ABSTRACT
RIG-I is a cytosolic sensor critically involved in the activation of the innate immune response to RNA virus infection. In the present study, we evaluated the inhibitory effect of a RIG-I agonist on the replication of two emerging arthropod-borne viral pathogens, dengue virus (DENV) and chikungunya virus (CHIKV), for which no therapeutic options currently exist. We demonstrate that when a low, noncytotoxic dose of an optimized 5′triphosphorylated RNA (5′pppRNA) molecule was administered, RIG-I stimulation generated a robust antiviral response against these two viruses. Strikingly, 5′pppRNA treatment before or after challenge with DENV or CHIKV provided protection against infection. In primary human monocytes and monocyte-derived dendritic cells, the RIG-I agonist blocked both primary infection and antibody-dependent enhancement of DENV infection. The protective response against DENV and CHIKV induced by 5′pppRNA was dependent on an intact RIG-I/MAVS/TBK1/IRF3 axis and was largely independent of the type I IFN response. Altogether, this in vitro analysis of the antiviral efficacy of 5′pppRNA highlights the therapeutic potential of RIG-I agonists against emerging viruses such as DENV and CHIKV.
IMPORTANCE DENV and CHIKV are two reemerging mosquito-borne viruses for which no therapeutic options currently exist. Both viruses overlap geographically in tropical regions of the world, produce similar fever-like symptoms, and are difficult to diagnose. This study investigated the inhibitory effect of a RIG-I agonist on the replication of these two viruses. RIG-I stimulation using 5′pppRNA before or after DENV or CHIKV infection generated a protective antiviral response against both pathogens in immune and nonimmune cells; interestingly, the protective response against the viruses was largely independent of the classical type I interferon response. The antiviral efficacy of 5′pppRNA highlights the therapeutic potential of RIG-I agonists against emerging viruses such as DENV and CHIKV.
INTRODUCTION
During infection, viral nucleic acids are the main pathogen-associated molecular patterns (PAMPs) recognized by the innate immune system (1). Sensing of PAMPs results in the control of the first waves of viral infection through the production of antiviral effector molecules and contributes to the mobilization of the adaptive arm of the immune response (2–4). Double-stranded RNA (dsRNA), generated during the replicative cycle of many viruses, is sensed by receptors such as Toll-like receptor 3 (TLR3) and different members of the RIG-I-like receptor (RLR) family, including RIG-I (retinoic acid-inducible gene I), MDA5 (melanoma differentiation factor 5), and LGP-2 (laboratory of genetics and physiology-2). RIG-I and MDA5 consist of two N-terminal caspase activation and recruitment domains (CARD), a DExD/H-box RNA helicase-sensing domain, and a C-terminal regulatory domain (RD). LGP-2 contains the RNA helicase-sensing domain and the RD but lacks the CARD (4–8).
Viral RNA extracted from infected cells has been shown to potently activate RIG-I (9, 10). Chemically or enzymatically synthesized dsRNA molecules bearing an exposed 5′-triphosphate end (5′ppp) were first identified as RIG-I inducers (11, 12), with the presence of the 5′ppp moiety being essential for RIG-I activation. Further characterization of a potent RIG-I ligand demonstrated that the presence of a blunt base pairing at the 5′ end, as well as a minimum length of 20 nucleotides, were essential for optimal RIG-I recognition of the molecule (11, 12). While short dsRNAs bearing a 5′ppp end are preferentially recognized by RIG-I, long dsRNA lacking the triphosphate moiety, such as poly(I·C), are recognized by TLR3 and MDA5 (13). More recently, a SELEX technology identified RNA aptamers that specifically target the RIG-I protein. The selected aptamers contained poly(U) motifs that were crucial for RIG-I activation of the immune response but, unexpectedly, activated RIG-I in a 5′-triphosphate-independent manner (14).
Binding of 5′ppp dsRNA to RIG-I leads to a conformational alteration, resulting in dissociation of the CARD from the helicase domain and exposure of the CARD (15, 16). This conformational change results in the generation of an active state characterized by ATP hydrolysis and ATP-driven translocation of RNA along the RIG-I molecule (15–18). RIG-I first forms a small binding unit upon recognition of the 5′ppp dsRNA, which occurs independently of ATP binding (19). In a second step, RIG-I oligomerizes on the 5′ppp dsRNA in an ATP hydrolysis-dependent manner, and the length of dsRNA dictates the strength of the type I interferon (IFN) response (19). Activated RIG-I is then able to interact with its mitochondrial adaptor MAVS via a CARD-CARD interaction. MAVS triggers the activation of IRF3, IRF7, and NF-κB through the IKK-related kinases TBK1 and IKKε, leading to the induction of type I IFN (IFN-β and IFN-α), proinflammatory cytokines, and selected antiviral genes, such as IFN-stimulated gene 15 (ISG15), ISG54, and ISG56 (20). Expansion of the antiviral response is then driven by the binding of type I IFN on its receptor, which activates the induction of hundreds of ISGs through the JAK-STAT pathway (3, 4, 21–24).
Given the importance of the innate immune response for host survival, TLR and RLR agonists have been the subject of intense study. Treatment with agonists of TLRs 2, 3, 4, 5, 7, and 9 inhibited hepatitis B virus as well as herpes simplex virus-2 replication in a type I IFN-dependent manner (25–27). Furthermore, pretreatment of cells with poly(I·C) also inhibited the replication of hepatitis C virus (HCV), human immunodeficiency virus (HIV), influenza virus, respiratory syncytial virus (RSV), DENV, and CHIKV (28–34). More recently, an RNA-based agonist of RIG-I was shown to block the replication of multiple viruses, including influenza virus, HIV, HCV, vesicular stomatitis virus (VSV), and vaccinia virus in vitro, as well as influenza virus in vivo (35). This broad-spectrum antiviral activity was in part attributed to the potent and specific stimulation of antiviral and inflammatory genes through IRF3/7, STAT1, and NF-κB transcription factors (35).
DENV and CHIKV are arthropod-borne viruses belonging to the Flavivirus and Alphavirus genera, respectively. Illness caused by CHIKV is usually diagnosed based on febrile symptoms and arthralgia and is often confused with dengue fever, given the similarities in clinical signs. CHIKV causes a severe arthralgia that may persist for many months but is not associated with the hemorrhagic fever that develops in a small proportion of severe dengue cases. Most DENV infections are asymptomatic or cause a self-limiting dengue fever, while a small proportion of infections leads to severe and potentially lethal manifestations, such as dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), which are associated with antibody-dependent enhanced infections (36–38). Dengue fever, with millions of cases reported each year (36, 37), is already a leading infectious disease in tropical areas, while chikungunya fever is a lesser-known disease also affecting the same subtropical regions of the world. After a 50-year period of relative quiescence (39), CHIKV has reemerged with millions of estimated cases since 2005 (40, 41). The dramatic geographic expansion and increased incidence of DENV infections, as well as the emergence of CHIKV strains with an increased epidemic potential, highlight the increased burden of both viruses in tropical regions. The current lack of vaccines and effective antivirals stresses the importance of investigating new strategies to combat these serious human pathogens. Ideally, such strategies should target both viruses, as they cause similar symptoms, have an overlapping geographic distribution, and occur in regions in which the capacity to perform (differential) diagnosis is often limited.
The present study describes the protective innate immune response against DENV and CHIKV infection triggered by a well-characterized 5′ triphosphorylated RIG-I agonist. The current study demonstrates that treatment with 5′pppRNA triggers a protective antiviral response sufficient to prevent DENV and CHIKV infection in both immune and nonimmune cells. The protective antiviral response was largely independent of the IFN-α/β receptor (IFNAR)/STAT1 axis but dependent on an intact RIG-I/MAVS/TBK1/IRF3 axis.
MATERIALS AND METHODS
In vitro synthesis of 5′pppRNA.
The sequence of 5′pppRNA was derived from the 5′ and 3′ untranslated regions (UTR) of the VSV genome as previously described (15). In vitro-transcribed RNA was prepared as previously described (35). Briefly, RNA was prepared using the Ambion MEGAscript T7 kit according to the manufacturer's guidelines (Invitrogen, NY, USA). 5′pppRNA was purified using the Qiagen miRNA minikit (Qiagen, Valencia, CA). An RNA with the same sequence but lacking the 5′ppp moiety was purchased from IDT (Integrated DNA Technologies Inc., IA, USA). This RNA generated results identical to those obtained with 5′pppRNA that was dephosphorylated enzymatically with calf intestinal alkaline phosphatase (Invitrogen, NY, USA).
Cell culture and transfections.
A549 cells were grown in F12K medium (ATCC, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) and antibiotics. C6/36 insect cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS and antibiotics. Lipofectamine RNAiMax (Invitrogen, NY, USA) was used for transfections of 5′pppRNA in A549 cells according to the manufacturer's instructions. For short interfering RNA (siRNA) knockdown, A549 cells were transfected with 50 nM (30 pmol) human RIG-I (sc-6180), IFN-α/βR α chain (sc-35637) and β chain (sc-40091), STING (sc-92042), TLR3 (sc-36685), MDA5 (sc-61010), MAVS (sc-75755), interleukin-28R (IL-28R; sc-62497), IL-10Rβ (sc-75331), STAT1 p844/91 (sc-44123), IRF1 (sc-35706), IRF3 (sc-35710), IRF7 (sc-38011), and control siRNA (sc-37007) (Santa Cruz Biotechnology, Dallas, T) using Lipofectamine RNAiMax according to the manufacturer's guidelines.
MRC-5 cells (ATCC CCL-171) were grown in Earle's minimum essential medium (EMEM) supplemented with 10% FBS, 2 mM l-glutamine, 1% nonessential amino acids (PAA), and antibiotics. For siRNA-mediated knockdown of gene expression, MRC-5 cells were transfected with 16.7 nM (10 pmol) siRNA using Dharmafect1 (Dharmacon) according to the manufacturer's guidelines.
Mouse embryonic fibroblast cells (MEFs) were grown in DMEM with 10% FBS and antibiotics.
Primary cell isolation.
Human peripheral blood mononuclear cells (PBMC) were isolated from the blood of healthy volunteers in a study approved by the institutional review board and by the VGTI-FL Institutional Biosafety Committee (2011-6-JH1). Written informed consent, approved by the VGTI-FL Inc. ethics review board (FWA number 161), was provided and signed by study participants. Research conformed to ethical guidelines established by the ethics committee of the OHSU VGTI and Martin Health System. Briefly, PBMC were isolated from freshly collected blood using Ficoll-Paque plus medium (GE Healthcare Bio, Uppsala, Sweden) per the manufacturer's instructions. Monocytes were then isolated using the negative selection human monocyte enrichment kit (Stem Cell, Vancouver, Canada) per the kit's instructions and used for further experiments. To obtain monocyte-derived dendritic cells (MDDC), monocytes were allowed to adhere to 100-mm dishes for 1 h in serum-free RPMI at 37°C. After adherence, remaining platelets and nonadherent cells were removed by two washes with serum-free RPMI. The cells were differentiated into MDDC by culturing for 7 days in MDDC differentiation medium (Miltenyi Biotec, Auburn, GA). Medium was replenished after 3 days of differentiation.
Virus production, quantification, and infection.
Confluent monolayers of C6/36 insect cells were infected with DENV serotype 2 strain New Guinea C (DENV NGC) at a multiplicity of infection (MOI) of 0.5. Virus was allowed to adsorb for 1 h at 28°C in a minimal volume of serum-free DMEM. After adsorption, the monolayer was washed once with serum-free medium and covered with DMEM containing 2% FBS. After 7 days of infection, medium was harvested, cleared by centrifugation (500 × g, 5 min), and concentrated down by centrifugation (2,000 × g, 8 min) through a 15-ml Millipore Amicon centrifugal filter unit (Millipore, Billerica, MA). The virus was concentrated by ultracentrifugation on a sucrose density gradient (20% sucrose cushion) using a Sorvall WX 100 ultracentrifuge (ThermoScientific, Rockford, IL) for 2 h at 134,000 × g and 10°C with the brake turned off. Concentrated virus was then washed to remove sucrose using a 15-ml Amicon tube. After 2 washes, the virus was resuspended in DMEM plus 0.1% bovine serum albumin (BSA) and stored at −80°C. Titers of DENV stocks were determined by fluorescence-activated cell sorting (FACS), infecting Vero cells with 10-fold serial dilutions of the stock, and then immunofluorescence staining of intracellular DENV E protein at 24 h postinfection (p.i.). Titers were expressed as IU/ml. DENV titers in cell culture supernatants from 5′ppp-treated and control cells were determined by plaque assay on confluent Vero cells. Cells in 6-well clusters were incubated with 10-fold serial dilutions of the sample in a total volume of 500 μl of DMEM without serum. After 1 h of infection, the inoculum was removed and cells were overlaid with 3 ml of 2% agarose in complete DMEM. The cells were fixed and stained, and plaques were counted 5 days postinfection.
In infection experiments, A549 cells, monocytes, or MDDC were infected in a small volume of medium without FBS for 1 h at 37°C and then incubated with complete medium for 24 to 72 h prior to analysis. All procedures with live DENV were performed in a biosafety level 2+ facility at the Vaccine and Gene Therapy Institute–Florida.
CHIKV strain LS3 and enhanced green fluorescent protein (EGFP)-expressing reporter virus CHIKV LS3-GFP have been described (42). Virus production, titration, and infection were performed essentially as described previously (42). Working stocks of CHIKV were routinely produced in Vero E6 cells at 37°C, and infections were performed in EMEM with 25 mM HEPES (Lonza) supplemented with 2% fetal calf serum (FCS), l-glutamine, and antibiotics. After 1 h, the inoculum was replaced with fresh culture medium. All procedures with live CHIKV were performed in a biosafety level 3 facility at the Leiden University Medical Center.
Flow cytometry analysis.
The percentage of cells infected with DENV was determined by standard intracellular staining (ICS) with a mouse IgG2a monoclonal antibody (MAb) specific for DENV-E protein (clone 4G2), followed by staining with a secondary anti-mouse antibody coupled to phycoerythrin (PE) (BioLegend, San Diego, CA). Cells were analyzed on an LSRII flow cytometer (Becton, Dickinson, New Jersey, USA). Calculations as well as population analyses were done using FACS Diva software.
Cell viability analysis.
Cell surface expression of phosphatidylserine was measured using an allophycocyanin (APC)-conjugated annexin V antibody, as recommended by the manufacturer (BioLegend, San Diego, CA). Briefly, specific annexin V binding was achieved by incubating A549 cells in annexin V binding buffer (Becton, Dickinson, NJ, USA) containing a saturating concentration of APC-annexin V antibody and 7-amino-actinomycin D (7-AAD) (Becton, Dickinson, New Jersey, USA) for 15 min in the dark. APC-annexin V and 7-AAD binding to the cells was analyzed by flow cytometry, as described previously, using an LSRII flow cytometer and FACS Diva software. Alternatively, the viability of siRNA- or 5′pppRNA-transfected cells was assessed using the CellTiter 96 aqueous nonradioactive cell proliferation assay (Promega). Absorbance was measured using a Berthold Mithras LB 940 96-well plate reader.
Protein extraction and immunoblot analysis.
DENV-infected cells were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8, 1% sodium deoxycholate, 1% NP-40, 5 mM EDTA, 150 mM NaCl, 0.1% sodium dodecyl sulfate), and the insoluble fraction was removed by centrifugation at 17,000 × g for 15 min (4°C). Protein concentration was determined using the Pierce bicinchoninic (BCA) protein assay kit (Thermo Scientific, Rockford, IL). Protein extracts were resolved by SDS-PAGE on 4 to 20% acrylamide Mini-Protean TGX precast gels (Bio-Rad, Hercules, CA) in a 1× Tris-glycine-SDS buffer (Bio-Rad, Hercules, CA). Proteins were electrophoretically transferred to an Immobilon-PSQ polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA) for 1 h at 100 V in a buffer containing 30 mM Tris, 200 mM glycine, and 20% methanol. Membranes were blocked for 1 h at room temperature in Odyssey blocking buffer (Odyssey, USA) and then probed with the following primary antibodies: anti-IRF1 (Santa Cruz Biotechnology, Dallas, TX), anti-pIRF3 at Ser 396 (EMD Millipore, MA, USA), anti-IRF3 (IBL, Japan), anti-IRF7 (Cell Signaling, MA, USA), anti-RIG-I (EMD Millipore, MA, USA), anti-IFIT1 (Thermo Fisher Scientific, Rockford, IL, USA), anti-ISG15 (Cell Signaling Technology, Danvers, MA), anti-pSTAT1 at Tyr701 (Cell Signaling, MA, USA), anti-STAT1 (Cell Signaling, MA, USA), anti-STING (Novus Biologicals, Littleton, CO), anti-DENV (Santa Cruz Biotechnology, USA), and anti-β-actin (Odyssey, USA). Antibody signals were detected by immunofluorescence using the IRDye 800CW and IRDye 680RD secondary antibodies (Odyssey, USA) and the LiCor imager (Odyssey, USA). Protein expression levels were determined and normalized to β-actin using ImageJ software (National Institutes of Health, Bethesda, MD).
CHIKV-infected cells were lysed and proteins were analyzed by Western blotting as described previously (42). CHIKV proteins were detected with rabbit antisera against nsP1 (a generous gift of Andres Merits, University of Tartu, Estonia) and E2 (43). Mouse monoclonal antibodies against β-actin (Sigma), the transferrin receptor (Zymed), cyclophilin A (Abcam), and cyclophilin B (Abcam) were used for detection of loading controls. Biotin-conjugated swine α-rabbit (Dako), goat α-mouse (Dako), and Cy3-conjugated mouse α-biotin (Jackson) were used for fluorescent detection of the primary antibodies with a Typhoon-9410 scanner (GE Healthcare).
RT-qPCR.
Total RNA was isolated from cells using an RNeasy kit (Qiagen, Valencia, CA) per the manufacturer's instructions. RNA was reverse transcribed using the SuperScript VILO cDNA synthesis kit according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). PCR primers were designed using Roche's Universal Probe Library Assay Design Center (Roche). Quantitative reverse transcription-PCR (RT-qPCR) was performed on a LightCycler 480 system using LightCycler 480 probes master (Roche, Penzberg, Germany). All data are presented as a relative quantification with efficiency correction based on the relative expression of target gene versus glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the invariant control. The N-fold differential mRNA expression of genes in samples was expressed as 2ΔΔCT. Primers used in this study were the following: DENV2 (probe 5) forward, 5′-ATCCTCCTATGGTACGCACAAA-3′; reverse, 5′-CTCCAGTATTATTGAAGCTGCTATCC-3′; GAPDH (probe 60) forward, 5′-AGCCACATCGCTCAGACAC-3′; reverse, 5′-GCCCAATACGACCAAATCC-3′; IFNA2 (probe 49) forward, 5′-AATGGCCTTGACCTTTGCTT-3′; reverse, 5′-CACAGAGCAGCTTGACTTGC-3′; IFNAR1 (probe 65) forward, 5′-ATTTACACCATTTCGCAAAGC-3′; reverse, 5′-CACTATTGCCTTATCTTCAGCTTCTA-3′; IFNAR2 (probe 87) forward, 5′-TAGCCTCCCCAAAGTCTTGA-3′; reverse, 5′-AAATGACCTCCACCATATCCA-3′; IFNB1 (probe 20) forward, 5′-CTTTGCTATTTTCAGACAAGATTCA-3′; reverse, 5′-GCCAGGAGGTTCTCAACAAT-3′; ILA (probe 66) forward, 5′-TGACGCCCTCAATCAAAGTA-3′; reverse, 5′-TGACTTATAAGCACCCATGTCAA-3′; IL-6 (probe 7) forward, 5′-CAGGAGCCCAGCTATGAACT-3′; reverse, 5′-GAAGGCAGCAGGCAACAC-3′; IL28RA (probe 12) forward, 5′-CCCCCACTGGATCTGAAGTA-3′; reverse, 5′-GAGTGACTGGAAATAGGGTCTTG-3′; IL-29 (probe 75) forward, 5′-CCTGAGGCTTCTCCAGGTG-3′; reverse, 5′-CCAGGACCTTCAGCGTCA-3′; TNFA (probe 79) forward, 5′-GACAAGCCTGTAGCCCATGT-3′; reverse, 5′-TCTCAGCTCCACGCCATT-3′.
RNA isolation, denaturing agarose electrophoresis, and in-gel hybridization.
CHIKV RNA isolation and analysis were performed essentially as described previously (42). Briefly, total RNA was isolated by lysis in 20 mM Tris-HCl (pH 7.4), 100 mM LiCl, 2 mM EDTA, 5 mM dithiothreitol (DTT), 5% (wt/vol) lithium dodecyl sulfate, and 100 μg/ml proteinase K. After acid phenol (Ambion) extraction, RNA was precipitated with isopropanol, washed with 75% ethanol, and dissolved in 1 mM sodium citrate (pH 6.4). RNA samples were separated in 1.5% denaturing formaldehyde-agarose gels using the morpholinepropanesulfonic acid (MOPS) buffer system. RNA molecules were detected by direct hybridization of the dried gel with 32P-labeled oligonucleotides. CHIKV genomic and subgenomic RNAs (sgRNAs) were visualized with probe CHIKV-hyb4 (5′-TGTGGGTTCGGAGAATCGTGGAAGAGTT-3′), and negative-stranded RNA was detected with probe CHIKV-hyb2 (5′-AACCCATCATGGATCCTGTGTACGTGGA-3′). Probes (10 pmol) were labeled with 10 μCi [γ-32P]ATP (PerkinElmer). Prehybridization (1 h) and hybridization (overnight) were done at 55°C in 5× SSPE (0.9 M NaCl, 50 mM NaH2PO4, 5 mM EDTA, pH 7.4), 5× Denhardt's solution, 0.05% SDS, and 0.1 mg/ml homomix I. Storage Phosphor screens were exposed to hybridized gels and scanned with a Typhoon-9410 scanner (GE Healthcare), and data were quantified with Quantity One v4.5.1 (Bio-Rad).
Statistical analysis.
Values were expressed as the means ± standard errors of the means (SEM), and statistical analysis was performed with Microsoft Excel using an unpaired, two-tailed Student's t test to determine significance. Differences were considered significant at P < 0.05.
RESULTS
5′pppRNA inhibits DENV infection.
To determine the capacity of the 5′pppRNA RIG-I agonist to induce a protective antiviral response to DENV infection, A549 cells were challenged with DENV at different multiplicities of infection (MOI); infection and replication were monitored by flow cytometry, RT-qPCR, plaque assay, and immunoblotting (Fig. 1A to F). DENV established infection in A549 cells which was completely abrogated in cells pretreated with 1 ng/ml of 5′pppRNA (Fig. 1A). A similar antiviral effect was observed at higher concentrations of 5′pppRNA (10 ng/ml). Importantly, the antiviral effect was strictly dependent on the 5′ppp moiety, as transfection of cells with the identical RNA sequence lacking the 5′ppp did not prevent DENV infection (Fig. 1B). Pretreatment of cells with 5′pppRNA also led to an 8.5-fold decrease in DENV RNA synthesis (Fig. 1C). Release of infectious DENV was completely suppressed by 5′pppRNA treatment (4.3 × 106 PFU/ml in untreated cells versus undetectable in 5′pppRNA-treated cells) (Fig. 1D), leading to a complete inhibition of DENV E protein expression (Fig. 1D, lane 3). To compare the effect of 5′pppRNA to that of the dsRNA ligand poly(I·C), A549 cells were pretreated with 5′pppRNA or poly(I·C) (0.1 to 1 ng/ml) and subsequently challenged with DENV (Fig. 1E). Treatment with 1 ng/ml of 5′pppRNA almost completely suppressed DENV infection, whereas at the same concentration, only a 1.8-fold decrease of the number of DENV-infected cells was observed upon poly(I·C) treatment (Fig. 1E). Cytosolic delivery of dsRNA by transfection was required in A549 cells, as demonstrated by the absence of a protective antiviral effect in cells in medium to which 5 μg/ml of 5′pppRNA or poly(I·C) had just been added (Fig. 1E). To determine whether pretreatment with 5′pppRNA maintained a protective effect, A549 cells were transfected with 5′pppRNA prior to DENV challenge and the virus was allowed to replicate up to 72 h p.i. (Fig. 1F). The combination treatment completely inhibited DENV infection at all time points for up to 72 h p.i. (Fig. 1F). The viability of uninfected cells and cells protected from infection by 5′pppRNA was indistinguishable (Fig. 1G). Altogether, these results demonstrate the antiviral potential of 5′pppRNA against DENV infection in nonimmune cells.
FIG 1.
Pretreatment with 5′pppRNA inhibits DENV replication in vitro. (A and B) A549 cells were pretreated with various concentrations of 5′pppRNA (0.01 to 10 ng/ml) or control (Ctrl) RNA lacking the 5′ppp at the same concentrations for 24 h prior to DENV challenge. The percentage of DENV-infected cells was determined by intracellular staining (ICS) of DENV E protein expression using flow cytometry. Data are from two independent experiments performed in triplicate and represent the means ± SEM. *, P < 0.05. FSC, forward scatter. (C and D) A549 cells were pretreated with 5′pppRNA (1 ng/ml) for 24 h prior to DENV challenge (MOI, 0.1). DENV RNA level (C), viral titers (D), and DENV E protein expression level (D) were determined by RT-qPCR, plaque assay, and Western blotting, respectively. Error bars represent SEM from three independent samples. *, P < 0.05. One representative DENV E protein Western blot out of three independent triplicates is shown. (E) A549 cells were transfected using Lipofectamine (Lipo.) RNAiMax with increasing concentrations of 5′pppRNA and poly(I·C) (0.1 to 1 ng/ml) or treated with the same dsRNA sequences (5,000 ng/ml) in the absence of transfection reagent. Cells were then challenged with DENV (MOI, 1), and the percentage of infected cells was determined by FACS 24 h after infection. Data are the means ± SEM from two independent experiments performed in triplicate. *, P < 0.05. (F and G) The percentage of A549 DENV-infected cells and cell viability were assessed by flow cytometry and determined at 24 h (black bars), 48 h (gray bars), and 72 h (white bars) after DENV challenge (MOI, 0.01). Cells were pretreated with 5′pppRNA (1 ng/ml) for 24 h before DENV challenge. Data are the means ± SEM from a representative experiment performed in triplicate. *, P < 0.05.
To assess the potential of 5′pppRNA as a postinfection treatment, A549 cells were first infected with DENV, subsequently treated with the RIG-I agonist at 4 h and 8 h after infection, and analyzed 48 h later to detect DENV infection. Interestingly, infection was almost completely inhibited even when cells were treated at 8 h p.i., as shown by the 12.4-fold reduction of the number of DENV-infected cells (Fig. 2A). This suggests that as DENV replicates over time, 5′pppRNA prevents further spread of the virus by protecting uninfected cells and clearing virus from infected cells. The observed effects of 5′pppRNA on DENV infection were confirmed by RT-qPCR, yielding a 3.6-fold (+4 h) and 10.8-fold (+8 h) decrease in DENV viral RNA levels at 48 h p.i. (Fig. 2B). Cell viability was not significantly affected by a 24-h 5′pppRNA treatment, and an ∼20% decrease in viability was observed at 48 h p.i. in cells protected from infection by 5′pppRNA (Fig. 2C and D).
FIG 2.
Postinfection treatment with 5′pppRNA inhibits de novo DENV infection. (A) A549 cells were treated with 5′pppRNA (1 ng/ml) 4 h (black bars) or 8 h (gray bars) following DENV challenge (MOI, 0.01). The percentage of DENV-infected cells was determined by intracellular staining (ICS) of DENV E protein expression using flow cytometry at 48 h after infection. Data represent the means ± SEM from a representative experiment performed in triplicate. *, P < 0.05. (B) DENV RNA levels were determined by RT-qPCR (48 h after infection) on A549 cells treated with 5′pppRNA (1 ng/ml) 4 h (black bars) and 8 h (gray bars) after infection. *, P < 0.05. (C and D) Cell viability of A549 cells was measured by flow cytometry 24 h (black bars) and 48 h (gray bars) after infection. Cells were treated with 5′pppRNA 4 h after DENV infection. Data are the means ± SEM from a representative experiment performed in triplicate. (E) A549 cells were challenged with DENV (MOI, 0.1) for 4 h and transfected with 5′pppRNA (0.1 to 10 ng/ml) and incubated for an additional 20 h. Whole-cell extracts (WCEs) were prepared and subjected to immunoblot analysis 24 h postinfection. Data are from one representative experiment. (F) A549 cells were infected with DENV at different MOI and were transfected with 5′pppRNA (1 ng/ml) 4 h after infection. The expression level of genes was determined by RT-qPCR 24 h after DENV challenge. Data are the means ± SEM from a representative experiment performed in triplicate. *, P < 0.05.
To investigate the antiviral response triggered by 5′pppRNA, various signaling parameters were monitored by immunoblotting and RT-qPCR in cells treated with increasing doses of 5′pppRNA in the presence or absence of DENV infection (Fig. 2E and F). Interferon signaling was detected by immunoblotting in 5′pppRNA-treated cells, both in the presence or absence of DENV, as demonstrated by increased STAT1 Tyr701 phosphorylation and ISG expression of STAT1, RIG-I, and IFIT1 (Fig. 2E, lanes 2 to 8). Although DENV can evade the host innate response (44–46), we did not observe a significant inhibition of IFN signaling based on the expression of antiviral markers STAT1, RIG-I, and IFIT1 in infected or uninfected cells (Fig. 2E, lanes 2 to 8). 5′pppRNA treatment elicited a strong antiviral response in uninfected and DENV-infected A549 cells (Fig. 2E), and delivery of 5′pppRNA at 4 h p.i. potently stimulated type I IFN and inflammatory responses via the upregulation of genes, such as those of IFN-α, IFN-β, IL-6, and IL-1α (Fig. 2F).
5′pppRNA-restricted DENV infection requires an intact RIG-I pathway.
Introduction of RIG-I siRNA (10 and 30 pmol) into A549 cells severely reduced RIG-I as well as IFIT1 induction in response to 5′pppRNA treatment (Fig. 3A, lanes 5 to 8). Induction of the type I and type III IFNs, as well as the inflammatory response, were all dependent on intact RIG-I signaling, since the mRNA levels of IFN-β, IFN-α, IL-29, and tumor necrosis factor alpha (TNF-α) were drastically decreased in the absence of a functional RIG-I sensor (Fig. 3B). To explore the respective involvement of RIG-I, TLR3, and MDA5 in the 5′pppRNA-mediated anti-DENV effect, the expression of these immune sensors was knocked down in A549 cells by siRNA (Fig. 3C). While impairing RIG-I expression completely suppressed the 5′pppRNA-mediated antiviral effect, this was not the case upon knockdown of TLR3/MDA5 (Fig. 3C). The efficacy of poly(I·C) in preventing DENV infection was reduced to a larger extent in the absence of TLR3/MDA5 than in the absence of RIG-I, suggesting a predominant role for TLR3/MDA5 in mediating poly(I·C) antiviral effect in A549 cells (Fig. 3C). To demonstrate that the antiviral activity of 5′pppRNA against DENV relies on a functional RIG-I axis, the expression of RIG-I, STING, MAVS, and TBK1 was depleted in A549 cells using specific siRNAs. In addition, suitable knockout MEFs were used (Fig. 3D to F). Following 5′pppRNA treatment, DENV viral replication was assessed by flow cytometry. Whereas ∼35% of A549 cells were infected with DENV in the untreated population, the absence of RIG-I led to an ∼1.5-fold increase in the number of infected cells (Fig. 3D). Transient knockdown of RIG-I resulted in the abrogation of the protective response induced by 5′pppRNA in control cells (Fig. 3D), whereas the absence of STING did not affect DENV infection and did not significantly reduce the 5′pppRNA-induced antiviral response (Fig. 3D). Similar results were observed with A549 cells depleted for the mitochondrial adaptor MAVS; depletion of MAVS strongly reduced the 5′pppRNA-mediated protective antiviral response (Fig. 3E). Finally, TBK1-deficient MEFs were more susceptible to DENV infection than wild-type MEFs and were not responsive to the 5′pppRNA treatment, as demonstrated by the high level of DENV infection (Fig. 3F). In conclusion, 5′pppRNA treatment efficiently generates a RIG-I/MAVS/TBK1-dependent antiviral response that limits DENV infection in vitro.
FIG 3.
5′pppRNA inhibits DENV infection in vitro in a RIG-I/MAVS/TBK1-dependent manner. (A) A549 cells were transfected with control or RIG-I siRNA (10 or 30 pmol), and 48 h later they were treated with 5′pppRNA (10 ng/ml) for 24 h. Expression of IFIT1, RIG-I, and β-actin was evaluated by Western blotting. RIG-I knockdown and impairment of the 5′ppp-induced immune response is representative of at least 3 independent experiments. (B) A549 cells were transfected with control siRNA or RIG-I siRNA (30 pmol), and 48 h later they were treated with 5′pppRNA (10 ng/ml) for 24 h. mRNA expression level of IFN-β, IFN-α, TNF-α, and IL-29 was evaluated by RT-qPCR. Data are from a representative experiment performed in triplicate and show the means ± SEM. *, P < 0.05. (C) A549 cells were transfected with control (black bars), RIG-I (gray bars), or a combination of TLR3/MDA5 (white bars) siRNA (30 pmol each), and 48 h later they were treated with 5′pppRNA (10 ng/ml) or poly(I·C) (1 ng/ml). Cells were then infected with DENV (MOI, 0.5), and at 24 h p.i. the percentage of infected cells was assessed by intracellular staining of DENV E protein using flow cytometry. Data are from a representative experiment performed in triplicate and show the means ± SEM. *, P < 0.05. (D and E) A549 cells were treated with 5′pppRNA (0.1 to 10 ng/ml) for 24 h 2 days after transfection with 30 pmol of control (black bars), RIG-I (gray bars), or STING (white bars) siRNA (D) or with 30 pmol of control (black bars) or MAVS (gray bars) siRNA (E). Cells were then challenged with DENV (MOI, 0.1) for 24 h. The percentage of DENV-infected cells was determined by intracellular staining of DENV E protein and flow cytometry 24 h after infection. Data are the means ± SEM from a representative experiment performed in triplicate. *, P < 0.05. (F) TBK1+/+ (black bars) and TBK1−/− (gray bars) MEF cells were treated with 10 ng/ml of 5′pppRNA 24 h before DENV challenge at an MOI of 5. The percentage of DENV-infected cells was evaluated by flow cytometry. Data are the means ± SEM of a representative experiment performed in triplicate. *, P < 0.05.
5′pppRNA generates an IRF3-dependent and IFNAR/STAT1-independent antiviral protective effect.
To determine whether the potent RIG-I activation could compensate for the type I and type III IFN response, expression of the type I IFN receptor (IFN-α/βR) as well as the type III IFN receptor (IL-28R plus IL-10Rβ) was knocked down using siRNA in A549 cells (Fig. 4A to C). Expression of both type I and III IFN receptor was efficiently reduced, as shown by the downregulation of IFNAR1 (IFN-α/βR α chain), IFNAR2 (IFN-α/βR β chain), and IL-28R mRNA expression levels (Fig. 4A). Furthermore, knockdown of type I IFN signaling was highly efficient, as demonstrated by the reduction of IFIT1 and RIG-I induction following IFN-α2b stimulation (6.2-fold reduction of IFIT1 versus control siRNA [siCTRL]; Fig. 4B, lane 3 versus lane 6). Knocking down the type III IFN receptor did not interfere with the ability of 5′pppRNA and IFN-α2b to induce IFIT1 and RIG-I expression (Fig. 4B, lanes 2 and 3 versus lanes 8 and 9). Interestingly, induction of IFIT1 but not RIG-I was only partially reduced following 5′pppRNA treatment in the absence of type I IFN receptor (1.6-fold reduction of IFIT1 versus siCTRL; Fig. 4B, lane 2 versus lane 5), suggesting that certain ISGs were upregulated by 5′pppRNA in an IFN-independent manner. Knocking down expression of both type I and type III IFN receptors did not limit IFIT1 induction by 5′pppRNA, as the increase of IFIT1 was only reduced 1.9 times compared to the siRNA control (Fig. 4B). This type I and III IFN-independent activation of the innate system was sufficient to suppress DENV infection in A549 cells stimulated with a higher (10 ng/ml) but not a low dose (0.1 to 1 ng/ml) of 5′pppRNA (Fig. 4C). To further confirm that type I IFN signaling was not necessarily required to mediate an immune response to 5′pppRNA, STAT1 was depleted in A549 cells using siRNA (Fig. 4D, lanes 5 to 8). The increased expression of IFIT1 following 5′pppRNA treatment was not impacted by the absence of the STAT1 transcription factor (Fig. 4D, lanes 2 to 4 versus lanes 6 to 8). The STAT1-independent induction of the antiviral response was sufficient to block DENV infection in A549 cells stimulated with a high 5′pppRNA concentration (Fig. 4E). Finally, to determine which IRF transcription factor downstream of RIG-I was involved in the antiviral protective effect, IRF1, IRF3, and IRF7 expression was knocked down using siRNA (Fig. 4F). Depletion of these different transcription factors was highly efficient, as shown in Fig. 4F. Only IRF3 knockdown resulted in inhibition of the protective antiviral response generated by 5′pppRNA treatment. Indeed, the absence of either IRF1 or IRF7 did not impair 5′pppRNA-mediated antiviral protection (Fig. 4G). Altogether, these data demonstrate that the 5′pppRNA-mediated anti-DENV effect in vitro is largely independent of the type I or type III IFN responses but requires the activation of a functional RIG-I/IRF3 axis to mediate its protective effect.
FIG 4.
5′pppRNA-induced antiviral effect is IRF3 dependent but IFNAR and STAT1 independent. (A) A549 cells were transfected with control, IFN-α/βR α chain (IFNAR1), IFN-α/βR β chain (IFNAR2), or IL-28R siRNA, and 48 h later mRNA levels of IFNAR1, IFNAR2, and IL-28R were evaluated by RT-qPCR. Data are from a representative experiment performed in triplicate. *, P < 0.05 (B) A549 cells were transfected with the control siRNA, IFN-α/βR or IL-28R siRNA, or a combination of both. After 48 h, cells were treated with 5′pppRNA (10 ng/ml) or IFN-α2b (100 UI/ml) for 24 h. Expression of IFIT1, RIG-I, and β-actin was evaluated by Western blotting. The evaluation of 5′ppp-induced immune response by Western blotting in the absence of type I IFN receptor, representative of three independent experiments, and in the absence of type III IFN receptor, representative of one experiment. (C) After siRNA knockdown of IFN-α/βR as described for panel B, cells were treated with increasing concentrations of 5′pppRNA (0.1 to 10 ng/ml) and then infected with DENV (MOI, 0.1). The percentage of DENV-infected cells was evaluated by flow cytometry. Data are the means ± SEM of a representative experiment performed in triplicate. *, P < 0.05. (D) A549 cells were transfected with control and STAT1 siRNA, and 48 h later they were treated with 5′pppRNA (0.01 to 1 ng/ml) for 24 h. Expression of STAT1, IFIT1, and β-actin was evaluated by Western blotting. The induction of 5′ppp-induced immune response in the absence of STAT is representative of two independent experiments. (E) A549 cells were transfected with control or STAT1 siRNA and incubated for 48 h. Cells were treated with increasing concentrations of 5′pppRNA (0.1 to 10 ng/ml) and then infected with DENV (MOI, 0.1). The percentage of DENV-infected cells was evaluated by flow cytometry. Data are the means ± SEM from a representative experiment performed in triplicate. *, P < 0.05. (F) A549 cells were transfected with control, IRF1, IRF3, or IRF7 siRNA for 48 h, and the protein expression level of these transcription factors was evaluated by Western blotting. This panel is representative of one experiment. (G) A549 cells were transfected with control IRF1, IRF3, or IRF7 and then treated as described for panel E. The percentage of DENV-infected cells was evaluated by flow cytometry. Data are the means ± SEM from a representative experiment performed in triplicate. *, P < 0.05.
A protective antiviral response against DENV in primary human myeloid cells.
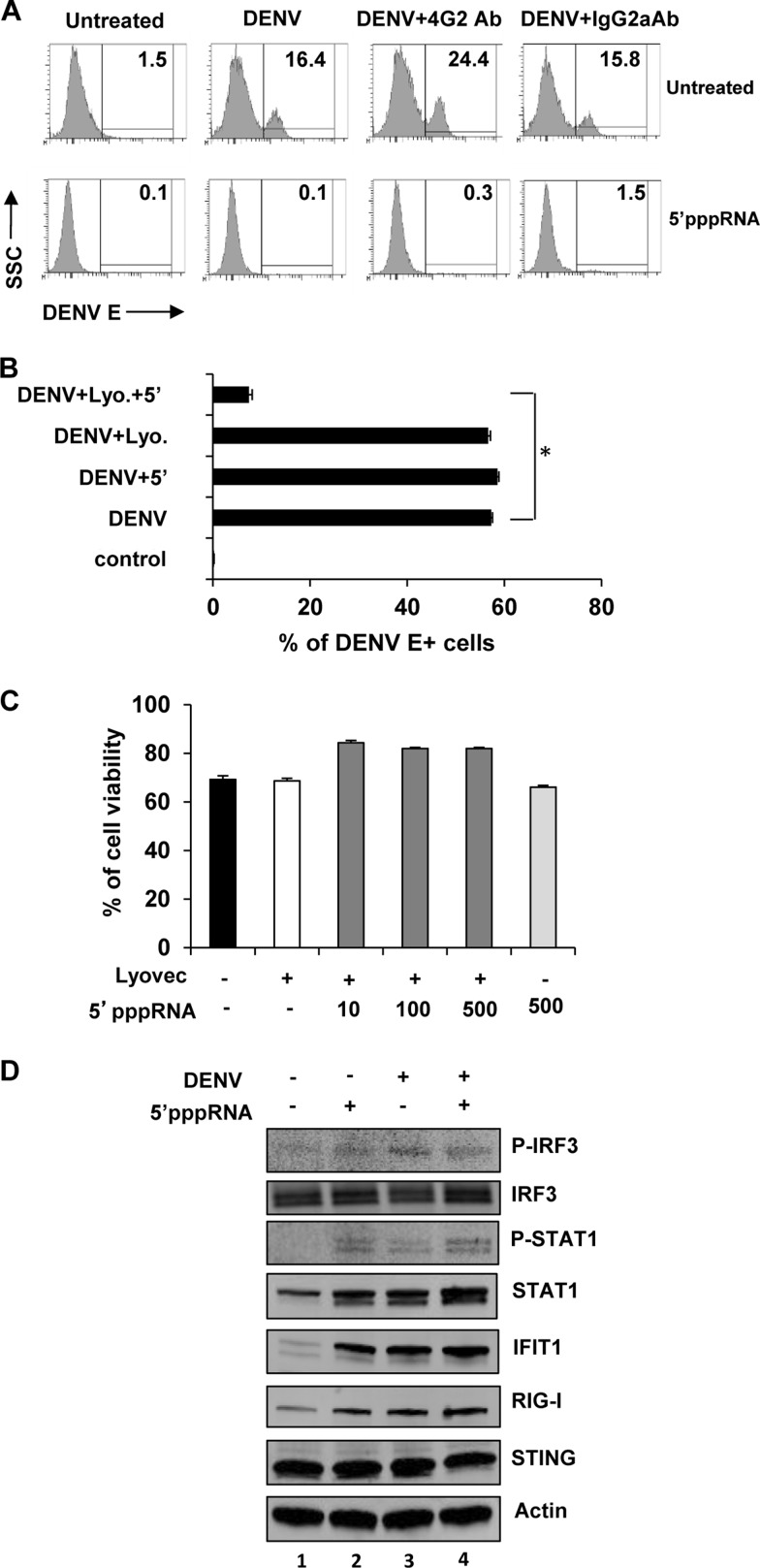
Cells of the myeloid lineage, including monocyte/macrophages and dendritic cells, are the primary target cells for DENV infection among human peripheral blood mononuclear immune cells (47, 48). Severe and potentially lethal manifestations associated with secondary DENV infection are often related to antibody-dependent enhancement (ADE) of infection (36–38). To address the impact of 5′pppRNA on ADE-mediated DENV infection, we demonstrated, using isolated human monocytes, that anti-DENV E 4G2 antibody increased DENV infectivity from 16.4% to 24.4% (Fig. 5A), whereas a control isotype IgG2a antibody did not significantly increase viral infectivity (Fig. 5A). Both primary and ADE DENV infections were completely suppressed by 5′pppRNA treatment (16.4% and 24.4% in untreated cells versus 0.1% and 0.3% in 5′pppRNA-treated cells, respectively). Similarly, in primary human MDDC, which are highly permissive to DENV, infection decreased 8.4-fold in the presence of 5′pppRNA in combination with Lyovec (Fig. 5B), and cell viability was not affected by increasing concentrations of 5′pppRNA (Fig. 5C). MDDC treated with 5′pppRNA at 4 h p.i. were assessed for markers of activation of the innate immune response (Fig. 5D). Increased levels of phosphorylated IRF3 and STAT1 were observed, and a 2- to 10-fold increase in the expression of ISGs RIG-I and IFIT1 following 5′pppRNA treatment were observed (Fig. 5D, lane 2). A similar response was observed with DENV infection alone (Fig. 5D, lane 3). The innate DNA sensor STING was previously shown to be cleaved and inactivated by DENV NS2/3 protease (44); in the current experiments, STING expression was not modulated by 5′pppRNA or DENV infection alone (Fig. 5D, lane 2 and 3). Also, postinfection treatment with 5′pppRNA moderately increased the levels of the following markers of the innate immune response compared to virus alone: phospho-STAT1 (3-fold increase), STAT1 (1.4-fold increase), IFIT1 (1.3-fold increase), and RIG-I (1.3-fold increase) (Fig. 5D, lanes 3 and 4). Surprisingly, 5′pppRNA did not further increase the level of phospho-IRF3 compared to DENV infection alone (Fig. 5D, lane 3 and 4), an observation that is in part attributable to the early and transient kinetics of IRF3 phosphorylation. These data demonstrate that RIG-I activation by 5′pppRNA triggers an immune response capable of inhibiting DENV in both primary and ADE models of infection.
FIG 5.
5′pppRNA treatment protects human myeloid cells from primary and ADE DENV infection. (A) Negatively selected monocytes were challenged with DENV (MOI, 20) in the presence or absence of the enhancing antibody 4G2 (0.5 μg/ml) for 4 h. They were subsequently transfected with 5′pppRNA (100 ng/ml) using Lyovec and incubated for 20 h. An IgG2a antibody (0.5 μg/ml) served as a negative control. The percentage of DENV-infected cells was determined by flow cytometry 24 h after infection. (B) CD14− MDDCs were challenged with DENV (MOI, 10) for 4 h, followed by transfection with 5′pppRNA (100 ng/ml) and incubation for an additional 20 h. Data represent the means ± SEM of an experiment performed in triplicate. *, P < 0.05. (C) Cell viability was assessed by flow cytometry on CD14− MDDC and determined 24 h after 5′pppRNA treatment (10 to 500 ng/ml) in the presence of Lyovec. Data are the means ± SEM of a representative experiment performed in triplicate. (D) CD14− MDDCs were challenged with DENV (MOI, 10) for 4 h and then were treated with 5′pppRNA (100 ng/ml) for an additional 20 h. WCEs were resolved by SDS-PAGE and analyzed by immunoblotting for phospo-IRF3, IRF3, phospho-STAT1, STAT1, IFIT1, RIG-I, STING, and β-actin. Results are from one representative experiment that was repeated once.
5′pppRNA treatment inhibits CHIKV replication in a RIG-I-dependent manner.
To explore the potential of the 5′pppRNA agonist to prevent CHIKV infection, human fibroblast MRC-5 cells were pretreated with increasing concentrations of 5′pppRNA prior to challenge with a CHIKV LS3-GFP reporter virus (Fig. 6A). CHIKV replication was strongly inhibited in a dose-dependent manner in cells treated with 5′pppRNA 1 h prior to infection (Fig. 6A); as little as 1 ng/ml completely blocked CHIKV EGFP reporter gene expression, and the 5′pppRNA concentration required to completely block CHIKV replication in MRC-5 cells was 10-fold lower than that required to inhibit DENV in A549 cells. It is currently unclear whether this is due to virus-specific immune evasion or cell type-specific differences, as CHIKV does not replicate in A549 cells. Also, introduction of control RNA lacking the 5′ triphosphate moiety only led to a minor reduction of GFP reporter gene expression in CHIKV LS3-GFP-infected cells (Fig. 6A). Cell viability, monitored in parallel, was not significantly affected by transfection of either 5′pppRNA or control RNA lacking the 5′ triphosphate (Fig. 6B). Analysis of intracellular RNA of CHIKV-infected cells pretreated with 5′pppRNA or control RNA showed that treatment with 0.1 ng/ml 5′pppRNA reduced CHIKV positive- and negative-strand RNA accumulation to minimally detectable levels (Fig. 6C), and at higher doses of 5′pppRNA viral RNA was undetectable. Transfection of cells with control RNA prior to infection had no significant effect on the accumulation of CHIKV RNA (Fig. 6C). To determine the effect of RIG-I agonist treatment on the expression of CHIKV nonstructural proteins (translated from genomic RNA) and structural proteins (translated from the sgRNA), cells were pretreated with 5′pppRNA or control RNA and infected with CHIKV, and nsP1 and E2 expression was analyzed by Western blotting (Fig. 6D). Transfection of 0.1 ng/ml 5′pppRNA led to a 4-fold reduction in nsP1 expression and an 8-fold reduction in E2 expression. Higher doses of 5′pppRNA reduced nsP1 and E2 expression over 30-fold (Fig. 6D). Transfection of control RNA lacking the 5′ triphosphate had no noticeable effect on CHIKV protein expression (Fig. 6D). Finally, the effect of 5′pppRNA treatment on the production of infectious progeny was determined. Compared to untreated cells, transfection of MRC-5 cells with 0.1 ng/ml of 5′pppRNA 1 h prior to CHIKV infection led to an ∼1 log reduction in virus titer, while transfection with 1 ng/ml and 10 ng/ml 5′pppRNA reduced viral progeny titers by ∼2 and ∼3 logs, respectively (Fig. 6E). Transfection of control RNA lacking the 5′ triphosphate did not significantly affect CHIKV progeny titers (Fig. 6E).
FIG 6.
Treatment with 5′pppRNA inhibits CHIKV replication in a RIG-I-dependent manner. (A) MRC-5 cells were treated with 0.015 to 4 ng/ml of control RNA or 5′pppRNA from 1 h prior to infection to 24 h postinfection with CHIKV LS3-GFP (MOI, 0.1). At 24 h p.i., cells were fixed and EGFP reporter gene expression was quantified. *, P < 0.05. cntrl, control. (B) To assess potential cytotoxicity, MRC-5 cell viability was measured 24 h posttransfection of 5′pppRNA or control RNA lacking the 5′ triphosphate. Data are represented as the means ± SEM from a representative experiment performed in quadruplicate. (C) The intracellular accumulation of CHIKV positive- and negative-strand RNA was determined by in-gel hybridization of RNA isolated from MRC-5 cells that were treated with 5′pppRNA (0.1 to 10 ng/ml) 1 h prior to infection (MOI, 0.1). (D) CHIKV E2, E3E2, and nsP1 protein expression was assessed by Western blotting of lysates of MRC-5 cells that were treated with various concentrations of control RNA or 5′pppRNA 1 h prior to infection with CHIKV. Data are representative of at least two independent experiments. (E) The effect of 5′pppRNA and control RNA treatment on CHIKV progeny titers as assessed by plaque assay. (F) siRNA-transfected MRC-5 cells were either left untreated or were transfected with 5′pppRNA, after which they were infected with CHIKV LS3-GFP (MOI, 0.1). CHIKV-driven EGFP reporter gene expression was measured at 24 h p.i. and was normalized to the expression level in CHIKV-infected cells that had been transfected with a nontargeting scrambled siRNA (scr). *, P < 0.05. (G) MRC-5 cells were transfected with 10 pmol of scrambled siRNA (siScr) or siRNA targeting RIG-I, STAT1, or STING 48 h prior to treatment with 1 ng/ml of 5′pppRNA. Expression levels of RIG-I, STAT1, STING, and IFIT1 were monitored by Western blotting. Cyclophilin A or B was used as a loading control. Data are representative of at least two independent experiments.
To determine which innate immune pathways are involved in the 5′pppRNA-mediated inhibition of CHIKV replication, several key proteins of the IFN signaling pathway (RIG-I, STAT1, and STING) were depleted in MRC-5 cells using siRNAs. Knockdown levels were assessed by Western blotting (Fig. 6G). Subsequently, cells depleted for RIG-I, STAT1, or STING were treated with 5′pppRNA and infected 1 h later with CHIKV LS3-GFP (Fig. 6F). CHIKV-driven GFP reporter gene activity was reduced to almost background levels in 5′pppRNA-treated cells that were depleted for STAT1 and STING, suggesting these proteins are not involved in the 5′pppRNA-mediated antiviral response to CHIKV. In contrast, CHIKV replication was observed in cells depleted of RIG-I and treated with 5′pppRNA, although EGFP reporter gene expression was ∼30% of that in untreated cells transfected with scrambled (or RIG-I-targeting) siRNAs (Fig. 6F). This partial recovery of replication might be due to incomplete knockdown of RIG-I in a fraction of the cells and/or paracrine IFN signaling of those cells, which could affect CHIKV replication of RIG-I-depleted cells. CHIKV replication in cells depleted for RIG-I, STAT1, or STING, but not treated with 5′pppRNA, was similar or slightly increased compared to that of cells transfected with a scrambled control siRNA. In parallel, the siRNA-treated cells were transfected with 1 ng/ml 5′pppRNA, and 24 h later the IFN signaling response was analyzed by monitoring the upregulation of IFITI or STAT1 (Fig. 6G). Knockdown of RIG-I expression resulted in a strong reduction of 5′pppRNA-induced IFITI upregulation, whereas the 5′pppRNA-induced upregulation of IFITI was not affected by STAT1 depletion. siRNA-mediated knockdown of STING also did not block the 5′pppRNA-induced upregulation of STAT1, indicating that STAT1 and STING are dispensable for the response to 5′pppRNA, whereas RIG-I is required.
Postinfection treatment with 5′pppRNA inhibits CHIKV replication and stimulates the RIG-I pathway in both uninfected and CHIKV-infected cells.
To explore the antiviral potential of 5′pppRNA against CHIKV, MRC-5 cells were first infected with CHIKV LS3-GFP at an MOI of 0.1, followed by transfection with 5′pppRNA (1 ng/ml) or control RNA at several time points postinfection. Measurement of EGFP expression by the reporter virus in infected MRC-5 cells that were fixed at 24 h p.i. indicated that treatment with 5′pppRNA at 1 or 3 h p.i. reduced reporter gene expression to less than 20% of that in untreated infected control cells (Fig. 7A). Even when treatment was initiated as late as 5 h p.i., a more than 50% reduction in EGFP expression was observed (Fig. 7A). Transfection of control RNA merely led to an ∼20% reduction in EGFP reporter gene expression, largely independent of the time of addition. Postinfection treatment of CHIKV-infected cells with 5′pppRNA also reduced viral progeny titers at 24 h p.i., depending on the time of addition (Fig. 7B). CHIKV titers in the medium of untreated infected cells were 6 × 106 PFU/ml at 24 h p.i., while treatment from 1 h p.i. onward led to a more than 2-log reduction in infectious progeny, i.e., 5 × 104 PFU/ml. When treatment was initiated at 3, 5, or 8 h p.i., CHIKV titers of 2 × 105, 7 × 105, and 1 × 106, respectively, were measured at 24 h p.i. Transfection of CHIKV-infected cells with control RNA resulted in a less than 1-log reduction in infectious progeny titer (Fig. 7B).
FIG 7.
Postinfection treatment with 5′pppRNA inhibits CHIKV replication in an addition-dependent manner. MRC-5 cells were infected with CHIKV LS3-GFP at an MOI of 0.1, and at the indicated time points postinfection they were transfected with 1 ng/ml 5′pppRNA or control RNA. (A) Cells were fixed at 24 h p.i., and EGFP reporter gene expression was quantified and normalized to that in untreated cells. *, P < 0.05. (B) CHIKV progeny titers 24 h p.i. and after 5′pppRNA or control RNA treatment were determined by plaque assay. (C) MRC-5 cells were transfected with 0.1, 1, or 10 ng/ml 5′pppRNA or control RNA 1 h prior to infection with CHIKV LS3-GFP (MOI, 0.1). At 24 h p.i., cell lysates were prepared and STAT1, RIG-I, and IFIT-I protein levels were determined by Western blotting. Actin or the transferrin receptor were used as loading controls. Data are representative of at least two independent experiments.
To assess the activation of the RIG-I signaling pathway in MRC-5 cells after 5′pppRNA treatment in the presence or absence of CHIKV infection, the expression levels of STAT1, RIG-I, and IFIT1 were analyzed by immunoblotting (Fig. 7C). Both in mock-infected and CHIKV-infected cells, transfection of 0.1 ng/ml 5′pppRNA induced a strong upregulation of STAT1, RIG-I, and IFITI (Fig. 7C), an effect that was more pronounced with treatment of 1 or 10 ng/ml of 5′pppRNA. In contrast, introduction of control RNA had no effect on expression of these proteins. CHIKV infection alone did not lead to increased STAT1, RIG-I, and IFIT1 expression, and CHIKV infection did not inhibit the 5′pppRNA-induced upregulation of RIG-I or downstream IFN signaling (Fig. 7C).
DISCUSSION
The absence of directly acting antivirals and registered vaccines for the treatment of important human pathogens, such as DENV and CHIKV, has emphasized the need for the development of therapeutic strategies. Initiated within minutes of virus binding to target cells, the innate immune response is the first natural barrier against viral infection. The innate response triggers an array of protective processes, resulting in the production of antiviral effectors and the induction of adaptive immune responses (1–4). Mimicking the early steps of the host antiviral response through stimulation of innate recognition receptors represents a novel therapeutic strategy for the treatment of emerging diseases, such as those caused by DENV and CHIKV.
We previously demonstrated that a 5′pppRNA derived from the 5′ and 3′ UTRs of VSV blocked the replication of multiple viruses both in vitro and in vivo (35). In the present study, we further characterized the antiviral potential of 5′pppRNA and demonstrated that 5′pppRNA treatment restricted DENV and CHIKV infection in human myeloid, epithelial, and fibroblastic cells. The antiviral effect was observed when 5′pppRNA was administered both pre- and postinfection, with demonstrated inhibition of DENV and CHIKV protein expression, RNA levels, and production of infectious progeny. The in vitro protective antiviral effect was dependent on an intact RIG-I/MAVS/TBK1/IRF3 axis but largely independent of IFNAR and STAT1. The protective effect was sustained over time, as cells remained free from infection even 72 h after infection, and 5′pppRNA also blocked antibody-dependent enhancement of DENV infectivity, a phenomenon that is associated with complications and disease severity in dengue patients. The present data on the 5′pppRNA immune-stimulating effect on human cells, combined with previous observations demonstrating that 5′pppRNA triggered a full range of antiviral and inflammatory responses in serum, lungs, and spleens of treated mice (35), highlights the importance of evaluating the efficacy of RIG-I agonists as a potential immune stimulator in humans.
The type I IFN system represents an important innate antiviral response to DENV and CHIKV. Indeed, type I IFN has been shown to restrict the propagation of DENV and CHIKV in both in vitro and in vivo models. Treatment with either IFN-α or IFN-β suppressed the replication of both DENV (49) and CHIKV (50) in cell culture. Moreover, infection of mice lacking IFNAR led to a significantly higher lethality after DENV (51) or CHIKV infection (52, 53). Among the viral RNA sensors, RIG-I, MDA5, and TLR3 are activated upon DENV infection and are essential for host defense against the virus (54). RIG-I appears to play a major role in the response to CHIKV infection, as CHIKV-induced IFN-β expression was more strongly reduced in RIG-I-deficient MEFs than in those lacking MDA5 (53). The antiviral activity of 5′pppRNA observed in previous studies has been attributed to the potent activation of inflammatory and antiviral programs driven by transcription factors such as IRF3, IRF7, STAT1, and NF-κB (35). In this study, the antiviral protective response was dependent on a functional RIG-I pathway but was largely independent of STAT signaling. We also demonstrated that 5′pppRNA treatment triggered a robust host antiviral response associated with the expression of several IFN-stimulated genes (ISGs), including RSAD2 (Viperin), IFIT, and IFITM proteins MX1 and OAS. Although hundreds of ISGs have been identified, only a few of them have been fully characterized with respect to their inhibitory function. Recently, Schoggins et al. identified a panel of broadly active antiviral molecules that facilitate inhibition of viral infection (24), with IFITM proteins and Viperin characterized as important ISGs required for inhibition of CHIKV (55–57). Although most of these essential antiviral genes were upregulated following RIG-I stimulation (35), an in-depth analysis is now required to identify which IRF3-driven ISGs mediate the 5′pppRNA antiviral effect observed against DENV and CHIKV infection in vitro.
Poly(I·C), another dsRNA innate immune stimulator, has been extensively studied in vitro and in vivo and has demonstrated a broad range of efficacy against many viral infections, including those by DENV and CHIKV (30, 31, 58–61). In vitro studies have shown that the poly(I·C)-mediated DENV antiviral response was dependent primarily on IFN-β induction and was reversed by IKK inhibitors (31). Poly(I·C), in combination with TLR7/8 agonists, not only prevented DENV infection in vitro but also decreased DENV viremia in vivo through increased inflammatory and humoral responses (31, 60). The immune response generated by poly(I·C) was previously evaluated in a phase 1 clinical trial with healthy volunteers, and whole-genome transcriptional analysis of peripheral blood mononuclear cells demonstrated upregulation of genes involved in multiple innate pathways, including antiviral and inflammasome signaling (62). This observation underscores the fact that synthetic dsRNA remains an attractive innate immune stimulator in humans.
Vaccination is the primary approach to prevent viral infection. Increasing the immunogenicity of vaccines with molecular immune modulators that elicit immune responses is crucial to enhance vaccine efficacy. Poly(I·C) and, more recently, 5′pppRNA have been used as adjuvants and were shown in combination with influenza vaccine to improve protection in mice (26, 63–65). Interestingly, we observed that RIG-I activation using 5′pppRNA not only induced antiviral effectors but also mobilized cytokines and chemokines involved in trafficking processes of immune cells, including CXCL10, CCL5, and CCL3. The proinflammatory cytokine IL-6, which was potentiated by the 5′pppRNA treatment, also is crucial to maintain and potentiate CD8+ T cell survival and killing in vivo (66). A significant type III IFN response was observed in cells challenged with 5′pppRNA. Recent studies have shown that type I and III IFNs activate similar components of the JAK-STAT pathways, although type III IFNs induced a delayed and stronger induction of ISGs than type I IFNs (67). We did not find a significant antiviral role for type III IFN in DENV-infected A549 cells, although the induction of type III IFN may have biological relevance in vivo by bridging the innate and adaptive immune responses. Indeed, IL-28A stimulation presented antiviral immunostimulatory effects by increasing the total number of lymphocytes and CD4+ T cells in lung lymphocyte preparations of vaccinia virus-infected mice (68). The present study demonstrates the antiviral effect of 5′pppRNA but also suggests new perspectives on the use of RIG-I agonists as vaccine adjuvants in the context of DENV and CHIKV infection.
Taken together, our study provides compelling evidence that stimulation of the natural host defense with 5′pppRNA represents a valuable alternative strategy to conventional antiviral drugs directed against specific viral targets. RIG-I activation mimics and stimulates evolutionarily conserved immune responses to infection and induces multiple antiviral factors that block viral infection at different steps, reducing the possible development of antiviral resistance. These novel approaches to boost host antiviral innate and inflammatory immune responses are broad spectrum in nature rather than virus specific. Such a broadly acting antiviral molecule may be desirable in tropical areas where the population is exposed to multiple pathogens, e.g., where both DENV and CHIKV are endemic and cause coinfections and where the facilities for (differential) diagnosis are limited.
ACKNOWLEDGMENTS
We thank Vladimir Beljanski for critical reading of the manuscript. We are grateful to Andres Merits (University of Tartu, Estonia) for his generous gift of CHIKV antisera.
This work was supported by funding from VGTI Florida, the Canadian Institutes of Health and Research (grant CCI-117954), the European Union Seventh Framework Programme under SILVER grant agreement 260644 and the Marie Curie Initial Training Network EUVIRNA (grant agreement 264286).
We declare no conflict of interest.
Footnotes
Published ahead of print 29 January 2014
REFERENCES
- 1.Goubau D, Deddouche S, Reis ESC. 2013. Cytosolic sensing of viruses. Immunity 38:855–869. 10.1016/j.immuni.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- 3.Liu SY, Sanchez DJ, Cheng G. 2011. New developments in the induction and antiviral effectors of type I interferon. Curr. Opin. Immunol. 23:57–64. 10.1016/j.coi.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- 5.Belgnaoui SM, Paz S, Hiscott J. 2011. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr. Opin. Immunol. 23:564–572. 10.1016/j.coi.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 6.Kumar H, Kawai T, Akira S. 2011. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30:16–34. 10.3109/08830185.2010.529976 [DOI] [PubMed] [Google Scholar]
- 7.Loo YM, Gale M., Jr 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692. 10.1016/j.immuni.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkins C, Gale M., Jr 2010. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22:41–47. 10.1016/j.coi.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehwinkel J, Reis e Sousa C. 2010. RIGorous detection: exposing virus through RNA sensing. Science 327:284–286. 10.1126/science.1185068 [DOI] [PubMed] [Google Scholar]
- 10.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397–408. 10.1016/j.cell.2010.01.020 [DOI] [PubMed] [Google Scholar]
- 11.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- 12.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- 13.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610. 10.1084/jem.20080091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang SY, Sun HY, Lee KH, Oh BH, Cha YJ, Kim BH, Yoo JY. 2012. 5′-Triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res. 40:2724–2733. 10.1093/nar/gkr1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, Hartmann G. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34. 10.1016/j.immuni.2009.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737. 10.1038/ni1087 [DOI] [PubMed] [Google Scholar]
- 17.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. 2011. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147:423–435. 10.1016/j.cell.2011.09.039 [DOI] [PubMed] [Google Scholar]
- 18.Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. 2009. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323:1070–1074. 10.1126/science.1168352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel JR, Jain A, Chou YY, Baum A, Ha T, Garcia-Sastre A. 2013. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 14:780–787. 10.1038/embor.2013.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandvaux N, Servant MJ, ten Oever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532–5539. 10.1128/JVI.76.11.5532-5539.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawai T, Akira S. 2007. SnapShot: pattern-recognition receptors. Cell 129:1024. 10.1016/j.cell.2007.05.017 [DOI] [PubMed] [Google Scholar]
- 22.Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650. 10.1016/j.immuni.2011.05.006 [DOI] [PubMed] [Google Scholar]
- 23.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568. 10.1038/nri2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Lu M, Meng Z, Trippler M, Broering R, Szczeponek A, Krux F, Dittmer U, Roggendorf M, Gerken G, Schlaak JF. 2007. Toll-like receptor-mediated control of HBV replication by nonparenchymal liver cells in mice. Hepatology 46:1769–1778. 10.1002/hep.21897 [DOI] [PubMed] [Google Scholar]
- 26.Isogawa M, Robek MD, Furuichi Y, Chisari FV. 2005. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J. Virol. 79:7269–7272. 10.1128/JVI.79.11.7269-7272.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svensson A, Bellner L, Magnusson M, Eriksson K. 2007. Role of IFN-alpha/beta signaling in the prevention of genital herpes virus type 2 infection. J. Reprod. Immunol. 74:114–123. 10.1016/j.jri.2006.09.002 [DOI] [PubMed] [Google Scholar]
- 28.Boukhvalova MS, Sotomayor TB, Point RC, Pletneva LM, Prince GA, Blanco JC. 2010. Activation of interferon response through toll-like receptor 3 impacts viral pathogenesis and pulmonary toll-like receptor expression during respiratory syncytial virus and influenza infections in the cotton rat Sigmodon hispidus model. J. Interferon Cytokine Res. 30:229–242. 10.1089/jir.2009.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau YF, Tang LH, Ooi EE, Subbarao K. 2010. Activation of the innate immune system provides broad-spectrum protection against influenza A viruses with pandemic potential in mice. Virology 406:80–87. 10.1016/j.virol.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li YG, Siripanyaphinyo U, Tumkosit U, Noranate N, A-Nuegoonpipat A, Pan Y, Kameoka M, Kurosu T, Ikuta K, Takeda N, Anantapreecha S. 2012. Poly (I:C), an agonist of toll-like receptor-3, inhibits replication of the chikungunya virus in BEAS-2B cells. Virol. J. 9:114. 10.1186/1743-422X-9-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang Z, Wu S, Li Y, He L, Wu M, Jiang L, Feng L, Zhang P, Huang X. 2011. Activation of Toll-like receptor 3 impairs the dengue virus serotype 2 replication through induction of IFN-beta in cultured hepatoma cells. PLoS One 6:e23346. 10.1371/journal.pone.0023346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trapp S, Derby NR, Singer R, Shaw A, Williams VG, Turville SG, Bess JW, Jr, Lifson JD, Robbiani M. 2009. Double-stranded RNA analog poly(I:C) inhibits human immunodeficiency virus amplification in dendritic cells via type I interferon-mediated activation of APOBEC3G. J. Virol. 83:884–895. 10.1128/JVI.00023-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang N, Liang Y, Devaraj S, Wang J, Lemon SM, Li K. 2009. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. J. Virol. 83:9824–9834. 10.1128/JVI.01125-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Wang X, Liu M, Hu Q, Song L, Ye L, Zhou D, Ho W. 2010. A critical function of toll-like receptor-3 in the induction of anti-human immunodeficiency virus activities in macrophages. Immunology 131:40–49. 10.1111/j.1365-2567.2010.03270.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goulet ML, Olagnier D, Xu Z, Paz S, Belgnaoui SM, Lafferty EI, Janelle V, Arguello M, Paquet M, Ghneim K, Richards S, Smith A, Wilkinson P, Cameron M, Kalinke U, Qureshi S, Lamarre A, Haddad EK, Sekaly RP, Peri S, Balachandran S, Lin R, Hiscott J. 2013. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog. 9:e1003298. 10.1371/journal.ppat.1003298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halstead SB. 2007. Dengue. Lancet 370:1644–1652. 10.1016/S0140-6736(07)61687-0 [DOI] [PubMed] [Google Scholar]
- 37.Halstead SB, Suaya JA, Shepard DS. 2007. The burden of dengue infection. Lancet 369:1410–1411. 10.1016/S0140-6736(07)60645-X [DOI] [PubMed] [Google Scholar]
- 38.Rothman AL. 2011. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 11:532–543. 10.1038/nri3014 [DOI] [PubMed] [Google Scholar]
- 39.Schwartz O, Albert ML. 2010. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 8:491–500. 10.1038/nrmicro2368 [DOI] [PubMed] [Google Scholar]
- 40.Her Z, Kam YW, Lin RT, Ng LF. 2009. Chikungunya: a bending reality. Microbes Infect. 11:1165–1176. 10.1016/j.micinf.2009.09.004 [DOI] [PubMed] [Google Scholar]
- 41.Powers AM, Logue CH. 2007. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J. Gen. Virol. 88:2363–2377. 10.1099/vir.0.82858-0 [DOI] [PubMed] [Google Scholar]
- 42.Scholte FE, Tas A, Martina BE, Cordioli P, Narayanan K, Makino S, Snijder EJ, van Hemert MJ. 2013. Characterization of synthetic Chikungunya viruses based on the consensus sequence of recent E1-226V isolates. PLoS One 8:e71047. 10.1371/journal.pone.0071047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metz SW, Geertsema C, Martina BE, Andrade P, Heldens JG, van Oers MM, Goldbach RW, Vlak JM, Pijlman GP. 2011. Functional processing and secretion of Chikungunya virus E1 and E2 glycoproteins in insect cells. Virol. J. 8:353. 10.1186/1743-422X-8-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, Maringer K, Bernal-Rubio D, Shabman RS, Simon V, Rodriguez-Madoz JR, Mulder LC, Barber GN, Fernandez-Sesma A. 2012. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 8:e1002934. 10.1371/journal.ppat.1002934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrison J, Aguirre S, Fernandez-Sesma A. 2012. Innate immunity evasion by dengue virus. Viruses 4:397–413. 10.3390/v4030397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez-Madoz JR, Belicha-Villanueva A, Bernal-Rubio D, Ashour J, Ayllon J, Fernandez-Sesma A. 2010. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. J. Virol. 84:9760–9774. 10.1128/JVI.01051-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blackley S, Kou Z, Chen H, Quinn M, Rose RC, Schlesinger JJ, Coppage M, Jin X. 2007. Primary human splenic macrophages, but not T or B cells, are the principal target cells for dengue virus infection in vitro. J. Virol. 81:13325–13334. 10.1128/JVI.01568-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kou Z, Quinn M, Chen H, Rodrigo WW, Rose RC, Schlesinger JJ, Jin X. 2008. Monocytes, but not T or B cells, are the principal target cells for dengue virus (DV) infection among human peripheral blood mononuclear cells. J. Med. Virol. 80:134–146. 10.1002/jmv.21051 [DOI] [PubMed] [Google Scholar]
- 49.Diamond MS, Edgil D, Roberts TG, Lu B, Harris E. 2000. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J. Virol. 74:7814–7823. 10.1128/JVI.74.17.7814-7823.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sourisseau M, Schilte C, Casartelli N, Trouillet C, Guivel-Benhassine F, Rudnicka D, Sol-Foulon N, Le Roux K, Prevost MC, Fsihi H, Frenkiel MP, Blanchet F, Afonso PV, Ceccaldi PE, Ozden S, Gessain A, Schuffenecker I, Verhasselt B, Zamborlini A, Saib A, Rey FA, Arenzana-Seisdedos F, Despres P, Michault A, Albert ML, Schwartz O. 2007. Characterization of reemerging chikungunya virus. PLoS Pathog. 3:e89. 10.1371/journal.ppat.0030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson AJ, Roehrig JT. 1999. New mouse model for dengue virus vaccine testing. J. Virol. 73:783–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Couderc T, Chretien F, Schilte C, Disson O, Brigitte M, Guivel-Benhassine F, Touret Y, Barau G, Cayet N, Schuffenecker I, Despres P, Arenzana-Seisdedos F, Michault A, Albert ML, Lecuit M. 2008. A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog. 4:e29. 10.1371/journal.ppat.0040029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schilte C, Couderc T, Chretien F, Sourisseau M, Gangneux N, Guivel-Benhassine F, Kraxner A, Tschopp J, Higgs S, Michault A, Arenzana-Seisdedos F, Colonna M, Peduto L, Schwartz O, Lecuit M, Albert ML. 2010. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J. Exp. Med. 207:429–442. 10.1084/jem.20090851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. 2011. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl. Trop. Dis. 5:e926. 10.1371/journal.pntd.0000926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. 10.1016/j.cell.2009.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teng TS, Foo SS, Simamarta D, Lum FM, Teo TH, Lulla A, Yeo NK, Koh EG, Chow A, Leo YS, Merits A, Chin KC, Ng LF. 2012. Viperin restricts chikungunya virus replication and pathology. J. Clin. Investig. 122:4447–4460. 10.1172/JCI63120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helbig KJ, Carr JM, Calvert JK, Wati S, Clarke JN, Eyre NS, Narayana SK, Fiches GN, McCartney EM, Beard MR. 2013. Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl. Trop. Dis. 7:e2178. 10.1371/journal.pntd.0002178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaajetaan GR, Geelen TH, Grauls GE, Bruggeman CA, Stassen FR. 2012. CpG and poly(I:C) stimulation of dendritic cells and fibroblasts limits herpes simplex virus type 1 infection in an IFNbeta-dependent and -independent way. Antiviral Res. 93:39–47. 10.1016/j.antiviral.2011.10.015 [DOI] [PubMed] [Google Scholar]
- 59.Mazaleuskaya L, Veltrop R, Ikpeze N, Martin-Garcia J, Navas-Martin S. 2012. Protective role of Toll-like receptor 3-induced type I interferon in murine coronavirus infection of macrophages. Viruses 4:901–923. 10.3390/v4050901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sariol CA, Martinez MI, Rivera F, Rodriguez IV, Pantoja P, Abel K, Arana T, Giavedoni L, Hodara V, White LJ, Anglero YI, Montaner LJ, Kraiselburd EN. 2011. Decreased dengue replication and an increased anti-viral humoral response with the use of combined Toll-like receptor 3 and 7/8 agonists in macaques. PLoS One 6:e19323. 10.1371/journal.pone.0019323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao J, Wohlford-Lenane C, Zhao J, Fleming E, Lane TE, McCray PB, Jr, Perlman S. 2012. Intranasal treatment with poly(I · C) protects aged mice from lethal respiratory virus infections. J. Virol. 86:11416–11424. 10.1128/JVI.01410-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caskey M, Lefebvre F, Filali-Mouhim A, Cameron MJ, Goulet JP, Haddad EK, Breton G, Trumpfheller C, Pollak S, Shimeliovich I, Duque-Alarcon A, Pan L, Nelkenbaum A, Salazar AM, Schlesinger SJ, Steinman RM, Sekaly RP. 2011. Synthetic double-stranded RNA induces innate immune responses similar to a live viral vaccine in humans. J. Exp. Med. 208:2357–2366. 10.1084/jem.20111171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ichinohe T, Kawaguchi A, Tamura S, Takahashi H, Sawa H, Ninomiya A, Imai M, Itamura S, Odagiri T, Tashiro M, Chiba J, Sata T, Kurata T, Hasegawa H. 2007. Intranasal immunization with H5N1 vaccine plus poly I:poly C12U, a Toll-like receptor agonist, protects mice against homologous and heterologous virus challenge. Microbes Infect. 9:1333–1340. 10.1016/j.micinf.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 64.Ichinohe T, Watanabe I, Ito S, Fujii H, Moriyama M, Tamura S, Takahashi H, Sawa H, Chiba J, Kurata T, Sata T, Hasegawa H. 2005. Synthetic double-stranded RNA poly(I:C) combined with mucosal vaccine protects against influenza virus infection. J. Virol. 79:2910–2919. 10.1128/JVI.79.5.2910-2919.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez-Gil L, Goff PH, Hai R, Garcia-Sastre A, Shaw ML, Palese P. 2013. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. J. Virol. 87:1290–1300. 10.1128/JVI.02338-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dejean AS, Beisner DR, Ch'en IL, Kerdiles YM, Babour A, Arden KC, Castrillon DH, DePinho RA, Hedrick SM. 2009. Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat. Immunol. 10:504–513. 10.1038/ni.1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maher SG, Sheikh F, Scarzello AJ, Romero-Weaver AL, Baker DP, Donnelly RP, Gamero AM. 2008. IFNalpha and IFNlambda differ in their antiproliferative effects and duration of JAK/STAT signaling activity. Cancer Biol. Ther. 7:1109–1115. 10.4161/cbt.7.7.6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bartlett NW, Buttigieg K, Kotenko SV, Smith GL. 2005. Murine interferon lambdas (type III interferons) exhibit potent antiviral activity in vivo in a poxvirus infection model. J. Gen. Virol. 86:1589–1596. 10.1099/vir.0.80904-0 [DOI] [PubMed] [Google Scholar]