ABSTRACT
The Gag protein of the murine retrovirus mouse mammary tumor virus (MMTV) orchestrates the assembly of immature virus particles in the cytoplasm which are subsequently transported to the plasma membrane for release from the cell. The morphogenetic pathway of MMTV assembly is similar to that of Saccharomyces cerevisiae retrotransposons Ty1 and Ty3, which assemble virus-like particles (VLPs) in intracytoplasmic ribonucleoprotein (RNP) complexes. Assembly of Ty1 and Ty3 VLPs depends upon cellular mRNA processing factors, prompting us to examine whether MMTV utilizes a similar set of host proteins to facilitate viral capsid assembly. Our data revealed that MMTV Gag colocalized with YB-1, a translational regulator found in stress granules and P bodies, in intracytoplasmic foci. The association of MMTV Gag and YB-1 in cytoplasmic granules was not disrupted by cycloheximide treatment, suggesting that these sites were not typical stress granules. However, the association of MMTV Gag and YB-1 was RNA dependent, and an MMTV RNA reporter construct colocalized with Gag and YB-1 in cytoplasmic RNP complexes. Knockdown of YB-1 resulted in a significant decrease in MMTV particle production, indicating that YB-1 plays a role in MMTV capsid formation. Analysis by live-cell imaging with fluorescence recovery after photobleaching (FRAP) revealed that the population of Gag proteins localized within YB-1 complexes was relatively immobile, suggesting that Gag forms stable complexes in association with YB-1. Together, our data imply that the formation of intracytoplasmic Gag-RNA complexes is facilitated by YB-1, which promotes MMTV virus assembly.
IMPORTANCE Cellular mRNA processing factors regulate the posttranscriptional fates of mRNAs, affecting localization and utilization of mRNAs under normal conditions and in response to stress. RNA viruses such as retroviruses interact with cellular mRNA processing factors that accumulate in ribonucleoprotein complexes known as P bodies and stress granules. This report shows for the first time that mouse mammary tumor virus (MMTV), a mammalian retrovirus that assembles intracytoplasmic virus particles, commandeers the cellular factor YB-1, a key regulator of translation involved in the cellular stress response. YB-1 is essential for the efficient production of MMTV particles, a process directed by the viral Gag protein. We found that Gag and YB-1 localize together in cytoplasmic granules. Functional studies of Gag/YB-1 granules suggest that they may be sites where virus particles assemble. These studies provide significant insights into the interplay between mRNA processing factors and retroviruses.
INTRODUCTION
Retroviruses are single-stranded, positive-sense RNA viruses that encode a limited genome and therefore depend on cellular factors to facilitate virus replication. During early infection, retroviral genomic RNA is reverse transcribed into double-stranded DNA, which is integrated into the host chromosome. Synthesis of viral RNA is mediated by RNA polymerase II, and viral transcripts are processed just like cellular mRNAs, with the addition of a 5′ methyl guanosine cap and a 3′ poly(A) tail. A subset of the nascent viral RNA is spliced, but the majority remains unspliced and performs two essential roles, serving as an mRNA for translation of the viral structural proteins Gag and Gag-Pol and as genomic RNA for encapsidation into virus particles. Retroviruses efficiently package full-length RNAs that are not translated, suggesting that there might be a mechanism in which unspliced genomic RNA is sequestered from translation machinery for encapsidation into virions (reviewed in reference 1).
Mouse mammary tumor virus (MMTV) is an oncogenic betaretrovirus that exhibits B/D-type morphogenesis, assembling immature viral capsids in the cytoplasm of infected cells (2). Once assembled, immature capsids are transported to the plasma membrane where they are enveloped and released from the cell. The genesis of MMTV virions is directed by the Gag polyprotein, which is composed of seven distinct domains that are proteolytically cleaved: MA, or matrix, the plasma membrane-targeting domain (3); pp21, a phosphoprotein of unknown function; p3, p8, and n, involved in Gag-Gag interactions required for immature capsid assembly (4, 5); CA, or capsid, which forms the protein core of the mature virion; and NC, or nucleocapsid, which binds to the viral genomic RNA for encapsidation into virions (reviewed in reference 6).
Most mammalian retroviruses, including human immunodeficiency virus (HIV), follow the C-type morphogenesis pathway in which the virus core is formed at the plasma membrane late in the assembly process (reviewed in reference 7). In C-type retroviral morphogenesis, cytoplasmic Gag-RNA complexes are lower-order oligomers, making viral RNP complexes difficult to study using conventional microscopy (8). However, B/D-type retroviruses such as MMTV provide an opportunity to study the properties of early viral RNP complexes because complete immature capsids form within cytoplasmic granules that are readily visualized (2). The B/D-type retrovirus assembly pathway shares common features with the Saccharomyces cerevisiae long terminal repeat (LTR) retrotransposons Ty1 and Ty3, which assemble virus-like particles (VLPs) composed of Gag and RNA in the cytoplasm. To identity cellular factors that contribute to the formation of retrotransposon RNP complexes, genome-wide studies of proteins affecting Ty1 and Ty3 replication were performed in yeast. In these studies, several mRNA processing factors with roles in posttranscriptional regulation were found that colocalized with Gag, modulated retrotransposition, and were required for VLP formation (9–11). Many of the host factors essential for Ty1 and Ty3 replication are mRNA binding proteins that accumulate in P bodies and stress granules, cytoplasmic RNP complexes that sequester nontranslating mRNAs (12–23).
P bodies exist constitutively in cells and are proposed to be sites where translationally repressed mRNAs are sorted for storage, reassociation with polysomes, or degradation (12, 16–18, 23–26). In contrast, stress granules form only in response to cellular stressors, including heat shock (HS), oxidative stress, UV irradiation, and virus infection (27–29). Core constituents of stress granules include T-cell restricted intracellular antigen (TIA-1) and Ras-GAP SH3 domain binding protein 1 (G3BP1), with overexpression of either inducing stress granule formation (27, 30, 31). TIA-1 is an mRNA binding protein that recruits nontranslating poly(A)-containing RNAs to stress granules and initiates stress granule formation through a homo-oligomerization domain (30). G3BP1 contains two RNA recognition motifs and possesses RNase activity; multimerization of G3BP1 is a key step in stress granule formation in response to cellular stress (31). Cellular mRNAs and proteins traffic transiently in and out of stress granules, indicating that mRNAs are not stored there; instead, mRNPs in stress granules undergo dynamic exchange with polysomes (22). Specific stress granule and P-body factors that play a positive role in Ty1 and Ty3 assembly and retrotransposition include Dhh1/Rck/p54, Edc3, Dcp2, Pat1, Kem1/Xrn1, and Lsm1 (9–11). Ty3 interacts with P-body and stress granule proteins in so-called “retrosomes,” cytoplasmic sites that are distinct from stress granules or P bodies but nonetheless contain many of the mRNA processing factors typically found in stress granules and P bodies (11).
YB-1, a major component of cellular mRNPs, serves as a key translational regulator in mammalian cells and accumulates in stress granules following cellular stress (32). YB-1 is essential for viability in mice, but because there is no ortholog in Saccharomyces cerevisiae, it does not play a role in Ty1 or Ty3 assembly (17, 33). YB-1 is not considered a core component of stress granules, and accordingly, overexpression of YB-1 does not typically induce stress granule formation. Although YB-1 moves into stress granules during cellular stress, it is also a component of P bodies (alternatively called GW bodies) (17, 34); therefore small YB-1 RNPs are constitutive and play a central role in mRNA utilization in nonstressed cells (35). Although YB-1 does have antiviral activity in some RNA viruses (36), the retroviruses HIV-1 and murine leukemia virus (MLV) were recently reported to utilize YB-1 to stabilize viral RNA and increase virus production (37, 38).
Here, we report that YB-1 interacts with MMTV Gag in an RNA-dependent manner within small cytoplasmic RNP complexes distinct from stress granules. The biological relevance of the Gag–YB-1 interaction was demonstrated by knockdown of YB-1, which significantly impaired virus production without affecting intracellular Gag protein levels. Using a fluorescence recovery after photobleaching (FRAP)-based assay to study Gag dynamics, we identified two functionally distinct Gag-containing cytoplasmic complexes: (i) G3BP1-induced stress granules that contained highly mobile Gag molecules and (ii) YB-1 foci containing complexes of immobile Gag proteins, conceivably located within assembled immature capsids. Finally, in contrast to HIV-1 and human T-cell leukemia virus type 1 (HTLV-1), MMTV infection did not inhibit stress granule formation. Instead, MMTV-infected cells appeared to be “primed” to form large RNPs more readily as overexpression of YB-1 induced formation of cytoplasmic granules at a significantly higher frequency in infected cells than in uninfected cells. Together, our results suggest that MMTV has taken advantage of the mRNA regulatory pathway by co-opting YB-1 to facilitate virus production.
MATERIALS AND METHODS
Cells, plasmids, and transfections.
Uninfected NMuMG (normal murine mammary gland) cells (ATCC CRL-1636), MMTV strain C3H [MMTV(C3H)]-infected NMuMG (NMuMG.C3H) cells (kindly provided by Tatyana Golovkina, University of Chicago) (39, 40), and QT6 (quail fibroblast) (41) cells were maintained in culture as previously described (2, 42). Rous sarcoma virus (RSV)-infected QT6 cells were established by transfection of the pRC.V8 molecular clone (42). Plasmids expressing myc–YB-1, myc-HuR, and myc-PABPc1 were a gift from Michael Malim (Kings College, London, England) (43). The yellow fluorescent protein (YFP)–TIA-1 plasmid was kindly provided by Nancy Kedersha (Harvard University) (43) and pGFP-G3BP1 (where GFP is green fluorescent protein) was a gift from Jamal Tazi (Institut Génétique Moléculaire, Montpellier, France) (31). The pSrprb-mRFP-4BoxB (where mRFP is monomeric red fluorescent protein and 4BoxB is four BoxB stem-loops) and λN-enhanced GFP (eGFP) constructs were a gift from Jan Ellenberg (European Molecular Biology Laboratory, Heidelberg, Germany). The MMTV pGag-GFP and pGag-mCherry constructs were described previously (2).
The pGagΔNC-mCherry plasmid was created by PCR amplification of the mCherry gene and replacing the GFP sequence in pMA-CA.Gag-GFP (2) using BamHI-NotI. The CREB1 leucine zipper domain coding sequence was PCR amplified from RSV pGag.Zip-GFP (44) and inserted into pGagΔNC-mCherry using EcoRI-BamHI to create pGag.Zip-mCherry. To create the pGag.pp21.imCherry construct, we first created pGag-4×CTE (with four copies of the constitutive transport element [CTE]) by inserting MMTV Gag in place of HIV-1 Gag in pGPV-4×CTE (45) (a kind gift from Michael Malim, Kings College, London) using EcoRI-BamHI. The PflMI site within mCherry was deleted through a silent mutation using site-directed mutagenesis, and the resultant mCherry gene was inserted between the seventh and eighth codons of pp21 using splicing by overlap extension (46). The resulting fragment was then inserted into pGag-4×CTE using SacI-PflMI sites present within MMTV Gag. PCR primer sequences are available upon request. NMuMG cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, whereas QT6 cells were transfected using the calcium phosphate method (42).
The subviral RNA (svRNA) construct was based on our previously described MMTV Gag-GFP construct (2). The R-U5 region of the MMTV 5′ untranslated region (UTR) was PCR amplified from NMuMG.C3H total cellular DNA using the following primers: 5′-ATC GAT AGA TCT GCA ACA GTC CTA ACA TTC ACC and 5′-ATC GAT GAT ATC TAC CTC TTC TCC GTA GGC G. The PCR-amplified fragment was inserted into BglIII and EcoRV sites in Gag-GFP to create 5′ UTR Gag-GFP intermediate 1. The EcoRV cloning site between the 5′ UTR and the Gag initiation codon was eliminated by QuikChange mutagenesis using primers 5′-CCG CCT ACG GAG AAG AGG TAG GTT ACG GTG A and 5′-TCA CCG TAA CCT ACC TCT TCT CCG TAG GCG to create 5′ UTR Gag-GFP intermediate 2.
The MMTV Rem response element (RemRE) was PCR amplified from NMuMG.C3H total cellular DNA using primers 5′-ATC GAT GCG GCC GCA GAT CTT AAC GTG CTT CTT TTA AAA and 5′-ATC GAT GCG GCC GCG GAT CCA GTA CTA AAA GAT AAT GAT TCA TTT C and inserted into the NotI site in 5′ UTR Gag-GFP intermediate 2 to create CMV-R-U5-Gag-GFP-RemRE (where CMV is cytomegalovirus). This construct was digested with BsrGI and EcoNI, blunted, and religated to create CMV-R-U5-Δgag-RemRE. To insert BoxB loops into CMV-R-U5-Δgag-RemRE, both this construct and Srprb-mRFP-4BoxB were first transformed into GM272 (dam-3 mutant dcm-6 mutant) E. coli (a gift from David Spector, Penn State College of Medicine) (47). Srprb-mRFP-4BoxB (a gift from Jan Ellenberg, EMBL) was digested with BsrGI, blunted using a Klenow reaction, and digested with XbaI to isolate the BoxB loops. The isolated BoxB loops were inserted into CMV-R-U5-Δgag-RemRE digested with ScaI and XbaI to create MMTV svRNA.
Immunofluorescence and confocal microscopy.
Following transfection, cells were washed twice with phosphate-buffered saline (PBS), fixed in 3.7% paraformaldehyde, permeabilized in 0.25% Triton X-100, and blocked in 10% bovine serum albumin. Samples were immunostained using rabbit anti-myc (Ab9106; Abcam), rabbit anti-YB-1 (Ab12148; Abcam), rabbit anti-TIA-1 (Ab40693; Abcam), and/or mouse monoclonal anti-CA (a gift from Tatyana Golovkina, University of Chicago) primary antibodies followed by appropriate anti-rabbit-Cy3 (C2306; Sigma), anti-rabbit-Cy5 (Ab97077; Abcam), anti-mouse-Cy3 (Sigma C2818), or anti-mouse-fluorescein isothiocyanate (FITC) (F9137; Sigma) secondary antibodies. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) (EMD Chemicals, Inc.) and mounted on glass slides using SlowFade reagent (Invitrogen).
Samples were imaged using a Leica TCS SP2-ABOS confocal microscope (Leica Microsystems, Wetzlar, Germany). Images were obtained using sequential scanning with laser excitations of 405 nm (DAPI), 488 nm (eGFP), 514 nm (YFP), 543 nm (Cy3), or 633 nm (Cy5). Colocalization analysis was conducted using Colocalization Colormap (48) and/or Just Another Colocalization Plugin (JACoP) (49) for ImageJ (National Institutes of Health, Bethesda, MD; http://imagej.nih.gov/ij/). Thresholds were set to approximately the same levels of fluorescence present in the original confocal image. Images were prepared for publication using CorelDrawX3 (Corel Corp., Ottawa, Canada). Any adjustments to intensity were applied to all images in a series.
siRNA knockdowns.
YB-1 or TIA-1 expression was knocked down in NMuMG.C3H cells by transfection with 5 pmol of YB-1-specific small interfering RNAs (siRNAs) (5′-CUGAGUAAAUGCCGGCUUAdTdT and 5′-CGAUCCACCAGCUGAGAAUdTdT [Sigma-Aldrich]) or TIA-1-specific siRNA (5′-CAGCACACAGCGUUCACAAdTdT [Sigma-Aldrich]) for 16 h using Lipofectamine 2000. Control cells were transfected with scrambled siRNA (siRNA Universal Negative Control 1) (SIC001; Sigma-Aldrich). Transfection medium was replaced with primary growth medium for 8 h, and cells were transfected with an additional 5 pmol of YB-1, TIA-1, or control siRNA. To reconstitute YB-1 expression, cells were cotransfected with 2 μg of siRNA-resistant pmyc–YB-1 during the initial siRNA transfection.
Virus budding assays.
The MMTV budding assay was described previously (50). Briefly, MMTV(C3H)-infected NMuMG (NMuMG.C3H) cells were transfected with pYFP-TIA-1, p.EGFP.N2 (mock), YB-1-specific siRNA, TIA-1-specific siRNA, or control siRNA, as described above. Virus particles released into the medium were collected for 3 h, pelleted through a 20% sucrose cushion, and resuspended in SDS-PAGE sample buffer. Proteins present in the corresponding cell lysates and supernatants were separated by SDS-PAGE, and the Gag/CA protein was detected by Western blotting using anti-CA antibody with quantitative imaging of chemiluminescence using an EC3 Chemi HR 410 imaging system (UVP; Upland, CA). Virus budding efficiency was calculated as the amount of CA protein detected in the supernatant (virus, V) divided by the sum of the Gag/CA protein present in the lysate (L) and supernatant [V/(V+L)] (50, 51). Expression levels of YB-1 and TIA-1 (endogenous and epitope-tagged proteins), as determined by Western blotting using anti-YB-1 or anti-TIA-1 antibodies, were compared to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) loading controls using goat and-GAPDH antibody (A00191; GenScript). Western blots were developed using Amersham ECL-Plus reagent (GE Healthcare Life Sciences) or SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and quantitated using the UVP imaging system described above. Budding assays were performed at least eight times in separate transfections conducted on two different days. The mean, standard error of the mean, and P values were calculated using two-tailed paired Student's t tests (GraphPad Prism, version 5; GraphPad Software, Inc.).
Co-IPs.
Coimmunoprecipitations (co-IPs) were performed essentially as described previously (50), with the following changes. Cells were lysed in 50 mM Tris, pH 7.05, 1% Triton X-100, 0.5% deoxycholic acid, and 150 mM NaCl on ice for 15 min; cellular debris was removed by centrifugation, and half of each lysate was treated with 25 μg of RNase A (Invitrogen) for 15 min. Five micrograms of rabbit anti-YB-1, rabbit anti-G3BP1 (Bethyl Laboratories), or rabbit anti-lamin A/C (Santa Cruz Biotechnology) antibody was added to cleared lysates for 1 h at room temperature. Protein A-Sepharose beads (Invitrogen) were washed twice in coimmunoprecipitation buffer (50 mM Tris, pH 8.0, 1% Triton X-100, 0.1% SDS, 1% deoxycholic acid, and 150 mM NaCl), added to each lysate sample, and incubated overnight at 4°C with gentle end-over-end mixing. After being pelleted by centrifugation, beads were washed twice with co-IP buffer and twice with TE buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, pH 8.0), resuspended in SDS-PAGE sample buffer (125 mM Tris-HCl, pH 6.8, 20% glycerol, 0.5% bromophenol blue, 4% SDS, and 10% β-mercaptoethanol), and boiled. Eluted proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Western blotting was performed using anti-CA antibody at a 1:4,000 dilution, followed by goat anti-mouse-horseradish peroxidase conjugate antibody diluted 1:10,000. Chemiluminescent signals were developed using SuperSignal West Pico Substrate (Thermo Scientific).
Fluorescence recovery after photobleaching (FRAP).
Cells were seeded onto 35-mm glass bottom dishes (MatTek Corp.), transfected as above, and imaged 15 to 18 h after transfection using a Leica TCS SP8-ABOS confocal microscope fitted with a 37°C, 5% CO2 live-cell chamber. Samples were imaged using a white light laser tuned to 489 nm for GFP or 580 nm for mCherry. Cells were imaged for approximately 20 s (prebleach); then two circular, 5-μm-diameter regions of interest (ROIs) per cell were subjected to photobleaching for approximately 10 s using a 580-nm white light laser at 100% power. Cells were imaged at 1.91 frames per second for 15 min after bleaching.
Individual foci were selected for analysis in ImageJ by drawing 1.5-μm-diameter ROIs around foci and monitoring the integrated signal intensity over the course of the experiment. To ensure that foci were bleached to at least 30% of the initial fluorescence intensity, data were first normalized by the method of Phair et al. (52) using the following formula in Microsoft Excel:
where is the normalized relative intensity (in arbitrary units [AU]) at time t, is the average total cell prebleach intensity, ACt is the total cell fluorescence intensity at time t, BGt is the background fluorescence intensity at time t, is the average experimental ROI prebleach intensity, and It is the experimental ROI fluorescence intensity at time t. Foci that were not bleached to ≤30% of the normalized initial intensity were discarded from the analysis. To facilitate fitting of the data by GraphPad Prism, the data were subjected to additional full-scale normalization (53) using the following formula:
where is the full-scale normalized value at time t, and is the first postbleach value. Average signal intensity and standard error of the mean for each time point from at least four foci were calculated using Microsoft Excel and fit using single or double exponential association curves using GraphPad Prism.
RESULTS
Constitutive colocalization of MMTV Gag with YB-1.
Previous studies of HIV-1 and HTLV-1 demonstrated that stress granule formation was inhibited in infected cells (54, 55). In contrast, yeast LTR retrotransposons Ty1 and Ty3 assemble RNP complexes in the cytoplasm in conjunction with mRNA processing factors that include orthologs of mammalian stress granule and P-body proteins (10, 11). Therefore, we reasoned that MMTV might utilize a similar set of host factors as Ty1 and Ty3 because, like the yeast retrotransposons, MMTV assembles viral RNPs and complete immature capsids at intracytoplasmic sites. To test this possibility, we began our studies with two constituents of stress granules, YB-1 and TIA-1, using antibodies that could reliably detect these endogenous proteins in normal mouse cells derived from mammary glands (NMuMG cells).
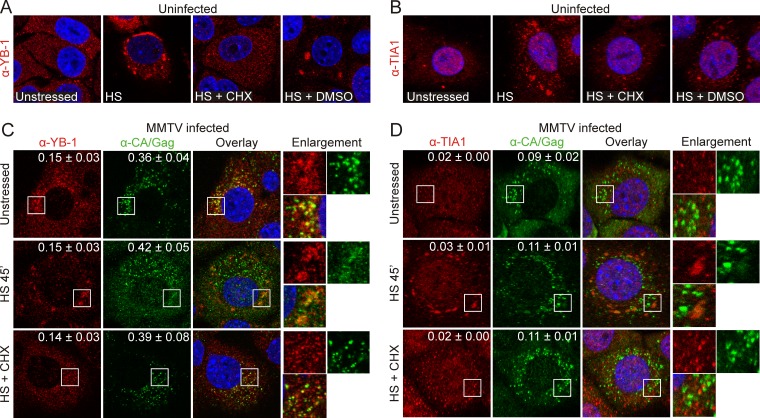
NMuMG cells were immunostained with YB-1 or TIA-1 antibodies and examined using confocal microscopy. In uninfected cells, YB-1 had a speckled cytoplasmic distribution characteristic of its localization within cellular mRNPs (Fig. 1A, unstressed) (17, 33), whereas TIA-1 was predominantly nuclear (Fig. 1B, unstressed) with a slightly punctate cytoplasmic appearance. When cells were subjected to heat shock at 44°C for 45 min to induce stress granules, YB-1 and TIA-1 accumulated in large cytoplasmic aggregates, as previously reported in other cell types (Fig. 1A and B, HS). Stress granule formation depends on the presence of mRNAs that cycle dynamically between stress granules and polysomes (22). Therefore, treating stressed cells with the translational inhibitor cycloheximide (CHX) causes stress granules to disassemble even with continued stress (17). As shown in Fig. 1, when NMuMG cells were heat shocked for 30 min at 44°C and subsequently treated with CHX for 15 min at 44°C, YB-1 and TIA-1 no longer accumulated in cytosolic granules, consistent with CHX-induced disassembly of stress granules (Fig. 1A and B, HS + CHX). In contrast, in cells subjected to the same conditions but treated with vehicle control (dimethyl sulfoxide [DMSO]), YB-1 and TIA-1 remained in cytoplasmic aggregates characteristic of stress granules (Fig. 1A and B, HS + DMSO).
FIG 1.
MMTV Gag colocalizes with endogenous YB-1 in cycloheximide-resistant cytoplasmic complexes. (A) Uninfected NMuMG cells were unstressed, subjected to heat shock at 44°C for 45 min (HS) or heat shocked for 30 min at 44°C and treated with cycloheximide (HS + CHX) or vehicle control (HS + DMSO) for 15 min at 44°C, and then fixed and stained for YB-1. (B) Uninfected NMuMG cells were treated as in described for panel A and immunostained for TIA-1. (C) MMTV(C3H)-infected NMuMG cells were treated as described above and immunostained for YB-1 and Gag. White boxes indicate the areas of enlargement. M1 and M2 coefficients are shown in the upper right-hand corner and were calculated for ≥6 cells using Just Another Colocalization Plugin (JACoP) (49) for ImageJ (http://imagej.nih.gov/ij/). Values are shown as the means ± standard errors of the means. (D) MMTV(C3H)-infected cells treated as described above were immunostained for TIA-1 and Gag. M1 and M2 values were calculated for 10 cells under each condition. α, anti.
We next examined the distribution of endogenous YB-1 and TIA-1 in NMuMG cells chronically infected with the MMTV strain C3H (NMuMG.C3H cells) (39, 40). MMTV capsids were stained using anti-CA antibody. In NMuMG.C3H cells, a population of YB-1 colocalized with a portion of MMTV Gag in small cytoplasmic foci (Fig. 1C, unstressed). Overlap between Gag and YB-1 immunofluorescence was quantitatively assessed using Mander's colocalization analysis, which measures the overall fraction of colocalized pixels in each channel (M1 and M2) of a two-color confocal image (56, 57). Mander's analysis revealed that 15% of the total YB-1 signal overlapped that of Gag (M1 = 0.15 ± 0.03, n = 10 cells) and that 36% of the total Gag fluorescence overlapped that of YB-1 (M2 = 0.36 ± 0.04). These data indicate that Gag colocalizes in small granules with a sizeable portion of the cytosolic YB-1 population. In contrast, TIA-1 and Gag colocalization was markedly lower under identical conditions (Fig. 1D, unstressed) (M1 = 0.02; M2 = 0.09 ± 0.02; n = 10 cells).
Because Gag was associated with YB-1 in small foci in infected cells, we examined whether Gag and YB-1 would traffic into stress granules when cells were stressed with HS. In HS-treated NMuMG.C3H cells, YB-1 and Gag accumulated in stress granules that were much larger than the Gag–YB-1 granules present constitutively in unstressed cells (Fig. 1C, HS 45 min). However, the overall fraction of Gag molecules associated with YB-1 in HS-treated cells was the same as in unstressed cells (M1, 0.16 ± 0.03; M2, 0.40 ± 0.05; n = 10 cells). From this experiment, it was not possible to determine whether preexisting cytoplasmic Gag–YB-1 complexes or free Gag and YB-1 moved into HS-induced stress granules. Treatment of cells with HS also did not change the overall degree of colocalization between Gag and TIA-1 (Fig. 1D, HS 45 min) (M1 = 0.03 ± 0.01; M2 = 0.11 ± 0.01; n = 10 cells), which was again lower than the amount of Gag–YB-1 colocalization. However, Gag did accumulate to a small degree in TIA-1-containing HS-induced stress granules (Fig. 1D, HS 45 min, enlargement). Additionally, we noted an increase in the Gag signal in the nucleus during HS (Fig. 1C, HS 45 min). We previously reported that a subpopulation of MMTV(C3H) Gag localizes to the nucleus and nucleolus under steady-state conditions (2) although the function of MMTV Gag in the nucleus, if any, remains unknown.
Treatment of heat-shocked NMuMG.C3H cells with CHX resulted in dissolution of stress granules, as expected, because CHX treatment traps mRNAs on polysomes and prevents them from cycling into stress granules (17, 22). Similarly, large stress granules that contained Gag and YB-1 dissolved in CHX-treated cells; unexpectedly, however, small cytoplasmic Gag–YB-1 granules remained intact, and the degree of colocalization was nearly identical to that in untreated cells (Fig. 1C, HS + CHX) (M1 = 0.14 ± 0.03 and M2 = 0.39 ± 0.08; n = 10 cells). CHX treatment did not alter the low degree of colocalization between Gag and TIA-1 (Fig. 1D, HS + CHX) (M1 = 0.02 ± 0.00 and M2 = 0.11 ± 0.01; n = 10 cells), which remained unchanged under all conditions tested. The results of the experiments shown in Fig. 1 demonstrate that MMTV Gag associated specifically with YB-1 under steady-state conditions in small cytoplasmic granules that were distinct from HS-induced stress granules, based on the failure of the constitutive Gag–YB-1 granules to disassemble with CHX treatment. These findings raise the possibility that constitutive Gag–YB-1 cytoplasmic granules contain translationally repressed mRNAs that do not cycle through polysomes. Alternatively, it is possible that these Gag–YB-1 granules form through protein-protein interactions independently of RNA.
Overexpression of stress granule-associated proteins recruits Gag into stress granules in MMTV-infected cells.
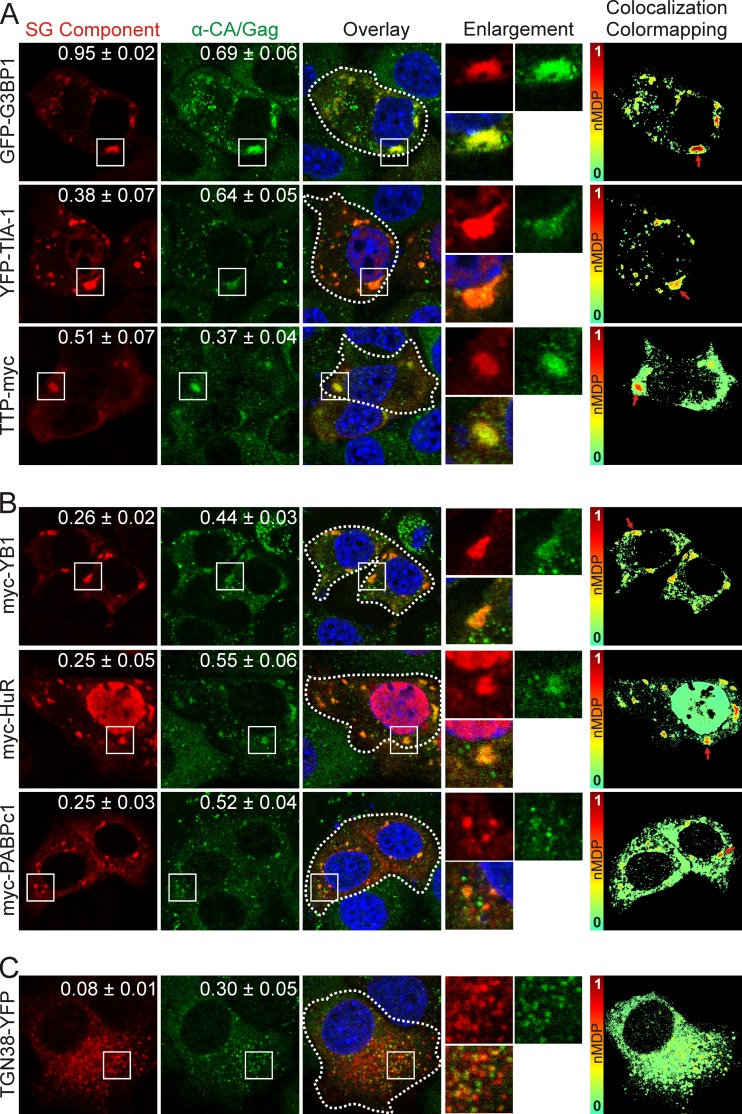
Cells tolerate HS only for relatively short periods of time (17). Therefore, to determine whether more MMTV Gag protein would move into stress granules over a longer period of time, we induced stress granule formation by transient overexpression of several epitope-tagged core stress granule proteins (17). TIA-1, G3BP1, and tristetraprolin (TTP) serve a scaffolding role in stress granules, and overexpression of these proteins reliably causes the nucleation of large cytoplasmic complexes that disassemble with CHX treatment, similarly to authentic stress granules (30, 31, 58). As expected, expression of GFP-G3BP1 and YFP–TIA-1 resulted in formation of numerous large stress granules, whereas stress granules formed by TTP-myc overexpression tended to be smaller and less numerous (Fig. 2A). MMTV Gag accumulated to a high degree in stress granules induced by GFP-G3BP1 expression, with Mander's analysis demonstrating that 95% of the intracellular GFP-G3BP1 signal overlapped that of Gag (M1 = 0.95 ± 0.02), whereas approximately 69% of the overall Gag fluorescence colocalized with GFP-G3BP1 (Fig. 2A, top row) (M2 = 0.69 ± 0.06; n = 10 cells). In YFP–TIA-1-expressing cells, 38% of total YFP–TIA-1 fluorescence overlapped that of Gag (Fig. 2A, middle row) (M2 = 0.38 ± 0.07), while 64% of the total Gag signal overlapped that of YFP–TIA-1 (M1 = 0.64 ± 0.05, n = 10 cells). The overall overlap of TTP-myc and Gag was 51%, and that of the converse was 37% (Fig. 2A, bottom row) (M1 = 0.51 ± 0.07; M2 = 0.37 ± 0.04, n = 10 cells). Whereas Mander's colocalization analysis quantifies the fraction of colocalization of each fluorescently tagged protein throughout an image, we also examined whether the fluorescence intensities of each tagged protein were similar to one another within the regions of colocalization. To this end, we analyzed the images using ImageJ colocalization color mapping, in which the normalized mean deviation product (nMDP) is represented as a thermal map of the original image, with “hot” colors (red-orange) indicating areas of high signal correlation between the intensities of each channel and “cool” colors (blue-green) representing low correlation (48; http://imagej.nih.gov/ij/). This analysis revealed a high correlation (red-orange) between Gag and the core stress granule proteins G3BP1, TIA-1, and TTP within large stress granules (Fig. 2A, colocalization color mapping, red arrows), indicating that the local concentrations of both Gag and these core stress granule proteins were very high.
FIG 2.
Expression of stress granule-associated proteins relocalizes MMTV Gag to stress granules. Confocal microscopy images show MMTV Gag localization with core stress granule proteins that nucleate assembly (A), proteins that localize to stress granules but have not been reported to nucleate stress granule assembly (B), and a control protein that localizes to the trans-Golgi network and does not induce stress granule formation (C). White boxes indicate the areas shown in the enlargements. To the right, colocalization color maps were generated of the cells outlined by white dashed lines using the colocalization color map plugin (48) for ImageJ (http://imagej.nih.gov/ij/). Red arrows point to discrete regions of high colocalization on the color-mapping images. SG, stress granule.
We next examined whether MMTV Gag would accumulate in cytoplasmic granules resulting from overexpression of YB-1, HuR, and PABPc1 (14, 19, 27) (Fig. 2B). MMTV Gag colocalized with YB-1 in cytosolic foci, with an M1 of 0.26 ± 0.02 and an M2 of 0.44 ± 0.03 (n = 10 cells) (Fig. 2B, top row). Similarly, Gag and myc-HuR colocalized in cytoplasmic aggregates, with an M1 of 0.25 ± 0.05 and an M2 of 0.55 ± 0.06 (n = 10 cells) (Fig. 2B, middle row). With expression of myc-PABPc1, 25% of the total cellular Gag colocalized with PABP (Fig. 2B, bottom row) (M1 = 0.25 ± 0.03) and 53% of myc-PABPc1 overlapped Gag (M2 = 0.53 ± 0.04; n = 10 cells). For myc–YB-1, myc-HuR, and myc-PABPc1, colocalization color mapping revealed that Gag accumulated within prominent cytoplasmic granules at locally high concentrations (Fig. 2C, colocalization color mapping, red arrows). In these experiments, the portion of Gag that did not accumulate in large granules remained primarily within small intracytoplasmic foci.
To discern whether the accumulation of Gag within granules was specifically due to overexpression of stress granule-associated proteins, we transfected NMuMG.C3H cells with YFP-tagged trans-Golgi network 38 (TGN38), an integral membrane protein that cycles between the Golgi complex and the plasma membrane (59). Although Gag overlapped with TGN38-YFP as indicated by Mander's analysis (Fig. 2C) (M1 = 0.08 ± 0.1; M2 = 0.30 ± 0.05; n = 10), Gag did not accumulate in large foci typical of stress granules, as shown by colocalization color mapping (Fig. 2C). Together, these data indicate that expression of stress granule-related proteins specifically induced Gag to accumulate in large cytoplasmic granules. Finding that Gag accumulates in stress granules induced overexpression of stress granule-associated proteins raised three testable hypotheses: (i) sequestration of Gag in stress granules interferes with virus assembly, and therefore reducing expression of stress granule proteins would increase virion production; (ii) MMTV takes advantage of stress granule proteins to promote virus assembly, and thus knocking down stress granule-associated proteins would decrease capsid production; or (iii) accumulation of Gag in stress granules is irrelevant with respect to the capsid assembly process.
Functional role of stress granule-associated proteins in MMTV particle assembly.
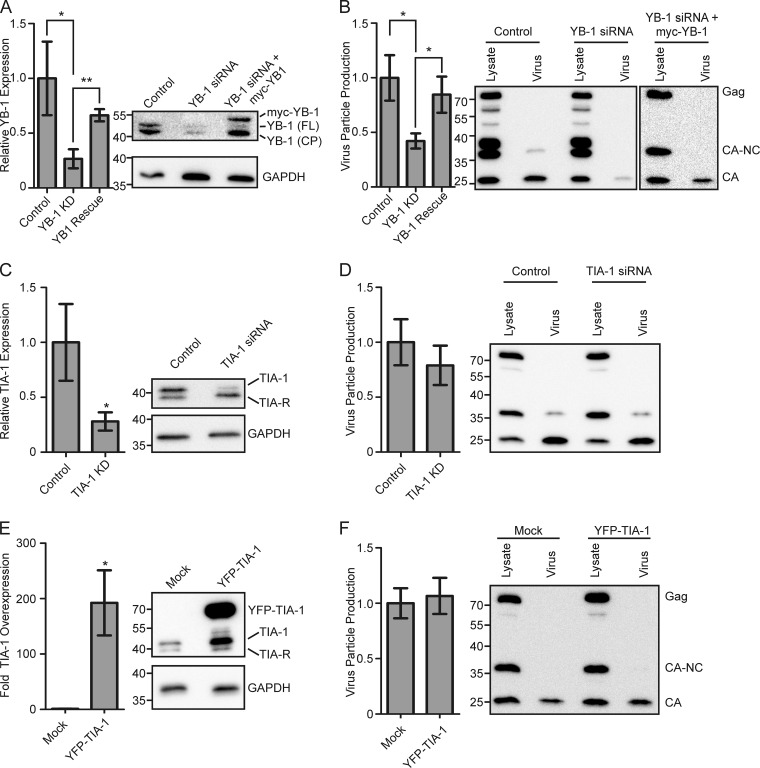
To differentiate between these possibilities, we tested the effect of altering the expression levels of two stress granule-related proteins, YB-1 and TIA-1. YB-1 expression was knocked down using an siRNA targeted against the 3′ untranslated region of YB-1, and the effect on MMTV virus production was assessed. Although YB-1 knockout is embryonic lethal in mice (60), the expression level of YB-1 was reduced by 77% ± 7% in MMTV-infected cells transfected with a YB-1 siRNA (Fig. 3A, YB-1 KD) (P = 0.04) without any apparent toxicity. The amount of YB-1 protein in cells expressing the YB-1 siRNA was compared to that in cells transfected with a scrambled siRNA, and YB-1 levels in each case were internally controlled for GAPDH expression. Expression of the YB-1 siRNA reduced the amount of both full-length YB-1 [YB-1 (FL)] and a YB-1 cleavage product[YB-1 (CP)] (Fig. 3A) (61). The decrease in YB-1 expression was associated with a reduction in MMTV particle production by 58% ± 7% compared to that of the scrambled siRNA control (P = 0.02) (Fig. 3B). Virus particle production (measured as the amount of Gag protein released into the culture supernatant after pelleting through a sucrose cushion) was normalized to intracellular Gag expression levels in each replicate of the experiment, as described in Materials and Methods. Although there was a significant decrease in MMTV particle production, it is possible that the inhibitory effect of YB-1 on virus assembly was underestimated, considering that every cell was producing virus particles whereas only a subset of cells was transfected with YB-1 siRNA (estimated transfection efficiency of NMuMG cells, 15 to 20%).
FIG 3.
Functional role of YB-1 in MMTV assembly. All graphs show the mean ± standard error of the mean calculated over at least four repetitions. Statistical significance (*, P < 0.05; **, P < 0.005) was determined by a two-tailed Student's t test (GraphPad Prism). (A) Graph showing mean relative YB-1 expression in NMuMG.C3H cells transfected with scrambled siRNA control, YB-1 siRNA (YB1 knockdown [KD]), or YB-1 siRNA and siRNA-resistant myc–YB-1 (YB1 Rescue). YB-1 expression was determined by Western blotting and standardized to the amount of GAPDH within each lane. YB-1 expression in cells treated with scrambled control siRNA was set to 100%. A representative Western blot is shown at right. YB-1 (FL) denotes full-length YB-1, while YB-1 (CP) is a previously described YB-1 cleavage product (61). Bands corresponding to YB-1 (FL) and myc–YB-1, but not YB-1 (CP), were quantitated, and results are shown in the graph. (B) Graph depicting virus production from cells used in the experiments described in panel A. Virus production was measured by quantitating the amount of Gag protein present in the medium (pelleted through a sucrose cushion) divided by the sum of the Gag protein present in the medium and lysates (amount in medium/amount in medium + amount in lysates) (2). A representative Western blot is shown to the right of the graph. Relative TIA-1 expression (C) and virus production (D) from NMuMG.C3H cells treated with TIA-1 siRNA or a control siRNA are shown. Western blots from representative experiments are shown to the right of the graphs. Overexpression of TIA-1 (E) and the effect on virus production (F) relative to cells transfected with a GFP control plasmid are shown. Representative Western blots are shown to the right of each graph.
To examine whether the decrease in MMTV virus production was directly related to YB-1 protein levels, we restored YB-1 expression by cotransfecting an siRNA-resistant myc–YB-1 plasmid lacking the native 3′ untranslated region of YB-1 (YB-1 rescue). In cells cotransfected with YB-1 siRNA and myc–YB-1, we detected myc-tagged YB-1 and YB-1 (CP), but not full-length, endogenous YB-1 (Fig. 3A). In YB-1 siRNA-treated cells, expression of myc–YB-1 restored YB-1 (FL) expression to 66% ± 5% of pretreatment levels, using GAPDH levels as an internal standard (Fig. 3A), and MMTV budding increased to 84% ± 16% of control levels (P = 0.02) (Fig. 3B, YB-1 siRNA+myc–YB-1), indicating a clear correlation between intracellular YB-1 levels and virus particle production. We examined the effect of YB-1 overexpression on virus production, but there was no effect (data not shown). One problem in interpreting the YB-1 overexpression experiment was our observation that ectopic expression of myc–YB-1 did not consistently increase intracellular YB-1 levels as measured by Western blotting, ostensibly due to the tight autoregulation of YB-1 expression (62, 63).
To determine whether the decrease in virus production with YB-1 knockdown was a common feature of inhibiting stress granule-associated proteins, we examined the effects of targeting the core stress granule protein TIA-1 on MMTV assembly. Depletion of TIA-1 by approximately 80% relative to cells treated with a scrambled siRNA (Fig. 3C, TIA-1 KD) had no significant effect on virus production (Fig. 3D) (P = 0.46). Furthermore, overexpression of TIA-1 to a level 200-fold higher than that of mock-transfected cells induced stress granule formation but did not alter MMTV virus particle production (Fig. 3E and F). Similarly, nucleation of stress granules by ectopic expression of G3BP1 had no effect on virus production (data not show). Together, these data suggest that induction of stress granule formation per se does not interfere with MMTV virus production, nor does the recruitment of Gag to stress granules enhance virus assembly. Instead, our data demonstrate that that the stress granule-associated protein YB-1 specifically facilitates MMTV particle assembly, possibly due the interaction of Gag and YB-1 in cytoplasmic granules.
MMTV Gag colocalizes with stress granule-associated proteins in the absence of virus infection.
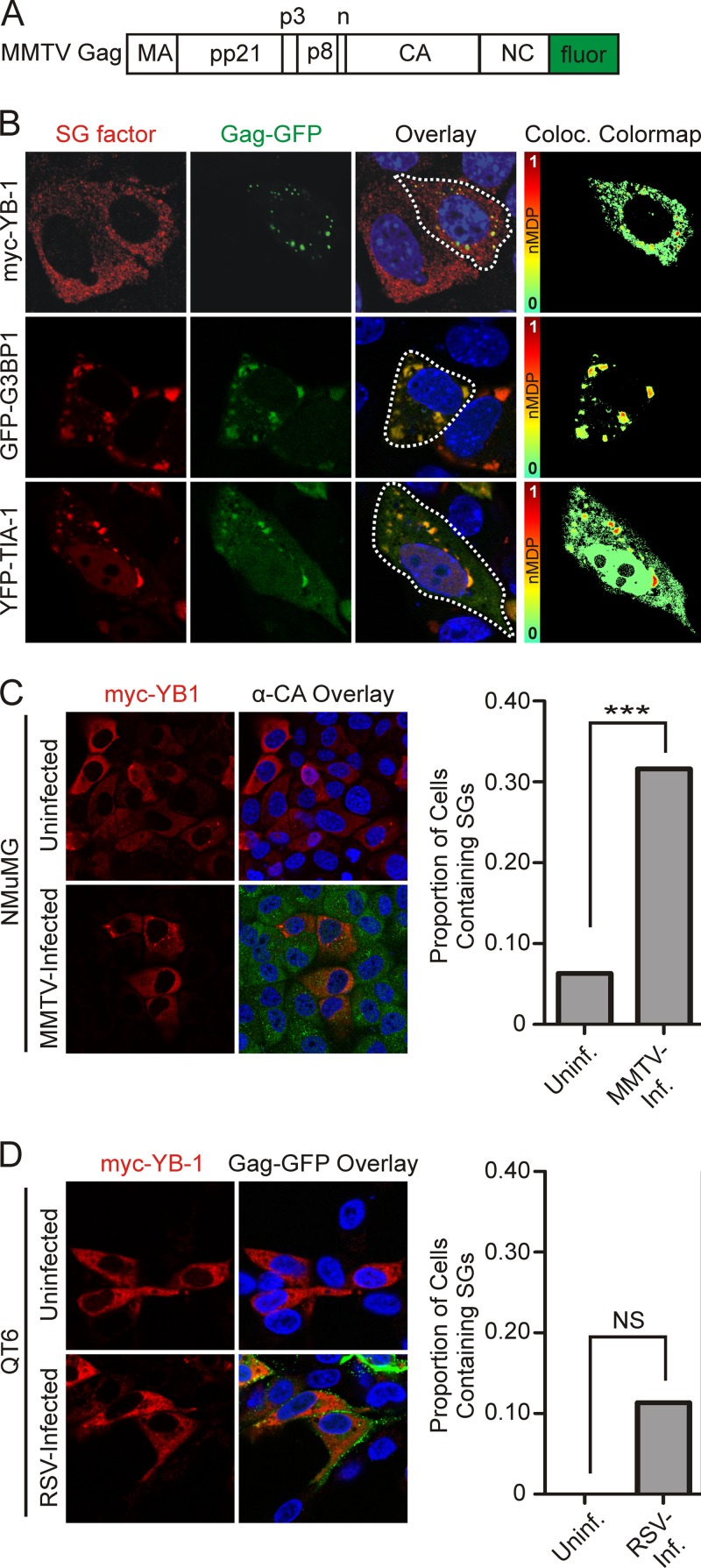
Next, we examined whether MMTV Gag would accumulate in stress granules when expressed in the absence of additional viral constituents. Expression of a Gag-GFP or Gag-mCherry fusion protein (Fig. 4A) (2) in uninfected NMuMG cells revealed that fluorophore-tagged Gag colocalized with YB-1 in cytoplasmic granules, as demonstrated by the punctate yellow signal in the overlay images and the red-orange foci in the colocalization color-mapping image analysis (Fig. 4B). However, large YB-1-induced stress granules were not observed even though they had been present in MMTV-infected cells overexpressing myc–YB-1 (compare top rows of Fig. 2B and 4B). Instead, YB-1 remained dispersed in smaller granules in the cytoplasm, and colocalization with Gag-GFP was less robust than in MMTV-infected cells. In contrast, expression of GFP-G3BP1 or YFP–TIA-1 induced the formation of large stress granules that strongly colocalized with Gag (Fig. 4B, middle and bottom rows).
FIG 4.
Gag is sufficient to traffic to stress granules in uninfected cells. MMTV Gag containing a C-terminal GFP tag (2) (A) was coexpressed with myc–YB-1, GFP-G3BP1, or YFP–TIA-1 (B) in uninfected NMuMG cells. Colocalization color maps were generated as described above and are shown in the far right column. A dashed white line drawn around the perimeter indicates the outline of the cell shown in the colocalization color-mapping image. (C) Uninfected or MMTV-infected NMuMG cells were transfected with myc–YB-1 and fixed for immunofluorescence 18 h after transfection. MMTV Gag was detected using anti-CA antibody (green), and YB-1 was detected using anti-YB-1 antiserum (red). Cells were scored for the presence or absence of YB-1-containing cytoplasmic granules. (D) Uninfected or RSV-infected QT6 cells were transfected with plasmids expressing myc–YB-1 (uninfected) or myc–YB-1 and Gag-GFP (RSV infected). Cells were evaluated for YB-1 staining in cytoplasmic granules. Statistical analysis of the data shown in panels C and D was determined using Fisher's exact test (GraphPad Prism). NS, nonsignificant (P > 0.05); ***, P < 0.0001. Data were derived from >30 cells from at least two experiments conducted on separate days.
Because YB-1 expression did not induce the formation of large stress granules in uninfected cells (Fig. 4B) but did induce stress granules in MMTV-infected cells (Fig. 2B), we quantitated the effect of MMTV infection on the formation of YB-1-induced stress granules. Expression of myc–YB-1 induced the formation of cytoplasmic granules in 31.6% of MMTV-infected cells (n = 98 cells) but in only 6.3% of uninfected cells (Fig. 4C) (P < 0.0001; n = 190 cells). These results suggest that MMTV infection creates an environment favoring YB-1-induced stress granule formation that requires a viral component in addition to the Gag protein, possibly the viral RNA. The predilection of YB-1 expression to induce stress granules was not observed in cells infected with another retrovirus, Rous sarcoma virus (RSV). In fact, there was a complete absence of stress granules observed in RSV-infected, myc–YB-1 transfected quail cells (n = 34 cells), and only 11% of uninfected, transfected quail cells contained stress granules (n = 44 cells) (Fig. 4D). Accordingly, we conclude that retrovirus infection does not generally promote the formation of large YB-1-induced cytoplasmic granules.
The interaction between Gag and YB-1 is RNase sensitive and dependent on the Gag NC domain.
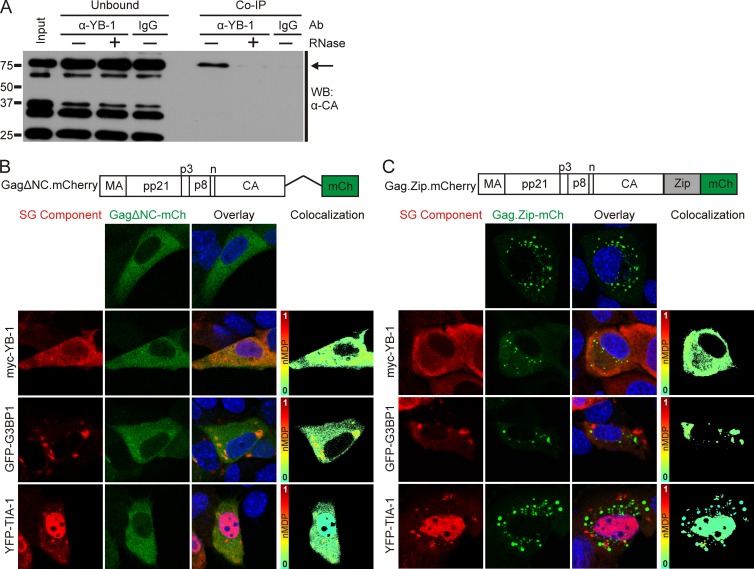
Because YB-1 binds mRNA, regulates translation, and is present in mRNP-containing stress granules, we reasoned that RNA might bridge the association of Gag with YB-1. To test this idea, we performed coimmunoprecipitation (co-IP) experiments in the absence or presence of RNase A in MMTV-infected cells. Using an antibody to endogenous YB-1 for the pulldown and anti-CA antibody to detect Gag by immunoblotting, we found that YB-1 and the full-length Gag polyprotein were coimmunoprecipitated (Fig. 5A) and that this interaction was disrupted by RNase treatment. In contrast, MMTV Gag was very inefficiently coimmunoprecipitated with endogenous G3BP1 under similar conditions (data not shown).
FIG 5.
RNA-dependent interaction of Gag with stress granule proteins. (A) Endogenous YB-1 was immunoprecipitated from MMTV(C3H)-infected NMuMG whole-cell lysates using 5 μg of YB-1 antibody or an irrelevant IgG antibody as a control. Prior to the immunoprecipitation, samples were treated with (+) or without (−) RNase A. Gag (indicated by the arrow) was detected by Western blotting (WB) using anti-CA monoclonal antibody (Ab). (B) Schematic diagram showing the NC deletion mutant Gag.ΔNC-mCherry with images of cells expressing the protein (false colored green in the images) expressed alone or coexpressed with myc–YB-1, GFP-G3BP1, or YFP–TIA-1 in uninfected NMuMG cells. Colocalization color maps were generated as described in the legend of Fig. 4 and are shown to the right. (C) Schematic representation of the Gag.Zip-mCherry construct, in which the NC domain of Gag was replaced with the CREB1 leucine zipper domain, with images of cells expressing the construct (false colored green) alone or with myc–YB-1, GFP-G3BP1, or YFP–TIA-1 in NMuMG cells. Colocalization color maps are shown to the right. mCH, mCherry.
The NC domain of Gag is required for both Gag-RNA and Gag-Gag interactions. We previously reported that deletion of NC abrogates the formation of distinct cytoplasmic foci of Gag (50). Therefore, we examined whether NC was required for association of Gag and YB-1 in cells. Gag.ΔNC-mCherry did not colocalize with myc–YB-1 and YFP–TIA-1 in uninfected NMuMG cells; however, there was a low level of colocalization observed between GFP-G3BP1 and Gag.ΔNC despite the deletion of the nucleic acid binding region of Gag (Fig. 5B). To determine whether the decrease in Gag association with YB-1, TIA-1, and G3BP1 was due to a defect in Gag multimerization or a loss of RNA binding activity, we replaced NC with the CREB1 leucine zipper domain, which is a nucleic acid-independent protein-protein interaction motif (Fig. 5E) (MMTV Gag.Zip-mCherry). For other retroviral Gag proteins, substitution of the CREB1 leucine zipper for NC promotes Gag-Gag interactions that are sufficient to drive the assembly of virus-like particles (44, 64). In our experiments, MMTV Gag.Zip-mCherry formed small cytoplasmic aggregates in uninfected NMuMG cells (Fig. 5C). However, these Gag.Zip-mCherry foci did not colocalize with YB-1, and they were distinct from the stress granules formed by GFP-G3BP1 and YFP–TIA-1 (Fig. 5C). Therefore, either the NC sequence itself or the RNA binding activity of the Gag NC domain is required for MMTV Gag to colocalize with YB-1, G3BP1, and TIA-1. It is possible that MMTV RNA facilitates the interaction of Gag and YB-1, given that (i) MMTV infection promotes YB-1-induced granule formation (Fig. 2B and 4C) and (ii) the association of Gag, when expressed alone, has reduced colocalization with YB-1 in cytoplasmic granules compared to Gag expressed in the context of an authentic viral infection (Fig. 1B, 2B, and 4C, compare YB-1 panels). However, based on these experiments, we cannot rule out the possibility that a cellular RNA or another factor promotes Gag–YB-1 association in cytoplasmic granules.
Colocalization of an MMTV RNA reporter with Gag, YB-1, and G3BP1.
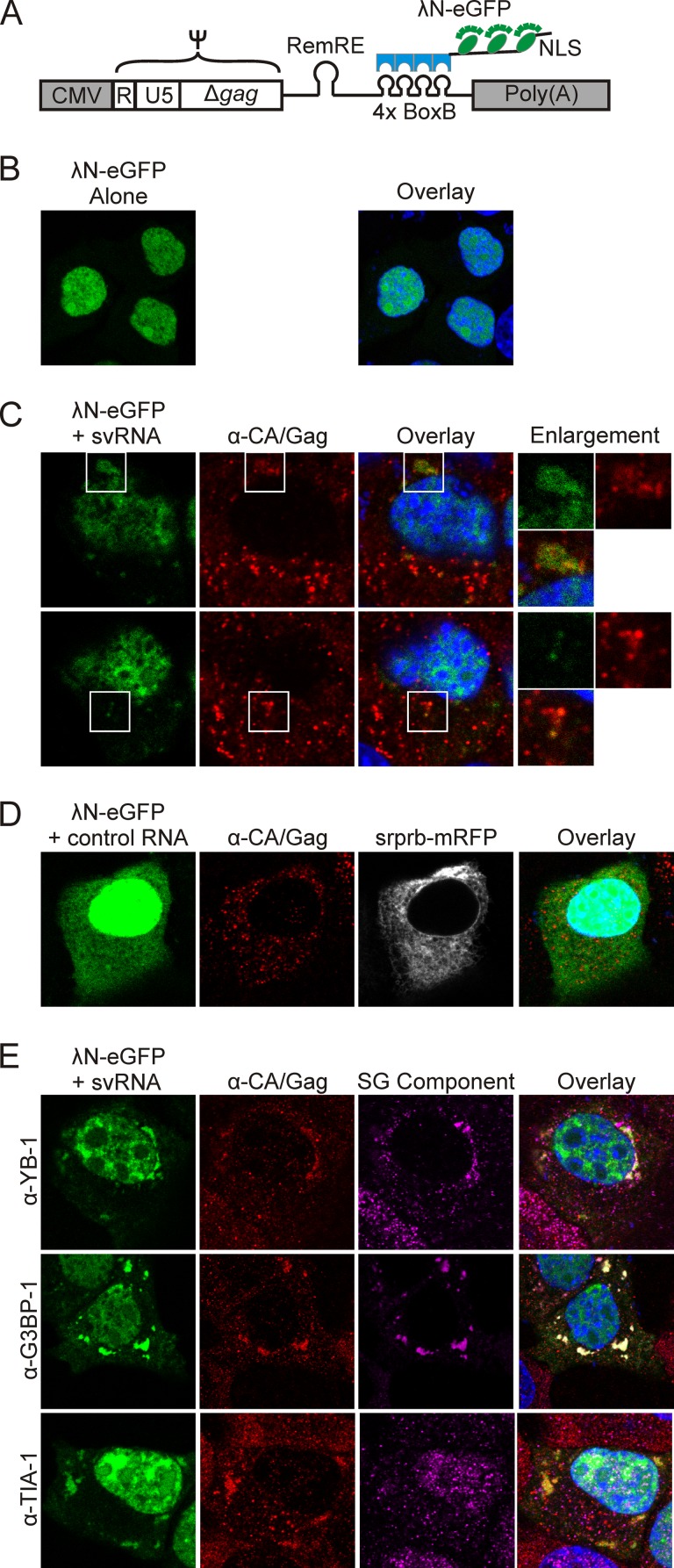
To examine whether viral RNA is present within Gag–YB-1 granules, we used a previously described subviral RNA (svRNA) construct (65) as a surrogate marker for the localization of MMTV genomic RNA in infected cells (Fig. 6A). We used this approach because of the concern that endogenous retrovirus sequences (e.g., Mtv1) highly homologous to the variant MMTV(C3H) could make it difficult to reliably distinguish between endogenous and exogenous viral sequences using probes designed for fluorescence in situ hybridization, especially if Mtv1 and MMTV(C3H) viral RNAS were colocalized in the cell (66–68). The svRNA construct contains the MMTV Ψ packaging signal encompassed within R, U5, and the first 746 nucleotides of the gag coding region (69, 70), the Rem response element (RemRE) (71, 72), and four tandem repeats of the λ bacteriophage BoxB stem-loop, which are bound with high affinity by the λN protein (73, 74). The λN-eGFP reporter protein contains three copies of eGFP (enhanced GFP), four copies of λN, and a nuclear localization signal (NLS) (Fig. 6A and B). The MMTV svRNA contains the splice donor site but not the splice acceptor; therefore nucleocytoplasmic transport of the svRNA is dependent on coexpression of the MMTV Rem protein, which mediates nuclear export of unspliced MMTV RNAs through its interaction with the RemRE (65). As we reported previously, when the λN-eGFP reporter binds to svRNA, the complex is exported into the cytoplasm in a Rem-dependent manner (65).
FIG 6.
Localization of MMTV subviral RNA in infected cells. (A) Schematic diagram depicting the MMTV subviral RNA (svRNA) construct, which is driven by the CMV promoter and contains R, U5, the first 746 nucleotides of gag (Ψ), the Rem response element (RemRE), and four BoxB stem-loops, which are bound by the λN-eGFP reporter. The λN-eGFP reporter contains an M9 nuclear localization signal (NLS) (74). (B) Expression of the λN-eGFP reporter alone in MMTV(C3H)-infected NMuMG (NMuMG.C3H) cells. (C) Two planes through the same NMuMG.C3H cell transfected with svRNA and λN-eGFP and immunostained for Gag. The svRNA and Gag colocalize in cytoplasmic complexes. White boxes indicate the areas shown in enlargements. (D) MMTV-infected NMuMG cells were transfected with λN-eGFP and immunostained for Gag and YB-1, G3BP1, or TIA-1.
To visualize the localization of svRNA in MMTV-infected NMuMG cells, we transfected plasmids expressing svRNA and the λN-eGFP reporter. Because these cells are chronically infected with MMTV, Rem is expressed from the integrated provirus. The Rem-dependent svRNA was exported into the cytoplasm, where it colocalized with Gag in cytoplasmic granules of various sizes (Fig. 6C, white boxes). Of note, not all of the cytoplasmic Gag foci (detected by anti-CA antibody and Cy3 secondary antibody) contained detectable svRNA (eGFP signal). There are several possible explanations for this finding: Gag-svRNA binding obscures the eGFP signal, the presence of unlabeled full-length MMTV RNA in infected cells outcompetes the svRNA for binding to Gag, or not all Gag foci contain viral RNA. The large granules of Gag seen in the top panel of Fig. 6C were observed only in MMTV-infected cells coexpressing the svRNA, suggesting that overexpression of the svRNA caused Gag to accumulate in these aggregates. As a control, we expressed a nonviral control RNA (Srprb-mRFP-4BoxB), which binds the λN-eGFP reporter and expresses the Srprb-mRFP protein (74). We found that Srprb-mRFP-4BoxB did not accumulate in granules with Gag in NMuMG.C3H cells, and its expression did not alter Gag localization (Fig. 6D).
To assess whether stress granule-associated proteins were associated with viral RNP complexes, MMTV-infected cells expressing the svRNA and the λN-eGFP reporter were immunostained for Gag and endogenous YB-1, G3BP1, or TIA-1. YB-1 and G3BP1 colocalized with Gag and svRNA in cytoplasmic granules (Fig. 6E, overlay, top and middle rows). In contrast, the svRNA and Gag were localized together in cytoplasmic aggregates but were separate from TIA-1 (Fig. 6E, overlay, bottom row). Finding that Gag-svRNA complexes do not contain the core stress granule component TIA-1 indicates that the viral RNP complexes are distinct from stress granules.
Kinetic analysis of cytoplasmic granules formed by MMTV Gag with YB-1 or G3BP1.
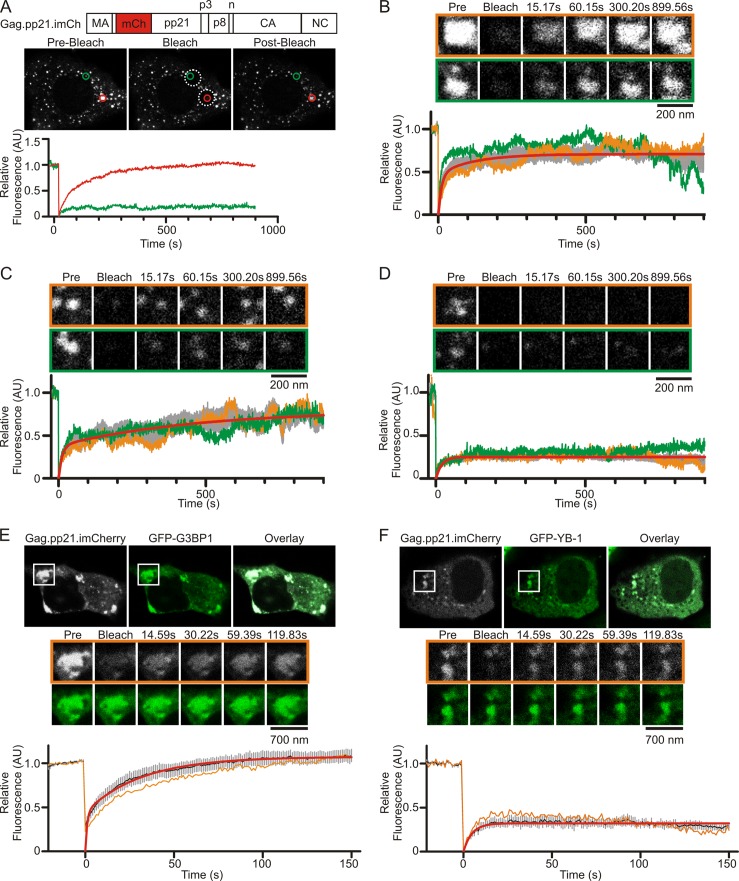
Up to this point, our data suggested that cytoplasmic viral RNP complexes containing Gag, YB-1, and svRNA could be sites of immature capsid assembly based on the finding that virus production was dependent on YB-1 (Fig. 3A and B). To test this possibility, we developed a fluorescence recovery after photobleaching (FRAP) assay to study the biophysical properties of Gag in living MMTV-infected cells. MMTV Gag was fluorescently labeled by inserting mCherry between the seventh and eighth codons of the pp21 domain (Fig. 7A). The resulting Gag.pp21.imCh protein was expressed in NMuMG.C3H cells, where it was processed by the MMTV protease and incorporated into virus particles (data not shown). Live-cell imaging of NMuMG.C3H cells revealed that Gag.pp21.imCh localized within small cytoplasmic foci, as previously observed for the wild-type Gag protein (compare Fig. 7A to 1C and D, unstressed cells).
FIG 7.
FRAP analysis of MMTV Gag in living MMTV(C3H)-infected cells. (A) Parameters used to define different populations of MMTV Gag complexes in living MMTV(C3H)-infected NMuMG cells. MMTV Gag was internally labeled with mCherry by inserting the fluorophore into the pp21 domain. The Gag.pp21.imCherry construct was expressed in NMuMG.C3H cells, and live cells were imaged by confocal microscopy. Individual foci were chosen for analysis, and the relative fluorescence of each focus was measured. Foci were monitored for 20 s (prebleach), subjected to photobleaching, and then monitored for fluorescence recovery for 15 min. The graphs show the relative fluorescence intensities of the foci indicated by the red and green circles. Analysis of 19 foci revealed discrete populations of foci with rapid (B), gradual (C), and limited (D) fluorescence recovery after photobleaching. Individual foci are shown in the orange and green boxes, with the relative fluorescence of each focus shown as the orange or green curve on the graph. The gray curves on each graph represent the mean fluorescence intensity from at least four foci ± standard errors of the means. The red line is the curve fit to the mean fluorescence intensity using GraphPad Prism using either a double (B and C) or single (D) exponential association curve. (E) FRAP analysis of Gag.pp21.imCherry within stress granules induced by expression of GFP-G3BP1 or GFP–YB-1 (F) in NMuMG.C3H cells. For each condition, a representative cell is shown at the initial prebleach time point, and a single cytoplasmic granule chosen for analysis is indicated by the white box. Enlarged images of the white boxed granule before (prebleach), during (bleach), and at various time points after photobleaching are shown for both the Gag.pp21.imCherry channel (grayscale, outlined by an orange line) and the GFP channel (green). Graphs show the mean fluorescence intensity for at least five granules (black line), with standard errors of the means (gray bars). The fluorescence intensity of the representative focus is show as an orange line. Red lines are curves fit to the averaged data with double (E) or single (F) exponential association equations using GraphPad Prism. Time is indicated in seconds (s). Pre, prebleach image. AU, arbitrary units.
We reasoned that if Gag proteins in cytoplasmic foci were highly mobile, cycling rapidly between the foci and the cytoplasm, then Gag was unlikely to be tightly bound within a higher-order complex. In contrast, if Gag proteins were relatively immobile within the foci, with little to no exchange with Gag proteins in the surrounding cytoplasm, that behavior would be consistent with the formation of a stable multimeric complex, possibly in an immature virus particle. Similar kinetic properties have been described for Gag proteins within assembled particles for retroviruses that form virus particles at the plasma membrane (e.g., HIV-1, equine infectious anemia virus [EIAV], and MLV) (75–78).
To assess the mobility of Gag.pp21.imCh, individual foci (Fig. 7A, green and orange circles) were subjected to FRAP (fluorescence recovery after photobleaching) analysis. Gag-containing foci were imaged for approximately 20 s to measure baseline fluorescence, and then 5-μm-diameter regions of interest (ROIs) (Fig. 7A, white dashed circles) containing Gag foci were photobleached. Foci within each ROI were monitored for fluorescence recovery at 1.91 frames per second for 15 min, based on the time required for assembly of HIV-1, EIAV, and MLV particles (75–78). The fluorescence intensity for each Gag focus was measured every 15 s for 15 min, normalized for loss of fluorescence due to imaging, and analyzed as described in Materials and Methods.
Analysis of 19 individual Gag.pp21.imCh foci revealed the presence of three categories of Gag foci that had distinct kinetics (Fig. 7B, C, and D). The first group of foci (n = 4) (Fig. 7B) exhibited rapid, two-phase recovery with a fast-phase recovery half-life (t1/2) of 7.8 s (95% confidence interval [CI], 6.7 to 9.3 s) and a slow-phase recovery t1/2 of 84.2 s (95% CI, 72.4 to 100.6 s). Examples of two foci in this category are shown in the images in Fig. 7B outlined by orange and green boxes, with relative fluorescence for each (orange and green lines, respectively) shown in the graph below and the average fluorescence ± standard error of the mean (SEM) shown in gray. The average mobile fraction for these foci was 0.691 (95% CI, 0.688 to 0.694), indicating that the majority of Gag proteins (∼70%) within these complexes underwent rapid exchange with Gag proteins outside the bleached region (Fig. 7B). Accordingly, we concluded that foci with rapid and complete fluorescence recovery after photobleaching represented high local concentrations of free (monomers or small oligomers) Gag proteins that had not assembled into virus particles.
In contrast, the second population of foci (n = 5) (Fig. 7C) exhibited two-phase recovery following bleaching, with a fast-phase recovery t1/2 of 9.1 s (95% CI, 7.4 to 11.7 s), a slow-phase t1/2 of 346.5 s (95% CI, 285.7 to 440.3 s), and a mobile fraction of 0.662 (95% CI, 0.657 to 0.668) (Fig. 7C). These kinetics indicated that early after bleaching, Gag proteins within these foci are rapidly exchanged with cytoplasmic Gag proteins outside the region of interest, but at later time points there was a slower addition of Gag molecules. Therefore, complexes characterized by rapid, partial fluorescence recovery followed by a prolonged phase of additional fluorescence recovery indicate the transition of Gag proteins from higher to lower mobility, which may occur in an assembling virus particle that continues to grow slowly over time. Indeed, the gradual increase in Gag fluorescence over the 15-min period was consistent with previous reports describing the kinetics of HIV-1, EIAV, and MLV virus particles assembling at the plasma membrane (75–78).
The third population of foci (n = 11) (Fig. 7D) exhibited limited, single-phase recovery after photobleaching with a t1/2 of 13.4 s (95% CI, 12.5 to 14.5) and a mobile fraction of 0.25 (95% CI, 0.249 to 0.252). The limited fluorescence recovery within these foci suggests that they represent assembled MMTV particles with very limited addition of new Gag molecules after photobleaching. Although we grouped these populations of MMTV Gag foci into three separate categories, it is likely these populations represent points along a continuum of immature capsid assembly, during which Gag monomers accumulate in distinct cytoplasmic locations, interact with the viral RNA, multimerize, and assemble into virus particles that are transported through the cytoplasm to the plasma membrane.
We next performed FRAP analysis to determine whether Gag proteins located with stress granules induced by GFP-G3BP1 overexpression exhibited kinetic properties characteristic of one of the populations we had defined (Fig. 7E). We found that Gag.pp21.imCh present in association with GFP-G3BP1 in stress granules exhibited rapid and complete two-phase recovery after photobleaching with a fast phase t1/2 of 6.6 s (95% CI, 3.6 to 39.9 s), a slow phase t1/2 of 35.1 s (95% CI, 20.0 to 142.6 s), and a mobile fraction of 1.10 (95% CI, 1.08 to 1.13; n = 5 foci). The kinetics of Gag recovery within G3BP1 granules is similar to that of the unassembled Gag complexes described in Fig. 7B, indicating that Gag diffused rapidly in and out of G3BP1 granules. These findings imply that the majority of Gag proteins within G3BP1 granules have not assembled into a stable complex. In support of this conclusion, the slow phase of Gag recovery within G3BP1 granules was markedly faster than that of the “assembling” complexes shown in Fig. 7C, suggesting that Gag recovery in G3BP1 granules is more rapid than would be expected for the ordered addition of Gag molecules to virus particles. However, we cannot rule out the possibility that a small population of Gag may be in the process of assembling into a higher-order complex within G3BP1 granules. The mobile fraction of >1.00 suggests that the rate of Gag trafficking into G3BP1 granules exceeds the rate of Gag movement out of these complexes, consistent with our observations that Gag accumulates in G3BP1 granules over time (data not shown). In interpreting these data, it is important to remember that the fluorescence intensities used to calculate fluorescence recovery are corrected for fluorescence loss incurred during photobleaching and imaging using the Leica SP8 imaging software. However, the raw images showing Gag.pp21.imCh fluorescence in Fig. 7E (and subsequently in Fig. 7F) were not corrected for fluorescence loss, and therefore these images cannot be used to accurately estimate fluorescence recovery “by eye.”
Interestingly, recovery of Gag.pp21.imCh fluorescence within YB-1-induced granules exhibited markedly slower kinetics (Fig. 7F) than Gag in G3BP1 complexes. The curve showing the average recovery of Gag fluorescence within YB-1 granules (n = 5 foci) was best fit by a single exponential equation, with a t1/2 of 3.3 s (95% CI, 2.7 to 4.5 s) and a mobile fraction of 0.32 (95% CI, 0.31 to 0.33). Accordingly, these data indicate that Gag mobility within YB-1-induced granules was dictated by a single binding event. Furthermore, the small mobile fraction signifies that Gag proteins within YB-1-induced granules underwent very limited exchange with free Gag in the cytoplasm, indicating that Gag forms a stable complex within YB-1-induced granules. Together with data showing the RNase sensitivity of Gag-YB-1 association, these data raise the possibility that Gag may bind viral or cellular RNA in association with YB-1 in cytoplasmic granules to nucleate immature capsid assembly.
DISCUSSION
Stress granules form in response to unfavorable cellular conditions, including viral infection. Although this stress response can create a hostile environment for pathogens, some viruses have evolved mechanisms to commandeer proteins involved in stress granule formation to promote virus replication (28). Here, we report that the betaretrovirus MMTV, unlike human retroviral pathogens HIV-1 and HTLV-I, does not disable the stress response by preventing stress granule formation. Instead, our data support the conclusion that MMTV Gag co-opts YB-1, a stress granule-associated protein and master regulator of translation, to facilitate virus assembly.
YB-1, a major component of cellular messenger ribonucleoprotein complexes (mRNPs), is present in both translating and nontranslating mRNPs (79, 80; for a review, see reference 81). In the cytosol, YB-1 is present in small cytoplasmic granules that coalesce into larger stress granules during unfavorable cellular conditions (18, 82). The mechanism by which YB-1 exerts posttranscriptional control is regulated by the amount of YB-1 that binds to an mRNA. At low YB-1:RNA mass ratios, the cold shock domain and C-terminal domain of YB-1 bind RNA, promoting an extended conformation of the mRNA, which favors translation (83, 84). Conversely, at high concentrations YB-1 multimerizes, compacting the mRNA and repressing translation (80, 84). In stress granules, YB-1 is thought to package translationally repressed cellular mRNAs to prevent their degradation, allowing the mRNAs to return to polysomes for translation when conditions improve. In addition to regulating cellular translation, YB-1 modulates expression of some viral RNAs. For example, YB-1 represses dengue virus replication by suppressing translation of viral RNA (36). YB-1 plays a different role in hepatitis C virus, where its recruitment to sites of virus assembly influences the balance between viral RNA replication and encapsidation (85). Recently, it was reported that YB-1 stabilizes intracellular levels of HIV-1 and murine leukemia virus RNAs, resulting in an increase in virus particle production (37, 38).
As reported here, we found that YB-1 colocalized with MMTV Gag in small cytoplasmic foci in unstressed cells. We speculate that YB-1 may associate with Gag and viral RNA in these granules and repress MMTV RNA translation, based on the failure of these granules to disassociate in the presence of cycloheximide. Our reasoning is based on the assumption that MMTV Gag and YB-1 granules would disassemble with cycloheximide treatment like “traditional” stress granules and P bodies if these granules contained RNAs that were recycling through polysomes. Further experiments will need to be done to directly test this idea. Alternatively, it is possible that MMTV Gag–YB-1 granules do not contain RNA. This explanation seems less likely given that MMTV Gag initiates immature capsid assembly in the cytoplasm in association with its RNA genome, and the data in Fig. 6 demonstrate that MMTV svRNA is present in Gag–YB-1 cytoplasmic granules.
The observation that MMTV Gag–YB-1 granules are constitutively present in infected cells is characteristic of GW or P bodies, which are present in unstressed cells and are involved in silencing, decay, transport, and stabilization of mRNAs (17, 23, 24, 26, 86–89). However, Gag does accumulate in stress granules induced by heat shock and with overexpression of stress granule-associated proteins, including YB-1, TIA-1, G3BP1, TTP, HuR, and PABPc1. Nonetheless, it does not appear that the accumulation of Gag in stress granules plays an important role in particle assembly because knocking down or overexpressing TIA-1, a core stress granule component, did not have any effect on virus production. Furthermore, functional analysis of stress granules induced by G3BP1 overexpression revealed that Gag was highly mobile in these complexes, arguing against stress granules as a site for Gag-driven immature capsid assembly. There is a close, dynamic interrelationship between stress granules and P bodies, and many proteins, including YB-1, are constituents of both types of mRNA regulatory granules (12, 14, 17–20, 89, 90). It is possible that the constitutive Gag–YB-1 granules contain components of P bodies, and this question is currently under study in our laboratory. The association of MMTV Gag with mRNA processing bodies (stress granules and P bodies) is complex and will require more detailed analysis to determine if there is a functional relationship between MMTV assembly and cellular factors in addition to the central role of YB-1 that was demonstrated in the manuscript. Based on our data, we propose that MMTV particles do not assemble in stress granules, nor is virus assembly inhibited by stress granule formation. Instead, our results favor the interpretation that MMTV co-opts YB-1, and possibly other stress granule- and P body-associated proteins, during the process of immature capsid formation.
Knockdown of YB-1 expression interfered with MMTV assembly, suggesting that YB-1 exerts a positive effect on virus particle formation, transport, or release. However, we did not observe a difference in Gag localization in cells expressing reduced levels of YB-1 (data not shown), implying that the defect did not alter the cytoplasmic localization of Gag but, rather, disrupted immature particle formation. The precise mechanism by which YB-1 facilitates MMTV particle formation is unclear, but our results have provided some clues. The interaction between Gag and YB-1 in MMTV-infected cells was RNase sensitive, suggesting that an RNA bridges the interaction. The NC domain, which specifically binds to the viral genome, was required for localization of Gag to YB-1 granules, and dimerization of Gag through a nucleic acid-independent mechanism was not sufficient to mediate the Gag–YB-1 association. The MMTV subviral RNA colocalized with Gag and YB-1, suggesting that MMTV RNA is an integral part of Gag–YB-1 granules; however, one caveat to this interpretation is that the subviral RNA may not follow the same subcellular trafficking pathway taken by the full-length MMTV genomic RNA. Functional analysis of Gag–YB-1 granules using FRAP revealed that these granules contained stable complexes of Gag that underwent very limited exchange with free Gag in the cytoplasm. The best model fitting the kinetic data was a single exponential equation, indicating that Gag mobility was governed by a single binding event. Thus, the FRAP data define two functionally distinct populations cytoplasmic granules: (i) stress granules induced by G3BP1, which contain highly mobile Gag proteins and therefore do not likely represent sites of particle assembly, and (ii) Gag–YB-1 granules in which Gag proteins are relatively immobile, consistent with sites of immature capsid assembly.
Taken together, these data have led us to construct a working model in which YB-1 and possibly other cellular proteins associate with MMTV RNPs in cytoplasmic granules. We hypothesize that binding of YB-1 to viral RNA sequesters it away from translation machinery, essentially “marking” the viral RNA as a genome. The YB-1–viral RNP complex may serve as a landmark for Gag to identify sites to initiate capsid assembly. It is possible that Gag is recruited to YB-1 granules (through an unknown mechanism), displacing YB-1 from the viral RNA due to high-affinity binding between Gag and the genomic RNA Ψ packaging signal. YB-1 also binds cellular mRNAs in small cytoplasmic granules, so in the absence of viral RNA, MMTV, like other retroviruses, may encapsidate cellular RNAs. Alternatively, it is possible that Gag and YB-1 associate first and that the viral genomic RNA is recruited independently to Gag–YB-1 granules. Further experimentation is needed to refine our understanding of the mechanism by which YB-1 promotes Gag-mediated assembly of MMTV.
The recruitment of Gag to YB-1 granules where genomic viral RNA may be sequestered would represent a mechanism to circumvent cellular antiviral defenses. This strategy may be common among other retroviruses and retrovirus-like elements. For example, the mammalian long interspersed nuclear element 1 (LINE-1) retrotransposon forms large cytoplasmic ribonucleoprotein complexes (RNPs) thought to be replication intermediates (91). These LINE-1 RNPs contain LINE-1 RNA and the retrotransposon proteins Orf1p and Orf2p, as well as YB-1 and other stress granule-associated proteins (91, 92). Additionally, YB-1 has a stabilizing effect on the genomic RNA of HIV-1 and Ψ-containing RNAs of MLV-based vectors, and YB-1 promotes HIV and MLV particle production (38, 93). Considering these data together, we propose that targeting mammalian retroviral and retrotransposon structural proteins to YB-1-containing cytoplasmic granules where viral RNAs are sequestered may be a mechanism to counteract cellular defenses and promote replication of retroelements and retroviruses. Importantly, these findings are similar to data from HIV-1 and Ty3 indicating that assembly of retrovirus or retrotransposon particles occurs in conjunction with stress granule- or P body-associated proteins but not in stress granules or P bodies themselves (11, 54, 94).
ACKNOWLEDGMENTS
We acknowledge the members of our laboratory for their thoughtful comments and critiques of the manuscript. We are grateful for the generosity of the scientists who contributed reagents, as indicated in Materials and Methods. We thank Nancy Kedersha (Harvard University) for helpful advice. Images were acquired at the Microscopy Imaging Facility at the Penn State Hershey College of Medicine.
This work was funded by the National Cancer Institute at the National Institutes of Health (R01 CA76534 to L.J.P. and F30 CA165774 to D.V.B), the National Science Foundation (Graduate Research Fellowship to A.R.B.), and the Pennsylvania Department of Health Tobacco Settlement CURE Funds (L.J.P.).
The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions of this project.
Footnotes
Published ahead of print 5 February 2014
REFERENCES
- 1.Butsch M, Boris-Lawrie K. 2002. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 76:3089–3094. 10.1128/JVI.76.7.3089-3094.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyer AR, Bann DV, Rice B, Pultz IS, Kane M, Goff SP, Golovkina TV, Parent LJ. 2013. Nucleolar trafficking of the mouse mammary tumor virus gag protein induced by interaction with ribosomal protein L9. J. Virol. 87:1069–1082. 10.1128/JVI.02463-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hizi A, Henderson LE, Copeland TD, Sowder RC, Krutzsch HC, Oroszlan S. 1989. Analysis of gag proteins from mouse mammary tumor virus. J. Virol. 63:2543–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zabransky A, Hoboth P, Hadravova R, Stokrova J, Sakalian M, Pichova I. 2010. The noncanonical Gag domains p8 and n are critical for assembly and release of mouse mammary tumor virus. J. Virol. 84:11555–11559. 10.1128/JVI.00652-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zabransky A, Sakalian M, Pichova I. 2005. Localization of self-interacting domains within betaretrovirus Gag polyproteins. Virology 332:659–666. 10.1016/j.virol.2004.12.007 [DOI] [PubMed] [Google Scholar]
- 6.Mirambeau G, Lyonnais S, Gorelick RJ. 2010. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 7:724–734. 10.4161/rna.7.6.13777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogt VM. 1997. Retroviral virions and genomes, p 27–69 In Coffin JM, Hughes SH, Varmus HE. (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 8.Larson DR, Johnson MC, Webb WW, Vogt VM. 2005. Visualization of retrovirus budding with correlated light and electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 102:15453–15458. 10.1073/pnas.0504812102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beliakova-Bethell N, Beckham C, Giddings TH, Jr, Winey M, Parker R, Sandmeyer S. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94–101. 10.1261/rna.2264806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Checkley MA, Nagashima K, Lockett SJ, Nyswaner KM, Garfinkel DJ. 2010. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol. Cell. Biol. 30:382–398. 10.1128/MCB.00251-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandmeyer SB, Clemens KA. 2010. Function of a retrotransposon nucleocapsid protein. RNA Biol. 7:642–654. 10.4161/rna.7.6.14117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson P, Kedersha N. 2006. RNA granules. J. Cell Biol. 172:803–808. 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson P, Kedersha N. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell. Biol. 10:430–436. 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- 14.Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150. 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- 15.Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO. 2006. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870–32878. 10.1074/jbc.M606149200 [DOI] [PubMed] [Google Scholar]
- 16.Franks TM, Lykke-Andersen J. 2008. The control of mRNA decapping and P-body formation. Mol. Cell 32:605–615. 10.1016/j.molcel.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedersha N, Anderson P. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81. 10.1016/S0076-6879(07)31005-7 [DOI] [PubMed] [Google Scholar]
- 18.Kedersha N, Anderson P. 2009. Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci. 90:155–185. 10.1016/S1877-1173(09)90004-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kedersha N, Anderson P. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963–969 [DOI] [PubMed] [Google Scholar]
- 20.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884. 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. 284:C273–C284. 10.1152/ajpcell.00314.2002 [DOI] [PubMed] [Google Scholar]
- 22.Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D. 2008. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell 19:4469–4479. 10.1091/mbc.E08-05-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker R, Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25:635–646. 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- 24.Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805–808. 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheth U, Parker R. 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125:1095–1109. 10.1016/j.cell.2006.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371–382. 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montero H, Trujillo-Alonso V. 2011. Stress granules in the viral replication cycle. Viruses 3:2328–2338. 10.3390/v3112328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pothof J, Verkaik NS, Hoeijmakers JH, van Gent DC. 2009. MicroRNA responses and stress granule formation modulate the DNA damage response. Cell Cycle 8:3462–3468. 10.4161/cc.8.21.9835 [DOI] [PubMed] [Google Scholar]
- 30.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383–5398. 10.1091/mbc.E04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831. 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Davydova EK, Evdokimova VM, Ovchinnikov LP, Hershey JW. 1997. Overexpression in COS cells of p50, the major core protein associated with mRNA, results in translation inhibition. Nucleic Acids Res. 25:2911–2916. 10.1093/nar/25.14.2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evdokimova VM, Ovchinnikov LP. 1999. Translational regulation by Y-box transcription factor: involvement of the major mRNA-associated protein, p50. Int. J. Biochem. Cell Biol. 31:139–149. 10.1016/S1357-2725(98)00137-X [DOI] [PubMed] [Google Scholar]
- 34.Bloch DB, Nobre RA, Yang WH. 2013. GW/P-bodies and autoimmune disease. Adv. Exp. Med. Biol. 768:61–70. 10.1007/978-1-4614-5107-5_5 [DOI] [PubMed] [Google Scholar]
- 35.Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. 2011. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Mosc.) 76:1402–1433. 10.1134/S0006297911130049 [DOI] [PubMed] [Google Scholar]
- 36.Paranjape SM, Harris E. 2007. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 282:30497–30508. 10.1074/jbc.M705755200 [DOI] [PubMed] [Google Scholar]
- 37.Li W, Wang X, Gao G. 2012. Expression of YB-1 enhances production of murine leukemia virus vectors by stabilizing genomic viral RNA. Protein Cell 3:943–949. 10.1007/s13238-012-2090-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu X, Li W, Wang X, Gao G. 2013. YB-1 stabilizes HIV-1 genomic RNA and enhances viral production. Protein Cell 4:591–597. 10.1007/s13238-013-3011-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dzuris JL, Golovkina TV, Ross SR. 1997. Both T and B cells shed infectious mouse mammary tumor virus. J. Virol. 71:6044–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jude BA, Pobezinskaya Y, Bishop J, Parke S, Medzhitov RM, Chervonsky AV, Golovkina TV. 2003. Subversion of the innate immune system by a retrovirus. Nat. Immunol. 4:573–578. 10.1038/ni926 [DOI] [PubMed] [Google Scholar]
- 41.Moscovici C, Moscovici MG, Jimenez H, Lai MM, Hayman MJ, Vogt PK. 1977. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell 11:95–103. 10.1016/0092-8674(77)90320-8 [DOI] [PubMed] [Google Scholar]
- 42.Craven RC, Leure-duPree AE, Weldon RA, Jr, Wills JW. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, Malim MH. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81:2165–2178. 10.1128/JVI.02287-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson MC, Scobie HM, Ma YM, Vogt VM. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177–11185. 10.1128/JVI.76.22.11177-11185.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swanson CM, Puffer BA, Ahmad KM, Doms RW, Malim MH. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 23:2632–2640. 10.1038/sj.emboj.7600270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2:924–932. 10.1038/nprot.2007.132 [DOI] [PubMed] [Google Scholar]
- 47.Palmer BR, Marinus MG. 1994. The dam and dcm strains of Escherichia coli—a review. Gene 143:1–12. 10.1016/0378-1119(94)90597-5 [DOI] [PubMed] [Google Scholar]
- 48.Jaskolski F, Mulle C, Manzoni OJ. 2005. An automated method to quantify and visualize colocalized fluorescent signals. J. Neurosci. Methods 146:42–49. 10.1016/j.jneumeth.2005.01.012 [DOI] [PubMed] [Google Scholar]
- 49.Bolte S, Cordelieres FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- 50.Beyer AR, Bann DV, Rice B, Pultz IS, Kane M, Goff SP, Golovkina TV, Parent LJ. 2013. Nucleolar trafficking of the mouse mammary tumor virus Gag protein induced by interaction with ribosomal protein L9. J. Virol. 87:1069–1082. 10.1128/JVI.02463-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelle TD, Wills JW. 1996. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J. Virol. 70:2269–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phair RD, Gorski SA, Misteli T. 2004. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375:393–414. 10.1016/S0076-6879(03)75025-3 [DOI] [PubMed] [Google Scholar]
- 53.Rapsomaniki MA, Kotsantis P, Symeonidou IE, Giakoumakis NN, Taraviras S, Lygerou Z. 2012. easyFRAP: an interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics 28:1800–1801. 10.1093/bioinformatics/bts241 [DOI] [PubMed] [Google Scholar]
- 54.Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, Clement JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, Boccaccio G, Mouland AJ. 2010. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 123:369–383. 10.1242/jcs.055897 [DOI] [PubMed] [Google Scholar]
- 55.Legros S, Boxus M, Gatot JS, Van Lint C, Kruys V, Kettmann R, Twizere JC, Dequiedt F. 2011. The HTLV-1 Tax protein inhibits formation of stress granules by interacting with histone deacetylase 6. Oncogene 30:4050–4062. 10.1038/onc.2011.120 [DOI] [PubMed] [Google Scholar]
- 56.Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. 1992. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 103:857–862 [DOI] [PubMed] [Google Scholar]
- 57.Manders EMM, Verbeek FJ, Aten JA. 1993. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169:375–382. 10.1111/j.1365-2818.1993.tb03313.x [DOI] [PubMed] [Google Scholar]
- 58.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23:1313–1324. 10.1038/sj.emboj.7600163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Girotti M, Banting G. 1996. TGN38-green fluorescent protein hybrid proteins expressed in stably transfected eukaryotic cells provide a tool for the real-time, in vivo study of membrane traffic pathways and suggest a possible role for ratTGN38. J. Cell Sci. 109:2915–2926 [DOI] [PubMed] [Google Scholar]
- 60.Uchiumi T, Fotovati A, Sasaguri T, Shibahara K, Shimada T, Fukuda T, Nakamura T, Izumi H, Tsuzuki T, Kuwano M, Kohno K. 2006. YB-1 is important for an early stage embryonic development: neural tube formation and cell proliferation. J. Biol. Chem. 281:40440–40449. 10.1074/jbc.M605948200 [DOI] [PubMed] [Google Scholar]
- 61.Sorokin AV, Selyutina AA, Skabkin MA, Guryanov SG, Nazimov IV, Richard C, Th'ng J, Yau J, Sorensen PH, Ovchinnikov LP, Evdokimova V. 2005. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA-damage stress response. EMBO J. 24:3602–3612. 10.1038/sj.emboj.7600830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fukuda T, Ashizuka M, Nakamura T, Shibahara K, Maeda K, Izumi H, Kohno K, Kuwano M, Uchiumi T. 2004. Characterization of the 5′-untranslated region of YB-1 mRNA and autoregulation of translation by YB-1 protein. Nucleic Acids Res. 32:611–622. 10.1093/nar/gkh223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skabkina OV, Lyabin DN, Skabkin MA, Ovchinnikov LP. 2005. YB-1 autoregulates translation of its own mRNA at or prior to the step of 40S ribosomal subunit joining. Mol. Cell. Biol. 25:3317–3323. 10.1128/MCB.25.8.3317-3323.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chung HY, Morita E, von Schwedler U, Muller B, Krausslich HG, Sundquist WI. 2008. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 82:4884–4897. 10.1128/JVI.02667-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bann DV, Parent LJ. 2012. Application of live-cell RNA imaging techniques to the study of retroviral RNA trafficking. Viruses 4:963–979. 10.3390/v4060963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Golovkina TV, Chervonsky A, Prescott JA, Janeway CA, Jr, Ross SR. 1994. The mouse mammary tumor virus envelope gene product is required for superantigen presentation to T cells. J. Exp. Med. 179:439–446. 10.1084/jem.179.2.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Golovkina TV, Jaffe AB, Ross SR. 1994. Coexpression of exogenous and endogenous mouse mammary tumor virus RNA in vivo results in viral recombination and broadens the virus host range. J. Virol. 68:5019–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hook LM, Agafonova Y, Ross SR, Turner SJ, Golovkina TV. 2000. Genetics of mouse mammary tumor virus-induced mammary tumors: linkage of tumor induction to the gag gene. J. Virol. 74:8876–8883. 10.1128/JVI.74.19.8876-8883.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mustafa F, Al Amri D, Al Ali F, Al Sari N, Al Suwaidi S, Jayanth P, Philips PS, Rizvi TA. 2012. Sequences within both the 5′ UTR and Gag are required for optimal in vivo packaging and propagation of mouse mammary tumor virus (MMTV) genomic RNA. PLoS One 7:e47088. 10.1371/journal.pone.0047088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rizvi TA, Ali J, Phillip PS, Ghazawi A, Jayanth P, Mustafa F. 2009. Role of a heterologous retroviral transport element in the development of genetic complementation assay for mouse mammary tumor virus (MMTV) replication. Virology 385:464–472. 10.1016/j.virol.2008.12.027 [DOI] [PubMed] [Google Scholar]
- 71.Mertz JA, Chadee AB, Byun H, Russell R, Dudley JP. 2009. Mapping of the functional boundaries and secondary structure of the mouse mammary tumor virus Rem-responsive element. J. Biol. Chem. 284:25642–25652. 10.1074/jbc.M109.012476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mullner M, Salmons B, Gunzburg WH, Indik S. 2008. Identification of the Rem-responsive element of mouse mammary tumor virus. Nucleic Acids Res. 36:6284–6294. 10.1093/nar/gkn608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Austin RJ, Xia T, Ren J, Takahashi TT, Roberts RW. 2002. Designed arginine-rich RNA-binding peptides with picomolar affinity. J. Am. Chem. Soc. 124:10966–10967. 10.1021/ja026610b [DOI] [PubMed] [Google Scholar]
- 74.Daigle N, Ellenberg J. 2007. LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat. Methods 4:633–636. 10.1038/nmeth1065 [DOI] [PubMed] [Google Scholar]
- 75.Ivanchenko S, Godinez WJ, Lampe M, Krausslich HG, Eils R, Rohr K, Brauchle C, Muller B, Lamb DC. 2009. Dynamics of HIV-1 assembly and release. PLoS Pathog. 5:e1000652. 10.1371/journal.ppat.1000652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin J, Sherer NM, Heidecker G, Derse D, Mothes W. 2009. Assembly of the murine leukemia virus is directed towards sites of cell-cell contact. PLoS Biol. 7:e1000163. 10.1371/journal.pbio.1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jouvenet N, Bieniasz PD, Simon SM. 2008. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454:236–240. 10.1038/nature06998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 13:394–401. 10.1038/ncb2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Minich WB, Maidebura IP, Ovchinnikov LP. 1993. Purification and characterization of the major 50-kDa repressor protein from cytoplasmic mRNP of rabbit reticulocytes. Eur. J. Biochem. 212:633–638. 10.1111/j.1432-1033.1993.tb17701.x [DOI] [PubMed] [Google Scholar]
- 80.Minich WB, Ovchinnikov LP. 1992. Role of cytoplasmic mRNP proteins in translation. Biochimie 74:477–483. 10.1016/0300-9084(92)90088-V [DOI] [PubMed] [Google Scholar]
- 81.Lyabin DN, Eliseeva IA, Ovchinnikov LP. 2014. YB-1 protein: functions and regulation. Wiley Interdiscip. Rev. RNA 5:95–110. 10.1002/wrna.1200 [DOI] [PubMed] [Google Scholar]
- 82.Chernov KG, Barbet A, Hamon L, Ovchinnikov LP, Curmi PA, Pastre D. 2009. Role of microtubules in stress granule assembly: microtubule dynamical instability favors the formation of micrometric stress granules in cells. J. Biol. Chem. 284:36569–36580. 10.1074/jbc.M109.042879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Evdokimova VM, Kovrigina EA, Nashchekin DV, Davydova EK, Hershey JW, Ovchinnikov LP. 1998. The major core protein of messenger ribonucleoprotein particles (p50) promotes initiation of protein biosynthesis in vitro. J. Biol. Chem. 273:3574–3581. 10.1074/jbc.273.6.3574 [DOI] [PubMed] [Google Scholar]
- 84.Skabkin MA, Kiselyova OI, Chernov KG, Sorokin AV, Dubrovin EV, Yaminsky IV, Vasiliev VD, Ovchinnikov LP. 2004. Structural organization of mRNA complexes with major core mRNP protein YB-1. Nucleic Acids Res. 32:5621–5635. 10.1093/nar/gkh889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chatel-Chaix L, Melancon P, Racine ME, Baril M, Lamarre D. 2011. Y-box-binding protein 1 interacts with hepatitis C virus NS3/4A and influences the equilibrium between viral RNA replication and infectious particle production. J. Virol. 85:11022–11037. 10.1128/JVI.00719-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212. 10.1016/j.chom.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kedersha N, Tisdale S, Hickman T, Anderson P. 2008. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448:521–552. 10.1016/S0076-6879(08)02626-8 [DOI] [PubMed] [Google Scholar]
- 88.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719–723. 10.1038/ncb1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moser JJ, Fritzler MJ. 2013. Relationship of other cytoplasmic ribonucleoprotein bodies (cRNPB) to GW/P bodies. Adv. Exp. Med. Biol. 768:213–242. 10.1007/978-1-4614-5107-5_13 [DOI] [PubMed] [Google Scholar]
- 90.Moser JJ, Fritzler MJ. 2010. Cytoplasmic ribonucleoprotein (RNP) bodies and their relationship to GW/P bodies. Int. J. Biochem. Cell Biol. 42:828–843. 10.1016/j.biocel.2009.11.018 [DOI] [PubMed] [Google Scholar]
- 91.Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, Gilbert N. 2010. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 6:e1001150. 10.1371/journal.pgen.1001150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr 2007. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell Biol. 27:6469–6483. 10.1128/MCB.00332-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989–12000. 10.1128/JVI.76.23.11989-12000.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reed JC, Molter B, Geary CD, McNevin J, McElrath J, Giri S, Klein KC, Lingappa JR. 2012. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J. Cell Biol. 198:439–456. 10.1083/jcb.201111012 [DOI] [PMC free article] [PubMed] [Google Scholar]