and UL51-complementing cells, as indicated below the graph. (G to H) Same as panels A to C except that measurements were performed by using HEp-2 cells. Note that the y axis in panel F has a linear scale. For replication and release measurements (A, B, E, and F), each point represents the mean of three independent experiments, and the error bars represent the ranges of values obtained. Statistical significance for replication and release experiments, where noted in the text, was determined by using a Student t test, as implemented in Microsoft Excel. Panels C and F are each representative of three independent experiments. The differences in plaque sizes between the HSV-1(F) BAC and the UL51 deletion mutants shown in panel G are significant, with P values of <0.01 determined by using a Kolmogorov-Smirnov test.
As is normal in Vero cell infections, all viruses released only a small fraction of infectivity into the medium. The addition of a FLAG tag did not impair the efficiency of virus release, since WT and UL51-FLAG viruses released similar fractions of the infectivity produced (4.0% versus 2.7% at 24 h). The deletion viruses, however, showed an additional release defect. Even though they produced roughly the same peak titer as the UL51-FLAG virus, they released roughly 10-fold less virus (0.3% for deletion 1 and 0.4% for deletion 2) (Fig. 2B).
The plaques formed by the deletion viruses were almost 100-fold smaller than those formed by the wild-type virus (Fig. 2C). This difference in plaque size between the deletion and wild-type viruses might be due to a specific effect on CCS, or it might be a result of the single-step replication and release defects in the deletion viruses. However, the same difference in plaque area was observed between the UL51-FLAG virus and the deletion viruses despite the similar single-step replication of these viruses. This suggests that pUL51 plays a critical role in CCS in Vero cells and that this function can be partly uncoupled from its previously described role in virus replication and from the virus release function observed here. The defect in plaque formation was due specifically to the deletion in pUL51, since it was identical in the two independently constructed deletion recombinants and since it was completely corrected in the complementing cell line that expresses wild-type pUL51 (Fig. 2D).
In HEp-2 cells, there was no significant virus replication defect for any of the viruses compared to the wild type (Fig. 2E). The UL51-FLAG virus and the two deletion viruses showed a small but significant (P < 0.05) release defect compared to the wild type but were not significantly different from each other (Fig. 2F). The two deletion viruses did, however, show a CCS defect compared to both the wild-type and UL51-FLAG viruses (Fig. 2G). This defect was not as dramatic as that seen on Vero cells. Mutant virus plaques were about 6-fold smaller than the plaques formed by the wild-type and UL51-FLAG viruses. Since the deletion viruses and the UL51-FLAG virus did not differ from each other in single-step growth or virus release, this suggests that the difference in plaque size is due to the loss of a specific CCS function of pUL51 in the deletion viruses.
UL51 contains a highly conserved YXXΦ motif near the N terminus.
The UL51 protein is thought to localize to the cytoplasmic face of Golgi membranes, and this localization suggests a possible function in trafficking of viral proteins or virions in transport vesicles that bud from this compartment. We hypothesized that pUL51 contains sequence motifs for this function. A search of the UL51 protein sequence using the Eukaryotic Linear Motif online resource (24) revealed several membrane-trafficking motifs that might be expected to play a role in virion or virus glycoprotein sorting for CCS. Many of these motifs, however, have very low sequence complexity and thus might be expected to appear by chance, regardless of protein function. To identify likely functional motifs, an alignment of UL51 proteins was created from sequences of all herpesviruses for which a UL51 sequence is available. One motif, a YXXΦ sequence found at residues 19 to 22 in HSV-1 UL51, is found at a very similar position in all herpesvirus pUL51 homolog sequences from all subfamilies of the Herpesviridae (Fig. 3), with the single exception of PrV, suggesting that this motif might carry out a conserved function.
FIG 3.

Alignment of N-terminal sequences of UL51 homologs from human herpesviruses. Homologs of UL51 from all herpesviruses for which sequences are available were aligned by using the MUSCLE sequence alignment program (52). The alignment from the N terminus of the human herpesvirus homologs is shown. The positions of the conserved cysteine residue that is the palmitoylation site (26) and of the conserved YXXΦ motif are boxed. VZV, varicella-zoster virus; Kaposi's sarcoma-associated herpesvirus; HHV6, human herpesvirus 6.
Mutation of the YXXΦ motif results in a cell-specific defect in CCS.
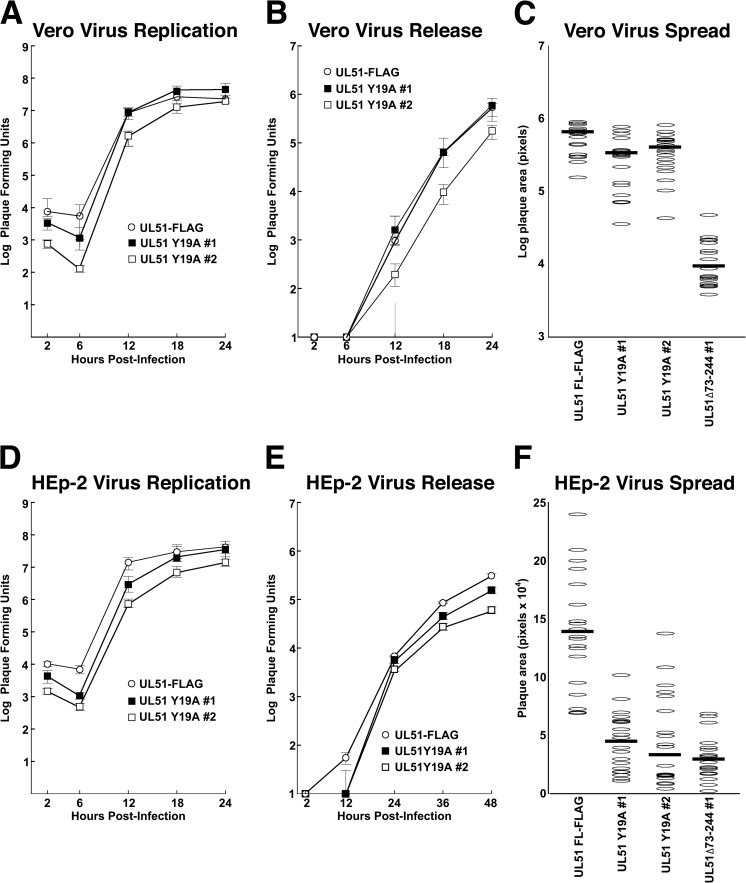
To test for the function of the YXXΦ motif in CCS, we constructed two independent recombinant viruses in which we mutated the tyrosine codon at position 19 to an alanine codon in the context of the UL51-FLAG recombinant virus (Fig. 1A). Both viruses expressed FLAG-tagged pUL51 at the same level as the parent UL51-FLAG virus (Fig. 1B). The Y19A recombinant showed no detectable defect in single-step growth (Fig. 4A and D) or the efficiency of virus release into the medium (Fig. 4B and E) on either Vero or HEp-2 cells, suggesting that the YXXΦ motif is not important for the virus replication or release functions of pUL51. Despite the strong effect of the pUL51 deletion on spread in Vero cells, the Y19A mutant showed no evident spread defect (Fig. 4C). The mutant did, however, have a spread defect in HEp-2 cells that was just as large as the defect induced by the UL51Δ73–244 virus. This suggests that the YXXΦ motif has a cell-specific function in CCS.
FIG 4.
Growth and spread of UL51(Y19A) mutants on Vero and HEp-2 cells. (A) Single-step growth of UL51-FLAG and two independently isolated UL51(Y19A) mutant viruses measured on Vero cells. Stocks were prepared from the total infected culture (cells and medium). (B) Virus released into the medium during the single-step growth experiment shown in panel A. (C) Sizes of plaques formed by control and mutant viruses. Twenty plaques were measured for each virus. Note that the y axis has a logarithmic scale. (D to F) Same as panels A to C except that measurements were performed with HEp-2 cells. Note that the y axis in panel F has a linear scale. For replication and release measurements (A, B, D, and E), each point represents the mean of three independent experiments, and the error bars represent the ranges of values obtained. Panels C and F are each representative of three independent experiments. The differences in plaque sizes between UL51-FLAG and the UL51(Y19A) mutants shown in panel F are significant, with P values of <0.01.
Expression of a pUL51-EGFP fusion specifically inhibits CCS and disrupts normal gE localization and function.
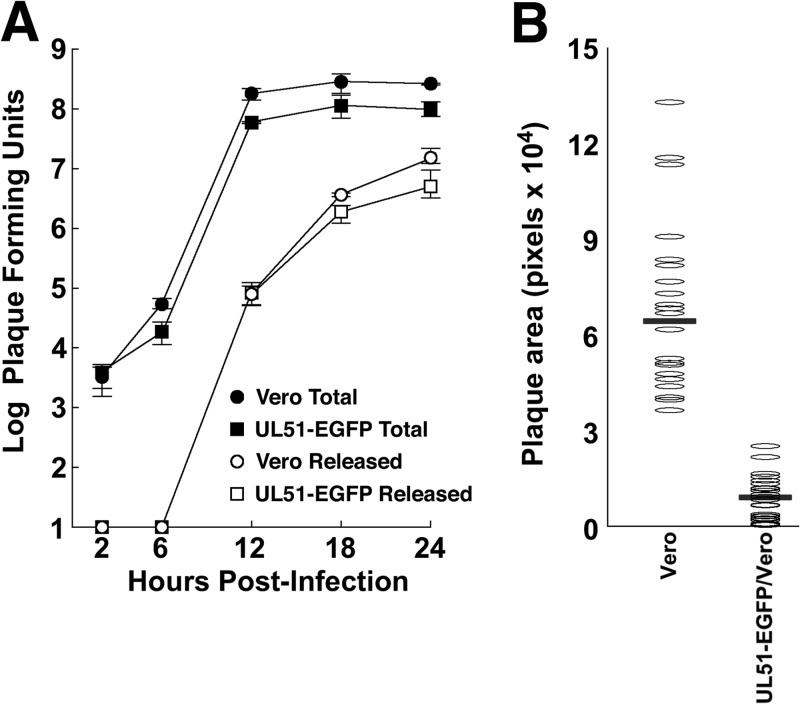
In an attempt to create a complementing cell line for propagation of a full UL51 deletion, we stably transfected Vero cells with a construct that expresses a pUL51-EGFP fusion under the control of pUL51 promoter-regulatory sequences. Stable transfectant clones were isolated, which did not express detectable pUL51-EGFP unless infected with HSV-1. Surprisingly, we noted that HSV-1(F) formed significantly smaller plaques in these cell lines than in untransfected Vero cells. We therefore characterized one of these lines with respect to the replication, release, and spread of wild-type HSV-1(F) (Fig. 5). We found that the pUL51-EGFP-expressing cells supported single-step replication and virus release as well as normal Vero cells (Fig. 5A). However, the wild-type virus formed only small plaques on the pUL51-EGFP-expressing cells (Fig. 5B). This effect is specific for the expression of the pUL51-EGFP fusion, since the expression of wild-type unfused pUL51 did not inhibit spread (Fig. 2D). This further shows that virus replication and spread functions for pUL51 can be distinguished genetically and suggests that the pUL51-EGFP construct is a specific dominant negative inhibitor of the CCS function of pUL51.
FIG 5.
Growth, release, and spread of HSV-1(F) on pUL51-EGFP-expressing cells. (A) Single-step growth and supernatant virus curves for HSV-1(F) on Vero cells (circles) and a stably transfected clonal Vero cell line that expresses pUL51-EGFP in response to infection. (B) Sizes of plaques formed by HSV-1(F) on Vero or pUL51-EGFP-expressing cells. Horizontal bars indicate the median plaque sizes. Data from one of three representative experiments are shown. The difference in plaque sizes is significant, with a P value of <0.001 determined by using a Kolmogorov-Smirnov test.
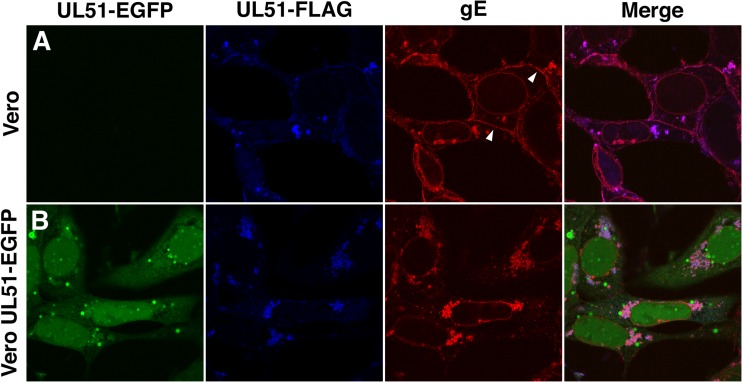
The degree of inhibition of spread seen in cells that express pUL51-EGFP is similar to that previously reported for deletions of the US8 gene, which encodes gE (4, 5, 25), suggesting that mutation of UL51 might interfere with gE function. We therefore tested for disruptions of two other correlates of gE function: localization at cell junctions and support of syncytium formation. gE function in epithelial cell spread is correlated with its ability to localize to cell junctions. To test the hypothesis that pUL51-EGFP might disrupt gE function, we determined the localization of pUL51-EGFP, pUL51-FLAG, and gE in Vero and pUL51-EGFP-expressing cells infected with the UL51-FLAG virus (Fig. 6). In normal Vero cells, gE is concentrated in several locations, including the nuclear envelope and cytoplasmic membrane aggregates, and at cell junctions (Fig. 6A, white arrowheads). pUL51-FLAG localizes in the same cytoplasmic membrane aggregates as gE, but it does not concentrate as gE does at either the nuclear membrane or cell junctions. This localization of pUL51 is consistent with its previously reported localization to Golgi membranes in transfected cells (26). In contrast to pUL51-FLAG, most pUL51-EGFP is found dispersed in both the cytoplasm and nucleoplasm and lining small spherical membranes in the cytoplasm, although some is found in cytoplasmic membrane aggregates, where it colocalizes with pUL51-FLAG and gE (Fig. 6B). Interestingly, while gE is still concentrated on the nuclear envelope and in cytoplasmic membranes in pUL51-EGFP-expressing cells, it no longer concentrates at cellular junctions (compare red staining in Fig. 6A and B), suggesting that the expression of pUL51-EGFP interferes with gE localization and thereby with the spread function of gE.
FIG 6.
Change in gE localization in pUL51-EGFP-expressing cells. Localizations of pUL51-EGFP, pUL51-FLAG, and gE were determined 16 h after infection of Vero (A) or pUL51-EGFP-expressing (B) cells with the UL51-FLAG virus. pUL51-FLAG was detected with anti-FLAG antibody (blue), and gE was detected with mouse monoclonal anti-gE (red). Arrowheads point to sites of gE staining at cell junctions.
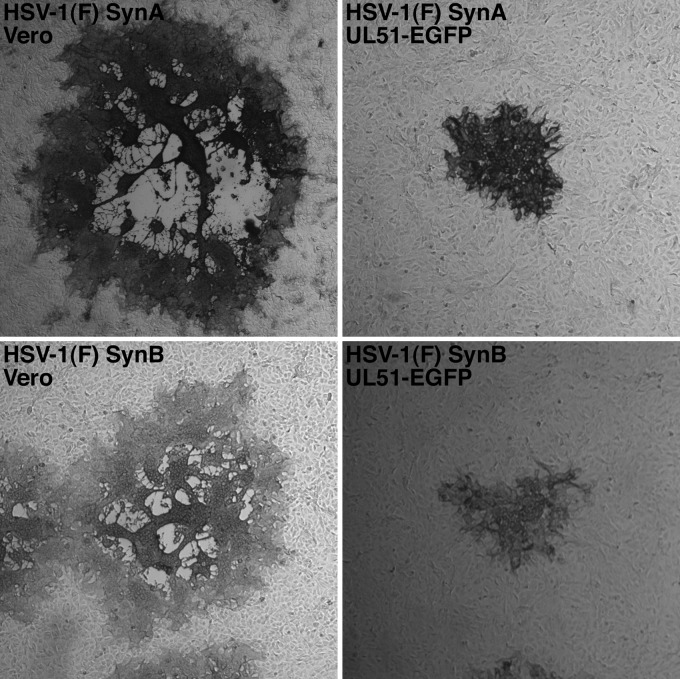
HSV-1 gE function is required for syncytium formation by viral syncytial mutants (3, 16). To determine whether this function of gE is disrupted in pUL51-EGFP-expressing cells, we isolated 12 syncytial variants of HSV-1(F) and tested for their ability to form syncytial plaques on Vero and UL51-EGFP-expressing cells. Two examples are shown in Fig. 7. On Vero cells, the 12 syncytial variants showed variable syncytial plaque morphology, ranging from plaques that were collections of small syncytia to plaques in which all of the cells were apparently fused into a single syncytium (Fig. 7, left). None of the syncytial variants were able to form a syncytial plaque on the UL51-EGFP-expressing cell line (Fig. 7, right), instead forming smaller plaques consisting of rounded cells only, suggesting that gE function in syncytium formation may also be impaired by the expression of pUL51-EGFP.
FIG 7.
Morphology of syncytial HSV-1(F) variants on Vero and pUL51-EGFP-expressing cells. Representative plaques immunostained by using an anti-gD monoclonal antibody are shown.
pUL51 interacts with gE.
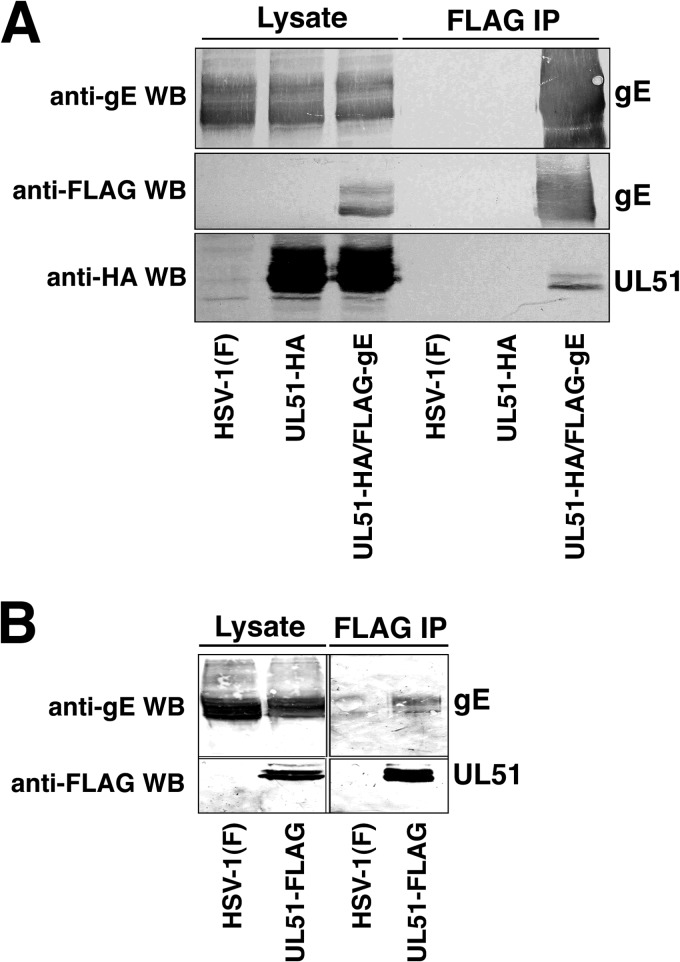
The observations that gE and pUL51 partially colocalize and that expression of a pUL51-EGFP fusion disrupts gE localization suggested that pUL51 and gE might physically interact. We constructed recombinant viruses carrying affinity purification tags on either gE, pUL51, or both to allow efficient purification and asked whether the proteins were copurified from infected cells (Fig. 8). gE was FLAG tagged by the insertion of a FLAG epitope-coding sequence immediately following the signal peptide cleavage site so that mature gE was tagged at its N terminus. We found that the addition of the tag did not change gE localization or the ability to support the formation of wild-type-sized plaques (data not shown). In the dually tagged virus, gE was FLAG tagged and pUL51 was HA tagged at the C terminus. Purification of FLAG-tagged gE resulted in the copurification of HA-tagged pUL51 (Fig. 8A), and in the reciprocal experiment, purification of FLAG-tagged pUL51 resulted in the copurification of untagged gE (Fig. 8B), suggesting that gE and pUL51 can form a complex in infected cells. Other abundant virion proteins, including VP16 and gD, did not copurify with pUL51 (not shown).
FIG 8.
Copurification of gE and pUL51. Images of Western blots are shown. (A) Flag-tagged gE was purified from lysates of Vero cells infected with the indicated viruses using anti-FLAG magnetic beads, and samples of the unfractionated lysates and of the purified proteins were separated by SDS-PAGE, blotted onto nitrocellulose, and probed as indicated at the left. (B) Same as panel A except that FLAG-tagged pUL51 was purified.
UL51 mutant spread phenotypes cannot be accounted for by defects in gE function.
Both gE and pUL51 are required for efficient CCS, and one hypothesis to explain the relationship might be that the sole function of pUL51 is to ensure the proper localization and function of gE. If so, the effect of mutations in UL51 on CCS could never be larger than that of a deletion of gE. To test this hypothesis, we generated two independently isolated recombinant viruses in which amino acids 1 to 335 of gE were deleted and compared their spread phenotype with that of our UL51Δ73–244 mutant. As expected, the gE-null viruses did not express detectable gE and could be amplified to high titers on noncomplementing cells (not shown). They also formed small plaques that were about one-fourth of the size of the plaques of the wild-type virus on Vero cells (Fig. 9). They formed much larger plaques, however, than those formed by the UL51Δ73–244 mutant, suggesting that pUL51 has one or more functions in CCS that do not rely on gE expression.
FIG 9.
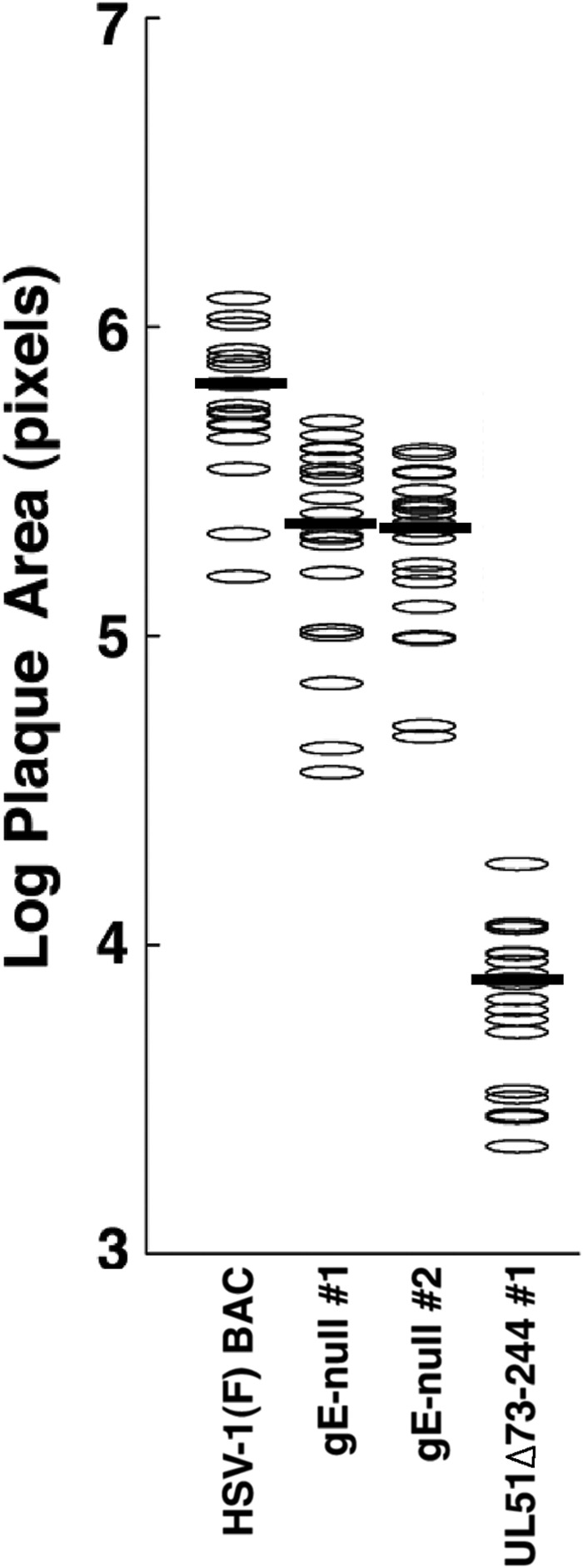
Comparison of spread phenotypes of gE and UL51 deletions. Plaques formed by each of the indicated viruses on Vero cells were measured and plotted as described in the legend of Fig. 2. Dark bars represent the median plaque size. The difference between the HSV-1(F) BAC and the gE-null viruses was significant, with a P value of <0.001.
DISCUSSION
UL51 is conserved in all herpesviruses, and its function has been examined in several herpesvirus systems. It is reported to be a virion tegument component and to localize to cellular membranes (26–28). In cells that transiently express pUL51 from a plasmid, pUL51 localizes to the Golgi apparatus, whereas in infected cells, pUL51 localizes to both Golgi and non-Golgi cytoplasmic membranes, suggesting that other factors in infected cells influence its localization (26). Membrane association of pUL51 requires its palmitoylation at a cysteine located at position 9 (26). Since there is no signal sequence, and since pUL51 is found in the tegument of the mature virion, pUL51 is likely displayed on the exterior of cytoplasmic membranes. From this position, it could participate in both virion assembly and vesicular trafficking interactions. In HSV-1, PrV, and HCMV, where recombinant viruses have been used to explore the function of pUL51 or its homolog pUL71, mutant phenotypes have indicated an important function in virus assembly at the point of secondary envelopment of capsids in the cytoplasm (14, 15, 17, 18). All of the mutant viruses previously studied showed small-plaque phenotypes as well, consistent with a role in CCS.
Here we show that partial deletion of HSV-1 UL51 results in a small-plaque phenotype that cannot be accounted for by single-step growth or release defects in two different cell lines. While the UL51Δ73–244 mutant does have both growth and release defects on Vero cells, it achieves final titers and release efficiencies similar to those obtained by a UL51-FLAG virus but forms plaques almost 100-fold smaller (Fig. 2). On HEp-2 cells, there is a smaller CCS defect but no significant growth or release defect. Furthermore, the CCS function of pUL51 can be specifically inhibited in Vero cells by the expression of a pUL51-EGFP fusion (Fig. 3).
While pUL51 evidently facilitates CCS in different cell types, the mechanism apparently differs to some extent. The highly conserved YXXΦ motif found near the N terminus of pUL51 is critical for CCS function in HEp-2 cells, since mutation of this motif results in a CCS defect comparable to that caused by a deletion of most of the protein. The same effect is not seen in Vero cells, where the plaques formed by the YXXΦ mutant are not significantly smaller than those formed by the UL51-FLAG control virus. YXXΦ motifs function by binding to the μ subunits of adapter protein (AP) complexes that direct coated-vesicle trafficking to a variety of cellular destinations, including the basolateral plasma membrane from the trans-Golgi network and endosomes (reviewed in reference 29). In HEp-2 cells, it is possible that the transport of virions to junctional surfaces of cells for CCS is mediated in part by the binding of one of the AP complexes to pUL51. Interestingly, a number of viral proteins have YXXΦ motifs, including the cytoplasmic domains of gE and gB (30, 31), raising the possibility that redundant signals can help mediate CCS.
The UL51Δ73–244 viruses exhibit better single-step growth than that seen by Nozawa et al. for a UL51 deletion that could express only the first 42 amino acids (15). The UL51Δ73–244 virus single-step growth kinetics are more similar to those shown by the PrV deletion described previously by Klupp et al., which allowed expression of the first 62 amino acids of pUL51 (14). Klupp et al. could not detect truncated pUL51 in Western blots, and we would not expect to have detected a truncated pUL51 because our antiserum was raised against C-terminal sequences. However, one possible explanation for the difference between our results and those of Nozawa et al. is that a truncated product is expressed and retains some of the UL51 function needed for single-step growth.
Studies of gE/gI function in HSV suggest that epithelial CCS has at least two specific components. Several lines of evidence suggest that one component is the engagement of cellular receptors at cell junctions by gE/gI. Mutations in the extracellular domain of gE can specifically inhibit CCS, consistent with a role in binding to cell surface factors (32). The extracellular domain of gE can concentrate at cellular junctions, suggesting that it binds to cellular factors there (10). Expression of truncated gE that lacked the cytoplasmic tail inhibited epithelial CCS, suggesting that this truncated construct might compete for binding to host cell receptors for CCS (33).
Trafficking of mature virions and perhaps other viral factors to junctional surfaces of cells probably forms a second component of CCS. Even in nonpolarized Vero cells, nascent virions are not released uniformly from the surface of the cell but rather are released from specialized sites on the basal surface where viral glycoproteins are concentrated (34), indicating that virions are specifically trafficked from the site of secondary envelopment. CCS probably relies on specific trafficking to junctional surfaces where the nascent virions are sterically protected from immune effectors, including neutralizing antibody. A gE deletion virus failed to deliver mature virions to the junctional surfaces of epithelial cells (35), and delivery of gE to cell junctions is necessary for CCS (8).
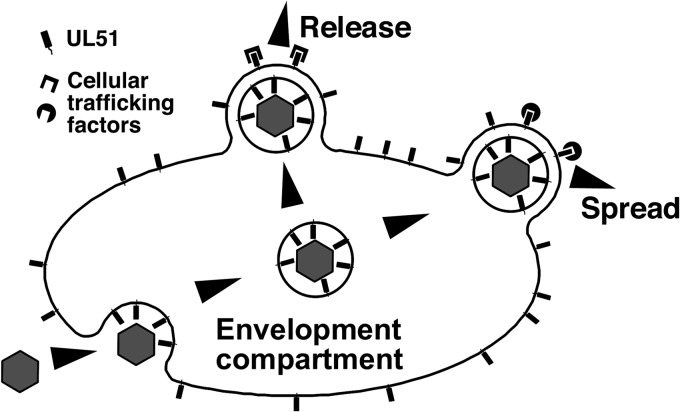
pUL51 is not displayed on the surface of cells or virions, making it unlikely that it participates directly in the engagement of specialized CCS receptors (26–28). Rather, the localization of pUL51 on the exterior face of the Golgi and other cytoplasmic membranes positions it to participate both in cytoplasmic envelopment and as a marker for trafficking on the exterior of transport vesicles that bud from the envelopment compartment (Fig. 10). Therefore, it seems likely that pUL51 is a critical mediator of trafficking of components for CCS. The effect of the pUL51-EGFP fusion on gE localization suggests that pUL51 participates in gE trafficking to cell junctions. However, the greater severity of the UL51 mutation phenotype on CCS suggests that it is responsible for the trafficking of other critical components as well, possibly including the viral particles themselves. Depending upon the cell type, pUL51 on the exterior of transport vesicles may interact with other viral or cellular cargo adapters to direct those vesicles along pathways for apical release, for delivery to junctional surfaces for CCS, or both.
FIG 10.
Schematic drawing of a possible mechanism for pUL51 function. Exposure of pUL51 on the exterior face of cytoplasmic membranes positions it to participate in multiple functions late in infection. It is positioned to interact with other tegument components to facilitate secondary envelopment. It may also mark the exterior of transport vesicles that bud from the envelopment compartment and interact with cell-specific cargo adapters to facilitate trafficking of virion proteins, including gE, or the virions themselves for CCS or for release.
Several conserved herpesvirus gene products have been implicated in CCS, including pUL34, pUL11, pUL16, pUL21, and now pUL51 (11, 13, 16). For all of these proteins, CCS function is correlated with their effects on gE function and localization. pUL11 binds directly to the gE cytoplasmic tail, where pUL11, pUL16, and pUL21 form a complex that is required for proper gE trafficking to cell junctions (16, 36). Our data suggest that the same may be true for pUL51, although whether all of these proteins form a single complex on the gE tail is not clear at this time.
Both epithelial CCS and spread from neurons apparently require specialized virion-sorting mechanisms, but the viral factors that are required differ. gE/gI is not essential for epithelial CCS in HSV but is apparently crucial for anterograde neuronal spread in both HSV and PrV (6, 37–39). The US9 gene is also required for efficient transneuronal spread but has little or no effect on epithelial CCS (40–45). Both proteins function in transneuronal spread by promoting trafficking of virion components into axons (44, 46–48). In PrV infection, both gE/gI and US9 promote interactions of virion components with transport motors via KIF1A (49). pUL51 may have a complementary trafficking function in epithelial CCS in that it is clearly crucial for CCS in some epithelial cell lines. While its function in neuronal spread has not been thoroughly explored, partial deletion of PrV UL51 has only a minor effect on neuroinvasiveness, suggesting that pUL51 does not play a critical role in spread between neurons (50).
The HCMV UL71 protein is required for efficient cytoplasmic virion assembly and may also be required for spread (17, 18). The secondary envelopment function of pUL71 is tied to leucine zipper-dependent oligomerization of the protein (51). The leucine zipper motif is not, however, well conserved among the herpesviruses and is not present in HSV or PrV pUL51 proteins, suggesting that either pUL51 oligomerization is unnecessary in alphaherpesviruses or it is mediated by other structural features of the protein.
ACKNOWLEDGMENTS
We are grateful to Harvey Friedman for the gift of anti-gE antiserum, to Gary Cohen and Roselyn Eisenberg for anti-gD monoclonal antibody, to Keith Jarosinski for helpful discussion and critical reading of the manuscript, and to students of the animal viruses laboratory course for assistance in recombinant BAC construction.
This work was supported by NIH grants AI097212 (R.J.R.) and AI52341 (J.D.B.).
Footnotes
Published ahead of print 22 January 2014
REFERENCES
- 1.Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325:417–470. 10.1007/978-3-540-77349-8_23 [DOI] [PubMed] [Google Scholar]
- 2.Perera RA, Samaranayake LP, Tsang CS. 2010. Shedding dynamics of Epstein-Barr virus: a type 1 carcinogen. Arch. Oral Biol. 55:639–647. 10.1016/j.archoralbio.2010.06.009 [DOI] [PubMed] [Google Scholar]
- 3.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gI or the putative gJ. J. Gen. Virol. 75:1245–1258. 10.1099/0022-1317-75-6-1245 [DOI] [PubMed] [Google Scholar]
- 4.Dingwell KS, Brunetti CR, Hendricks RL, Tang Q, Tang M, Rainbow AJ, Johnson DC. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dingwell KS, Johnson DC. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933–8942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGraw HM, Awasthi S, Wojcechowskyj JA, Friedman HM. 2009. Anterograde spread of herpes simplex virus type 1 requires glycoprotein E and glycoprotein I but not Us9. J. Virol. 83:8315–8326. 10.1128/JVI.00633-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson DC, Frame MC, Ligas MW, Cross AM, Stow ND. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J. Virol. 62:1347–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farnsworth A, Johnson DC. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J. Virol. 80:3167–3179. 10.1128/JVI.80.7.3167-3179.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMillan TN, Johnson DC. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928–1940. 10.1128/JVI.75.4.1928-1940.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wisner T, Brunetti C, Dingwell K, Johnson DC. 2000. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J. Virol. 74:2278–2287. 10.1128/JVI.74.5.2278-2287.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haugo AC, Szpara ML, Parsons L, Enquist LW, Roller RJ. 2011. Herpes simplex virus type 1 pUL34 plays a critical role in cell-to-cell spread of virus in addition to its role in virus replication. J. Virol. 85:7203–7215. 10.1128/JVI.00262-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duffy C, LaVail JH, Tauscher AN, Wills EG, Blaho JA, Baines JD. 2006. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J. Virol. 80:8664–8675. 10.1128/JVI.00498-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J, Chadha P, Meckes DG, Jr, Baird NL, Wills JW. 2011. Interaction and interdependent packaging of tegument protein UL11 and glycoprotein E of herpes simplex virus. J. Virol. 85:9437–9446. 10.1128/JVI.05207-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klupp BG, Granzow H, Klopfleisch R, Fuchs W, Kopp M, Lenk M, Mettenleiter TC. 2005. Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79:3831–3840. 10.1128/JVI.79.6.3831-3840.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nozawa N, Kawaguchi Y, Tanaka M, Kato A, Kato A, Kimura H, Nishiyama Y. 2005. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of virus particles. J. Virol. 79:6947–6956. 10.1128/JVI.79.11.6947-6956.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han J, Chadha P, Starkey JL, Wills JW. 2012. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc. Natl. Acad. Sci. U. S. A. 109:19798–19803. 10.1073/pnas.1212900109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schauflinger M, Fischer D, Schreiber A, Chevillotte M, Walther P, Mertens T, von Einem J. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J. Virol. 85:3821–3832. 10.1128/JVI.01540-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Womack A, Shenk T. 2010. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. mBio 1(5):e00282–10. 10.1128/mBio.00282-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roller RJ, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117–129. 10.1128/JVI.74.1.117-129.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ejercito PM, Kieff ED, Roizman B. 1968. Characteristics of herpes simplex virus strains differing in their effect on social behavior of infected cells. J. Gen. Virol. 2:357–364. 10.1099/0022-1317-2-3-357 [DOI] [PubMed] [Google Scholar]
- 21.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. 10.2144/000112096 [DOI] [PubMed] [Google Scholar]
- 22.Bjerke SL, Cowan JM, Kerr JK, Reynolds AE, Baines JD, Roller RJ. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601–7610. 10.1128/JVI.77.13.7601-7610.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803–8817. 10.1128/JVI.75.18.8803-8817.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, Jodicke L, Dammert MA, Schroeter C, Hammer M, Schmidt T, Jehl P, McGuigan C, Dymecka M, Chica C, Luck K, Via A, Chatr-Aryamontri A, Haslam N, Grebnev G, Edwards RJ, Steinmetz MO, Meiselbach H, Diella F, Gibson TJ. 2012. ELM—the database of eukaryotic linear motifs. Nucleic Acids Res. 40:D242–D251. 10.1093/nar/gkr1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weeks BS, Sundaresan P, Nagashunmugam T, Kang E, Friedman HM. 1997. The herpes simplex virus-1 glycoprotein E (gE) mediates IgG binding and cell-to-cell spread through distinct gE domains. Biochem. Biophys. Res. Commun. 235:31–35 [DOI] [PubMed] [Google Scholar]
- 26.Nozawa N, Daikoku T, Koshizuka T, Yamauchi Y, Yoshikawa T, Nishiyama Y. 2003. Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J. Virol. 77:3204–3216. 10.1128/JVI.77.5.3204-3216.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. 1998. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J. Gen. Virol. 79:3027–3031 [DOI] [PubMed] [Google Scholar]
- 28.Lenk M, Visser N, Mettenleiter TC. 1997. The pseudorabies virus UL51 gene product is a 30-kilodalton virion component. J. Virol. 71:5635–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Boulan E, Musch A. 2005. Protein sorting in the Golgi complex: shifting paradigms. Biochim. Biophys. Acta 1744:455–464. 10.1016/j.bbamcr.2005.04.007 [DOI] [PubMed] [Google Scholar]
- 30.Beitia Ortiz de Zarate I, Kaelin K, Rozenberg F. 2004. Effects of mutations in the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B on intracellular transport and infectivity. J. Virol. 78:1540–1551. 10.1128/JVI.78.3.1540-1551.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alconada A, Bauer U, Sodeik B, Hoflack B. 1999. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J. Virol. 73:377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polcicova K, Goldsmith K, Rainish BL, Wisner TW, Johnson DC. 2005. The extracellular domain of herpes simplex virus gE is indispensable for efficient cell-to-cell spread: evidence for gE/gI receptors. J. Virol. 79:11990–12001. 10.1128/JVI.79.18.11990-12001.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collins WJ, Johnson DC. 2003. Herpes simplex virus gE/gI expressed in epithelial cells interferes with cell-to-cell spread. J. Virol. 77:2686–2695. 10.1128/JVI.77.4.2686-2695.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mingo RM, Han J, Newcomb WW, Brown JC. 2012. Replication of herpes simplex virus: egress of progeny virus at specialized cell membrane sites. J. Virol. 86:7084–7097. 10.1128/JVI.00463-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson DC, Webb M, Wisner TW, Brunetti C. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 75:821–833. 10.1128/JVI.75.2.821-833.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farnsworth A, Wisner TW, Johnson DC. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81:319–331. 10.1128/JVI.01842-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dingwell KS, Doering LC, Johnson DC. 1995. Glycoproteins E and I facilitate neuron-to-neuron spread of herpes simplex virus. J. Virol. 69:7087–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Husak PJ, Kuo T, Enquist LW. 2000. Pseudorabies virus membrane proteins gI and gE facilitate anterograde spread of infection in projection-specific neurons in the rat. J. Virol. 74:10975–10983. 10.1128/JVI.74.23.10975-10983.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulder WA, Jacobs L, Priem J, Kok GL, Wagenaar F, Kimman TG, Pol JM. 1994. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and trigeminal routes in the porcine central nervous system. J. Gen. Virol. 75:3095–3106. 10.1099/0022-1317-75-11-3095 [DOI] [PubMed] [Google Scholar]
- 40.Brideau AD, Card JP, Enquist LW. 2000. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J. Virol. 74:834–845. 10.1128/JVI.74.2.834-845.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longnecker R, Roizman B. 1987. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science 236:573–576. 10.1126/science.3033823 [DOI] [PubMed] [Google Scholar]
- 42.Lyman MG, Demmin GL, Banfield BW. 2003. The attenuated pseudorabies virus strain Bartha fails to package the tegument proteins Us3 and VP2. J. Virol. 77:1403–1414. 10.1128/JVI.77.2.1403-1414.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishiyama Y, Kurachi R, Daikoku T, Umene K. 1993. The Us9, 10, 11, and 12 genes of herpes simplex virus type 1 are of no importance for its neurovirulence and latency in mice. Virology 194:419–423. 10.1006/viro.1993.1279 [DOI] [PubMed] [Google Scholar]
- 44.Snyder A, Polcicova K, Johnson DC. 2008. Herpes simplex virus gE/gI and US9 proteins promote transport of both capsids and virion glycoproteins in neuronal axons. J. Virol. 82:10613–10624. 10.1128/JVI.01241-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomishima MJ, Enquist LW. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741–752. 10.1083/jcb.200011146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ch'ng TH, Enquist LW. 2005. Efficient axonal localization of alphaherpesvirus structural proteins in cultured sympathetic neurons requires viral glycoprotein E. J. Virol. 79:8835–8846. 10.1128/JVI.79.14.8835-8846.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyman MG, Feierbach B, Curanovic D, Bisher M, Enquist LW. 2007. Pseudorabies virus Us9 directs axonal sorting of viral capsids. J. Virol. 81:11363–11371. 10.1128/JVI.01281-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howard PW, Howard TL, Johnson DC. 2013. Herpes simplex virus membrane proteins gE/gI and US9 act cooperatively to promote transport of capsids and glycoproteins from neuron cell bodies into initial axon segments. J. Virol. 87:403–414. 10.1128/JVI.02465-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kratchmarov R, Kramer T, Greco TM, Taylor MP, Ch'ng TH, Cristea IM, Enquist LW. 2013. Glycoproteins gE and gI are required for efficient KIF1A-dependent anterograde axonal transport of alphaherpesvirus particles in neurons. J. Virol. 87:9431–9440. 10.1128/JVI.01317-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klopfleisch R, Klupp BG, Fuchs W, Kopp M, Teifke JP, Mettenleiter TC. 2006. Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice. J. Virol. 80:5571–5576. 10.1128/JVI.02589-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meissner CS, Suffner S, Schauflinger M, von Einem J, Bogner E. 2012. A leucine zipper motif of a tegument protein triggers final envelopment of human cytomegalovirus. J. Virol. 86:3370–3382. 10.1128/JVI.06556-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]