ABSTRACT
The antiviral role of TRIM E3 ligases in vivo is not fully understood. To test the hypothesis that TRIM5α and TRIM22 have differential transcriptional regulation and distinct anti-HIV roles according to infection phase and compartment, we measured TRIM5α, TRIM22, and type I interferon (IFN-I)-inducible myxovirus resistance protein A (MxA) levels in peripheral blood mononuclear cells (PBMCs) during primary and chronic HIV-1 infection, with chronic infection samples being matched PBMCs and central nervous system (CNS)-derived cells. Associations with biomarkers of disease progression were explored. The impact of IFN-I, select proinflammatory cytokines, and HIV on TRIM E3 ligase-specific expression was investigated. PBMCs from individuals with primary and chronic HIV-1 infection had significantly higher levels of MxA and TRIM22 than did PBMCs from HIV-1-negative individuals (P < 0.05 for all comparisons). PBMCs from chronic infection had lower levels of TRIM5α than did PBMCs from primary infection or HIV-1-uninfected PBMCs (P = 0.0001 for both). In matched CNS-derived samples and PBMCs, higher levels of MxA (P = 0.001) and TRIM5α (P = 0.0001) in the CNS were noted. There was a negative correlation between TRIM22 levels in PBMCs and plasma viral load (r = −0.40; P = 0.04). In vitro, IFN-I and, rarely, proinflammatory cytokines induced TRIM5α and TRIM22 in a cell type-dependent manner, and the knockdown of either protein in CD4+ lymphocytes resulted in increased HIV-1 infection. These data suggest that there are infection-phase-specific and anatomically compartmentalized differences in TRIM5α and TRIM22 regulation involving primarily IFN-I and specific cell types and indicate subtle differences in the antiviral roles and transcriptional regulation of TRIM E3 ligases in vivo.
IMPORTANCE Type I interferon-inducible TRIM E3 ligases are a family of intracellular proteins with potent antiviral activities mediated through diverse mechanisms. However, little is known about the contribution of these proteins to antiviral immunity in vivo and how their expression is regulated. We show here that TRIM5α and TRIM22, two prominent members of the family, have different expression patterns in vivo and that the expression pattern depends on HIV-1 infection status and phase. Furthermore, expression differs in peripheral blood versus central nervous system anatomical sites of infection. Only TRIM22 expression correlated negatively with HIV-1 viral load, but gene silencing of both proteins enhances HIV-1 infection of target cells. We report subtle differences in TRIM5α and TRIM22 gene induction by IFN-I and proinflammatory cytokines in CD4+ lymphocytes, monocytes, and neuronal cells. This study enhances our understanding of antiviral immunity by intrinsic antiviral factors and how their expression is determined.
INTRODUCTION
Induction of the type I interferons (IFNs) IFN-α and IFN-β is a hallmark of and one of the earliest immune responses of mammalian cells to viral infection (1). The role of IFN-α/β in HIV-1 infection is controversial, as some studies have shown protective roles of IFN-I (2, 3), while others have highlighted the pathological roles of IFN-I (2). Nevertheless, administration of recombinant human IFN-α to patients in the asymptomatic phase of HIV-1 infection is beneficial, with attenuated CD4 T cell decline and reductions in the incidence of AIDS-defining events, although these effects were not observed in more advanced disease (4, 5). Transiently high levels of endogenous serum IFN-α have been described for primary HIV-1 infection (6, 7).
Type I interferons induce the expression of some members of the antiviral tripartite motif (TRIM) E3 ligase family, which consists of approximately 100 distinct proteins characterized by the presence of a RING domain, one or two B boxes, and a coiled-coil domain (8–11). TRIM5α, the best characterized of these proteins, blocks HIV-1 replication in Old World monkey cells through a direct interaction with the viral capsid (12, 13). TRIM5α is responsible for species-specific postentry restriction of retroviruses such as murine leukemia N-tropic virus (N-MLV) and HIV-1 in primate cells (13, 14). TRIM22 is also induced by IFN-I and inhibits viral replication by interfering with viral gene transcription and virion assembly (15–19). Genetic association studies have demonstrated that polymorphic variants of the human TRIM5α gene are associated with reduced susceptibility to HIV infection or are overrepresented among HIV-negative individuals compared to HIV-positive ones (20, 21), suggesting that human TRIM5α may have some protective role against HIV-1 infection. It has also been reported that human TRIM5α genetic variants can influence the rate of disease progression, although the effects appear to be dependent on the phase of infection and of modest magnitude (22, 23). Human TRIM5α may also select for escape mutants after a prolonged duration of HIV-1 infection (24), suggesting ongoing immune pressure during infection. In a prospective cohort study of HIV-1-negative individuals at high risk for HIV-1 infection, we showed that elevated expression levels of TRIM5α were associated with decreased susceptibility to HIV-1 infection (25). We subsequently found that TRIM22 but not TRIM5α, IFN-α, IFN-β, or myxovirus resistance protein A (MxA) expression correlated negatively with plasma viral load and positively with CD4+ T cell counts in primary HIV-1 infection, suggesting a protective, antiviral role in vivo (17).
The role of TRIM E3 ligases as an important component of innate defense against HIV-1 is therefore now well established. However, little is known about whether TRIM E3 ligases have significant antiviral activity in vivo or how they may be regulated and affected by HIV-1 infection. We hypothesized that TRIM E3 ligases contribute significantly to anti-HIV-1 immunity during the early phases of infection, as has been demonstrated with other components of antiviral innate immune mechanisms. Alternatively, the antiviral impact of these factors may become more pronounced as adaptive immune mechanisms become progressively dysfunctional in chronic HIV-1 infection. Furthermore, we reasoned that innate defenses such as TRIM E3 ligases may be more important in remote or immune-privileged sites without well-developed adaptive immune systems. We therefore studied the relationship between the expression of IFN-I and TRIM5α and TRIM22 in primary and chronic HIV-1 infection and in peripheral blood- and central nervous system (CNS)-derived cells. We explored the in vivo association between TRIM E3 ligase expression levels and biomarkers of HIV-1 disease progression. Finally, we explored in vitro the role of select proinflammatory cytokines in the regulation of TRIM E3 ligases and the impact of gene silencing of these antiviral factors on HIV-1 replication in CD4+ T cells.
MATERIALS AND METHODS
Subjects.
Study subjects were part of the CAPRISA 002 acute infection study, which is an observational natural history study of HIV-1 subtype C infection established in Durban, South Africa, in 2004 (25, 26). Briefly, the cohort consisted of 245 high-risk seronegative women who were followed up to identify acute or recent infections. Participants were enrolled into the primary infection phase if they were antibody positive within 5 months of a previous antibody-negative test or if they had evidence of virus replication without HIV-1 antibodies, as assessed by rapid tests and PCR testing. Women from other seroincident cohorts in Durban were enrolled into the CAPRISA 002 study if they met the above-described criteria. Time of infection was defined as the midpoint between the last HIV-1 antibody-negative test and the first HIV-1 antibody-positive test or 14 days prior to the first positive HIV-1 RNA PCR assay for those individuals identified as being antibody negative but HIV-1 RNA PCR positive. Peripheral blood mononuclear cells (PBMCs) from a total of 19 HIV-1-uninfected and 28 recently infected individuals from the CAPRISA study cohort were available for use in this study. We analyzed samples at a single time point within 12 months postinfection and defined this as primary infection.
We also analyzed matched PBMC and cerebrospinal fluid (CSF) samples from a separate cohort of individuals presenting with chronic meningitis, as described previously (27). Briefly, 150 consecutive patients with suspected tuberculous meningitis (TBM) were prospectively recruited between January 2008 and April 2009 at the Inkosi Albert Luthuli Central Hospital (IALCH) in Durban, South Africa. All patients were clinically assessed by a neurologist and had a computerized tomography (CT) scan done to exclude contraindications for a lumbar puncture (LP). A total of 26 HIV-1-positive samples were analyzed. All patients analyzed were antiretroviral naive. The study was approved by the Biomedical Research Ethics Committee of the University of KwaZulu-Natal, and participants provided written informed consent.
Sample processing, viral load quantification, and CD4 cell enumeration.
PBMCs were isolated by Ficoll-Histopaque (Sigma, St. Louis, MO) density gradient centrifugation from blood within 6 h of blood collection and frozen in liquid nitrogen until use. Six milliliters of whole CSF was spun at 1,800 rpm in an Eppendorf centrifuge (catalog no. 5430) for 10 min, and cell pellets were washed and resuspended in 1 ml RPMI 1640 (supplemented with 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum). Cells were immediately counted and lysed for RNA extractions. Viral load was determined by using the automated Cobas Amplicor HIV-1 Monitor Test v1.5 (Roche Molecular Systems, Inc., Branchburg, NJ) on plasma and CSF samples. CD4+ cells were enumerated by using the Multitest kit (CD4/CD3/CD8/CD45) on a four-parameter FACSCalibur flow cytometer (Becton, Dickinson, Franklin Lakes, NJ).
Cell sorting.
We employed magnetically activated cell sorting MACS (Miltenyi Biotec, Bergisch Gladbach, Germany, and Stemcell Technologies, Vancouver, Canada) to isolate the different cell populations from six fresh HIV-1-negative PBMC samples. Purity was assessed by fluorescence-activated cell sorter (FACS) analysis. CD4 cells were isolated by using negative-selection CD4+ T Cell Isolation kit II MACS (Miltenyi Biotec, Bergisch Gladbach, Germany), and monocytes were isolated by using negative-selection Monocytes Isolation kit II MACS (Miltenyi Biotec, Bergisch Gladbach, Germany). Natural killer (NK) cells were isolated by using the Easy Sep negative-selection human NK cell enrichment kit (Stemcell Technologies, Vancouver, Canada). The purity achieved ranged between 85% and 95%.
FACS analysis.
Nine matched PBMC or CSF pellets were surface stained on ice with the following murine anti-human monoclonal antibodies: anti-CD3-fluorescein isothiocyanate (FITC), anti-CD4-peridinin chlorophyll protein (PerCP), and anti-CD8-PerCP markers for T cells; anti-CD19-phycoerythrin (PE) for B cells; anti-CD14-allophycocyanin (APC) for monocytes; and anti-CD56-PE-Cy7 and anti-CD16-PE for NK cells (Becton, Dickinson). Stained cells were washed in phosphate-buffered saline (PBS) and fixed with 1% formaldehyde. Between 50,000 and 100,000 events were acquired per sample on an LSRII flow cytometer, and data analysis was performed by using FlowJo version 8.8.2 (TreeStar, Inc., Ashland, OR).
Cell lines and reagents.
Human astroglioma cells (U87.CD4.CCR5) (catalog no. 4035; NIH AIDS Reagent Repository) and neuroblastoma cells (ATCC CRL-2137; ATCC, Rockville, MD) were maintained in Dulbecco's modified Eagle's medium (DMEM) with 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (FBS). Recombinant human alpha 2b interferon (IFN-α2b) (catalog no. CYT-205), interleukin-2 (IL-2) (catalog no. CYT-209), and tumor necrosis factor alpha (TNF-α) (catalog no. CYT-223) were purchased from Prospec (East Brunswick, NJ).
IFN-α and cytokine stimulation of cells.
CD4 cells, monocytes, astrocytes, and neuroblasts in 24-well plates (1 × 106 cells/well) were stimulated with 1,000 U IFN-α, TNF-α (10 ng/μl), or IL-2 (83 ng/μl) (28) for 24 h, whereupon cells were washed and resuspended in cell lysis buffer from an RNA extraction kit (RNeasy kit; Qiagen, Hilden, Germany). For all stimulations, RNA was extracted, and gene expression was assessed for TRIM5α and TRIM22. Cells were also lysed with CytoBustor protein extraction reagent (catalog no. 71009-3; Novagen, Merck, Darmstadt, Germany) supplemented with a protease inhibitor cocktail (catalog no. P8340; Sigma) for Western blotting of TRIM5α and TRIM22. All experiments were performed in triplicate.
Short interfering RNA transfection and pseudotyped HIV-1 infection.
Three unique 27-mer short interfering RNA (siRNA) duplexes for human TRIM5α (TRIM5.1 [catalog no. SR31385A], TRIM5.2 [catalog no. SR31385B], TRIM5.3 [catalog no. SR31385C], and TRIM5.4 [a mixture of the 3 TRIM5α siRNAs]), three for TRIM22 (TRIM22.1 [catalog no. SR307012A], TRIM22.2 [catalog no. SR307012B], TRIM22.3 [catalog no. SR307012C], and TRIM22.4 [a mixture of the 3 TRIM22 siRNAs]), and a universal scrambled negative-control siRNA (catalog no. SR30004) were chemically synthesized by Origene (Origene Technologies, New York, NY).
Six million CD4 cells or neuroblasts were transduced with 20 μM TRIM5α- and TRIM22-specific siRNA or control siRNA by using the Bio-Rad Gene Pulser II apparatus (Bio-Rad, Berkeley, CA) at 100 V and 1.9 mA.
Forty-eight hours after transfection, cells were stimulated with or without 6,000 U IFN-α in a 24-well plate (6 × 106 cells/well) for 24 h. All experiments were performed in triplicate. Cells were harvested for TRIM5α and TRIM22 gene expression analysis and Western blotting.
Vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped HIV-1 virions of HIV laboratory-derived strain JCRSF were generated in HEK 293T cells by using JCRSF and VSV-G plasmids, respectively (generously provided by Warner Greene, UCSF), as described previously (29, 30). CD4+ T cells and neuroblasts were infected with a VSV-G-pseudotyped HIV-1 laboratory strain (JRCSF) by spinoculation (31). In brief, the virus was coincubated with 6 × 106 cells in each well of a 24-well plate and centrifuged for 90 min at 1,500 rpm at 21°C. Cells were washed twice and incubated for a further 48 h at 37°C in 5% CO2. The mean infection rate (based on intracellular detection of p24 by flow cytometry) was 48% (range, 15.4 to 74%) by 48 h after infection.
Cells transfected with the above-mentioned siRNAs were infected with a VSV-G-pseudotyped HIV-1 laboratory strain (JRCSF) (600 ng of p24/ml), as described above, and incubated for 48 h. Cell lysates were harvested and analyzed by using a p24 Vironostika HIV-1 Antigen Microelisa kit (bioMérieux, Durham, NC).
RNA isolation and analysis.
For all PBMCs and CNS-derived patient samples, RNA was extracted immediately after thawing, and counting of cells was performed without in vitro stimulation. RNA was extracted from 2 × 106 PBMCs and all available CSF cells by using the RNeasy kit (Qiagen) according to the manufacturer's instructions. RNA integrity was confirmed by using morpholinepropanesulfonic acid (MOPS) gels. The total RNA concentration was quantified, and samples were used only if the optical density at 260 nm (OD260)/OD280 ratio was 1.90 or higher. All RNA samples were DNase treated. One microgram of total RNA from each sample was reverse transcribed by using the iScript cDNA synthesis kit (Bio-Rad). For the CSF samples, all RNA extracted was reversed transcribed.
RNA quantitation by real-time PCR.
The PCR primers and cycling conditions used for MxA (to represent a universal IFN-I readout [32]), TRIM5α, and TRIM22 real-time quantitative PCR were validated in our laboratory and are shown in Table 1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was validated as the most suitable reference gene among 5 genes based on PCR efficiency. Each PCR mixture was comprised of 0.5 pmol/μl (for MxA, TRIM5α, and TRIM22) or 0.25 pmol/μl (for GAPDH) of each primer, 5 μl SYBR green I master mix (2×) (Roche), 1 μg cDNA, and water to 10 μl. Reactions were run in duplicate on a Roche LightCycler 480 version 1.5 instrument, with 1 cycle at 95°C (10 min) followed by 45 cycles consisting of denaturation, annealing, and extension steps (Table 1). Detection of the fluorescent products was carried out at the end of the 72°C extension period. To confirm amplification specificity, the PCR products were subjected to a melting-curve analysis and agarose gel electrophoresis. Serial dilutions of cDNA from total RNA were performed for each target gene. These dilutions served as standard curves for quantitative analysis.
TABLE 1.
Primers used in this study
| Gene | GenBank accession no. | Sequence (5′–3′)a | Cycling conditions (denaturation, annealing, and extension steps) |
|---|---|---|---|
| MxA | NM_0024462 | 5′-AAGCTGATCCGCCTCCACTT-3′ (F) | 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s |
| 5′-TGCAATGCACCCCTGTATACC-3′ (R) | |||
| TRIM5α | NM_033034 | 5′-AGGAGTTAAATGTAGTGCT-3′ (F) | 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s |
| 5′-ATAGATGAGAAATCCATGGT-3′ (R) | |||
| TRIM22 | NM_006074 | 5′-GGTTGAGGGGATCGTCAGTA-3′ (F) | 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s |
| 5′-TTGGAAACAGATTTTGGCTTC-3′ (R) | |||
| GAPDH | NM_002046 | 5′-AAGGTCGGAGTCAACGGATT-3′ (F) | 95°C for 15 s, 65°C for 15 s, and 72°C for 15 s |
| 5′-CTCCTGGAAGATGGTGATGG-3′ (R) |
F, forward; R, reverse.
Western blotting.
Briefly, cell lysates were mixed with 4× Laemmli sample buffer (catalog no. 161-0747; Bio-Rad) and boiled for 10 min. Samples were loaded onto 4 to 15% PAGE gels (catalog no. 456-1083; Bio-Rad) and electrophoresed for 90 min at 80 V in SDS running buffer. Separated proteins were transferred onto nitrocellulose (catalog no. 170-4270; Bio-Rad) for 30 min by using the Trans-Blot Turbo transfer system (catalog no. 170-4155; Bio-Rad). The nitrocellulose membrane was then washed three times in Tris-buffered saline with Tween 20 (TBST) (catalog no. 170-6435; Bio-Rad) for 10 min. The nitrocellulose membrane was blocked with 5% bovine serum albumin (BSA) (catalog no. 10735094001; Roche) in TBST. The membrane was then incubated with the primary antibody (TRIM5α, TRIM22, or tubulin) in 5% BSA (catalog no. 10735094001; Roche) in TBST at a 1:125, 1:500, or 1:5,000 dilution overnight, followed by three washes in TBST for 10 min. Antibodies used in this Western blot analysis were goat polyclonal anti-TRIM5α (catalog no. ab4389; Abcam), rabbit polyclonal anti-TRIM22 (catalog no. HPA003575; Sigma Prestige), and mouse polyclonal anti-alpha-tubulin (catalog no. ab7291; Abcam). The membrane was incubated with the secondary antibody (anti-goat, anti-rabbit, or anti-mouse) at a 1:20,000 dilution in 5% BSA in TBST for 1 h on a rocker, followed by three washes in TBST for 10 min. Antibody-antigen complexes were detected by using enhanced chemiluminescence reagents (SuperSignal West Dura extended-duration substrate, catalog no. 00034075; Thermo Scientific, Pierce Protein Research, Rockford, IL, USA). Proteins were visualized by using the ChemiDoc XRS+ system with Image Lab software (catalog no. 170-8265; Bio-Rad).
Statistical analysis.
All expression data were log transformed to ensure normality. Values are expressed as medians. Differences between matched PBMC and CSF samples were evaluated by using Student's t test for paired data. Differences between cohorts were compared by using analysis of variance (ANOVA) or a Kruskal-Wallis test with Tukey's or Dunn's multiple-comparison test, respectively. We correlated TRIM5α and TRIM22 to IFN-I gene expression values and all gene expression values to viral load and CD4 T cell counts using Pearson's correlation. Scatter plots and bar graphs were generated by using Instat Graphpad Prism V.5. All statistical analyses were performed by using Instat Graphpad Prism V.5 and SAS version 9.3 (SAS Institute, Inc., Cary, NC). P values of <0.05 were considered statistically significant.
RESULTS
Cohort characteristics.
The samples analyzed consisted of 19 HIV-1-uninfected participants, 28 HIV-1-infected participants with primary infection, and 26 chronically infected participants. The mean ages were 35 years (range, 19 to 54 years), 29 years (range, 18 to 59 years), and 33 years (range, 12 to 42 years), respectively, and percentages of females were 100% (19 participants), 100% (28 participants), and 73.1% (19 participants), respectively. Although the cohorts differed significantly in terms of gender (P < 0.01), gene expression values did not differ significantly between males and females in the chronic cohort (data not shown).
Expression of IFN-I (MxA), TRIM5α, and TRIM22 in compartments and according to HIV-1 infection status or phase.
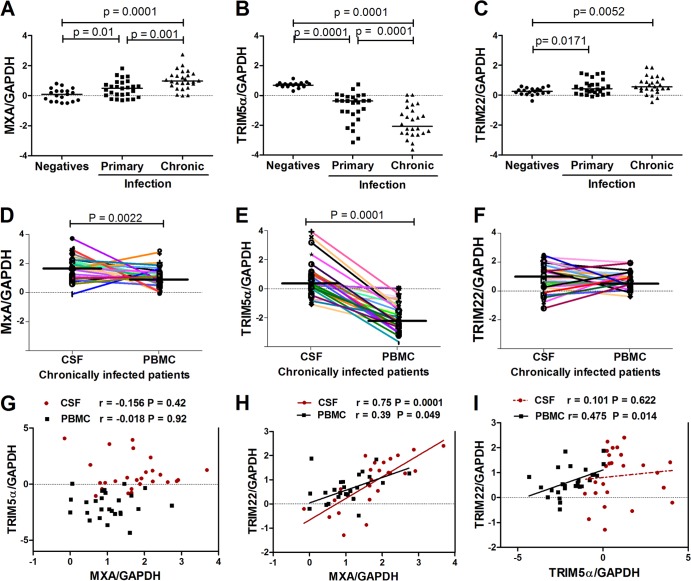
Here, we sought to better understand the relationship between the expression of TRIM5α, TRIM22, and a widely utilized universal correlate of IFN-I induction, myxovirus resistance protein A (MxA), in primary (or early) versus chronic HIV-1 infection. Furthermore, it is unknown whether TRIM E3 ligases are expressed in cells that reside or home to the CNS, a remote anatomical site that may represent a significant reservoir of HIV replication and where adaptive immune responses may be limited. mRNA expression levels were measured in PBMCs from 19 HIV-1-negative individuals and 28 HIV-1-infected samples from patients with primary infection. In addition, expression levels were measured in 26 matched CSF and PBMC samples obtained during the chronic phase of HIV-1 infection. Patients with chronic infection had significantly higher levels of MxA (P = 0.0001) and TRIM22 (P = 0.0052) (Fig. 1A and C) than did subjects in the HIV-1-negative group. Similarly, patients with primary infection also had higher levels of MxA (P < 0.05) and TRIM22 (P = 0.0171) than did subjects in the HIV-1-negative group (Fig. 1A and C). Both the primary and chronically infected patients had lower levels of TRIM5α (P = 0.0001) than did the HIV-1-negative group (Fig. 1B). We analyzed mRNA levels of MxA, TRIM5α, and TRIM22 in both PBMCs and CSF-derived cells. We found that there were higher levels of MxA (P = 0.0022) and TRIM5α (P = 0.0001) in CSF cells than in PBMCs (Fig. 1D and E); however, there was no significant difference in TRIM22 levels (Fig. 1F).
FIG 1.
Expression of antiviral factors according to HIV-1 infection status and compartment. (A to C) Expression of the IFN-I-responsive gene (MxA) (A), TRIM5α (B), and TRIM22 (C) in PBMCs from HIV-1-uninfected versus -infected subjects. (D to F) Expression of MxA (D), TRIM5α (E), and TRIM22 (F) in PBMCs versus CSF-derived cells from chronically infected patients. The samples from participants with primary infection were all collected within 12 months of infection. One time point, closest to 12 months postinfection, was used per participant (n = 28). For HIV-1-negative participants, only those who remained HIV-1 negative during the entire follow-up period (2 years) were included in this analysis (n = 19). The chronically infected group consisted of participants presenting with chronic meningitis (n = 26). Matched CSF samples and PBMCs from the same patients were analyzed (n = 26). Data are depicted as normalized ratios of MxA, TRIM5α, or TRIM22 to GAPDH. Median expression levels between HIV-negative and HIV-positive samples were compared. (G to I) Correlations (Pearson) between MxA and TRIM5α (G), TRIM22 and MxA (H), and TRIM22 and TRIM5α (I) in both CSF samples and PBMCs from patients with chronic HIV-1 infection.
Although both TRIM5α and TRIM22 are interferon response genes in vitro, we previously found no correlation between TRIM5α and IFN-I in the primary HIV-1 infection phase, in contrast to TRIM22 levels, which strongly correlated with IFN-I (17). We therefore next explored the relationships between TRIM5α, TRIM22, and IFN-I (MxA) expression in the periphery and the CNS compartment during chronic HIV-1 infection. There was no correlation between TRIM5α and MxA in CSF cells or PBMCs (Fig. 1G). We found a significant positive correlation between TRIM22 and MxA in CSF cells (r = 0.75; P < 0.0001) and PBMCs (r = 0.39; P = 0.04) (Fig. 1H). In addition, there was a significant positive correlation between TRIM22 and TRIM5α in PBMCs (r = 0.475; P = 0.014) (Fig. 1I). Taken together, these data suggest that there is a greater correlation between IFN-I and TRIM22 expression in vivo, whereas TRIM5α expression dynamics are more unique and seemingly independent of IFN-I in different phases or compartments of HIV-1 infection.
Cell populations in matched CSF samples and PBMCs and baseline expression of TRIM5α and TRIM22 in immune cells.
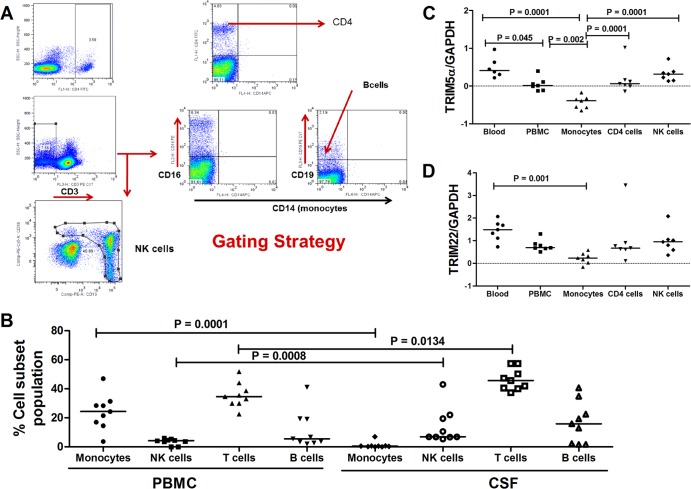
Differential TRIM E3 ligases and MxA expression in the blood versus CSF-derived cells raised the possibility that this may be determined by distinct immune cells or proportions that reside in these separate compartments. We therefore next investigated the type of immune cells that could be detected in matched PBMCs versus CSF samples from participants with chronic HIV-1 infection. Figure 2A shows representative flow cytometry plots with the gating strategy employed to define different cell populations.
FIG 2.
TRIM5α and TRIM22 expression in different immune cells. (A) Representative flow cytometry plots showing the gating strategy employed to define different cell populations. (Top) Cells were first gated for singlets (forward scatter height [FSC-H] versus forward scatter area [FSC-A]), and CD4+ T cells were defined from the lymphocyte gate (side scatter area [SSC-A] versus FSC-A) as CD3+ CD4+ events. (Middle) Flow cytometry plots illustrate the gating for B cells as CD3− CD19+ cells and monocytes as CD3− CD19− CD56− CD14+ cells. (Bottom) NK cells were defined as CD3− CD56+ CD16+ cells. (B) Data were compared between each subset of cells in the CSF and PBMCs only by using the paired t test. Magnetic cell sorting was employed to isolate CD4 cells and monocytes from 6 fresh HIV-1-negative PBMC samples. Natural killer cells were isolated by using the Easy Sep negative-selection human NK cell enrichment kit (Stemcell Technologies). (C and D) Different cell populations were analyzed for TRIM5α (C) and TRIM22 (D) mRNA expression. Data are depicted as normalized ratios of TRIM5α or TRIM22 to GAPDH.
We found that the CNS compartment had significantly higher proportions of T cells (P = 0.0134) and natural killer (NK) cells (P = 0.0008) (Fig. 2B) than did matched PBMCs. Lower numbers of monocytes were noted in CSF than in PBMCs (P = 0.0001) (Fig. 2B). These differences in specific cell subsets between the PBMC and CNS compartments led us to investigate mRNA levels of TRIM5α and TRIM22 in monocytes, CD4+ T cells, and NK cells isolated from HIV-1-negative participants. Higher levels of TRIM5α were noted in CD4+ T cells (P = 0.0001) and NK cells (P = 0.0001) than in monocytes (Fig. 2C). No significant differences in TRIM22 expression in these cell populations was noted (Fig. 2D). These data suggest that, at least during CNS inflammation, immune cells, particularly T cells, can traffic to and accumulate in this compartment compared to the periphery. Accumulation of CD4 T cells in this compartment probably contributed to the higher level of TRIM5α expression observed in CSF cells.
Association of antiviral gene expression with markers of disease progression in primary and chronic HIV-1 infection.
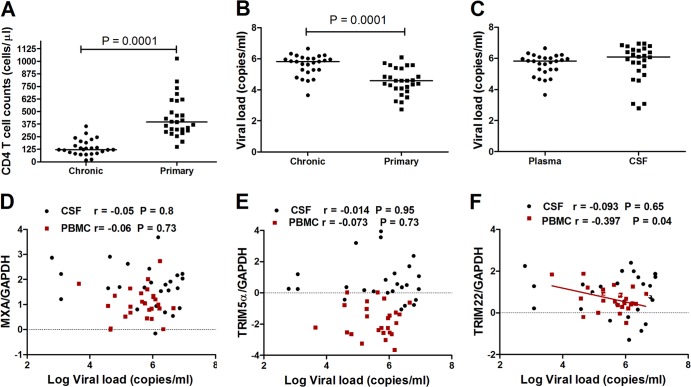
As expected, patients with primary HIV-1 infection had significantly higher CD4+ T cell counts and lower plasma viral loads than did patients chronically infected with HIV (P = 0.0001 for both) (Fig. 3A and B). There were no significant differences in viral load between plasma and CSF in the chronically infected group (Fig. 3C).
FIG 3.
Association of antiviral gene expression with markers of disease progression in primary and chronic HIV-1 infection. (A and B) Differences between samples from participants with primary infection closest to 12 months postinfection (n = 28) and samples from chronically infected patients (n = 28) were compared in terms of CD4 T cell counts and viral loads. (C) Viral loads in plasma and CSF samples from the chronically infected group were compared. (D to F) Pearson correlations were performed for MxA (D), TRIM5α (E), and TRIM22 (F) expression levels and viral loads in the CSF or peripheral blood compartments in the chronically HIV-1-infected group.
To determine if MxA, TRIM5α, or TRIM22 gene expression was associated with viral control during chronic infection, we investigated the correlation between viral loads and gene expression levels. No significant correlation for MxA or TRIM5α and viral load was noted for both the periphery and the CNS (Fig. 3D and E). However, there was a negative correlation between TRIM22 mRNA levels in PBMCs and plasma viral load (r = −0.397; P = 0.04) (Fig. 3F). These data are consistent with our previous findings for acute HIV-1 infection in which TRIM22 but not TRIM5α levels showed a weak negative correlation with plasma viral load (17).
Regulation of TRIM5α and TRIM22 by IFN-α and proinflammatory cytokines TNF-α and IL-2.
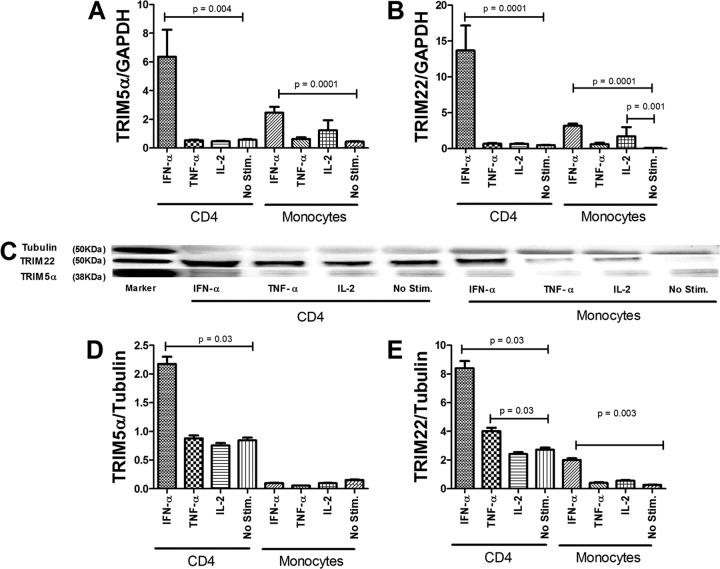
Our findings of differential expression kinetics in vivo between TRIM5α and TRIM22 and the lack of association between TRIM5α and MxA expression levels suggested that there may be subtle differences in the transcriptional regulation of these proteins besides IFN-I. We hypothesized that proinflammatory cytokines may play a role in the regulation of TRIM E3 ligases. To better understand how TRIM5α and TRIM22 are regulated in immune cells targeted by HIV-1, CD4 lymphocytes and monocytes purified by negative selection were stimulated with IFN-α, TNF-α, or IL-2, and expression of the TRIM E3 ligases was assessed by reverse transcription real-time PCR and Western blotting (Fig. 4A to E). IFN-α significantly but differentially induced TRIM5α mRNA expression in CD4 cells and monocytes (P < 0.05 for both), with higher levels of upregulation noted for the former (Fig. 4A). Likewise, TRIM22 mRNA levels increased significantly (P < 0.05) in both CD4 lymphocytes and monocytes in response to IFN-I stimulation, with higher levels of induction noted for CD4 cells (Fig. 4B). A modest upregulation of TRIM5α and TRIM22 mRNA levels in monocytes following IL-2 stimulation was noted. Next, we assessed whether stimulation with IFN-α, TNF-α, or IL-2 modulated TRIM5α and TRIM22 protein levels (Fig. 4C). CD4 cells stimulated with IFN-α expressed higher protein levels of TRIM5α than did unstimulated cells (P = 0.03), and there was no TRIM5α protein induction noted following stimulation with either TNF-α or IL-2 (Fig. 4C and D). Monocytes stimulated with IFN-α showed no significant increase in TRIM5α protein levels compared to unstimulated cells (Fig. 4C and D). CD4 cells and monocytes stimulated with IFN-α had higher protein levels of TRIM22 than did unstimulated cells (P = 0.03 and P = 0.003, respectively). CD4 T cells also showed increased TRIM22 protein expression levels following TNF-α stimulation (Fig. 4E).
FIG 4.
Regulation of TRIM5α and TRIM22 by IFN-α and select proinflammatory cytokines in immune cells. Shown is the effect of IFN-α stimulation and proinflammatory cytokine stimulation of CD4 cells and monocytes on antiviral gene expression. Cells from healthy donors were stimulated for 24 h. (A and B) Total cellular RNA from cells was then subjected to real-time RT-PCR to measure mRNA levels using primers specific for TRIM5α (A) and TRIM22 (B) and GAPDH. (C to E) Protein expression analyses of TRIM5α (C and D) and TRIM22 (C and E) are shown. Results shown in panels A, B, D, and E are the means of three independent experiments (with bars indicating ranges), each performed in duplicate.
We further tested whether IFN-α, IL-2, or TNF-α could modulate TRIM5α and TRIM22 in astrocytes and neuroblasts, cell lines similar to HIV target cells in the CNS compartment. IFN-α significantly but differentially induced TRIM5α and TRIM22 mRNA and protein levels in astrocytes and neuroblasts (P < 0.05 for all), whereas neither IL-2 nor TNF-α had a significant effect on TRIM E3 ligases expression (data not shown).
These data show that although both TRIM5α and TRIM22 are generally IFN-α inducible in most cells, some subtle differences may also exist between diverse immune cells, and some immunoregulatory cytokines may also play a role in the regulation of TRIM E3 ligases.
siRNA-mediated silencing of TRIM5α or TRIM22 in CD4 cells enhances HIV-1 infection.
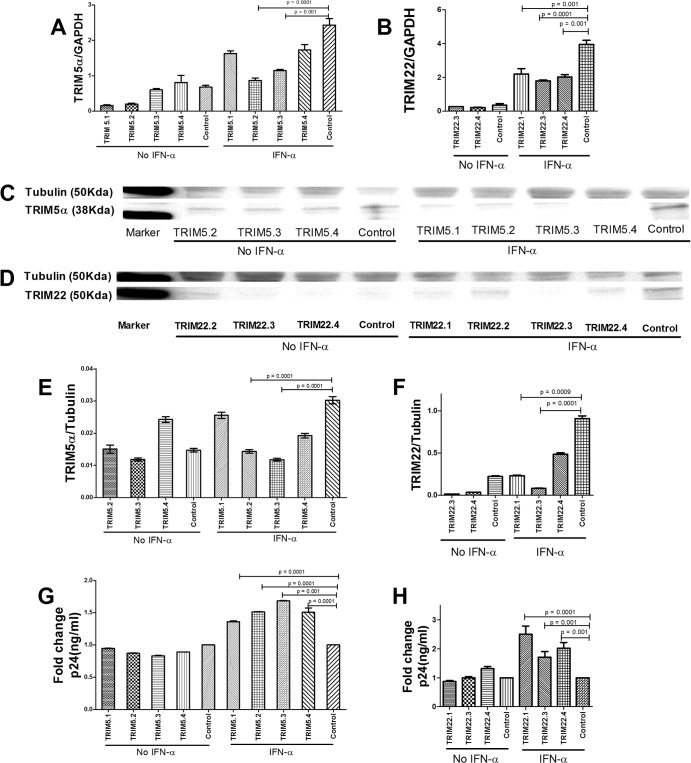
To further explore the impact of TRIM5α and TRIM22 on HIV-1 infection in primary immune cells targeted by HIV-1, we performed transient knockdown of these genes in CD4 lymphocytes, which were previously demonstrated to upregulate the expression of the TRIM E3 ligases in response to IFN-α (15, 17). Gene knockdown by 3 different siRNAs, each corresponding to TRIM5α and TRIM22 coding sequences, significantly reduced mRNA and protein levels of both genes in the presence of IFN-α, as verified by RT-PCR and Western blotting (Fig. 5A to F). Under IFN-α stimulation, CD4 cells treated with TRIM5.1, TRIM5.2, TRIM5.3, and TRIM5.4 (a mixture of the four TRIM5α siRNAs) showed a 28 to 64% reduction in mRNA levels and a 15 to 61% reduction in protein levels compared to control treated cells (Fig. 5A, C, and E). TRIM22 knockdown with the siRNA TRIM22.1, TRIM22.3, or TRIM22.4 (a mixture of the 3 TRIM22 siRNAs) resulted in a 44 to 54% reduction in mRNA levels and a 47 to 91% reduction in protein levels (Fig. 5B, D, and F), while TRIM22.2 siRNA knockdown data were inconsistent (data not shown).
FIG 5.
siRNA-mediated silencing of TRIM5α or TRIM22 in CD4 cells. To determine the functional impact of TRIM5α and TRIM22 on HIV-1 replication in CD4 cells, gene knockdown experiments were performed by transducing the cells with siRNA against TRIM5α or TRIM22 or a nontargeting (scramble) control siRNA, with or without IFN-α stimulation for 24 h. (A to F) Knockdown of TRIM5α or TRIM22 by siRNA in the absence or presence of IFN-α was validated by mRNA RT-PCR (A and B) and Western blotting (C to F). Cells were then challenged with a VSV-G-pseudotyped HIV-1 laboratory strain (JRCSF) (600 ng of p24/ml) for 48 h. (G and H) Culture lysates were collected and assessed for HIV p24 antigen levels by an enzyme-linked immunosorbent assay. Data are depicted as fold changes, where the levels of p24 antigen in knockdown cells were divided by the levels in control cells. TRIM22.2 siRNA knockdown efficiency was inconsistent, and the data are thus excluded from the graphs.
Next, we assessed the impact of TRIM5α and TRIM22 knockdown on HIV-1 infection in CD4 cells. Following IFN-α stimulation and infection with VSV-G-pseudotyped HIV-1, TRIM5α or TRIM22 knockdown cells showed higher levels (at least 1.3-fold and 1.7-fold higher, respectively) of intracellular p24 antigen than did control cells (P = 0.0001) (Fig. 5G and H).
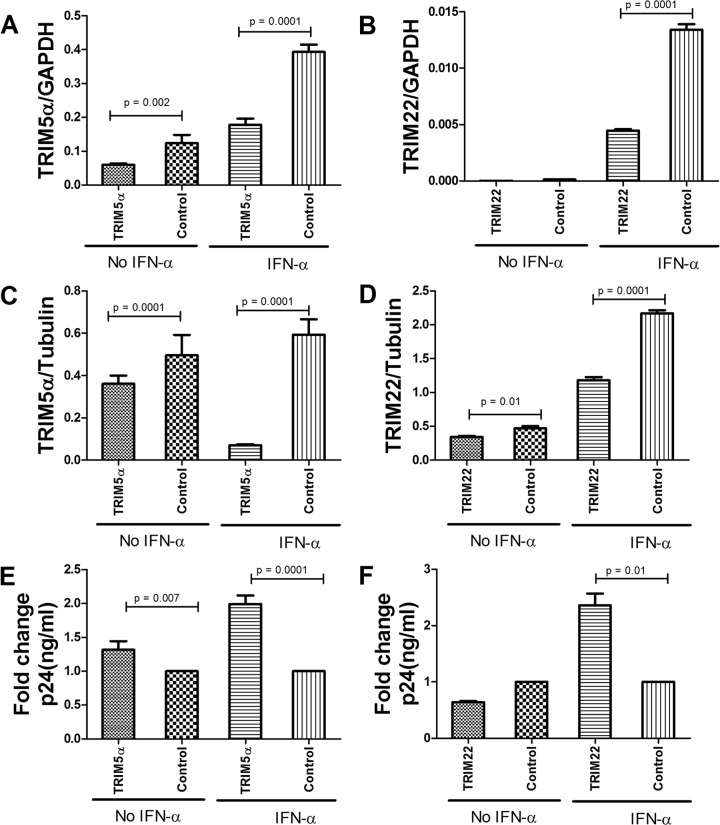
siRNA-mediated silencing of TRIM5α or TRIM22 in neuroblasts enhances HIV-1 infection.
HIV-1 is known to productively replicate in macrophages and microglia of the CNS, but it also infects astrocytes and other CNS cells via a CD4-independent pathway (33–35). To explore the possible differential antiviral role of TRIM E3 ligases in CNS-derived cells compared to CD4 lymphocytes, we performed transient knockdown of TRIM5α and TRIM22 and assessed the impact on IFN-α-induced anti-HIV-1 activity in neuroblasts (ATCC CRL-2137). Cell lines were used in lieu of patient-derived primary neuronal cells. Significantly lower levels of TRIM5α and TRIM22 mRNA and protein levels were noted in the knockdown cell lines than in the control cells transfected with nontargeting siRNA (all P ≤ 0.01), (Fig. 6A to D). It should be noted that knockdown of the TRIM E3 ligases appeared to be more efficient in the neuronal cell line than in primary CD4 cells, possibly because this cell line is more homogenous and easier to transfect. Furthermore, differences in expression were more pronounced with IFN-α stimulation than without. Upon infection with VSV-pseudotyped HIV, both the TRIM5α and TRIM22 knockdown cell lines showed higher (1.3- and 2.3-fold increases, respectively) intracellular p24 antigen levels than those of the control (scrambled siRNA-treated) cell line after IFN-α stimulation (P = 0.0001 and P = 0.001, respectively) (Fig. 6E and F). These data confirm the overall inhibitory effect of these TRIM E3 ligases on diverse cells despite subtle differences in the level or efficiency of the viral blockade.
FIG 6.
siRNA-mediated silencing of TRIM5α or TRIM22 in neuroblasts. To determine the functional impacts of TRIM5α and TRIM22 on HIV-1 infection in neuroblasts, gene knockdown experiments were performed by transducing the cells with siRNA against TRIM5α or TRIM22 or a nontargeting (scrambled) control siRNA, with or without IFN-α stimulation for 24 h. (A to D) Knockdown of TRIM5α or TRIM22 by siRNA in the absence or presence of IFN-α was validated by mRNA RT-PCR (A and B) and Western blotting (C and D). Cells were then challenged with a VSV-G-pseudotyped HIV-1 laboratory strain (JRCSF) (600 ng of p24/ml) for 48 h. (E and F) Culture lysates were collected and assessed for HIV p24 antigen levels by an enzyme-linked immunosorbent assay. Data are depicted as fold changes, where levels of p24 antigen in knockdown cells were divided by levels in control cells.
DISCUSSION
TRIM E3 ligases represent an important component of the innate immune response against retroviral infections (36). However, TRIM E3 ligases are a heterogeneous group of proteins, and little is known about the contribution of specific members of this family to antiviral activity in vivo and the factors that may contribute to differential expression or effectiveness in blocking viral replication (15, 36, 37). We hypothesized that TRIM E3 ligases, as a component of the innate immune system, may play a significant role in blocking retroviral infection during the early phases of infection and in a remote anatomical site likely deficient in a robust adaptive immune system. We demonstrate here that TRIM5α and TRIM22 are differentially regulated according to infection status or phase of infection. We also show that TRIM5α and TRIM22 have different or compartmentalized kinetics of expression and show some compartment-specific heterogeneity in association with HIV-1 viral load, a marker of viral replication. Furthermore, we show that while both TRIM5α and TRIM22 mRNA and protein expressions are strongly induced in CD4 T cells, monocytes, and neuronal cell lines by IFN-α, there are some cell-specific differences, with apparently stronger induction in CD4 T cells, and additionally, cytokines such as IL-2 and TNF-α may also subtly and differentially impact the expression of these antiviral proteins.
The majority of studies of host restriction factors in antiviral immunity have been performed in vitro, devoid of the complexity that the pathogen encounters in vivo in the form of different arms of the immune system and viral replication cofactors that all coexist and contribute to disease outcome. Here, we characterized the in vivo expression patterns of two TRIM E3 ligases, TRIM5 and TRIM22, in PBMCs. Both proteins were previously shown to be type I interferon inducible (15, 38, 39), and considering that TRIM22 is located downstream of the TRIM5 gene (40, 41), we anticipated similar expression patterns. However, our data show subtle differences in the expression of these TRIM E3 ligases at the transcriptional level, with TRIM22 more closely mimicking the expression of the IFN-I surrogate MxA in uninfected subjects as well as in early and chronic HIV-1 infection phases than TRIM5α (Fig. 1A to C). Likewise, in comparing MxA, TRIM5α, and TRIM22 expression levels in PBMC versus CNS compartments, MxA and TRIM22 had similar (but not identical) expression patterns, whereas there were marked differences in the expression levels of TRIM5α between the two compartments. Our findings of higher levels of MxA in the CNS compartment than in the periphery are consistent with observations of IFN-I reported by other groups (42–44). There was a significant positive correlation between MxA and TRIM22 but not between MxA and TRIM5α mRNA levels, even though TRIM22 and TRIM5α expression levels correlated positively. These data are consistent with a model where, in vivo, there is tighter regulation of TRIM22 than of TRIM5α by type I interferon but also offer the suggestion that other independent regulatory mechanisms are involved. Further studies will be needed to elucidate these non-type I interferon mechanisms of TRIM E3 ligase regulation in HIV-uninfected individuals but also in the context of HIV infection and in different phases and compartments of viral infection.
In this study, we found that MxA and TRIM22 levels were significantly elevated during chronic HIV-1 infection and in CNS-derived cells compared to PBMCs. Levels of type I interferons are known to be elevated due to generalized immune activation, which in turn has been associated with accelerated HIV/AIDS progression (45, 46). Therefore, elevated levels of MxA and TRIM22 in this context may reflect generalized immune activation as opposed to a robust antiviral immune response. Interestingly, TRIM5α levels were low in chronic HIV-1 infection but were elevated in the CNS compartment compared to the periphery, which is probably attributable to an accumulation of activated CD4 T cells in the CNS in our study cohort. TRIM5α has been reported to positively modulate innate immune responses and has been demonstrated to be a pattern recognition receptor (47, 48). Our data also revealed distinct compartmentalization of immune cells between the CNS and periphery, with enrichment of NK and T cells in the CNS compartment compared to matched samples from the periphery. These findings are consistent with other phenotypic characterizations of CNS samples from patients with meningitis (49–51); however, further work is needed to address whether this distinct sublocalization has an impact on disease pathogenesis in these locations.
We also investigated the association of antiviral gene expression and viral load, a commonly used marker of disease progression, extending our previous work performed exclusively with recently infected individuals (25). In agreement with previous studies, there was no correlation between viral loads and TRIM5α expression in patients with either primary or chronic HIV-1 infection. Interestingly, TRIM22 expression in PBMCs from patients with chronic HIV-1 infection showed a moderate negative correlation with viral loads. We have previously shown that TRIM22 has a negative correlation with plasma viral load and a positive correlation with CD4+ T cell counts in primary HIV-1 infection (17). Collectively, the in vitro and in vivo data provide strong evidence of immune pressure by TRIM22 against HIV-1.
In this study, we also sought to better understand how TRIM E3 ligases are regulated in CD4 cells and monocytes, primary HIV-1 target cells which were also found in high proportions in the CNS samples analyzed in this study. We also explored the expression patterns and antiviral activities of TRIM5α and TRIM22 in neuronal cell lines in an attempt to begin to unravel the role of intrinsic immunity in CNS-derived cells. Specifically, we investigated how IFN-α and select proinflammatory cytokines affect the expression of TRIM5α and TRIM22 in primary and immortalized neuronal cells. We found that TRIM5α and TRIM22 were IFN inducible in CD4 cells, monocytes, and neuronal cell lines. Most previous studies on immune regulation of TRIM E3 ligases have focused on IFN-α, but we reasoned that due to the discordance in the relationship between IFN-I and TRIM5α versus IFN-I and TRIM22 that we sometimes found in vivo, other regulatory cytokines may be involved.
We found that the pattern of expression of TRIM5α and TRIM22 was cell dependent, with higher IFN-α induction of TRIM E3 ligases in CD4 lymphocytes than in monocytes. Proinflammatory cytokines generally had a modest impact on TRIM E3 ligase expression, with TNF-α induction of TRIM22 in CD4 cells being the most notable effect. Yu and colleagues previously showed that TRIM22 overexpression could significantly induce the secretion of proinflammatory cytokines in the human macrophage cell line U937 in an NF-κB-dependent manner (52). These data suggest that some proinflammatory cytokines can induce TRIM E3 ligase production in some cells. One of the main features of advanced HIV-1 infection and associated chronic immune activation is the dysregulation of cytokine production, including elevated TNF-α levels (53, 54). This proinflammatory cytokine dysregulation may also have contributed to elevated TRIM E3 ligase expression levels in chronic HIV infection and in the CNS. Collectively, these data suggest that while IFN-α appears to be primarily responsible for the induction of TRIM E3 ligases, subtle cell-specific differences exist, and proinflammatory cytokines may also play a minor role in the regulation of these proteins in primary cells.
Finally, in this study, we investigated whether human TRIM5α or TRIM22 has any functional effects on HIV-1 infection in CD4 cells and neuroblasts. Knockdown of either TRIM5α or TRIM22 in either cell type revealed that both proteins had antiviral activity. These findings are consistent with previous studies suggesting that human TRIM5α and TRIM22 have some anti-HIV-1 activity, especially in saturating doses (13, 15, 17, 47). Clearly, the level of antiviral activity of human TRIM5α does not match that of its rhesus counterpart, and therefore, unsurprisingly, there was no correlation between human TRIM5α levels and viral load in any compartment. Based on these findings, we speculate that targeted enhancement of the expression of TRIM22 in HIV-1-infected individuals may be beneficial in reducing viral load and could be employed as a novel antiviral strategy. Whether one can achieve targeted enhancement of a host restriction factor without inducing pathological generalized immune activation will require further studies, but our data are suggestive that under certain circumstances, TRIM E3 ligases, and in particular TRIM22, have antiviral effects.
Some limitations of the current study should be noted. First, due to sample limitation and to allow comparison of in vivo data, we assessed the expression of the antiviral factors here at the transcriptional level only. Future studies should aim at analyzing protein expression levels of these factors in patient samples. Second, we used bulk PBMCs or CSF-derived cells rather than specific subsets, although the latter limitation was then partially addressed by performing in vitro experiments using sorted subsets and/or cell lines. Clearly, more studies using well-characterized human samples are needed to better understand the role of intrinsic immunity in HIV control and to assess whether there is a plausible pathway to harness these factors for immunotherapeutic or other novel antiviral approaches. It is also worth noting that TRIM E3 ligase knockdown using siRNA showed variable efficiency, with apparently high antiviral factor expression levels under IFN-α stimulation in some cases. However, even under these circumstances, it was clear that the knockdown of either factor limited VSV-G-pseudotyped HIV infection of target cells.
In conclusion, in this study, we have characterized the expression of the interferon-responsive gene MxA and the TRIM E3 ligases TRIM5α and TRIM22 in peripheral blood mononuclear cells from HIV-1-uninfected individuals and from patients during the early or chronic phase of HIV-1 infection. We demonstrate that in vivo, there may be some subtle differences in the expressions of TRIM5α and TRIM22, despite the two being IFN-I-inducible genes in vitro. Furthermore, differences in expression profiles between the PBMC and CNS compartments were also noted, which are likely due to differences in cellular composition between the compartments. TRIM22 but not TRIM5α expression levels correlated negatively with HIV-1 viral loads, suggesting that TRIM22 may be exerting significant antiviral activity in vivo. We have demonstrated that in addition to type I interferons, some immunoregulatory cytokines may modestly regulate the expression of TRIM5α and TRIM22 in different HIV-1-susceptible immune cells; however, the impact of these factors is cell type specific. Our study provides evidence for the role of TRIM E3 ligases in intrinsic antiviral immunity in peripheral blood and in a remote anatomical site of infection. These data have implications for a better understanding of HIV pathogenesis and provide a basis for further work to explore the contribution of host restriction factors to immunopathogenesis and novel therapeutic targets.
ACKNOWLEDGMENTS
We thank the study participants and CAPRISA clinical and laboratory staff for providing specimens. We give special acknowledgments to the following members of the CAPRISA acute infection study team: Carolyn Williamson, Lynn Morris, Clive Gray, and Francois van Loggerenberg. We are extremely grateful to the patients and registrars in the department of neurology and nurses for facilitating the collection of samples from chronically HIV-1-infected patients with meningitis.
This study was supported by South African Department of Science and Technology/National Research Foundation Research Chairs Initiative (SARChI) grants to T.N. and K.D., a grant from the Swiss South Africa Joint Research Programme (SSAJRP) to T.N. and J.L., and NIDA/NIH grant DP1DA034990 to J.L. The research of T.N. was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute. R.S. was supported by the Columbia University-Southern African Fogarty AIDS International Training and Research Program (AITRP) through the Fogarty International Center, National Institutes of Health (grant no. D43TW000231).
Footnotes
Published ahead of print 29 January 2014
REFERENCES
- 1.Sen GC. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255–281. 10.1146/annurev.micro.55.1.255 [DOI] [PubMed] [Google Scholar]
- 2.Hardy GA, Sieg SF, Rodriguez B, Jiang W, Asaad R, Lederman MM, Harding CV. 2009. Desensitization to type I interferon in HIV-1 infection correlates with markers of immune activation and disease progression. Blood 113:5497–5505. 10.1182/blood-2008-11-190231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheney KM, McKnight A. 2010. Interferon-alpha mediates restriction of human immunodeficiency virus type-1 replication in primary human macrophages at an early stage of replication. PLoS One 5:e13521. 10.1371/journal.pone.0013521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lane HC, Davey V, Kovacs JA, Feinberg J, Metcalf JA, Herpin B, Walker R, Deyton L, Davey RT, Jr, Falloon J, Polis MA, Salzman NP, Baseler M, Masur H, Fauci AS. 1990. Interferon-alpha in patients with asymptomatic human immunodeficiency virus (HIV) infection. A randomized, placebo-controlled trial. Ann. Intern. Med. 112:805–811 [DOI] [PubMed] [Google Scholar]
- 5.Kovacs JA, Bechtel C, Davey RT, Jr, Falloon J, Polis MA, Walker RE, Metcalf JA, Davey V, Piscitelli SC, Baseler M, Dewar R, Salzman NP, Masur H, Lane HC. 1996. Combination therapy with didanosine and interferon-alpha in human immunodeficiency virus-infected patients: results of a phase I/II trial. J. Infect. Dis. 173:840–848. 10.1093/infdis/173.4.840 [DOI] [PubMed] [Google Scholar]
- 6.von Sydow M, Sonnerborg A, Gaines H, Strannegard O. 1991. Interferon-alpha and tumor necrosis factor-alpha in serum of patients in various stages of HIV-1 infection. AIDS Res. Hum. Retroviruses 7:375–380. 10.1089/aid.1991.7.375 [DOI] [PubMed] [Google Scholar]
- 7.Khatissian E, Tovey MG, Cumont MC, Monceaux V, Lebon P, Montagnier L, Hurtrel B, Chakrabarti L. 1996. The relationship between the interferon alpha response and viral burden in primary SIV infection. AIDS Res. Hum. Retroviruses 12:1273–1278. 10.1089/aid.1996.12.1273 [DOI] [PubMed] [Google Scholar]
- 8.Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W. 2008. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 4:e16. 10.1371/journal.ppat.0040016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140–2151. 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meroni G, Diez-Roux G. 2005. TRIM/RBCC, a novel class of ‘single protein RING finger' E3 ubiquitin ligases. Bioessays 27:1147–1157. 10.1002/bies.20304 [DOI] [PubMed] [Google Scholar]
- 11.Han K, Lou DI, Sawyer SL. 2011. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 7:e1002388. 10.1371/journal.pgen.1002388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayah DM, Sokolskaja E, Berthoux L, Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573. 10.1038/nature02777 [DOI] [PubMed] [Google Scholar]
- 13.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. 10.1038/nature02343 [DOI] [PubMed] [Google Scholar]
- 14.Yap MW, Nisole S, Lynch C, Stoye JP. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. U. S. A. 101:10786–10791. 10.1073/pnas.0402876101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barr SD, Smiley JR, Bushman FD. 2008. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 4:e1000007. 10.1371/journal.ppat.1000007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouazzaoui A, Kreutz M, Eisert V, Dinauer N, Heinzelmann A, Hallenberger S, Strayle J, Walker R, Rubsamen-Waigmann H, Andreesen R, von Briesen H. 2006. Stimulated trans-acting factor of 50 kDa (Staf50) inhibits HIV-1 replication in human monocyte-derived macrophages. Virology 356:79–94. 10.1016/j.virol.2006.07.025 [DOI] [PubMed] [Google Scholar]
- 17.Singh R, Gaiha G, Werner L, McKim K, Mlisana K, Luban J, Walker BD, Karim SS, Brass AL, Ndung'u T. 2011. Association of TRIM22 with the type 1 interferon response and viral control during primary HIV-1 infection. J. Virol. 85:208–216. 10.1128/JVI.01810-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tissot C, Mechti N. 1995. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J. Biol. Chem. 270:14891–14898. 10.1074/jbc.270.25.14891 [DOI] [PubMed] [Google Scholar]
- 19.Kajaste-Rudnitski A, Marelli SS, Pultrone C, Pertel T, Uchil PD, Mechti N, Mothes W, Poli G, Luban J, Vicenzi E. 2011. TRIM22 inhibits HIV-1 transcription independently of its E3 ubiquitin ligase activity, Tat, and NF-kappaB-responsive long terminal repeat elements. J. Virol. 85:5183–5196. 10.1128/JVI.02302-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javanbakht H, Yuan W, Yeung DF, Song B, Diaz-Griffero F, Li Y, Li X, Stremlau M, Sodroski J. 2006. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353:234–246. 10.1016/j.virol.2006.05.017 [DOI] [PubMed] [Google Scholar]
- 21.Speelmon EC, Livingston-Rosanoff D, Li SS, Vu Q, Bui J, Geraghty DE, Zhao LP, McElrath MJ. 2006. Genetic association of the antiviral restriction factor TRIM5alpha with human immunodeficiency virus type 1 infection. J. Virol. 80:2463–2471. 10.1128/JVI.80.5.2463-2471.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldschmidt V, Bleiber G, May M, Martinez R, Ortiz M, Telenti A. 2006. Role of common human TRIM5alpha variants in HIV-1 disease progression. Retrovirology 3:54. 10.1186/1742-4690-3-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Manen D, Rits MA, Beugeling C, van Dort K, Schuitemaker H, Kootstra NA. 2008. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 4:e18. 10.1371/journal.ppat.0040018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kootstra NA, Navis M, Beugeling C, van Dort KA, Schuitemaker H. 2007. The presence of the Trim5alpha escape mutation H87Q in the capsid of late stage HIV-1 variants is preceded by a prolonged asymptomatic infection phase. AIDS 21:2015–2023. 10.1097/QAD.0b013e3282effa87 [DOI] [PubMed] [Google Scholar]
- 25.Sewram S, Singh R, Kormuth E, Werner L, Mlisana K, Karim SS, Ndung'u T. 2009. Human TRIM5alpha expression levels and reduced susceptibility to HIV-1 infection. J. Infect. Dis. 199:1657–1663. 10.1086/598861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Loggerenberg F, Mlisana K, Williamson C, Auld SC, Morris L, Gray CM, Abdool Karim Q, Grobler A, Barnabas N, Iriogbe I, Abdool Karim SS. 2008. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One 3:e1954. 10.1371/journal.pone.0001954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel VB, Singh R, Connolly C, Coovadia Y, Peer AK, Parag P, Kasprowicz V, Zumla A, Ndung'u T, Dheda K. 2010. Cerebrospinal T-cell responses aid in the diagnosis of tuberculous meningitis in a human immunodeficiency virus- and tuberculosis-endemic population. Am. J. Respir. Crit. Care Med. 182:569–577. 10.1164/rccm.200912-1931OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obad S, Olofsson T, Mechti N, Gullberg U, Drott K. 2007. Regulation of the interferon-inducible p53 target gene TRIM22 (Staf50) in human T lymphocyte activation. J. Interferon Cytokine Res. 27:857–864. 10.1089/jir.2006.0180 [DOI] [PubMed] [Google Scholar]
- 29.Lo HL, Yee JK. 2007. Production of vesicular stomatitis virus G glycoprotein (VSV-G) pseudotyped retroviral vectors. Curr. Protoc. Hum. Genet. Chapter 12:Unit 12.7. 10.1002/0471142905.hg1207s52 [DOI] [PubMed] [Google Scholar]
- 30.Burns JC, Friedmann T, Driever W, Burrascano M, Yee JK. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. U. S. A. 90:8033–8037. 10.1073/pnas.90.17.8033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Doherty U, Swiggard WJ, Malim MH. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074–10080. 10.1128/JVI.74.21.10074-10080.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Wussow P, Jakschies D, Hochkeppel HK, Fibich C, Penner L, Deicher H. 1990. The human intracellular Mx-homologous protein is specifically induced by type I interferons. Eur. J. Immunol. 20:2015–2019. 10.1002/eji.1830200920 [DOI] [PubMed] [Google Scholar]
- 33.Argyris EG, Acheampong E, Nunnari G, Mukhtar M, Williams KJ, Pomerantz RJ. 2003. Human immunodeficiency virus type 1 enters primary human brain microvascular endothelial cells by a mechanism involving cell surface proteoglycans independent of lipid rafts. J. Virol. 77:12140–12151. 10.1128/JVI.77.22.12140-12151.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bissel SJ, Wiley CA. 2004. Human immunodeficiency virus infection of the brain: pitfalls in evaluating infected/affected cell populations. Brain Pathol. 14:97–108 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1449744/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu NQ, Lossinsky AS, Popik W, Li X, Gujuluva C, Kriederman B, Roberts J, Pushkarsky T, Bukrinsky M, Witte M, Weinand M, Fiala M. 2002. Human immunodeficiency virus type 1 enters brain microvascular endothelia by macropinocytosis dependent on lipid rafts and the mitogen-activated protein kinase signaling pathway. J. Virol. 76:6689–6700. 10.1128/JVI.76.13.6689-6700.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ndung'u T. 2011. TRIM E3 ligases in HIV infection: can these intrinsic immunity factors be harnessed for novel vaccines or therapies? Virulence 2:360–366. 10.4161/viru.2.4.16372 [DOI] [PubMed] [Google Scholar]
- 37.Hattlmann CJ, Kelly JN, Barr SD. 2012. TRIM22: a diverse and dynamic antiviral protein. Mol. Biol. Int. 2012:153415. 10.1155/2012/153415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakuma R, Mael AA, Ikeda Y. 2007. Alpha interferon enhances TRIM5alpha-mediated antiviral activities in human and rhesus monkey cells. J. Virol. 81:10201–10206. 10.1128/JVI.00419-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carthagena L, Bergamaschi A, Luna JM, David A, Uchil PD, Margottin-Goguet F, Mothes W, Hazan U, Transy C, Pancino G, Nisole S. 2009. Human TRIM gene expression in response to interferons. PLoS One 4:e4894. 10.1371/journal.pone.0004894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawyer SL, Wu LI, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U. S. A. 102:2832–2837. 10.1073/pnas.0409853102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawyer SL, Emerman M, Malik HS. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 3:e197. 10.1371/journal.ppat.0030197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fink J, Gu F, Ling L, Tolfvenstam T, Olfat F, Chin KC, Aw P, George J, Kuznetsov VA, Schreiber M, Vasudevan SG, Hibberd ML. 2007. Host gene expression profiling of dengue virus infection in cell lines and patients. PLoS Negl. Trop. Dis. 1:e86. 10.1371/journal.pntd.0000086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouse BT, Sehrawat S. 2010. Immunity and immunopathology to viruses: what decides the outcome? Nat. Rev. Immunol. 10:514–526. 10.1038/nri2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glimaker M, Olcen P, Andersson B. 1994. Interferon-gamma in cerebrospinal fluid from patients with viral and bacterial meningitis. Scand. J. Infect. Dis. 26:141–147. 10.3109/00365549409011777 [DOI] [PubMed] [Google Scholar]
- 45.Mandl JN, Barry AP, Vanderford TH, Kozyr N, Chavan R, Klucking S, Barrat FJ, Coffman RL, Staprans SI, Feinberg MB. 2008. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat. Med. 14:1077–1087. 10.1038/nm.1871 [DOI] [PubMed] [Google Scholar]
- 46.Lehmann C, Taubert D, Jung N, Fatkenheuer G, van Lunzen J, Hartmann P, Romerio F. 2009. Preferential upregulation of interferon-alpha subtype 2 expression in HIV-1 patients. AIDS Res. Hum. Retroviruses 25:577–581. 10.1089/aid.2008.0238 [DOI] [PubMed] [Google Scholar]
- 47.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grutter MG, Luban J. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472:361–365. 10.1038/nature09976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn K-S, Fernandez-Sesma A, Jung J, Garcia-Sastre A. 2013. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 38:384–398. 10.1016/j.immuni.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moench TR, Griffin DE. 1984. Immunocytochemical identification and quantitation of the mononuclear cells in the cerebrospinal fluid, meninges, and brain during acute viral meningoencephalitis. J. Exp. Med. 159:77–88. 10.1084/jem.159.1.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cepok S, Jacobsen M, Schock S, Omer B, Jaekel S, Boddeker I, Oertel WH, Sommer N, Hemmer B. 2001. Patterns of cerebrospinal fluid pathology correlate with disease progression in multiple sclerosis. Brain 124:2169–2176. 10.1093/brain/124.11.2169 [DOI] [PubMed] [Google Scholar]
- 51.Tan DB, Yong YK, Tan HY, Kamarulzaman A, Tan LH, Lim A, James I, French M, Price P. 2008. Immunological profiles of immune restoration disease presenting as mycobacterial lymphadenitis and cryptococcal meningitis. HIV Med. 9:307–316. 10.1111/j.1468-1293.2008.00565.x [DOI] [PubMed] [Google Scholar]
- 52.Yu S, Gao B, Duan Z, Xu W, Xiong S. 2011. Identification of tripartite motif-containing 22 (TRIM22) as a novel NF-kappaB activator. Biochem. Biophys. Res. Commun. 410:247–251. 10.1016/j.bbrc.2011.05.124 [DOI] [PubMed] [Google Scholar]
- 53.Clerici M, Shearer GM. 1993. A TH1→TH2 switch is a critical step in the etiology of HIV infection. Immunol. Today 14:107–111. 10.1016/0167-5699(93)90208-3 [DOI] [PubMed] [Google Scholar]
- 54.Fauci AS. 1993. Multifactorial nature of human immunodeficiency virus disease: implications for therapy. Science 262:1011–1018. 10.1126/science.8235617 [DOI] [PubMed] [Google Scholar]