ABSTRACT
Human papillomavirus 45 (HPV45) is a member of the HPV18-related alpha-7 species and accounts for approximately 5% of all cervical cancer cases worldwide. This study evaluated the genetic diversity of HPV45 and the association of HPV45 variants with the risk of cervical cancer by sequencing the entire E6 and E7 open reading frames of 300 HPV45-positive cervical samples from 36 countries. A total of 43 HPV45 sequence variants were identified that formed 5 phylogenetic sublineages, A1, A2, A3, B1, and B2, the distribution of which varied by geographical region. Among 192 cases of cervical cancer and 101 controls, the B2 sublineage was significantly overrepresented in cervical cancer, both overall and in Africa and Europe separately. We show that the sequence analysis of E6 and E7 allows the classification of HPV45 variants and that the risk of cervical cancer may differ by HPV45 variant sublineage.
IMPORTANCE This work describes the largest study to date of human papillomavirus 45 (HPV45)-positive cervical samples and provides a comprehensive reference for phylogenetic classification for use in epidemiological studies of the carcinogenicity of HPV45 genetic variants, particularly as our findings suggest that the B2 sublineage of HPV45 is associated with a higher risk of cervical cancer.
INTRODUCTION
There are over 100 types of human papillomavirus (HPV), of which 12 have been classified as “carcinogenic to humans,” or group 1, by a working group of the International Agency for Research on Cancer (IARC) Monographs (1). While most HPV infections are asymptomatic and eventually cleared by the immune system, in some cases the infection will persist and, in rare cases, lead to cancer (reviewed in reference 2). Evidence suggests that not only HPV type but also sequence variations within high-risk HPVs may influence viral persistence and clinical outcome (3–8).
HPV45 is a high-risk HPV type that was first described in 1987 when it was cloned from a recurring cervical lesion found in a woman in the United States (9). In addition to being a member of the same phylogenetic species (alpha-7) as HPV18 (10, 11), HPV45 is similarly more common in adenocarcinoma than in squamous cell carcinoma of the cervix (12, 13). Approximately 5% of cervical cancers worldwide are positive for HPV45, although this proportion was reported to vary from 3% in Eastern Asia up to 9% in Africa (14). Based upon its level of enrichment in cervical cancer compared to cytologically normal women, HPV45 has been suggested to be the third most carcinogenic type after HPV16 and -18 (15).
Genetic variants of HPV45 have been classified into two major lineages, A and B, and five sublineages, A1, A2, A3, B1, and B2 (16). The whole-genome sequence of a variant lineage differs by approximately 1.0% from another variant lineage of the same HPV type, and differences of 0.5 to 0.9% define sublineages (17).
In contrast to other high-risk HPV types (e.g., HPV16 [18]), no studies exist on the association of HPV45 variants with cervical cancer risk. The aims of the current study, therefore, were to characterize the genetic diversity of HPV45 worldwide and to explore the association of HPV45 variant sublineages with the risk for cervical cancer.
MATERIALS AND METHODS
Origin of clinical specimens.
The IARC has coordinated cervical cancer case series, cervical cancer case-control studies, and population-based HPV prevalence surveys in a large number of countries around the world (19–35; also as-yet-unpublished studies from Fiji and Bhutan). The collection of samples has spanned a period of over 20 years from 1989 until 2012 and predates the introduction of HPV vaccines. Informed consent was obtained from all participants, and the studies were approved by the IARC Ethical Review Committee. Cervical samples (exfoliated cells or tissue biopsy specimens) derived from these studies have been comprehensively genotyped for HPV type by using a standardized and well-validated protocol (general primer GP5+/6+ PCR-enzyme immunoassay [EIA] followed by reverse line blot assay) (36) in one centralized laboratory (Molecular Pathology Unit, Department of Pathology, VU University Medical Center, Amsterdam, The Netherlands). All HPV45-positive cervical samples in the IARC biobank were selected for the current analysis, without exclusion. Forty-seven of these specimens were used in the context of a previous study (37). All specimens were categorized into the following regions: Africa, Asia and Oceania, Europe, North America, and South America. Country-specific details are noted in Table 1.
TABLE 1.
Geographic distribution of 300 HPV45-positive cervical samplesa
| Region and country | No. of samples |
|---|---|
| Africa | 108 |
| Algeria | 9 |
| Guinea | 17 |
| Kenya | 19 |
| Mali | 12 |
| Morocco | 6 |
| Nigeria | 13 |
| Senegal | 9 |
| South Africa | 17 |
| Tanzania | 4 |
| Uganda | 2 |
| Asia/Oceania | 104 |
| Bhutan | 10 |
| China | 4 |
| Fiji | 10 |
| India | 12 |
| Indonesia | 4 |
| Iran | 3 |
| South Korea | 1 |
| Mongolia | 14 |
| Nepal | 1 |
| Philippines | 36 |
| Thailand | 7 |
| Vanuatu | 2 |
| Europe | 43 |
| Georgia | 22 |
| Italy | 8 |
| Poland | 11 |
| Spain | 2 |
| North America | 5 |
| Canada | 3 |
| USA | 2 |
| South America | 40 |
| Argentina | 3 |
| Bolivia | 4 |
| Brazil | 8 |
| Chile | 4 |
| Cuba | 2 |
| Panama | 6 |
| Paraguay | 10 |
| Peru | 3 |
The regions and regional subtotals are in boldface type.
PCR and DNA sequencing.
DNA extraction from stored samples was performed using the High Pure PCR template preparation kit (Roche, Mannheim, Germany), and DNA isolates were subjected to β-globin PCR to assess sample quality, as described previously (38). Sequencing of the entire HPV45 E6 and E7 region (nucleotides 102 to 907) was performed as described previously (37) using a series of HPV45-specific primer pairs that were designed to amplify overlapping regions of the HPV45 E6 and E7 open reading frames in order to cover the entire E6 and E7 region.
To reveal single nucleotide polymorphisms (SNPs), sequences of the specimens were aligned with the prototype HPV45 sequence (NCBI accession number X74479) using multalin software (http://multalin.toulouse.inra.fr/multalin/). SNPs that were observed in only one sample were confirmed by reexamination of the sequence traces. Isolates that did not fall into existing lineage categories were confirmed by manual reexamination of the sequencing traces and with additional sequencing, where necessary. Multiple sequence traces for each sample were compiled to provide one sequence encompassing the entire HPV45 E6 and E7 region.
Phylogenetic analysis.
Unrooted consensus trees were built using the Phylogeny Inference Package (PHYLIP), version 3.69 (39). This included generating 10,000 bootstraps using the F84 model of DNA distances, clustering with the unweighted pair group method with arithmetic mean (UPGMA), and applying the majority rule extended, or greedy, method of consensus. Trees created with a maximum-likelihood method showed similar results and are not described further. All unique sequence variants found in the IARC samples, as well as two unique variants reported in the literature (40), were included in the trees.
Case-control analysis.
Samples were classified as either controls (including normal [n = 79], atypical squamous or glandular cells of undetermined significance [ASCUS; n = 2], or low-grade intraepithelial lesion [LSIL; n = 7]) or cases (squamous cell carcinoma [n = 138], adenocarcinoma [n = 11], adenosquamous cell carcinoma [n = 7], or unspecified invasive cervical cancer [n = 36]). Samples from population-based HPV prevalence studies for which histology and cytology were unavailable were also classified as controls (n = 13). Samples reported as cervical intraepithelial neoplasia (CIN) grade 3 or high-grade squamous intraepithelial lesion (HSIL) were excluded from the case-control analysis (n = 7) but were included in the previously described phylogenetic analysis. There were no samples reported as CIN1 or CIN2. Region-specific associations between variant sublineage and case-control status were assessed by 2-sided P values arising from Fisher's exact test without combining sublineages. Region-specific odds ratios (ORs) and 95% exact confidence intervals (CIs) were calculated for the B2 sublineage versus the combination of all other sublineages. A conditional logistic model stratified by region was used for the calculation of the worldwide OR and exact CI, comparing the B2 sublineage against all other sublineages combined. All statistics were calculated with SAS version 9.3 (SAS Institute, Cary, NC, USA).
Nucleotide sequence accession numbers.
All specimen sequences are available in GenBank (accession numbers KF591342 to KF591384).
RESULTS
Sequencing.
The entire E6 and E7 genes were sequenced in a total of 300 HPV45-positive cervical samples from 36 countries, including 10 countries in Africa, 12 countries in Asia/Oceania, 4 countries in Europe, 2 countries in North America, and 8 countries in South America (Table 1).
A total of 44 SNPs were identified across the E6 and E7 open reading frames. The combinations of these SNPs resulted in 43 unique sequences, which will be called variants (Table 2). Two additional variants (including 4 additional SNPs) were identified from the literature (40) and were included in the phylogenetic analysis. In E6, there were 28 SNPs (5.9% nucleotide variation), 15 resulting in amino acid changes. In E7, there were 20 SNPs (6.2% nucleotide variation), 12 resulting in amino acid changes. There were no SNPs observed in the 8-nucleotide region between the E6 and E7 open reading frames. The maximum pairwise difference of the E6 and E7 sequence between any two variants was approximately 2%.
TABLE 2.
HPV45 variants based on the sequence of the E6 and E7 regions of HPV45-positive cervical samplesa
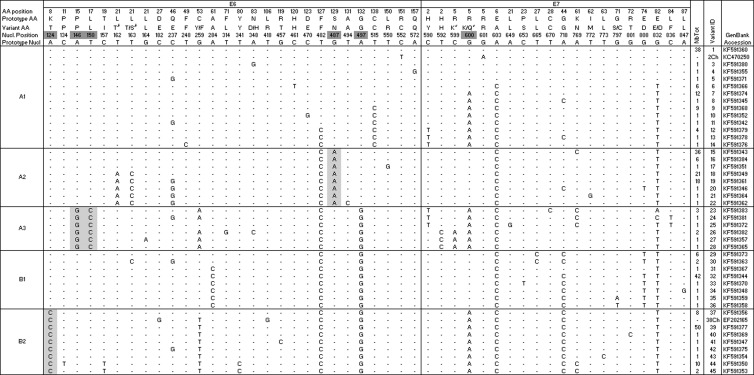
Light gray shading highlights the SNPs that are diagnostic for one lineage or sublineage. Nucleotide positions with dark gray shading are able to discriminate at least one (sub)lineage from one other (sub)lineage. Variant identifiers correspond to those in Fig. 1. AA, amino acid; Nucl., nucleotide; Ch, additional unique E6/E7 variant identified from the literature (40).
The SNP at nucleotide 162 was always found in conjunction with the SNP at nucleotide 163. With both changes, the amino acid at position 21 is threonine (T).
The SNP at nucleotide 599 was always found in conjunction with the SNP at nucleotide 600. With both changes, the amino acid at position 5 is lysine (K).
Phylogenetic analysis.
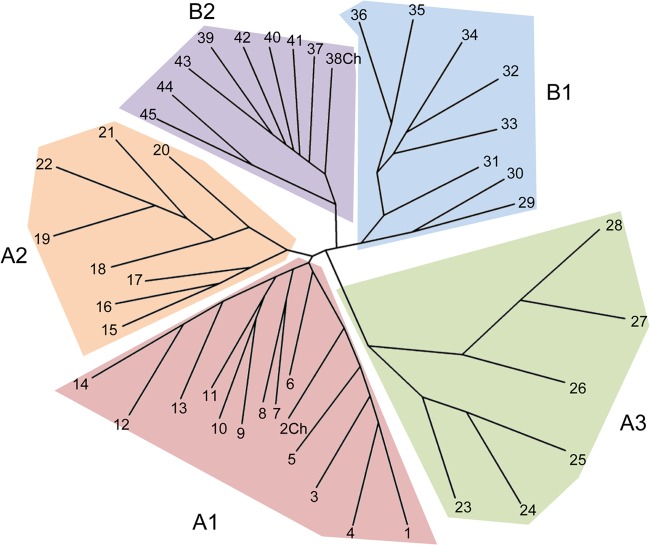
The 45 unique variants clustered into 5 groups in the phylogenetic tree (Fig. 1) that corresponded to the previously described sublineages A1, A2, A3, B1, and B2 (16). Twelve variants, representing 39 samples (variants ID 3 to 14 in Table 2), plus one variant from the literature (variant ID 2Ch [40]), were of the same A1 sublineage as the prototype variant (NCBI accession number X74479, variant ID 1, n = 38). Eight variants, representing 85 samples (variant IDs 15 to 22), corresponded to the previously reported A2 sublineage, and 6 variants, representing 9 samples (variant IDs 23 to 28), corresponded to the previously reported A3 sublineage. In the B lineage, we observed 8 variants, representing 55 samples (variant IDs 29 to 36), that corresponded to the previously reported B1 sublineage and 8 variants, representing 74 samples (variant IDs 37 to 45), plus one variant from the literature (variant ID 38Ch [40]) that corresponded to the previously described B2 sublineage.
FIG 1.
Phylogenetic tree of HPV45 variants based on E6 and E7. The numbers at the end of the branches correspond to the variant identifiers (IDs) listed in Table 2. The prototype sequence is variant 1 in sublineage A1.
There were 5 and 1 nucleotide positions in E6 and E7, respectively, which discriminated at least one sublineage from another (dark gray background for nucleotide positions in Table 2). Furthermore, 4 of these SNPs in E6 were “diagnostic” (i.e., consistently present and unique) for one sublineage only (light gray background for nucleotide bases in Table 2), but no such diagnostic SNP existed in E6 or E7 for the A1 or B1 sublineages. Based on our data, by genotyping a minimum of four nucleotide positions (e.g., 124, 150, 487, and 497), it is possible to correctly classify an isolate into one of the five sublineages.
The distribution of HPV45 sublineages varied by geographical region (Fig. 2), with a predominance of the A1 sublineage in Africa, the A2 sublineage in Asia/Oceania, and the B1 sublineage in Europe. The number of North American samples was low, but 3 out of 5 samples were of the A2 sublineage. The A3 sublineage was rare and was seen in Africa and South America only.
FIG 2.
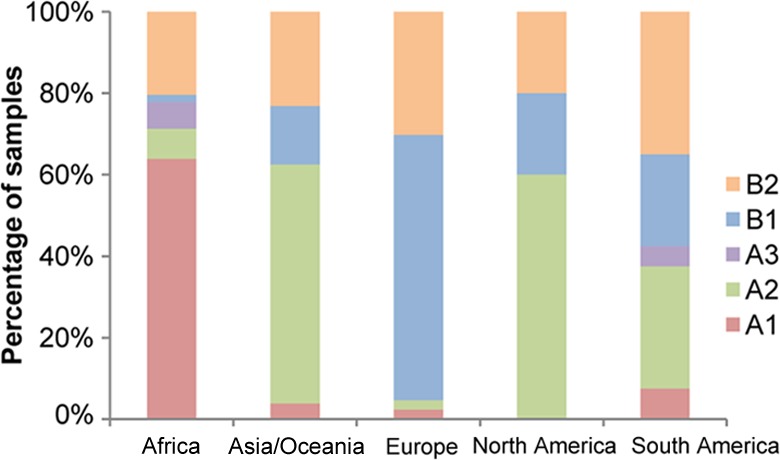
Geographical distribution of HPV45 sublineages shown as a proportion of the total number of HPV45-positive samples collected from each region, irrespective of case-control status.
Case-control analysis.
The distribution of HPV45 variant sublineages was compared between cases (n = 192) and controls (n = 101) (Table 3). To avoid misclassification, samples diagnosed as HSIL or CIN3 (n = 7) were excluded (although their inclusion as cases in a sensitivity analysis did not change the results; data not shown). The distribution of the variant sublineages differed significantly between the cases and controls among the samples from Africa (P = 0.01) and Europe (P = 0.02). In both regions, it appears that this difference was driven by the overrepresentation of the B2 lineage in cases with a relative risk of 6.2 (95% CI = 1.3 to 57.1) for African samples and 5.7 (95% CI = 0.9 to 60.0) for European samples. Although not statistically significant, Asia/Oceania showed a similar pattern, with a relative risk of 2.4 (95% CI = 0.8 to 7.6). The absence of controls from the Americas precluded a similar analysis. When the cases and controls from all regions were combined, the relative risk of cervical cancer for the B2 sublineage was 3.7 (95% CI = 1.8 to 8.5) compared to all other sublineages.
TABLE 3.
Distribution and statistical comparison of HPV45 sublineages between cases and controls
| Region | No. of samples by lineage |
Fisher's exact test P valuea | OR (95% CI),B2 vs non-B2b | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases |
Controls |
|||||||||||||
| A1 | A2 | A3 | B1 | B2 | Total | A1 | A2 | A3 | B1 | B2 | Total | |||
| Africa | 40 | 7 | 3 | 2 | 20 | 72 | 27 | 1 | 4 | 0 | 2 | 34 | 0.01 | 6.2 (1.3, 57.1) |
| Asia/Oceania | 2 | 31 | 0 | 4 | 15 | 52 | 2 | 29 | 0 | 10 | 7 | 48 | 0.13 | 2.4 (0.8, 7.6) |
| Europe | 1 | 1 | 0 | 12 | 10 | 24 | 0 | 0 | 0 | 16 | 2 | 18 | 0.02 | 5.7 (0.9, 60.0) |
| North America | 0 | 3 | 0 | 1 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| South America | 3 | 12 | 2 | 8 | 14 | 39 | 0 | 0 | 0 | 1 | 0 | 1 | 0.35 | |
| Total | 46 | 54 | 5 | 27 | 60 | 192 | 29 | 30 | 4 | 27 | 11 | 101 | 3.7 (1.8, 8.5) | |
Comparing the distribution of the 5 sublineages in cases and controls.
Exact confidence intervals.
To be certain that the cases were associated with HPV45 and not another HPV type, the analyses were repeated excluding the cases with multiple high-risk HPV infections. This resulted in a loss of 12, 7, 3, 1, and 6 cases from Africa, Asia/Oceania, Europe, North America, and South America, respectively. Despite the reduction in the number of samples, the associations between the B2 sublineage and cervical cancer remained significant for Africa (P = 0.03) and Europe (P = 0.02).
Approximately 10% of the cases were adenocarcinomas (n = 18). Results were consistent when restricting the case-control analyses to only squamous cell carcinomas (results not shown).
DISCUSSION
Given the uniquely large and diverse collection of HPV-genotyped cervical samples at the IARC, we were able to evaluate the genetic diversity within high-risk HPV types and report on the geographic distribution of variants, as well as measure their association with cervical cancer. By sequencing the entire E6 and E7 open reading frames of 300 HPV45-positive cervical samples, we were able to confirm all previously reported HPV45 sublineages (A1, A2, A3, B1, and B2) (16, 37, 40, 41) and to further characterize the genetic diversity in the E6 and E7 genes of HPV45. The amount of genetic variation in E6 and E7 captured in the present study (5.9% and 6.2% of nucleotides, respectively) was twice that of the largest previous report (2.9% and 3.1%) (40). For example, we identified five new variants belonging to the A3 sublineage in addition to the single previously known variant (40).
As has been shown previously for HPV16 and HPV33 (42, 43), the distribution of HPV45 variant lineages varies around the world. The HPV45 A1 sublineage was largely specific for Africa, similar to the AFR1 and AFR2 lineages (now known as lineages B and C) of HPV16 and the B lineage of HPV33. In contrast, the B1 and B2 sublineages of HPV45 were present in all regions, similar to the Eur (A1/A2) sublineage of HPV16 and the A1 sublineage of HPV33. Thus, HPV45 phylogenetic separation may have been partly driven by forces similar to those for other HPV types during the coevolution with human populations.
Due to such geographic heterogeneity of the variant sublineages, we performed the case-control comparison stratified by region, as was done for previous similar studies of HPV variants. Using this approach, we were able to identify significant associations between HPV45 variant sublineages and cervical cancer risk. This difference appeared to be predominantly driven by a significant overrepresentation of the B2 sublineage in cervical cancer, notably in Africa and Europe. Furthermore, given that this effect was not heterogeneous by region, we also present the significant pooled estimate worldwide. The only other study of the outcome of infection with HPV45 variants, a cohort study in Costa Rica, reported B lineages to be associated, albeit nonsignificantly, with persistent HPV45 infection and development of CIN3+ (6). Unfortunately, our study was not able to compare cervical cancer risk specifically for the Americas due to a lack of HPV45-positive controls.
By comparing the amino acid sequence of HPV45 to that of the better-characterized HPV16 and -18, we can surmise that there are not any observed changes in amino acids at the critical positions in E6/E7 (Fig. 3). For example, the HPV45 E6 protein appears to have two zinc binding domains that begin at amino acid positions 32 and 105, a PDZ binding domain (at amino acid positions 154 to 158), and a tyrosine at amino acid position 56 and isoleucine at position 130 that may be part of the LXXLL binding motif critical for association with LXXLL proteins such as E6AP (similar to Y54 and I128 in HPV16 [44]). The HPV45 E7 protein appears to have an RB1 binding site at positions 26 to 30, a casein kinase II (CKII) recognition site with serines at positions 33 and 35, and a zinc binding domain that begins at position 66. The lack of mutations in these biologically relevant positions for HPV45 is similar to what was seen in the analysis of HPV16 E6 variants (42; I. Cornet, personal communication) and HPV33 E6 and E7 variants (43). Additionally, there were no SNPs observed at the E6* splice sites (45) at positions 226 to 230 (donor) and 413 to 417 (acceptor) or at the neighboring intronic nucleotides. Nonetheless, it is possible that there are significant biological effects caused by the changes that we observed in the amino acids of E6 and E7 that have not yet been mapped to a specific biological or oncogenic function. It is also possible that SNPs in other regions of the HPV genome linked to those we describe for E6 and E7 are responsible for differences in oncogenic potential.
FIG 3.
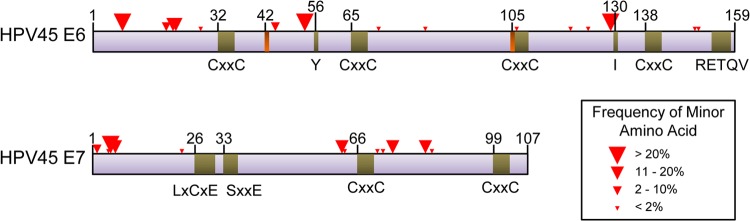
Positions of SNPs resulting in amino acid changes (red triangles) in HPV45 E6 and E7 open reading frames. Putative biologically relevant positions (green bars) based on sequence homology with HPV16 and HPV18 and E6* splicing sites (orange bars) are shown. CxxC is a zinc binding motif, RETQV is a PDZ binding motif, LxCxE is an RB1 binding motif, and SxxE is a casein kinase II recognition site. Y56 and I130 may be part of an LXXLL binding motif. Numbering of select amino acids is provided for reference.
Whole-genome sequencing remains the gold standard for complete phylogenetic characterization. However, it is not always feasible to do so. Importantly, our data suggest that it is possible to characterize the sublineage of HPV45 isolates through the genotyping of only four nucleotide positions in E6 (e.g., 124, 150, 487, and 497). This not only reduces the cost and complexity of phylogenetic studies but also allows one to include samples in which the DNA is not of sufficient quality or quantity for full-genome sequencing.
The major limitation of the current study was the relative rarity of HPV45 and, hence, the limited sample sizes within individual regions. The distribution of the lineages and the significant ORs that we report should, therefore, be interpreted with caution because of the broad CIs and the possibility that there is heterogeneity between the countries grouped together by region given that the distributions of cases and controls were not balanced by country (Table 4).
TABLE 4.
Country-specific distribution of HPV45-positive cases, controls, and samples excluded from case-control analyses (CIN3 and HSIL)
| Region and country | No. of samples |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case |
Control |
CIN3 or HSIL |
|||||||||||||
| A1 | A2 | A3 | B1 | B2 | A1 | A2 | A3 | B1 | B2 | A1 | A2 | A3 | B1 | B2 | |
| Africa | |||||||||||||||
| Algeria | 1 | 3 | 1 | 3 | 1 | ||||||||||
| Guinea | 6 | 10 | 1 | ||||||||||||
| Kenya | 8 | 3 | 8 | ||||||||||||
| Mali | 9 | 1 | 2 | ||||||||||||
| Morocco | 1 | 4 | 1 | ||||||||||||
| Nigeria | 10 | 1 | 2 | ||||||||||||
| Senegal | 6 | 3 | |||||||||||||
| South Africa | 10 | 2 | 1 | 3 | 1 | ||||||||||
| Tanzania | 4 | ||||||||||||||
| Uganda | 2 | ||||||||||||||
| Asia/Oceania | |||||||||||||||
| Bhutan | 1 | 1 | 3 | 4 | 1 | ||||||||||
| China | 1 | 1 | 2 | ||||||||||||
| Fiji | 6 | 3 | 1 | ||||||||||||
| India | 3 | 2 | 5 | 1 | 1 | ||||||||||
| Indonesia | 1 | 3 | |||||||||||||
| Iran | 1 | 1 | 1 | ||||||||||||
| South Korea | 1 | ||||||||||||||
| Mongolia | 12 | 1 | 1 | ||||||||||||
| Nepal | 1 | ||||||||||||||
| Philippines | 1 | 27 | 2 | 5 | 1 | ||||||||||
| Thailand | 7 | ||||||||||||||
| Vanuatu | 1 | 1 | |||||||||||||
| Europe | |||||||||||||||
| Georgia | 6 | 4 | 9 | 2 | 1 | ||||||||||
| Italy | 1 | 3 | 4 | ||||||||||||
| Poland | 3 | 1 | 7 | ||||||||||||
| Spain | 1 | 1 | |||||||||||||
| North America | |||||||||||||||
| Canada | 2 | 1 | |||||||||||||
| USA | 1 | 1 | |||||||||||||
| South America | |||||||||||||||
| Argentina | 1 | 1 | 1 | ||||||||||||
| Bolivia | 3 | 1 | |||||||||||||
| Brazil | 1 | 7 | |||||||||||||
| Chile | 1 | 1 | 2 | ||||||||||||
| Cuba | 1 | 1 | |||||||||||||
| Panama | 3 | 1 | 1 | 1 | |||||||||||
| Paraguay | 2 | 2 | 1 | 4 | 1 | ||||||||||
| Peru | 3 | ||||||||||||||
| Total | 46 | 54 | 5 | 27 | 60 | 29 | 30 | 4 | 27 | 11 | 2 | 1 | 0 | 1 | 3 |
In summary, the present study provides a practical approach for phylogenetic classification for use in epidemiological studies of the natural history and carcinogenicity of HPV45 genetic variants. The findings of this study suggest that the B2 sublineage may be associated with a higher risk of cervical cancer. Understanding the genetic basis of differences in the carcinogenicity of HPV45 variants may help us unravel the mechanisms of HPV infection and its malignant consequences.
ACKNOWLEDGMENTS
This work was supported by grants from The Association for International Cancer Research, United Kingdom (project grant 08-0213); the Institut National du Cancer, France (collaboration agreement 07/3D1514/PL-89-05/NG-LC); and the Fondation Innovations en Infectiologie (FINOVI) (project AO1-project 2). The work of A.A.C. was undertaken during the tenure of a Postdoctoral Fellowship from the International Agency for Research on Cancer, partially supported by the European Commission FP7 Marie Curie Actions—People—Co-funding of regional, national and international programs (COFUND).
The authors have no conflict of interest to declare.
We thank Vanessa Tenet and Jerome Vignat for technical assistance and Zigui Chen and Robert D. Burk for sharing data and scientific advice.
The members of the HPV Variant Study Group include the previous IARC staff (N. Muñoz, R. Herrero, and X. Bosch) and local study coordinators in the following countries: Algeria (D. Hammouda), Argentina (D. Loria and E. Matos), Bhutan (U. Tshomo and Dorji), Bolivia (J. L. Rios-Dalenz), Brazil (J. Eluf-Neto), Canada (P. Ghadirian), Chile (C. Ferreccio and J. M. Ojeda), China (M. Dai, L. K. Li, and R. F. Wu), Cuba (M. Torroella), Fiji (N. Pearce), Georgia (T. Alibegashvili and D. Kordzaia), Guinea (N. Keita and M. Koulibaly), India (T. Rajkumar and R. Rajkumar), Indonesia (Sarjadi), Iran (N. Khodakarami), Italy (M. Sideri), Kenya (P. Gichangi and H. De Vuyst), South Korea (D.-H. Lee and H. R. Shin), Mali (S. Bayo), Mongolia (B. Dondog), Morocco (N. Chaouki), Nepal (A. T. L. Sherpa), Nigeria (J. O. Thomas), Panama (E. de los Rios), Paraguay (P. A. Rolon), Peru (E. Caceres and C. Santos), Philippines (C. Ngelangel), Poland (A. Bardin and W. Zatonski), Senegal (C. S. Boye, C. Toure-Kane, E. S. Mbaye, and H. Diop-Ndiaye), South Africa (D. Moodley), Spain (S. de Sanjose and X. Castellsague), Tanzania (J. N. Kitinya), Thailand (S. Chichareon and S. Tunsakul), Uganda (H. R. Wabinga), and Vanuatu (B. Aruhuri and I. H. Frazer).
Footnotes
Published ahead of print 5 February 2014
REFERENCES
- 1.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V, WHO International Agency for Research on Cancer Monograph Working Group 2009. A review of human carcinogens—part B: biological agents. Lancet Oncol. 10:321–322. 10.1016/S1470-2045(09)70096-8 [DOI] [PubMed] [Google Scholar]
- 2.Baseman JG, Koutsky LA. 2005. The epidemiology of human papillomavirus infections. J. Clin. Virol. 32(Suppl 1):S16–S24. 10.1016/j.jcv.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 3.Berumen J, Ordonez RM, Lazcano E, Salmeron J, Galvan SC, Estrada RA, Yunes E, Garcia-Carranca A, Gonzalez-Lira G, Madrigal-de la Campa A. 2001. Asian-American variants of human papillomavirus 16 and risk for cervical cancer: a case-control study. J. Natl. Cancer Inst. 93:1325–1330. 10.1093/jnci/93.17.1325 [DOI] [PubMed] [Google Scholar]
- 4.Gheit T, Cornet I, Clifford GM, Iftner T, Munk C, Tommasino M, Kjaer SK. 2011. Risks for persistence and progression by human papillomavirus type 16 variant lineages among a population-based sample of Danish women. Cancer Epidemiol. Biomarkers Prev. 20:1315–1321. 10.1158/1055-9965.EPI-10-1187 [DOI] [PubMed] [Google Scholar]
- 5.Sathish N, Abraham P, Peedicayil A, Sridharan G, Chandy G. 2005. HPV 16 E6 sequence variations in Indian patients with cervical neoplasia. Cancer Lett. 229:93–99. 10.1016/j.canlet.2005.04.026 [DOI] [PubMed] [Google Scholar]
- 6.Schiffman M, Rodriguez AC, Chen Z, Wacholder S, Herrero R, Hildesheim A, Desalle R, Befano B, Yu K, Safaeian M, Sherman ME, Morales J, Guillen D, Alfaro M, Hutchinson M, Solomon D, Castle PE, Burk RD. 2010. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 70:3159–3169. 10.1158/0008-5472.CAN-09-4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villa LL, Sichero L, Rahal P, Caballero O, Ferenczy A, Rohan T, Franco EL. 2000. Molecular variants of human papillomavirus types 16 and 18 preferentially associated with cervical neoplasia. J. Gen. Virol. 81:2959–2968 [DOI] [PubMed] [Google Scholar]
- 8.Zuna RE, Moore WE, Shanesmith RP, Dunn ST, Wang SS, Schiffman M, Blakey GL, Teel T. 2009. Association of HPV16 E6 variants with diagnostic severity in cervical cytology samples of 354 women in a US population. Int. J. Cancer 125:2609–2613. 10.1002/ijc.24706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naghashfar ZS, Rosenshein NB, Lorincz AT, Buscema J, Shah KV. 1987. Characterization of human papillomavirus type 45, a new type 18-related virus of the genital tract. J. Gen. Virol. 68:3073–3079. 10.1099/0022-1317-68-12-3073 [DOI] [PubMed] [Google Scholar]
- 10.Bernard HU, Burk RD, Chen Z, van Doorslaer K, Hausen H, de Villiers EM. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79. 10.1016/j.virol.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. 2004. Classification of papillomaviruses. Virology 324:17–27. 10.1016/j.virol.2004.03.033 [DOI] [PubMed] [Google Scholar]
- 12.Clifford G, Franceschi S. 2008. Members of the human papillomavirus type 18 family (alpha-7 species) share a common association with adenocarcinoma of the cervix. Int. J. Cancer 122:1684–1685. 10.1002/ijc.23282 [DOI] [PubMed] [Google Scholar]
- 13.Guan P, Clifford GM, Franceschi S. 2013. Human papillomavirus types in glandular lesions of the cervix: a meta-analysis of published studies. Int. J. Cancer 132:248–250. 10.1002/ijc.27663 [DOI] [PubMed] [Google Scholar]
- 14.Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. 2011. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: variation by geographical region, histological type and year of publication. Int. J. Cancer 128:927–935. 10.1002/ijc.25396 [DOI] [PubMed] [Google Scholar]
- 15.Guan P, Howell-Jones R, Li N, Bruni L, de Sanjose S, Franceschi S, Clifford GM. 2012. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int. J. Cancer 131:2349–2359. 10.1002/ijc.27485 [DOI] [PubMed] [Google Scholar]
- 16.Chen Z, DeSalle R, Schiffman M, Herrero R, Burk RD. 2009. Evolutionary dynamics of variant genomes of human papillomavirus types 18, 45, and 97. J. Virol. 83:1443–1455. 10.1128/JVI.02068-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Schiffman M, Herrero R, Desalle R, Anastos K, Segondy M, Sahasrabuddhe VV, Gravitt PE, Hsing AW, Burk RD. 2011. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS One 6:e20183. 10.1371/journal.pone.0020183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornet I, Gheit T, Iannacone MR, Vignat J, Sylla BS, Del Mistro A, Franceschi S, Tommasino M, Clifford GM. 2013. HPV16 genetic variation and the development of cervical cancer worldwide. Br. J. Cancer 108:240–244. 10.1038/bjc.2012.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International Biological Study on Cervical Cancer (IBSCC) Study Group. J. Natl. Cancer Inst. 87:796–802 [DOI] [PubMed] [Google Scholar]
- 20.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518–527. 10.1056/NEJMoa021641 [DOI] [PubMed] [Google Scholar]
- 21.Franceschi S, Rajkumar T, Vaccarella S, Gajalakshmi V, Sharmila A, Snijders PJ, Munoz N, Meijer CJ, Herrero R. 2003. Human papillomavirus and risk factors for cervical cancer in Chennai, India: a case-control study. Int. J. Cancer 107:127–133. 10.1002/ijc.11350 [DOI] [PubMed] [Google Scholar]
- 22.Clifford GM, Gallus S, Herrero R, Munoz N, Snijders PJ, Vaccarella S, Anh PT, Ferreccio C, Hieu NT, Matos E, Molano M, Rajkumar R, Ronco G, de Sanjose S, Shin HR, Sukvirach S, Thomas JO, Tunsakul S, Meijer CJ, Franceschi S. 2005. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis. Lancet 366:991–998. 10.1016/S0140-6736(05)67069-9 [DOI] [PubMed] [Google Scholar]
- 23.Hammouda D, Munoz N, Herrero R, Arslan A, Bouhadef A, Oublil M, Djedeat B, Fontaniere B, Snijders P, Meijer C, Franceschi S. 2005. Cervical carcinoma in Algiers, Algeria: human papillomavirus and lifestyle risk factors. Int. J. Cancer 113:483–489. 10.1002/ijc.20600 [DOI] [PubMed] [Google Scholar]
- 24.Li LK, Dai M, Clifford GM, Yao WQ, Arslan A, Li N, Shi JF, Snijders PJ, Meijer CJ, Qiao YL, Franceschi S. 2006. Human papillomavirus infection in Shenyang City, People's Republic of China: a population-based study. Br. J. Cancer 95:1593–1597. 10.1038/sj.bjc.6603450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu RF, Dai M, Qiao YL, Clifford GM, Liu ZH, Arslan A, Li N, Shi JF, Snijders PJ, Meijer CJ, Franceschi S. 2007. Human papillomavirus infection in women in Shenzhen City, People's Republic of China, a population typical of recent Chinese urbanisation. Int. J. Cancer 121:1306–1311. 10.1002/ijc.22726 [DOI] [PubMed] [Google Scholar]
- 26.Bardin A, Vaccarella S, Clifford GM, Lissowska J, Rekosz M, Bobkiewicz P, Kupryjanczyk J, Krynicki R, Jonska-Gmyrek J, Danska-Bidzinska A, Snijders PJ, Meijer CJ, Zatonski W, Franceschi S. 2008. Human papillomavirus infection in women with and without cervical cancer in Warsaw, Poland. Eur. J. Cancer 44:557–564. 10.1016/j.ejca.2007.12.001 [DOI] [PubMed] [Google Scholar]
- 27.Dondog B, Clifford GM, Vaccarella S, Waterboer T, Unurjargal D, Avirmed D, Enkhtuya S, Kommoss F, Wentzensen N, Snijders PJ, Meijer CJ, Franceschi S, Pawlita M. 2008. Human papillomavirus infection in Ulaanbaatar, Mongolia: a population-based study. Cancer Epidemiol. Biomarkers Prev. 17:1731–1738. 10.1158/1055-9965.EPI-07-2796 [DOI] [PubMed] [Google Scholar]
- 28.Keita N, Clifford GM, Koulibaly M, Douno K, Kabba I, Haba M, Sylla BS, van Kemenade FJ, Snijders PJ, Meijer CJ, Franceschi S. 2009. HPV infection in women with and without cervical cancer in Conakry, Guinea. Br. J. Cancer 101:202–208. 10.1038/sj.bjc.6605140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sideri M, Cristoforoni P, Casadio C, Boveri S, Igidbashian S, Schmitt M, Gheit T, Tommasino M. 2009. Distribution of human papillomavirus genotypes in invasive cervical cancer in Italy: a representative, single institution case series. Vaccine 27(Suppl 1):A30–A33. 10.1016/j.vaccine.2008.12.028 [DOI] [PubMed] [Google Scholar]
- 30.Sherpa AT, Clifford GM, Vaccarella S, Shrestha S, Nygard M, Karki BS, Snijders PJ, Meijer CJ, Franceschi S. 2010. Human papillomavirus infection in women with and without cervical cancer in Nepal. Cancer Causes Control 21:323–330. 10.1007/s10552-009-9467-z [DOI] [PubMed] [Google Scholar]
- 31.Alibegashvili T, Clifford GM, Vaccarella S, Baidoshvili A, Gogiashvili L, Tsagareli Z, Kureli I, Snijders PJ, Heideman DA, van Kemenade FJ, Meijer CJ, Kordzaia D, Franceschi S. 2011. Human papillomavirus infection in women with and without cervical cancer in Tbilisi, Georgia. Cancer Epidemiol. 35:465–470. 10.1016/j.canep.2010.12.006 [DOI] [PubMed] [Google Scholar]
- 32.De Vuyst H, Ndirangu G, Moodley M, Tenet V, Estambale B, Meijer CJ, Snijders PJ, Clifford G, Franceschi S. 2012. Prevalence of human papillomavirus in women with invasive cervical carcinoma by HIV status in Kenya and South Africa. Int. J. Cancer 131:949–955. 10.1002/ijc.26470 [DOI] [PubMed] [Google Scholar]
- 33.Khodakarami N, Clifford GM, Yavari P, Farzaneh F, Salehpour S, Broutet N, Bathija H, Heideman DA, van Kemenade FJ, Meijer CJ, Hosseini SJ, Franceschi S. 2012. Human papillomavirus infection in women with and without cervical cancer in Tehran, Iran. Int. J. Cancer 131:E156–E161. 10.1002/ijc.26488 [DOI] [PubMed] [Google Scholar]
- 34.Aruhuri B, Tarivonda L, Tenet V, Sinha R, Snijders PJ, Clifford G, Pang J, McAdam M, Meijer CJ, Frazer IH, Franceschi S. 2012. Prevalence of cervical human papillomavirus (HPV) infection in Vanuatu. Cancer Prev. Res. (Phila.) 5:746–753. 10.1158/1940-6207.CAPR-11-0515 [DOI] [PubMed] [Google Scholar]
- 35.Mbaye EH, Gheit T, Dem A, McKay-Chopin S, Toure-Kane NC, Mboup S, Tommasino M, Sylla BS, Boye CS. 2014. Human papillomavirus infection in women in four regions of Senegal. J. Med. Virol. 86:248–256. 10.1002/jmv.23719 [DOI] [PubMed] [Google Scholar]
- 36.van den Brule AJ, Pol R, Fransen-Daalmeijer N, Schouls LM, Meijer CJ, Snijders PJ. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40:779–787. 10.1128/JCM.40.3.779-787.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Godinez JM, Heideman DA, Gheit T, Alemany L, Snijders PJ, Tommasino M, Meijer CJ, de Sanjose S, Bosch FX, Bravo IG. 2013. Differential presence of papillomavirus variants in cervical cancer: an analysis for HPV33, HPV45 and HPV58. Infect. Genet. Evol. 13:96–104. 10.1016/j.meegid.2012.09.011 [DOI] [PubMed] [Google Scholar]
- 38.Hesselink AT, Berkhof J, Heideman DA, Bulkmans NW, van Tellingen JE, Meijer CJ, Snijders PJ. 2009. High-risk human papillomavirus DNA load in a population-based cervical screening cohort in relation to the detection of high-grade cervical intraepithelial neoplasia and cervical cancer. Int. J. Cancer 124:381–386. 10.1002/ijc.23940 [DOI] [PubMed] [Google Scholar]
- 39.Felsenstein J. 1989. Mathematics vs. evolution: mathematical evolutionary theory. Science 246:941–942. 10.1126/science.246.4932.941 [DOI] [PubMed] [Google Scholar]
- 40.Chen Z, Schiffman M, Herrero R, DeSalle R, Anastos K, Segondy M, Sahasrabuddhe VV, Gravitt PE, Hsing AW, Burk RD. 2013. Evolution and taxonomic classification of alphapapillomavirus 7 complete genomes: HPV18, HPV39, HPV45, HPV59, HPV68 and HPV70. PLoS One 8:e72565. 10.1371/journal.pone.0072565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ntova CK, Kottaridi C, Chranioti A, Spathis A, Kassanos D, Paraskevaidis E, Karakitsos P. 2012. Genetic variability and phylogeny of high risk HPV type 16, 18, 31, 33 and 45 L1 gene in Greek women. Int. J. Mol. Sci. 13:1–17. 10.3390/ijms13010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornet I, Gheit T, Franceschi S, Vignat J, Burk RD, Sylla BS, Tommasino M, Clifford GM. 2012. Human papillomavirus type 16 genetic variants: phylogeny and classification based on E6 and LCR. J. Virol. 86:6855–6861. 10.1128/JVI.00483-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen AA, Heideman DAM, Boon D, Chen Z, Burk RD, De Vuyst H, Gheit T, Snijders PJF, Tommasino M, Franceschi S, Clifford GM. 2014. Human papillomavirus 33 worldwide genetic variation and associated risk of cervical cancer. Virology 448:356–362. 10.1016/j.virol.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Chen JJ, Gao Q, Dalal S, Hong Y, Mansur CP, Band V, Androphy EJ. 1999. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J. Virol. 73:7297–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sotlar K, Stubner A, Diemer D, Menton S, Menton M, Dietz K, Wallwiener D, Kandolf R, Bultmann B. 2004. Detection of high-risk human papillomavirus E6 and E7 oncogene transcripts in cervical scrapes by nested RT-polymerase chain reaction. J. Med. Virol. 74:107–116. 10.1002/jmv.20153 [DOI] [PubMed] [Google Scholar]