ABSTRACT
The coronavirus nucleocapsid (N) protein forms a helical ribonucleoprotein with the viral positive-strand RNA genome and binds to the principal constituent of the virion envelope, the membrane (M) protein, to facilitate assembly and budding. Besides these structural roles, N protein associates with a component of the replicase-transcriptase complex, nonstructural protein 3, at a critical early stage of infection. N protein has also been proposed to participate in the replication and selective packaging of genomic RNA and the transcription and translation of subgenomic mRNA. Coronavirus N proteins contain two structurally distinct RNA-binding domains, an unusual characteristic among RNA viruses. To probe the functions of these domains in the N protein of the model coronavirus mouse hepatitis virus (MHV), we constructed mutants in which each RNA-binding domain was replaced by its counterpart from the N protein of severe acute respiratory syndrome coronavirus (SARS-CoV). Mapping of revertants of the resulting chimeric viruses provided evidence for extensive intramolecular interactions between the two RNA-binding domains. Through analysis of viral RNA that was packaged into virions we identified the second of the two RNA-binding domains as a principal determinant of MHV packaging signal recognition. As expected, the interaction of N protein with M protein was not affected in either of the chimeric viruses. Moreover, the SARS-CoV N substitutions did not alter the fidelity of leader-body junction formation during subgenomic mRNA synthesis. These results more clearly delineate the functions of N protein and establish a basis for further exploration of the mechanism of genomic RNA packaging.
IMPORTANCE This work describes the interactions of the two RNA-binding domains of the nucleocapsid protein of a model coronavirus, mouse hepatitis virus. The main finding is that the second of the two domains plays an essential role in recognizing the RNA structure that allows the selective packaging of genomic RNA into assembled virions.
INTRODUCTION
Coronaviruses are a family of positive-strand RNA viruses that infect numerous mammalian and avian hosts (1, 2). Although they are generally species specific, the propensity of these viruses to cross from animal reservoirs to the human population has been dramatically demonstrated twice in just the past dozen years. In 2002, a previously unknown pathogen, severe acute respiratory syndrome coronavirus (SARS-CoV), initiated an epidemic that spread worldwide before the human chain of transmission was broken. The potential remains for SARS-CoV or very similar coronaviruses to reemerge from bat sources (3). More recently, there has emerged another lethal but less readily transmissible agent, Middle East respiratory syndrome coronavirus (MERS-CoV), which also appears to have originated in bats (4, 5).
The well-studied coronavirus mouse hepatitis virus (MHV) is a prototype for the molecular biology, genetics, and pathogenesis of this family of viruses. MHV is taxonomically classified within the betacoronaviruses, the same genus as includes SARS-CoV and MERS-CoV, and it can thus serve as a model for some of the fundamental attributes of these pathogens. A defining property of coronaviruses is that in addition to replicating their genomic RNA, they produce a nested set of transcripts, the latter serving as the mRNAs for the viral structural proteins and for additional accessory proteins. Viral RNA synthesis is carried out by a 16-subunit replicase-transcriptase complex that is generated by processing of two polyproteins encoded by the 5′ two-thirds of the genome. The virions of coronaviruses contain four structural proteins, three of which—the spike (S), membrane (M), and envelope (E) proteins—are embedded in the membrane envelope. The fourth structural component, the nucleocapsid (N) protein, resides in a helical ribonucleoprotein complex with the RNA genome in the virion interior.
N protein is thought to participate in a number of critical processes in coronavirus infection, in addition to its structural association with RNA and M protein in the assembled virion (1, 6). Various functions have been inferred for N protein in RNA replication and transcription (7–11), in the translation of viral mRNA (12, 13), and in the selective packaging of viral genomic RNA (14, 15). Some of these proposed roles of N are not clearly established; for others, the molecular details remain to be precisely defined. Despite this uncertainty about function, much has been learned about N protein structure in recent years. The coronavirus N protein has two distinct RNA-binding domains, a characteristic that is unusual and possibly unique among RNA virus capsid proteins. We have designated these domains N1b and N2b (16–18) (Fig. 1A), but they are also widely called the N-terminal domain (NTD) (10, 19–22) and the C-terminal domain (CTD) (21, 23–25), respectively. Domain N1b (or NTD) is a monomer in solution, whereas domain N2b (or CTD) forms dimers and higher-order oligomers that may be representative of the interactions driving helical nucleocapsid formation (24, 26–28). Structures for each of the RNA-binding domains of the N proteins of SARS-CoV, MHV, and infectious bronchitis virus (an avian gammacoronavirus) have been determined by X-ray crystallography or nuclear magnetic resonance (NMR). From these, RNA-binding surfaces have been deduced for grooves or platforms rich in basic and aromatic amino acid residues, but as yet, no structure has been obtained for an N protein-RNA complex.
FIG 1.
Construction of MHV mutants with chimeric N proteins containing RNA-binding domains from the SARS-CoV N protein. (A) Model of the MHV N protein showing domains as defined previously (16, 17). The RNA-binding domains are N1b (or NTD) and N2b (or CTD). Other segments of the molecule include the amino terminus N1a; the central spacer N2a, which harbors the SR region that interacts with nsp3; and the M protein-binding domain N3, which is linked to the molecule via spacer B. Numbering indicates amino acid residues. (B) Schematic of wild-type and mutant N proteins. Shading represents segments of SARS-CoV N sequence substituted within the MHV N molecule; the asterisk denotes a lethal substitution. (C) Western blots of equal amounts of immunopurified wild-type, S1b mutant, and S2b mutant virions probed with monoclonal anti-MHV N antibody J.3.3, which recognizes an epitope in domain N3 (31) (left) or with polyclonal anti-SARS-CoV N antibody (right). (D) Plaques of the S1b and S2b mutants at 33, 37, or 39°C compared with those of isogenic wild-type virus. Plaque titrations were carried out on L2 cells; monolayers were stained with neutral red at 49 h postinfection and were photographed 17 h later.
Adjacent to the RNA-binding domains are intrinsically disordered segments (29) falling at the amino terminus (domain N1a), the carboxy terminus (spacer B and domain N3), and the center (domain N2a) of the N molecule. No functional role has been ascribed to domain N1a. Domain N3 has been shown to interact with the carboxy terminus of the M protein (30–33), and it also appears to participate in one type of N-N interaction (16). Domain N2a contains a serine- and arginine-rich region (Fig. 1A) that interacts with the replicase subunit nonstructural protein 3 (nsp3) to mediate a crucial early step in infection (17, 18, 34).
In order to probe the functions of the two RNA-binding domains of the MHV N protein, we substituted each with its counterpart from SARS-CoV. A similar strategy, the replacement of the entire MHV N protein with that of bovine coronavirus (BCoV), led to our previous discovery of the domain N2a-nsp3 interaction (17). However, domains N1b and N2b of the MHV and BCoV N proteins are sufficiently homologous as to be functionally indistinguishable. We thus hypothesized that making substitutions from the more phylogenetically distant SARS-CoV N protein would have greater consequences. Analyses of the resulting chimeric N protein mutants provided evidence for extensive interactions between the two RNA-binding domains of N. We found no support for a role for either domain N1b or N2b in the interaction of N protein with M protein. Moreover, neither substitution altered the fidelity of viral transcript formation or had a significant effect on the N-nsp3 interaction. Strikingly, substitution of domain N2b, but not domain N1b, abolished the selective packaging of genomic RNA by MHV virions. The latter finding points to domain N2b as the principal determinant for the recognition of the MHV genomic RNA packaging signal (PS).
MATERIALS AND METHODS
Cells and viruses.
Stocks of wild-type MHV-A59 and mutants were grown in mouse 17 clone 1 (17Cl1) cells. Plaque titrations and plaque purifications were carried out with mouse L2 cells. The host range chimeric coronaviruses designated fMHV and fMHV.v2 (35, 36), which were used for reverse genetics, were grown in Felis catus whole-fetus (FCWF) cells.
MHV mutant construction.
All constructed mutants in this study were created by targeted RNA recombination, procedures for which have been described in detail previously (35, 36). Donor RNAs for targeted RNA recombination were synthesized from plasmids derived from the transcription vector pSG6X, which includes the 3′-most 8.6 kb of the MHV-A59 genome. pSG6X is identical to the previously described pMH54 (35), except for a coding-silent PspXI site (underlined) changing N gene codons 14 through 16 from AGCTCCTCT to AGCTCGAGT and a coding-silent BspEI site (underlined) changing N gene codons 444 through 446 from GTGCCAGAT to GTTCCGGAT. Each N gene mutation was first constructed in the plasmid pCK70XB (18), which is a template for MHV subgenomic RNA7, and then shuttled into pSG6X via the unique PspXI-BspEI fragment. Constructs made from pCK70XB took advantage of the unique PspXI site upstream of domain N1b, unique NheI and NgoMIV sites between domains N1b and N2b, and unique BstXI and BspEI sites downstream of domain N2b.
The S1b substitution was constructed by replacement of the PspXI-NheI segment of pCK70XB with a fragment synthesized by two-step PCR with partial products made from overlapping oligonucleotides and from PCR using a cloned SARS-CoV (strain Urbani) N gene cDNA as the template. An additional PspXI site was removed from the SARS-CoV N sequence in this process. Similarly, the S2b substitution was constructed by replacement of the NgoMIV-BstXI segment of pCK70XB, with concomitant removal of an additional BstXI site from the SARS-CoV N sequence. The S1b-S2b double substitution was made by shuttling the NheI-BspEI fragment of the S2b plasmid into the S1b plasmid. The S1bR1 and S1bR4 constructs were created by PCR-based mutagenesis of the S1b plasmid; the S1b2bR4 and S1b[2a]2b constructs were produced by PCR-based mutagenesis of the S1b-S2b plasmid. All of the partial S2b substitutions—S2bN, S2bC, S2b1, S2b2, and S2b3—were generated by replacement of the NgoMIV-BstXI segment of pCK70XB with fragments made by PCR from overlapping oligonucleotides.
For each viable viral mutant, two independent isolates were obtained: Alb733 and Alb735 for the S1b mutant, Alb737 and Alb739 for the S2b mutant, Alb774 and Alb775 for the S1bR1 mutant, Alb782 and Alb784 for the S2bC mutant, and Alb786 and Alb787 for the S2b2 mutant. In each case, when it was established that both isolates behaved identically in preliminary experiments, one of them was chosen for further analysis. Additionally, an isogenic wild-type recombinant, Alb741, was isolated by targeted RNA recombination with donor RNA from pSG6X.
gRNA infectivity assay.
The enhancement of the infectivity of MHV genomic RNA (gRNA) by wild-type or mutant N mRNA was measured by plaque assays. Purification of viral genomic RNA, in vitro transcription and purification of N mRNA, and conditions for cotransfection by electroporation were exactly as described in detail previously (18). The templates for synthesis of S1b and S2b chimeric N mRNA were the same pCK70XB-derived plasmids as were intermediates in the construction of donor RNA templates for targeted RNA recombination.
Virus purification.
Virus was grown in 17Cl1 cell monolayers that were infected at a multiplicity of 1 PFU/cell. Medium containing released virus was harvested at 12 to 16 h postinfection, at a point when monolayers exhibited maximal syncytium formation but only minimal lysis or detachment. Virus was precipitated from growth medium with polyethylene glycol, resuspended in magnesium- and calcium-free phosphate-buffered saline, pH 7.4 (PBS), and sedimented onto cushions of 60% sucrose in PBS by centrifugation at 151,000 × g for 2.5 h in a Beckman SW41 rotor at 4°C. After removal from cushions, virus samples were diluted with PBS to contain 10% sucrose and were layered onto step gradients of 10%-20%-40%-60% sucrose. Following centrifugation at 151,000 × g for 2.5 h in a Beckman SW41 rotor at 4°C, banded virions were collected from the 20% to 40% sucrose interface. Virions were then immunopurified with anti-M monoclonal antibody J.1.3 and nProtein A Sepharose beads (GE Healthcare) exactly as described previously (37).
Western blotting of purified virions was performed exactly as described previously (16, 37). Proteins were detected with anti-MHV-N monoclonal antibody J.3.3, anti-MHV-M monoclonal antibody J.1.3, or anti-SARS-CoV N rabbit polyclonal antibody. Both anti-MHV monoclonal antibodies were generously provided by John Fleming (University of Wisconsin, Madison). For normalization of immunopurified virions prior to RNA extraction, bound monoclonal antibodies were visualized by enhanced chemiluminescence detection (Pierce), which was quantitated with a Bio-Rad ChemiDoc XRS+ instrument.
Analysis of viral RNA.
RNA was extracted from purified virions or from infected cell monolayers with Ultraspec reagent (Biotecx) according to the manufacturer's instructions. Northern blotting of purified virion or intracellular RNA was carried out as detailed previously (38). RNA was probed with a PCR product corresponding to the 3′-most 539 nucleotides of the N gene and the entire 3′ untranslated region of the MHV genome. The probe was labeled with an AlkPhos Direct kit, and blots were visualized using CDP-Star detection reagent (GE Healthcare).
For verification of the composition of constructed mutants, reverse transcription of RNA was carried out with a random hexanucleotide primer and avian myeloblastosis virus reverse transcriptase (Life Sciences). PCR amplification of cDNA was performed with the Expand High Fidelity PCR system (Roche). Reverse transcription-PCR (RT-PCR) products were analyzed by agarose gel electrophoresis and were purified with QIAquick spin columns (Qiagen) prior to DNA sequencing.
To analyze the leader-body junctions of subgenomic mRNAs, random-primed cDNA made from total RNA purified from infected 17Cl1 cells was amplified by PCR using primer L (corresponding to nucleotides [nt] 16 to 39 at the 5′ end of the genome) paired with primer A (complementary to nucleotides 344 to 364 of gene 2a), primer B (complementary to nucleotides 247 to 264 of gene 5a), or primer C (complementary to nucleotides 626 to 645 of the N gene). PCR products were analyzed by electrophoresis in 1.5% agarose gels stained with ethidium bromide.
RESULTS
Construction of MHV N protein mutants containing SARS-CoV RNA-binding domains.
In order to probe the functions of the two RNA-binding domains of the MHV N protein, we exchanged each with its counterpart from the SARS-CoV N protein. In the S1b mutant, the MHV domain N1b (or NTD) was replaced by SARS-CoV domain N1b, with which it shares 44% amino acid identity (Fig. 1B). Similarly, in the S2b mutant, the MHV domain N2b (or CTD) was replaced by the even more divergent SARS-CoV domain N2b, with which it retains only 35% amino acid identity. The two mutants, as well as an otherwise isogenic wild-type virus, were constructed by targeted RNA recombination (35, 36). Additionally, we attempted to replace both domains of MHV N in an S1b-S2b construct, but this double substitution was found to be lethal.
For both the S1b mutant and the S2b mutant, the sequence of the entire N gene was determined and found to contain only the expected substitution and no extraneous mutations. To verify expression of the encoded N proteins, equal amounts of purified virions were analyzed by Western blotting (Fig. 1C). Both chimeric N proteins, as well as the wild type, reacted with an anti-MHV N monoclonal antibody that recognizes an epitope in domain N3 (31), which is common to all three N proteins. As expected, only the S1b and S2b N proteins were reactive with polyclonal antiserum that had been raised against bacterially expressed SARS-CoV N protein. Further sequencing was carried out to address the possibility that isolation of the mutants had been dependent on their acquisition of second-site mutations in proteins known to interact with N. It should be noted that in each of the chimeric N proteins, domain N3, which is the only clearly established M-interacting segment (30–33), and the SR region, which interacts with nsp3 (17, 18, 34), were both derived from the MHV N protein (Fig. 1B). For both the S1b mutant and the S2b mutant, the sequence of the first 600 codons of the replicase subunit nsp3 was confirmed to be unaltered; likewise, the entire M gene sequence of each mutant was found to be identical to that of the wild type. Therefore, no compensatory mutations in the Ubl1 domain of nsp3 or in the M protein had been required in order to allow either of the RNA-binding domain substitutions that were made in the N protein.
Although the S1b and S2b mutants were viable and grew to high titers, neither was as fit as wild-type virus. Both mutants formed plaques that were smaller than those of the wild type at 33, 37, and 39°C (Fig. 1D). The S1b mutant was markedly more impaired than the S2b mutant, and this difference became most pronounced at 39°C. These observations indicated that, compared to their MHV counterparts, the substituted SARS-CoV RNA-binding domains did not function optimally within the framework of the remainder of the MHV N molecule.
Analysis of revertants of the S1b and S2b mutants.
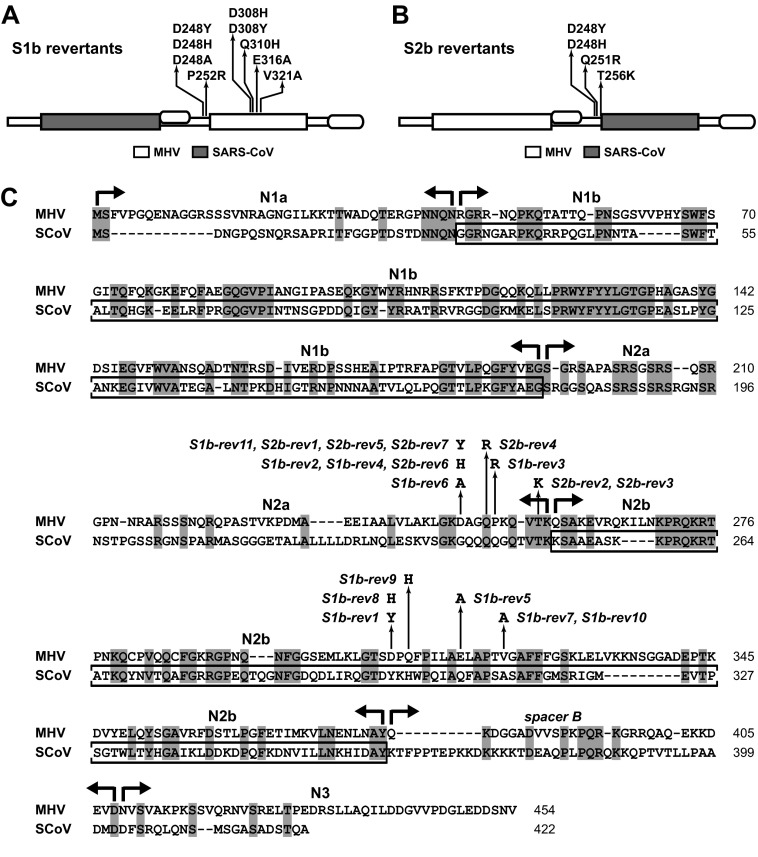
To identify genetic changes that could improve the growth of the N protein chimeras, we isolated adaptive or gain-of-function mutants (which, for simplicity, are hereafter referred to as revertants). For this purpose, cultures were started from multiple individual plaques of the S1b and S2b mutants and were serially passaged at 39°C in L2 cells at a low multiplicity of infection. Following passage six, by which time accelerated growth was usually noted, plaque titrations were carried out on L2 cells at 39°C, and a single plaque originating from each culture was purified for analysis. The plaques chosen ranged from slightly to substantially larger than those of the original mutant (see below), but none fully reached the size of plaques of wild-type MHV. In this manner, we obtained 18 independent revertants, 11 from the S1b mutant and 7 from the S2b mutant. The full N gene sequences of these viruses revealed a total of 11 unique reverting mutations (Fig. 2).
FIG 2.
Summaries of the loci of multiple independent growth-enhancing reverting mutations obtained for the S1b mutant (A) and the S2b mutant (B). (C) Alignment of the amino acid sequences of the MHV and SARS-CoV N proteins. Domain boundaries and the mutations of all 18 individual S1b and S2b revertants are shown. The SARS-CoV N1b and N2b domains that are contained in the S1b and S2b mutants, respectively, are boxed. GenBank accession numbers for the sequences shown are AY700211 for MHV-A59 and AY278741 for SARS-CoV strain Urbani.
Notably, each revertant had only a single mutation. All of these were coding mutations, and somewhat surprisingly, none of these mapped in the originally substituted SARS-CoV RNA-binding domain of its respective parent. All reverting mutations of the S1b mutant (summarized in Fig. 2A) clustered outside domain N1b, either in domain N2a near the N2b boundary or else in the center of (MHV) domain N2b. All reverting mutations of the S2b mutant (summarized in Fig. 2B) formed a single cluster at the same locus in domain N2a as the first set of S1b revertants. Moreover, two of the S2b reverting mutations, D248Y and D248H, were identical to S1b reverting mutations. Most, but not all, of the reverting mutations brought about a net increase of positive charge, either by changing a negatively charged residue to a neutral or polar residue or else by changing a neutral or polar residue to a positively charged residue (Fig. 2C). This may indicate that an important effect of most of the reverting mutations is to increase the general affinity of N protein for RNA. For a subset of five revertants, we sequenced the first 630 codons of the replicase subunit nsp3 and found no changes from the wild-type sequence. This finding was consistent with the absence of any S1b or S2b reverting mutations in the SR region of N. Additionally, for a subset of 10 revertants, we sequenced the entire M gene and found no changes from the wild type, except that two revertants (S1b-rev4 and S2b-rev1) harbored T228I, which altered the carboxy-terminal residue of M protein. This mutation is a common variant resulting from recombination between the genome and subgenomic RNA7 (30). Thus, none of the S1b or S2b revertants examined contained potential intergenic suppressor mutations either in the amino terminus of nsp3 or in the M protein.
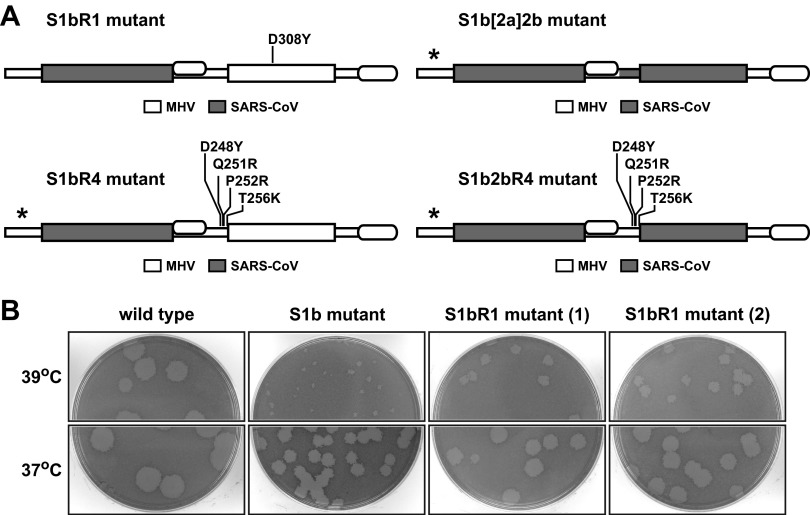
Our finding that five independent reverting mutations of the S1b substitution mapped in domain N2b points to the existence of a functional interaction between the two RNA-binding domains of the coronavirus N protein. Such an interaction is also suggested by the observation that three of these S1b reverting mutations (D308Y, Q310H, and V321A) change an MHV N2b residue to the corresponding aligned SARS-CoV residue (Fig. 2C). To more clearly test this genetic cross talk, we reconstructed one of the mutations, D308Y, in combination with the S1b substitution to determine whether it was sufficient for reversion (Fig. 3A). As shown in Fig. 3B, two independent isolates of the resulting S1bR1 mutant exhibited markedly larger plaques than the original S1b mutant at both 37 and 39°C. This established that the D308Y mutation alone is indeed capable of significantly enhancing the growth of the S1b mutant, thereby confirming that there is a genetic interaction between the N1b and N2b domains. We also attempted to learn the significance of the cluster of S1b and S2b reverting mutations at the downstream edge of domain N2a. To test whether these could have an additive effect, we designed constructs in which four of these mutations were combined either with the S1b substitution (in the S1bR4 mutant) or with both the S1b and S2b substitutions (in the S1b2bR4 mutant) (Fig. 2A). Additionally, we tested whether incorporation of the downstream end of SARS-CoV domain N2a could enable the recovery of the S1b2b double domain substitution (in the S1b[2a]2b mutant). However, we were unable to isolate any of the last three mutants in multiple independent targeted RNA recombination trials that included controls in which robust numbers of recombinants were obtained with wild-type donor RNA. These negative results are strong evidence that the S1bR4, S1b2bR4, and S1b[2a]2b mutations are lethal, suggesting that aggregation of multiple reverting mutations is not beneficial to chimeric N protein function.
FIG 3.
Chimeric N protein constructs incorporating one or multiple reverting mutations. (A) Schematic of wild-type and mutant N proteins. Shading represents segments of SARS-CoV N sequence substituted within the MHV N molecule; asterisks denote lethal substitutions. In the S1bR1 mutant, the S1b substitution was paired with the reverting mutation D308Y in domain N2b. In the S1bR4 mutant, the S1b substitution was paired with a set of four reverting mutations in domain N2a; likewise, in the S1b2bR4 mutant, the S1b and S2b substitutions were paired with the same set of four reverting mutations in domain N2a. In the S1b[2a]2b mutant, both the S1b and S2b substitutions were added to a partial substitution of domain N2a, in which residues 221 to 249 of SARS-CoV N replaced residues 234 to 257 of MHV N. (B) Plaques of two independent isolates of the S1bR1 mutant at 37 or 39°C compared with those of the S1b mutant and wild-type virus. Plaque titrations were carried out on L2 cells; monolayers were stained with neutral red at 72 h postinfection and were photographed 18 h later.
Characterization of a temperature-sensitive domain N2b mutant.
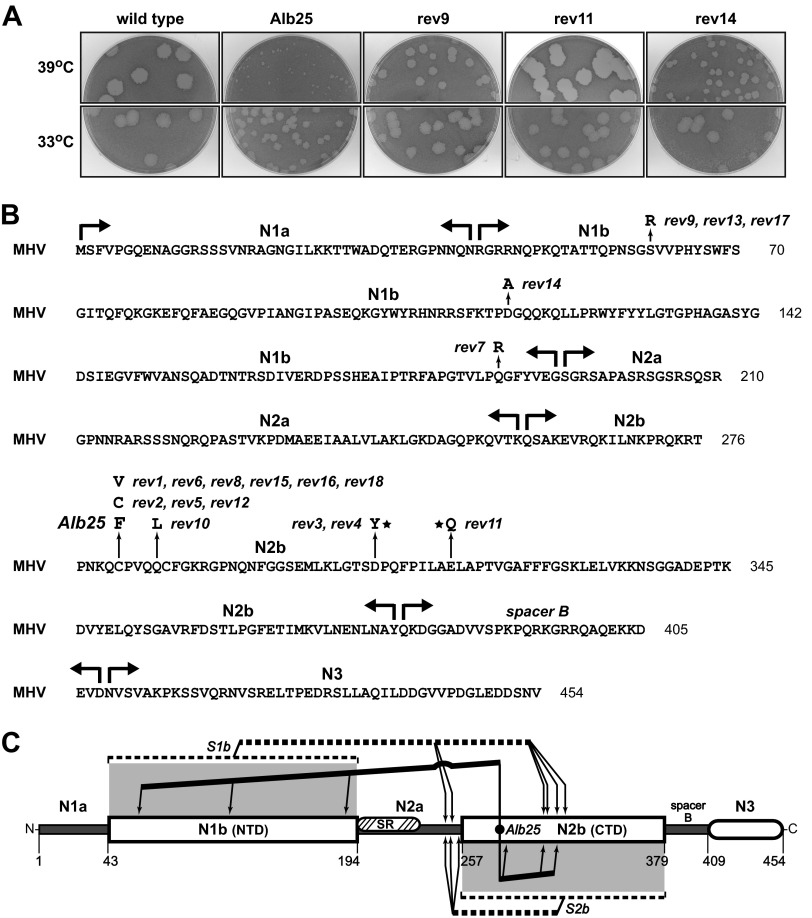
In a previous study, we identified a classical temperature-sensitive MHV mutant designated Alb25 (39). At the nonpermissive temperature, 39°C, Alb25 formed very tiny plaques compared to those of the wild type (Fig. 4A). Virions of Alb25 were also nearly 1,000-fold more thermolabile than wild-type virions. Because two other classical N gene mutants, Alb1 (39) and Alb4 (40), exhibited similarly extreme degrees of thermolability, we sequenced the N gene of Alb25 and found that it contained a single point mutation changing the first of two cysteine residues in domain N2b to phenylalanine (Fig. 4B). Eighteen independent revertants of Alb25 were isolated from passage 2 stocks on the basis of their ability to form plaques larger than those of the original mutant at 39°C, and the entire N gene of each was sequenced. Three revertants (rev2, rev5, and rev12) exactly returned phenylalanine back to cysteine at the site of the primary mutation. These viruses formed wild-type-sized plaques at the nonpermissive temperature (data not shown), demonstrating that the C281F mutation was responsible for the phenotype of Alb25. A second class of primary-site revertants, containing the mutation C281V, had plaques intermediate in size between those of the mutant and the wild type (data not shown). However, nine revertants retained the C281F mutation but had acquired an additional mutation elsewhere in the N molecule. Plaques formed by these second-site revertants at 39°C ranged from one-sixth the size of wild-type plaques (rev14) to equal in size to wild-type plaques (rev11) (Fig. 4A). The mutations in these revertants mapped to multiple sites in both domain N1b and domain N2b (Fig. 4B); notably, none were located in domain N1a, the SR region, spacer B, or domain N3. Like the S1b and S2b reverting mutations, most of the Alb25 second-site reverting mutations created a net increase of positive charge. Remarkably, three Alb25 revertants (rev3, rev4, and rev11) mapped to the same N2b residues as did three of the revertants of the S1b mutant (S1b-rev1, -rev5, and -rev8) (Fig. 2C). Collectively, the reverting mutations of the Alb25, S1b, and S2b mutants point to a broad set of intramolecular interactions between the two RNA-binding domains of the coronavirus N protein, as shown in Fig. 4C.
FIG 4.
Classical temperature-sensitive MHV N protein mutant Alb25 (39) and its revertants. (A) Plaques of representative examples of second-site Alb25 revertants at 33 or 39°C compared with those of the original Alb25 mutant and wild-type virus Alb240 (73). Plaque titrations were carried out on L2 cells; monolayers were stained with neutral red at 72 h postinfection and were photographed 18 h later. (B) MHV N protein amino acid sequence, showing domain boundaries and loci of the Alb25 mutation and 18 independent reverting mutations that partially or completely restore growth at the nonpermissive temperature. Stars denote reverting mutations at residues that were also mutated in particular S1b revertants. (C) Summary of N protein intramolecular genetic interactions revealed by revertants of the S1b, S2b, and Alb25 mutants. Lines running from the sites of the S1b or S2b substitutions or the Alb25 mutation point to the locations of compensating mutations.
Lack of effect of the S1b and S2b substitutions on subgenomic RNA leader-body junction formation.
A distinguishing characteristic of coronavirus infections is the transcription of a 3′ nested set of subgenomic RNAs (sgRNAs) that serve as the mRNAs for expression of all genes downstream of the replicase gene (1). Each sgRNA contains a 5′ leader (72 nucleotides [nt] for MHV), which is identical to the 5′ end of the genome, joined to a body sequence, which ranges from 1.7 to 9.6 kb identical to the 3′ end of the genome (Fig. 5A). The leader-body fusion occurs at a short transcription-regulating sequence (TRS) common to the 3′ end of the leader RNA and the 5′ end of each body RNA. This discontinuous transcription is brought about by a highly efficient strand-switching event that takes place during negative-strand RNA synthesis, and its frequency is influenced by RNA sequence and secondary structure flanking the TRS.
FIG 5.
Analysis of subgenomic mRNA leader-body junctions produced by the S1b and S2b mutants. (A) Positive-sense RNA species produced during MHV infection. At the top is the genome (gRNA), comprising the replicase genes (rep 1a and 1b) and downstream genes for the structural proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N), and for accessory proteins 2a, hemagglutinin-esterase (HE), 4, and 5a. Below the gRNA is the 3′ nested set of transcribed subgenomic mRNA species (sgRNA) 2 to 7. The leader RNA, which appears at the 5′ ends of the gRNA and all sgRNA species, is represented as a black rectangle. The positions and sizes of PCR primers L, A, B, and C are not drawn to scale; the exact positions of primers are given in Materials and Methods. The expected sizes of PCR products produced by primer pairs are indicated. (B) RT-PCR products crossing the leader-body junctions of sgRNA2, -4, -5, and -7 from total RNA purified from mock-infected cells (m) or from cells infected with wild-type MHV (wt) or the S1b or S2b mutants. PCR products were analyzed by agarose gel electrophoresis; std, DNA size standards. The star and circles indicate aberrant PCR products due to mispriming by downstream primers A and B, respectively. (C) Sequences of the PCR products for the leader-body junctions of sgRNA2 and sgRNA5. The sequence traces, obtained with downstream primers A and B, respectively, are negative sense. The corresponding positive-sense RNA sequences are given above each set of traces; the TRS for each is boxed.
N protein has been proposed to facilitate the coronavirus transcription process, both through sequence-specific binding to the TRS (7, 10, 13) and through helix-destabilizing and RNA chaperone activities (10, 11, 41, 42). For MHV and all other lineage A betacoronaviruses the consensus TRS is 5′-AAUCUAAAC-3′, whereas the SARS-CoV TRS is 5′-ACGAAC-3′ (43). Therefore, we reasoned that if transcription was dependent upon selective recognition of the TRS by N protein, then we would expect either or both of the SARS-CoV RNA-binding domain substitutions to adversely affect the fidelity of MHV sgRNA formation. To test this notion, we used RT-PCR to inspect the 5′ ends of sgRNAs formed in cells infected with the S1b and S2b mutants. We chose to analyze sgRNA2 and sgRNA5 as examples of lower-abundance transcripts, which might be more sensitive to misrecognition of the TRS (also, the TRS for sgRNA2, 5′-AAUCUAUAC-3′, deviates from the consensus). Additionally, we examined the highly abundant transcripts sgRNA4 and sgRNA7. Random-primed cDNA was amplified with a positive-sense leader primer (primer L) (Fig. 5A) paired with various negative-sense primers to cross the leader-body junctions of sgRNA2 (primer A), sgRNA4 and -5 (primer B), or sgRNA7 (primer C). The different primer pairs produced the same set of PCR products of the predicted sizes for RNA from cells infected with either the S1b mutant, the S2b mutant, or wild-type virus (Fig. 5B). Only two unexpected products were observed, a 300-bp band unique to the S1b mutant, which was generated with primers L and A, and a 200-bp band common to the S2b mutant and the wild type, which was generated with primers L and B. However, sequencing of these PCR products revealed that they did not reflect unusual leader-body junctions. Rather, they were caused, respectively, by mispriming of primer A within the SARS-CoV N1b sequence (which is absent in the S2b mutant and the wild type) and by mispriming of primer B within the MHV N1b sequence (which is absent in the S1b mutant). Sequences obtained for the main PCR products were identical for the two mutants and the wild type and corresponded exactly to the four expected leader-body junction sequences. Sequence histograms, partially shown for sgRNA2 and -5 in Fig. 5C, had no minor peaks that would have indicated a minor population of aberrant transcripts.
Additionally, we searched for evidence of transcription originating from fusion at the only occurrence in the MHV genome of the SARS-CoV TRS, 5′-ACGAAC-3′. This motif appears at nt 13588 to 13593 in the MHV nsp12 coding region. RT-PCR was carried out pairing primer L with negative-sense primers located either 266 or 584 nucleotides downstream of this potential TRS. However, like the wild type, neither the S1b or S2b mutant yielded a product of the expected size with either set of primer pairs; minor artifactual bands were the same for both mutants and the wild-type control. Overall, our results do not support the existence of a transcriptional requirement for sequence-specific TRS recognition by the N protein.
Ability of chimeric N proteins to support the infectivity of gRNA.
A critical early step in coronavirus infection is mediated by the interaction between the SR region of the N protein and the amino-terminal Ubl1 domain of nsp3 (17, 18, 34). Moreover, this interaction is directly related to the ability of N protein to enhance the infectivity of transfected coronavirus gRNA, which is only minimally infectious by itself (17). Since we and others have previously demonstrated that the latter function of N protein requires both of the RNA-binding domains of N in addition to the SR region (10, 18, 42), we examined the activities of the S1b and S2b mutant N proteins in gRNA-mRNA cotransfection assays. As observed before (17, 18), cotransfected wild-type MHV N mRNA stimulated the infectivity of wild-type gRNA more than 15-fold over that seen with no added N mRNA (Fig. 6). By comparison, the effectiveness of the chimeric N proteins was somewhat lower. Although the relative strengths of the S1b and S2b N mRNAs varied in multiple separate experiments, both consistently displayed 30 to 50% of the activity of wild-type MHV N mRNA. This less-than-optimal activity may account for some of the partial growth impairment exhibited by each of these mutants.
FIG 6.
Enhancement of the infectivity of gRNA by S1b or S2b mutant N mRNA. L2 cells were cotransfected by electroporation with wild-type MHV gRNA plus synthetic N mRNA or a water control (18). Transfected cells were seeded into dishes and overlaid with agar, and plaques were counted after 48 h of incubation at 37°C. Each histogram represents the mean infectious titer (±standard deviation) from four separate transfections.
Packaging defect of the S2b mutant.
Coronaviruses selectively package gRNA into assembled virions, generally excluding the large molar excess of sgRNA that is synthesized intracellularly during infection. For MHV and other lineage A betacoronaviruses (including BCoV and human coronavirus HKU1), this selectivity, at least in part, is due to an RNA packaging signal (PS) that is embedded in the coding region for the replicase subunit nsp15 (Fig. 7A) (14, 37, 44–49). Coronavirus PS identity and location are not universally conserved, though (50, 51). It is clear that SARS-CoV, a lineage B betacoronavirus, does not possess the nsp15 insertion that harbors the MHV PS (52), nor does it contain an RNA element resembling the MHV PS elsewhere in its genome (53). It was thus feasible that if N protein plays a role in the recognition of the PS, then that capability might have been lost in one or both of the SARS-CoV RNA-binding domain substitutions. To address this possibility, we performed Northern blot analysis of intracellular and packaged viral RNA. As shown in Fig. 7B, a probe specific for the 3′ end of the MHV genome detected the identical 3′ nested set of viral RNA species in total RNA extracted from cells that had been infected with the S1b mutant, the S2b mutant, or the wild type. In contrast to this pattern, RNA isolated from wild-type virions that were purified by sucrose step gradients and immunopurification consisted of gRNA almost entirely devoid of sgRNA (Fig. 7B). This result was in accord with previous work showing a high level of gRNA packaging selectivity by MHV (37, 44–46). Virions of the S1b mutant maintained a degree of packaging stringency equivalent to that of the wild type. Contrary to this, virions of the S2b mutant were found to have packaged substantial amounts of sgRNAs, in proportion to their relative abundance in infected cells. We had previously observed this same packaging-negative phenotype in constructed mutants in which the PS was either disrupted with silent mutations or else deleted (37). In the present case, however, the MHV PS was fully intact. We confirmed that the sequence of the genomic region of the PS (including a span from 200 nt upstream to 450 nt downstream) was identical for the S1b mutant, the S2b mutant, and the wild type. The failure of the S2b mutant to recognize the MHV PS was therefore a consequence of the substitution of the SARS-CoV N2b domain.
FIG 7.
RNA packaging phenotype of the S1b and S2b mutants. (A) Genomic schematic showing the relative positions of the MHV PS, which is located within the coding region for nsp15, and the wild-type or mutant N genes. (B) Northern blots of total RNA isolated from infected or mock-infected17Cl1 cells (left) or of RNA isolated from normalized amounts of immunopurified wild-type, S1b, or S2b virions (right). MHV RNA was detected with a probe specific for the 3′ end of the genome.
In prior work, we showed that the absence of the PS had only a minimal effect on viral growth, but its presence provided a selective advantage (37). Viruses containing the PS consistently outcompeted otherwise isogenic viruses lacking the PS over the course of multiple passages in tissue culture. This raised the question of whether revertants of the S2b mutant, which had been selected by serial passaging, had somehow regained packaging competence. Consequently, we chose to examine S2b-rev1 as a representative of the most frequent S2b reverting mutation, D248Y, which arose independently three times (Fig. 2). The plaque size of S2b-rev1 at 39°C, the temperature of revertant selection, was only slightly larger than that of the original S2b mutant (Fig. 8A). Nevertheless, despite the fact that S2b-rev1 had overtaken the S2b mutant following six serial passages, the packaging phenotype of S2b-rev1 remained identical to that of the S2b mutant (Fig. 8B). Thus, the D248Y mutation, which falls outside domain N2b, did not repair the packaging defect caused by the S2b substitution.
FIG 8.
RNA packaging phenotype of the most commonly arising S2b revertant. (A) Plaques of S2b-rev1 at 39°C compared with those of the original S2b mutant and wild-type virus. Plaque titrations were carried out on L2 cells; monolayers were stained with neutral red at 72 h postinfection and were photographed 18 h later. (B) Northern blot of RNA isolated from normalized amounts of immunopurified wild-type, S2b, or S2b-rev1 virions. MHV RNA was detected with a probe specific for the 3′ end of the genome.
We next sought to further localize the determinants of PS recognition in domain N2b. For this purpose, we designed a set of partial S2b chimeras (Fig. 9A and C). One pair of constructs, S2bN and S2bC, divided the original S2b substitution into two parts, based on structural work that suggested a critical role for SARS-CoV N2b residues 248 to 280 in RNA binding (28). An additional group of constructs, S2b1, S2b2, and S2b3, made smaller substitutions of three regions of SARS-CoV N2b that are colinear with their aligned MHV counterparts. Among these five constructs, only two could be isolated as viable viruses. The first of these, the S2bC mutant, had a robust growth phenotype, with plaques only slightly smaller than those of the wild type (Fig. 9B). The second chimera, the S2b2 mutant, was markedly defective, forming plaques that were tiny at 37°C and pinpoint at 39°C. In contrast, we failed to isolate the S2bN mutant following multiple independent targeted RNA recombination trials, which strongly indicated that the S2bN substitution is lethal. Likewise, we were unable to obtain S2b1 and S2b3 viruses, although our efforts to do so were less extensive in light of the minimal viability of the S2b2 mutant. Collectively, these results suggest that the N protein is much less tolerant to partial substitutions within domain N2b than to outright replacement of the entire domain.
FIG 9.
RNA packaging phenotype of partial S2b substitution mutants. (A) Schematic of wild-type and mutant N proteins. Shading represents segments of SARS-CoV domain N2b sequence substituted within MHV N domain N2b; asterisks denote lethal substitutions. (B) Plaques of the S2bC and S2b2 mutants and a revertant of the latter compared with wild-type plaques at 37 or 39°C. Plaque titrations were carried out on L2 cells; monolayers were stained with neutral red at 72 h postinfection and were photographed 18 h later. (C) Alignment of domain N2b amino acid sequences of the MHV and SARS-CoV N proteins. Brackets beneath the alignment indicate the extent of SARS-CoV N sequence substitution in each of the five partial chimeric mutants. Mutations in five independent revertants of the S2b2 mutant are shown above the alignment. (D) Northern blot of RNA isolated from normalized amounts of immunopurified wild-type, silPS, S2b, S2bC, and S2b2-rev2 virions. The silPS virus is a mutant in which the MHV PS was disrupted with 20 coding-silent mutations (18). MHV RNA was detected with a probe specific for the 3′ end of the genome.
For both the S2bC and S2b2 mutants, sequencing of the entire N and M genes revealed no changes other than the engineered mutations in N. Because the growth of the S2b2 mutant was too weak to examine its RNA packaging capability, we isolated revertants of this chimera following five serial passages at 39°C. Five independent revertants (one of which is shown in Fig. 9B) were obtained as viruses able to form large plaques at 37 and 39°C, and the entire N gene sequence of each was determined. Contrary to the pattern observed with the S1b and S2b revertants (Fig. 2), the mutations in the S2b2 revertants all fell within domain N2b, the domain in which the SARS-CoV sequence substitution had been made (Fig. 9C). Additionally, all but one of the revertants contained two mutations. These findings suggested that the reverting mutations compensated for folding defects that had been created by the juxtaposition of MHV and SARS-CoV amino acid residues within the domain N2b structure. One revertant, S2b2-rev2, which formed the largest plaques at 39°C, was selected as representative of the set.
Northern blot analysis of RNA isolated from highly purified virions revealed that like the original S2b mutant, the S2bC and S2b2-rev2 mutants packaged large amounts of sgRNAs in addition to gRNA (Fig. 9D). This contrasted sharply with the stringently selective incorporation of gRNA into wild-type virions. Moreover, the packaging defect of the three chimeric viruses was seen to be identical to that of the previously characterized silPS mutant (Fig. 9D). The silPS virus contains a set of 20 coding-silent mutations in nsp15 that disrupt both the primary and secondary structures of the MHV PS, but it has a wild-type N protein (37). The recapitulation of the silPS packaging-negative phenotype by viruses harboring only N protein mutations demonstrated that the RNA-binding domain N2b is the major, and perhaps sole, protein determinant of PS recognition for MHV. Our results also showed that, as with S2b-rev1 (Fig. 8), the reverting mutations in S2b2-rev2 corrected the overall function of N protein but did not reestablish recognition of the PS. The boundaries of the substitution in the S2b2 mutant allow us to conclude that alteration of a short interval of MHV domain N2b, residues 298 through 327, is sufficient to disrupt packaging selectivity. At this time, however, we cannot rule out the participation of other N2b residues in PS recognition.
DISCUSSION
Much has been learned previously about coronavirus protein interactions through the construction of interspecies chimeras that preserve or disrupt particular viral functions (17, 18, 35, 54–59). In the present study, we replaced the RNA-binding domains of the MHV N protein with those from a heterologous coronavirus N protein. We chose SARS-CoV N protein as the donor for these substitutions because we expected it to be sufficiently closely related to MHV to produce viable chimeric viruses. On the other hand, we anticipated that the extent of divergence between the MHV and SARS-CoV N proteins would be sufficient to uncover functions of domains N1b and N2b that are dependent upon their sequence-specific interactions with other N domains, other proteins, or RNA.
Role of domain N2b in PS recognition.
The most salient result of our study was the finding that entire or partial substitutions of MHV domain N2b abolished the selective packaging of gRNA into virions, a process which is mediated by the MHV PS. The MHV PS was originally discovered by the dissection of certain defective interfering RNAs that were able to be packaged in the presence of helper virus (44–46). The PS is a 95-nt stem-loop structure containing a 4-fold repeated motif, each copy of which displays a dipurine bulge (49). This element resides within the rep 1b gene, in a region that encodes a dispensable peptide loop on the surface of replicase subunit nsp15 (60). The role of the MHV PS in its native locus in the viral genome was only recently examined through the engineering of multiple point mutations (in the silPS mutant) or a total deletion (in the ΔPS mutant) (37). Surprisingly, knockout of the PS was not highly deleterious, although the silPS and ΔPS mutants were shown to be less fit than wild-type virus in tissue culture. Disruption of PS sequence and structure, nevertheless, was found to cause the indiscriminate packaging of sgRNAs along with gRNA into assembled virions. Thus, in the viral genome, in contrast to defective interfering RNAs, the PS is not absolutely required for gRNA packaging. Rather, it governs the selective incorporation of gRNA over sgRNA into progeny virions.
In the study presented here, we showed that the N protein mutants S2b, S2bC, and S2b2-rev2 had a packaging-negative phenotype identical to that of the silPS mutant (Fig. 9). The most straightforward conclusion to be drawn from this result is that MHV domain N2b specifically recognizes the MHV PS, which is unique to the lineage A betacoronaviruses. A homologous RNA structure does not occur in the nsp15 coding region of the SARS-CoV genome (37, 52, 53), despite unfounded assertions to the contrary (61, 62). It is therefore reasonable to assume that the SARS-CoV N protein did not evolve the capacity to recognize the MHV PS element and that the S2b substitution represents a loss-of-function mutation in this respect. The partial SARS-CoV N2b substitutions in the S2bC and S2b2-rev2 mutants provide us with a starting point for further localization of key residues for PS recognition. More finely targeted mutagenesis, as well as biochemical and structural studies, will be necessary to fully elucidate the molecular basis of gRNA packaging specificity.
Evidence has been presented previously for the specific binding of MHV N protein to PS RNA in vitro (14), and N would seem, a priori, to be the viral protein that acts in selective recognition of gRNA. However, it has been repeatedly demonstrated that not only gRNA but also all sgRNAs are coimmunoprecipitated from infected cell extracts by anti-N antibodies (15, 63, 64). Additionally, it has been shown that M protein associates with only the fraction of N protein that is bound to gRNA or to nonviral RNA containing the MHV PS (48, 64). Further, in a virus-like particle system it was found that M protein, in the absence of N protein, determined the selective packaging of nonviral RNA containing the PS (65). Our findings, which strongly argue that N protein is the major determinant of MHV packaging selectivity, are clearly incompatible with the latter result, but they do not exclude the participation of M protein in gRNA packaging. It may be possible to construct viruses with chimeric M proteins to address this question. Another caveat at this time is that we cannot generalize the role of domain N2b beyond the lineage A betacoronaviruses. Just as the locations and compositions of packaging signals do not appear to be conserved among all coronaviruses (50, 51, 53), so, too, their protein recognition elements may vary.
Viral functions not affected by N protein RNA-binding domain substitutions.
In contrast to the implication of domain N2b in PS recognition, we found no evidence to support a role for either domain N1b or N2b in specific recognition of the TRS during coronavirus transcription. MHV N protein (7, 13), and specifically domain N1b (10), has been shown to bind with high affinity to the MHV TRS, 5′-AAUCUAAAC-3′, and to the complement of this RNA motif. Such binding was envisioned to mediate the template-switching events that form sgRNA leader-body junctions during negative-strand RNA synthesis (10). We expected that if this were the case, then we would find a population of aberrant transcripts among the intracellular RNAs produced during infection with the S1b or the S2b mutant. However, for four different sgRNAs that were sampled, we detected only leader-body junctions identical to those of the wild type (Fig. 5). Correspondingly, Northern blots of RNA from S1b or S2b mutant-infected cells did not contain extra bands that might indicate the generation of leader-body junctions at alternative sites (Fig. 7B). Our results are consistent with recent findings indicating that high-affinity cognate TRS binding is not a general property of coronavirus N proteins (42). A requirement for sequence-specific TRS recognition would also seem to be ruled out by the demonstration of the “rewiring” of the SARS-CoV genome through replacement of all leader and body copies of the TRS with an alien hexanucleotide sequence (66). It is thus likely that the properties of N protein most pertinent to template strand-switching events in coronavirus transcription are its RNA helix-unwinding and annealing activities (10, 11, 41, 42). Consequently, it remains possible that aberrant transcription events could be caused by RNA-binding domain mutations other than those generated in the present study.
The substitution of RNA binding domains also did not affect the compatibility of the N and M proteins. Coronavirus assembly is stabilized by binding of the nucleocapsid to the carboxy-terminal endodomain of M protein (64, 67, 68). There is an abundance of genetic evidence that this essential N protein-M protein interaction maps to domain N3 and to the carboxy terminus of M (30–33). Such a localization accords well with cryo-electron microscopic (69) and cryo-electron tomographic (70) images of MHV and SARS-CoV virions that have shown thread-like connections between the M endodomain and the nucleocapsid. If there existed other critical N-M interactions that involved domains N1b or N2b, we would have expected them to have been perturbed in the S1b or S2b mutants, since the SARS-CoV M endodomain is equally as divergent from its MHV counterpart as are N domains N1b and N2b. However, sequence analysis of the S1b and S2b mutants indicated that no compensatory mutations in M were required for the viability of these chimeras. Likewise, there were no suppressor mutations found in the M genes of the S1b and S2b revertants or the partial S2b chimeras. This suggests that domain N3 is the only region of N protein that is necessary for its interaction with M protein.
N protein intramolecular interactions.
Each of the SARS-CoV RNA-binding domain substitutions was less than optimally functional when placed in the background of the MHV N protein. These deficiencies enabled the selection of multiple independent faster-growing revertants that arose upon passaging of the S1b and S2b mutants. All reversion events were found to be caused by second-site mutations in the N molecule, outside the originally substituted SARS-CoV RNA-binding domain. Many of the reverting mutations of the S1b mutant pointed to functional interactions between domains N1b and N2b (Fig. 2). Second-site revertants of Alb25, a classical domain N2b temperature-sensitive mutant, also exhibited genetic cross talk between domains N1b and N2b (Fig. 4). Moreover, some Alb25 reverting mutations were coincident with particular S1b reverting mutations in domain N2b; similarly, some S1b reverting mutations were coincident with some S2b reverting mutations in domain N2a.
This web of connections calls to mind the multiple interdomain interactions that have been shown to affect the function of MHV nsp5, the main protease of the replicase-transcriptase complex (71). In the case of the N protein, though, it is unclear whether the observed genetic interactions equate to direct physical contacts between the N1b and N2b domains. A study that used small-angle X-ray scattering to examine the shape of a SARS-CoV N1b-N2a-N2b construct in the absence of RNA found that domains N1b and N2b did not directly interact (29), but is not yet known if RNA can facilitate N protein interdomain contacts. It is intriguing that three of the S1b reverting mutations convert an MHV N2b residue to the corresponding SARS-CoV residue (Fig. 2C). However, if the SARS-CoV RNA-binding domains would prefer to interact with each other, rather than with their MHV equivalents, then it is perplexing that we were unable to isolate an S1b-S2b double substitution mutant. One possible explanation for this negative result is that there remains an unknown critical intermolecular interaction that involves both N1b and N2b, which would be abrogated by simultaneous substitution of both domains.
A related problem arises from consideration of the fact that most of the S1b and S2b reverting mutations increase the net positive charge of the region in which they appear. This suggests that the main effect of the reverting mutations is enhancement of nonspecific single-stranded RNA binding, which is principally characterized by electrostatic interactions between basic amino acid residues and the phosphate backbone of RNA (72). Consistent with this view, the most commonly arising S2b reverting mutation (D248Y) did not restore packaging competence to the S2b mutant (Fig. 8). This interpretation, however, does not explain why the nonspecific RNA-binding properties of SARS-CoV domains N1b and N2b should be different from those of the corresponding MHV N domains. These questions may remain unresolved until further information can be obtained about the structure of N protein in complex with RNA and about the ultrastructure of the coronavirus nucleocapsid. Even with a wealth of structural knowledge, we will likely also require a more detailed understanding of the biochemistry of RNA synthesis and recombination to fully apprehend why coronaviruses have evolved two independent RNA-binding domains.
ACKNOWLEDGMENTS
We thank the Applied Genomics Technology Core Facility of the Wadsworth Center for DNA sequencing.
This work was supported by National Institutes of Health (National Institute of Allergy and Infectious Diseases) grant R01 AI064603.
Footnotes
Published ahead of print 5 February 2014
REFERENCES
- 1.Masters PS, Perlman S. 2013. Coronaviridae, p 825–858 In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439–450. 10.1038/nrmicro2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang YJ, Luo CM, Tan B, Wang N, Zhu Y, Crameri G, Zhang SY, Wang LF, Daszak P, Shi ZL. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503:535–538. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annan A, Baldwin HJ, Corman VM, Klose SM, Owusu M, Nkrumah EE, Badu EK, Anti P, Agbenyega O, Meyer B, Oppong S, Sarkodie YA, Kalko EK, Lina PH, Godlevska EV, Reusken C, Seebens A, Gloza-Rausch F, Vallo P, Tschapka M, Drosten C, Drexler JF. 2013. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 19:456–459. 10.3201/eid1903.121503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, Alhakeem R, Durosinloun A, Al Asmari M, Islam A, Kapoor A, Briese T, Daszak P, Al Rabeeah AA, Lipkin WI. 2013. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 19:1819–1823. 10.3201/eid1911.131172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masters PS. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193–292. 10.1016/S0065-3527(06)66005-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stohlman SA, Baric RS, Nelson GN, Soe LH, Welter LM, Deans RJ. 1988. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J. Virol. 62:4288–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Almazán F, Galán C, Enjuanes L. 2004. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 78:12683–12688. 10.1128/JVI.78.22.12683-12688.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schelle B, Karl N, Ludewig B, Siddell SG, Thiel V. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 79:6620–6630. 10.1128/JVI.79.11.6620-6630.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossoehme NE, Li L, Keane SC, Liu P, Dann CE, III, Leibowitz JL, Giedroc DP. 2009. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 394:544–557. 10.1016/j.jmb.2009.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zúñiga S, Cruz JL, Sola I, Mateos-Gómez PA, Palacio L, Enjuanes L. 2010. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 84:2169–2175. 10.1128/JVI.02011-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tahara SM, Dietlin TA, Bergmann CC, Nelson GW, Kyuwa S, Anthony RP, Stohlman SA. 1994. Coronavirus translational regulation: leader affects mRNA efficiency. Virology 202:621–630. 10.1006/viro.1994.1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson GW, Stohlman SA, Tahara SM. 2000. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J. Gen. Virol. 81:181–188 http://vir.sgmjournals.org/content/81/1/181.long [DOI] [PubMed] [Google Scholar]
- 14.Molenkamp R, Spaan WJM. 1997. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology 239:78–86. 10.1006/viro.1997.8867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cologna R, Spagnolo JF, Hogue BG. 2000. Identification of nucleocapsid binding sites within coronavirus-defective genomes. Virology 277:235–249. 10.1006/viro.2000.0611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurst KR, Koetzner CA, Masters PS. 2009. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 83:7221–7234. 10.1128/JVI.00440-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurst KR, Ye R, Goebel SJ, Jayaraman P, Masters PS. 2010. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J. Virol. 84:10276–10288. 10.1128/JVI.01287-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurst KR, Koetzner CA, Masters PS. 2013. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 87:9159–9172. 10.1128/JVI.01275-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Q, Yu L, Petros AM, Gunasekera A, Liu Z, Xu N, Hajduk P, Mack J, Fesik SW, Olejniczak ET. 2004. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry 43:6059–6063. 10.1021/bi036155b [DOI] [PubMed] [Google Scholar]
- 20.Fan H, Ooi A, Tan YW, Wang S, Fang S, Liu DX, Lescar J. 2005. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure 13:1859–1868. 10.1016/j.str.2005.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayaram H, Fan H, Bowman BR, Ooi A, Jayaram J, Collisson EW, Lescar J, Prasad BV. 2006. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 80:6612–6620. 10.1128/JVI.00157-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saikatendu KS, Joseph JS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P. 2007. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 81:3913–3921. 10.1128/JVI.02236-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu IM, Oldham ML, Zhang J, Chen J. 2006. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between Corona- and Arteriviridae. J. Biol. Chem. 281:17134–17139. 10.1074/jbc.M602107200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeda M, Chang CK, Ikeya T, Güntert P, Chang YH, Hsu YL, Huang TH, Kainosho M. 2008. Solution structure of the C-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. J. Mol. Biol. 380:608–622. 10.1016/j.jmb.2007.11.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y, Tong X, Xu X, Li X, Lou Z, Rao Z. 2010. Structures of the N- and C-terminal domains of MHV-A59 nucleocapsid protein corroborate a conserved RNA-protein binding mechanism in coronavirus. Protein Cell 1:688–697. 10.1007/s13238-010-0079-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu IM, Gustafson CL, Diao J, Burgner JW, II, Li Z, Zhang J, Chen J. 2005. Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain. J. Biol. Chem. 280:23280–23286. 10.1074/jbc.M501015200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo H, Chen J, Chen K, Shen X, Jiang H. 2006. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization. Biochemistry 45:11827–11835. 10.1021/bi0609319 [DOI] [PubMed] [Google Scholar]
- 28.Chen CY, Chang CK, Chang YW, Sue SC, Bai HI, Riang L, Hsiao CD, Huang TH. 2007. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 368:1075–1086. 10.1016/j.jmb.2007.02.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang CK, Hsu YL, Chang YH, Chao FA, Wu MC, Huang YS, Hu CK, Huang TH. 2009. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 83:2255–2264. 10.1128/JVI.02001-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo L, Masters PS. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987–4999. 10.1128/JVI.76.10.4987-4999.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurst KR, Kuo L, Koetzner CA, Ye R, Hsue B, Masters PS. 2005. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 79:13285–13297. 10.1128/JVI.79.21.13285-13297.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verma S, Bednar V, Blount A, Hogue BG. 2006. Identification of functionally important negatively charged residues in the carboxy end of mouse hepatitis coronavirus A59 nucleocapsid protein. J. Virol. 80:4344–4355. 10.1128/JVI.80.9.4344-4355.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma S, Lopez LA, Bednar V, Hogue BG. 2007. Importance of the penultimate positive charge in mouse hepatitis coronavirus A59 membrane protein. J. Virol. 81:5339–5348. 10.1128/JVI.02427-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keane SC, Giedroc DP. 2013. Solution structure of mouse hepatitis virus (MHV) nsp3a and determinants of the interaction with MHV nucleocapsid (N) protein. J. Virol. 87:3502–3515. 10.1128/JVI.03112-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo L, Godeke GJ, Raamsman MJ, Masters PS, Rottier PJ. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393–1406. 10.1128/JVI.74.3.1393-1406.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goebel SJ, Hsue B, Dombrowski TF, Masters PS. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78:669–682. 10.1128/JVI.78.2.669-682.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo L, Masters PS. 2013. Functional analysis of the murine coronavirus genomic RNA packaging signal. J. Virol. 87:5182–5192. 10.1128/JVI.00100-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo L, Masters PS. 2010. Evolved variants of the membrane protein can partially replace the envelope protein in murine coronavirus assembly. J. Virol. 84:12872–12885. 10.1128/JVI.01850-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masters PS, Koetzner CA, Kerr CA, Heo Y. 1994. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J. Virol. 68:328–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koetzner CA, Parker MM, Ricard CS, Sturman LS, Masters PS. 1992. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 66:1841–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zúñiga S, Sola I, Moreno JL, Sabella P, Plana-Durán J, Enjuanes L. 2007. Coronavirus nucleocapsid protein is an RNA chaperone. Virology 357:215–227. 10.1016/j.virol.2006.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keane SC, Liu P, Leibowitz JL, Giedroc DP. 2012. Functional transcriptional regulatory sequence (TRS) RNA binding and helix destabilizing determinants of murine hepatitis virus (MHV) nucleocapsid (N) protein. J. Biol. Chem. 287:7063–7073. 10.1074/jbc.M111.287763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, Gorbalenya AE, Ziebuhr J. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305–2315. 10.1099/vir.0.19424-0 [DOI] [PubMed] [Google Scholar]
- 44.Makino S, Yokomori K, Lai MMC. 1990. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J. Virol. 64:6045–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Most RG, Bredenbeek PJ, Spaan WJM. 1991. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J. Virol. 65:3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fosmire JA, Hwang K, Makino S. 1992. Identification and characterization of a coronavirus packaging signal. J. Virol. 66:3522–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cologna R, Hogue BG. 2000. Identification of a bovine coronavirus packaging signal. J. Virol. 74:580–583. 10.1128/JVI.74.1.580-583.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narayanan K, Makino S. 2001. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 75:9059–9067. 10.1128/JVI.75.19.9059-9067.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen SC, van den Born E, van den Worm SH, Pleij CW, Snijder EJ, Olsthoorn RC. 2007. New structure model for the packaging signal in the genome of group IIa coronaviruses. J. Virol. 81:6771–6774. 10.1128/JVI.02231-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Escors D, Izeta A, Capiscol C, Enjuanes L. 2003. Transmissible gastroenteritis coronavirus packaging signal is located at the 5′ end of the virus genome. J. Virol. 77:7890–7902. 10.1128/JVI.77.14.7890-7902.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morales L, Mateos-Gomez PA, Capiscol C, del Palacio L, Enjuanes L, Sola I. 2013. Transmissible gastroenteritis coronavirus genome packaging signal is located at the 5′ end of the genome and promotes viral RNA incorporation into virions in a replication-independent process. J. Virol. 87:11579–11590. 10.1128/JVI.01836-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P. 2007. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 81:6700–6708. 10.1128/JVI.02817-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen SC, Olsthoorn RC. 2010. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology 401:29–41. 10.1016/j.virol.2010.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng D, Koetzner CA, McMahon T, Zhu Y, Masters PS. 1995. Construction of murine coronavirus mutants containing interspecies chimeric nucleocapsid proteins. J. Virol. 69:5475–5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Haan CA, Kuo L, Masters PS, Vennema H, Rottier PJ. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72:6838–6850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haijema BJ, Volders H, Rottier PJ. 2003. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 77:4528–4538. 10.1128/JVI.77.8.4528-4538.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuo L, Hurst KR, Masters PS. 2007. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J. Virol. 81:2249–2262. 10.1128/JVI.01577-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruch TR, Machamer CE. 2011. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J. Virol. 85:675–685. 10.1128/JVI.01570-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stobart CC, Sexton NR, Munjal H, Lu X, Molland KL, Tomar S, Mesecar AD, Denison MR. 2013. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J. Virol. 87:12611–12618. 10.1128/JVI.02050-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu X, Zhai Y, Sun F, Lou Z, Su D, Xu Y, Zhang R, Joachimiak A, Zhang XC, Bartlam M, Rao Z. 2006. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J. Virol. 80:7909–7917. 10.1128/JVI.00525-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qin L, Xiong B, Luo C, Guo ZM, Hao P, Su J, Nan P, Feng Y, Shi YX, Yu XJ, Luo XM, Chen KX, Shen X, Shen JH, Zou JP, Zhao GP, Shi TL, He WZ, Zhong Y, Jiang HL, Li YX. 2003. Identification of probable genomic packaging signal sequence from SARS-CoV genome by bioinformatics analysis. Acta Pharmacol. Sin. 24:489–496 [PubMed] [Google Scholar]
- 62.Hsieh PK, Chang SC, Huang CC, Lee TT, Hsiao CW, Kou YH, Chen IY, Chang CK, Huang TH, Chang MF. 2005. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 79:13848–13855. 10.1128/JVI.79.22.13848-13855.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baric RS, Nelson GW, Fleming JO, Deans RJ, Keck JG, Casteel N, Stohlman SA. 1988. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 62:4280–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Narayanan K, Maeda A, Maeda J, Makino S. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127–8134. 10.1128/JVI.74.17.8127-8134.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narayanan K, Chen CJ, Maeda J, Makino S. 2003. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 77:2922–2927. 10.1128/JVI.77.5.2922-2927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yount B, Roberts RS, Lindesmith L, Baric RS. 2006. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc. Natl. Acad. Sci. U. S. A. 103:12546–12551. 10.1073/pnas.0605438103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sturman LS, Holmes KV, Behnke J. 1980. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 33:449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Escors D, Ortego J, Laude H, Enjuanes L. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312–1324. 10.1128/JVI.75.3.1312-1324.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P, Milligan RA, Yeager M, Buchmeier MJ. 2006. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 80:7918–7928. 10.1128/JVI.00645-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bárcena M, Oostergetel GT, Bartelink W, Faas FG, Verkleij A, Rottier PJ, Koster AJ, Bosch BJ. 2009. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. U. S. A. 106:582–587. 10.1073/pnas.0805270106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stobart CC, Lee AS, Lu X, Denison MR. 2012. Temperature-sensitive mutants and revertants in the coronavirus nonstructural protein 5 protease (3CLpro) define residues involved in long-distance communication and regulation of protease activity. J. Virol. 86:4801–4810. 10.1128/JVI.06754-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Auweter SD, Oberstrass FC, Allain FH. 2006. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 34:4943–4959. 10.1093/nar/gkl620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuo L, Masters PS. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597–4608. 10.1128/JVI.77.8.4597-4608.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]