ABSTRACT
Avian influenza (AI) viruses of the H7 subtype have the potential to evolve into highly pathogenic (HP) viruses that represent a major economic problem for the poultry industry and a threat to global health. However, the emergence of HPAI viruses from low-pathogenic (LPAI) progenitor viruses currently is poorly understood. To investigate the origin and evolution of one of the most important avian influenza epidemics described in Europe, we investigated the evolutionary and spatial dynamics of the entire genome of 109 H7N1 (46 LPAI and 63 HPAI) viruses collected during Italian H7N1 outbreaks between March 1999 and February 2001. Phylogenetic analysis revealed that the LPAI and HPAI epidemics shared a single ancestor, that the HPAI strains evolved from the LPAI viruses in the absence of reassortment, and that there was a parallel emergence of mutations among HPAI and later LPAI lineages. Notably, an ultradeep-sequencing analysis demonstrated that some of the amino acid changes characterizing the HPAI virus cluster were already present with low frequency within several individual viral populations from the beginning of the LPAI H7N1 epidemic. A Bayesian phylogeographic analysis revealed stronger spatial structure during the LPAI outbreak, reflecting the more rapid spread of the virus following the emergence of HPAI. The data generated in this study provide the most complete evolutionary and phylogeographic analysis of epidemiologically intertwined high- and low-pathogenicity viruses undertaken to date and highlight the importance of implementing prompt eradication measures against LPAI to prevent the appearance of viruses with fitness advantages and unpredictable pathogenic properties.
IMPORTANCE The Italian H7 AI epidemic of 1999 to 2001 was one of the most important AI outbreaks described in Europe. H7 viruses have the ability to evolve into HP forms from LP precursors, although the mechanisms underlying this evolutionary transition are only poorly understood. We combined epidemiological information, whole-genome sequence data, and ultradeep sequencing approaches to provide the most complete characterization of the evolution of HPAI from LPAI viruses undertaken to date. Our analysis revealed that the LPAI viruses were the direct ancestors of the HPAI strains and identified low-frequency minority variants with HPAI mutations that were present in the LPAI samples. Spatial analysis provided key information for the design of effective control strategies for AI at both local and global scales. Overall, this work highlights the importance of implementing rapid eradication measures to prevent the emergence of novel influenza viruses with severe pathogenic properties.
INTRODUCTION
Influenza A viruses (IAVs) evolve rapidly due to the high error rate of the RNA polymerase, strong immune-driven natural selection, and the capacity for genome segments to be exchanged via reassortment (1). The rapidity of evolution, in combination with the increasing availability of genome-scale sequence data and new computational tools for phylogenetic analysis, facilitate the inference of the evolutionary history and spatial spread of IAVs, in turn providing a better understanding of the patterns and processes that underpin epidemics (2–8). Eighteen antigenically distinct hemagglutinin (HA) subtypes of influenza viruses have been identified to date (9, 10). In avian species, infections with the H5 and H7 subtypes are of greatest concern because of their potential to evolve into the highly pathogenic form of the virus that can devastate poultry populations and occasionally be transmitted to humans. Why the H5 and H7 subtypes are more prone to evolve into highly pathogenic forms than other subtypes remains poorly understood. Highly pathogenic avian influenza (HPAI) viruses can evolve directly from low-pathogenic (LPAI) virus precursors following introduction into domestic poultry (11–13). The emergence of HPAI viruses results from the insertion/substitution of basic amino acids at the HA cleavage site (14–16) or from nonhomologous recombination resulting in the insertion of a foreign nucleotide sequence (17–19). HPAI viruses replicate systemically in birds, damaging vital organs and tissues and frequently causing death (20). In addition to the HA, the virus polymerase gene complex (PB1, PB2, and PA) and nucleoprotein (NP) also modulate the pathogenicity of AI viruses, as demonstrated for H5N1 HPAI viruses (21–23). Importantly, the insertion of multiple basic amino acids at the cleavage site is not always sufficient to convert an LPAI virus phenotype into an HPAI one, as demonstrated in experimental studies (24) and sporadically in natural infections (25–27).
Although there are numerous phylogenetic studies of the emergence, spread, and evolution of HPAI viruses (3–6, 28–30), whole-genome sequence data from HPAI and the LPAI progenitor viruses collected during the same epidemic is limited. In most cases, the number of LPAI isolates collected prior to the emergence of the HPAI strain was insufficient to determine the genesis of the virulent phenotype, including whether this occurs via a stepwise evolutionary process (13–15, 17, 18, 31). Importantly, improved deep-sequencing technologies allow for greater insight into the emergence of new variants of influenza type A viruses (32, 33), potentially including HPAI strains.
From 1999 to 2001 in northern Italy, an LPAI H7N1 avian influenza virus evolved into an HPAI form that caused the death of over 16 million poultry and substantial economic losses to industry (34). From the end of March to December 1999, 199 outbreaks of LPAI were detected in northern Italy, which is home to more than 65% of Italian poultry production (35, 36). Despite control measures, an HPAI virus of the same subtype was isolated in December 1999 and rapidly spread to additional production facilities before being eradicated officially in April 2000. Following depopulation and restocking of the HPAI-infected areas, LPAI reemerged in August 2000, and 78 outbreaks were diagnosed and eradicated (37). To reduce the economic impact of the H7N1 infection in poultry (38), a vaccination program was initiated from November 2000 to May 2002.
The Italian HPAI H7N1 outbreak has been well characterized epidemiologically, including identifying risk factors and within-flock viral transmission dynamics (39–44). However, few pieces of genetic data have been published from this epidemic (12). Here, we describe the analysis of 109 whole genomes from H7N1 viruses collected between March 1999 and February 2001. In particular, we compared the evolutionary patterns and dynamics of the HPAI and LPAI viruses, including rates of nucleotide substitution, selection pressures, and the emergence of amino acid mutations that may facilitate evolution from low to high pathogenicity. In addition, we investigated the patterns of spatial spread of the LPAI and HPAI H7N1 strains among the affected Italian provinces. To complement the phylogenetic analysis and to help identify viral subpopulations harboring known molecular markers for viral pathogenicity, we utilized deep sequencing of representative clinical samples collected during the 1999-2001 H7N1 epidemic.
MATERIALS AND METHODS
Sanger sequencing.
We generated the complete genome sequences of 72 H7N1 avian influenza A viruses collected from poultry in northern Italy from March 1999 to February 2001, comprising 22 LPAI and 50 HPAI viruses. In addition, 24 HA segments were sequenced from LPAI viruses from which whole-genome sequences could not be obtained. Publicly available sequence data for this epidemic were downloaded from the Influenza Virus Resource at GenBank, and we also included in the analysis the complete genome of 37 H7N1 viruses and the HA gene of 11 viruses. Epidemiological information (collection date, city and province of collection, and pathotype) for all of the viruses included in this study (n = 144) is available in Table S1 in the supplemental material.
Viral RNA was extracted from the infected allantoic fluid of specific-pathogen-free fowls' eggs using the Nucleospin RNA II kit (Macherey-Nagel, Duren, Germany) and reverse transcribed with the SuperScript III reverse transcriptase kit (Invitrogen, Carlsbad, CA). PCR amplifications were performed with PfuTurbo DNA polymerase (Stratagene, La Jolla, CA) by using specific primers (sequences are available on request). The complete coding sequences were generated using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). The products of the sequencing reactions were cleaned up using the Performa DTR Ultra 96-well kit (Edge BioSystems, Gaithersburg, MD) and sequenced in a 16-capillary ABI Prism 3130xl genetic analyzer (Applied Biosystems, Foster City, CA). Sequence data were assembled and edited with SeqScape software v2.5 (Applied Biosystems). Sequences from all eight gene segments were aligned and compared to the most related sequences available in GenBank. In addition, the phylogenetic relationship between the Italian H7N1 viruses and all publicly available Eurasian H7 sequences (n = 443) was evaluated for the HA segment.
Library preparation, Illumina sequencing, and data analysis.
To assess virus population diversity, next-generation sequencing (NGS) was performed on all available H7N1 influenza A virus clinical samples: six LPAI and seven HPAI viruses. Full sample details are described in Table 1. Viral RNA was extracted directly from the infected clinical samples, such as trachea, lung, and intestine, using the Nucleospin RNA II kit (Macherey-Nagel, Duren, Germany). Complete influenza A virus genomes were prepared from the RNA using the SuperScript III one-step reverse transcription-PCR (RT-PCR) system with PlatinumTaq high fidelity (Invitrogen, Carlsbad, CA) by following the protocol of Zhou et al. (45). All amplified products were visualized on a 0.7% agarose gel stained with GelRed. High-throughput sequencing of the amplified genome was performed using an Illumina MiSeq at IGA Technology Services srl (UD), Italy. The amplified genomes were fragmented down to 600 to 800 bp and tagged with sequencing adapters with indexes in a single step, using the Nextera DNA sample preparation kit from Illumina. Libraries were run on the Caliper GX (PerkinElmer) for quality control, and a QBit fluorimeter (Invitrogen) was used to determine the molar concentration of the libraries. Finally, the indexed libraries were pooled in equimolar concentrations and sequenced in multiplex for 150-bp paired ends on Illumina MiSeq according to the manufacturer's instructions.
TABLE 1.
Identification number, epidemiological data, and total number of sequences obtained for each viral populationa
| Identity no. | Sample name and pathotype | Type of sample | Date of collection (day/mo/yr) | Location | Total no. of sequencesb | No. of mapped sequences |
|---|---|---|---|---|---|---|
| 4618/99 | A/turkey/Italy/4618/1999, HPAI | Trachea | 14/12/1999 | VR | 423,574 | 418,238 |
| 4828/99 | A/turkey/Italy/4828/1999, HPAI | Lung, trachea | 23/12/1999 | VR | 460,708 | 460,491 |
| 4827/99 | A/turkey/Italy/4827/1999, HPAI | Lung, trachea | 23/12/1999 | VR | 431,304 | 431,085 |
| 4749/99 | A/turkey/Italy/4749/1999, HPAI | Intestine | 21/12/1999 | VR | 544,596 | 541,929 |
| 4708/99 | A/turkey/Italy/4708/1999, HPAI | Trachea | 21/12/1999 | VR | 501,086 | 496,408 |
| 4911/99 | A/chicken/Italy/4911/1999, HPAI | Intestine | 27/12/1999 | RO | 386,926 | 378,252 |
| 4756/99 | A/turkey/Italy/4756/1999, HPAI | Intestine | 22/12/1999 | NAc | 463,704 | 413,277 |
| 2732/99 | A/turkey/Italy/2732/1999, LPAI | Lung, trachea | 03/08/1999 | VR | 533,706 | 533,293 |
| 3675/99 | A/turkey/Italy/3675/1999, LPAI | Trachea | 04/10/1999 | VR | 591,410 | 591,173 |
| 4295/99 | A/turkey/Italy/4295/1999, LPAI | Lung | 22/11/1999 | VR | 520,056 | 516,798 |
| 3283/99 | A/turkey/Italy/3283/1999, LPAI | Lung, trachea | 10/09/1999 | VR | 534,136 | 529,407 |
| 4829/99 | A/turkey/Italy/4829/1999, LPAI | Lung, trachea | 23/12/1999 | VR | 542,062 | 541,952 |
| 1744/99 | A/turkey/Italy/1744/1999, LPAI | Lung, trachea | 26/04/1999 | VR | 195,496 | 194,543 |
Data were derived from clinical specimens using the Illumina deep-sequencing platform.
After removal of adapter and low-quality sequences.
NA, not available.
Raw sequence reads were inspected using FASTQC to assess the quality of data coming from the high-throughput sequencing pipelines. Fastq files were cleaned with PRINSEQ and Trim Galore to remove adaptors and primers and exclude reads with a Phred quality score below 20. Only reads longer than 80 bp were considered and mapped to the reference sequences. Reads were aligned to both the A/Italy/4749/99 (HPAI) and A/Italy/3675/99 (LPAI) reference sequences using Stampy (46), an alignment tool particularly suited to map reads that contain sequence variation relative to the reference. This alignment protocol also enabled us to identify possible key mutations associated with the highly pathogenic virus in the low-pathogenic virus samples and vice versa. Unmapped reads were removed from the resulting BAM files using samtools version 0.1.14 (47). Depth of coverage of the alignment was visualized using BEDTools (48) and R. The BAM alignment files were parsed using a custom perl script to identify the frequency of polymorphisms at each site relative to the reference used for the alignment. A binomial test, using the error probability calculated from the base qualities at a site and by following the approach of Morelli et al. (49), was used to determine whether the frequency of each mutation was above the artifactual value expected from Illumina sequencing. All low-frequency polymorphisms that were likely to be miscalled bases during sequencing were removed from downstream analyses.
Phylogenetic analysis.
Nucleotide sequence alignments were manually constructed for each gene segment and for the concatenated whole genome using the Se-Al program (tree.bio.ed.ac.uk/software/seal/). To infer the evolutionary relationships for each gene segment, we employed the maximum likelihood (ML) method available in the PhyML program, incorporating a GTR model of nucleotide substitution with a gamma distribution of among-site rate variation (with four rate categories, Γ4) and an SPR branch-swapping search procedure (50). A bootstrap resampling process (1,000 replications), using the neighbor-joining (NJ) method and incorporating the ML substitution model defined above, was employed to assess the robustness of individual nodes of the phylogeny using PAUP* (51). Parameter values for the GTR substitution matrix, base composition, gamma distribution of among-site rate variation, and proportion of invariant sites (I) were estimated directly from the data using MODELTEST (52).
Analysis of selection pressures.
Gene- and site-specific selection pressures for all segments of the Italian H7N1 viruses were measured as the ratio of nonsynonymous (dN) to synonymous (dS) nucleotide substitutions per site. In all cases, dN/dS ratios were estimated using the single-likelihood ancestor counting (SLAC), fixed-effects likelihood (FEL), and internal fixed-effects likelihood (IFEL) methods available at the Datamonkey online version of the Hy-Phy package (53, 54). In addition, we employed the mixed-effects model of evolution (MEME), which is capable of identifying instances of both episodic and positive selection at individual sites (55). All analyses utilized the GTR model of nucleotide substitution and employed input NJ phylogenetic trees.
Evolutionary dynamics and spatial analysis.
Rates of nucleotide substitution per site per year (subs/site/year) and the time to the most recent common ancestor (tMRCA) of the sampled data were estimated using the Bayesian Markov chain Monte Carlo (MCMC) approach available in the BEAST program, version 1.6.2 (56). For each analysis, we employed a flexible Bayesian skyline coalescent tree prior (10 piece-wise constant groups), as this is typically the best descriptor of the complex population dynamics of influenza A virus (57). In addition, we utilized a HKY85 + Γ4 model of nucleotide substitution with two data partitions reflecting codon positions (1st plus 2nd positions, 3rd position) and with base frequencies unlinked across all codon positions (i.e., the SRD06 substitution model). Two molecular clock models, strict (constant) and relaxed (uncorrelated lognormal), were compared by analyzing values of the coefficient of variation (CoV) in Tracer (56), in which CoV values of >0 are evidence of non-clock-like evolutionary behavior. The molecular clock model utilized for each data set is indicated in Table 2. In all cases, statistical uncertainty is reflected in values of the 95% highest posterior density (HPD) for each parameter estimate, and in each case chain lengths were run for at least 50 million iterations to achieve convergence as assessed using the Tracer v1.5 program (56). Finally, maximum clade credibility (MCC) phylogenetic trees were summarized from the posterior distribution of trees using TreeAnnotator v1.6.1 (56) after the removal of 10% burn-in. The topologies of the MCC trees, visualized using FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/), were similar to those inferred using ML methods.
TABLE 2.
Estimated rates of nucleotide substitution and tMRCA for LPAI and HPAI H7N1 viruses collected during the Italian epidemic
| Gene and genetic group | Modela | Substitution rate (subs/site/year) |
tMRCA (mo/yr) |
||
|---|---|---|---|---|---|
| Mean (×10−3) | 95% HPD (×10−3) | Mean | 95% HPD | ||
| HA | |||||
| H7N1 | SRD06-SC-Skyline | 10.15 | 8.5–11.9 | Feb 1999 | Jan 1999-Mar 1999 |
| H7N1-LPAI | SRD06-SC-Skyline | 9.75 | 7.8–11.9 | Feb 1999 | Jan 1999-Mar 1999 |
| H7N1-HPAI | SRD06-SC-Skyline | 9.89 | 6.54–13.22 | Nov 1999 | Oct 1999-Dec 1999 |
| NA | |||||
| H7N1 | SRD06-SC-Skyline | 9.86 | 8.01–11.79 | Feb 1999 | Dec 1998-Mar1999 |
| H7N1-LPAI | SRD06-SC-Skyline | 10.06 | 7.89–12.37 | Feb 1999 | Jan 1999-Mar 1999 |
| H7N1-HPAI | SRD06-SC-Skyline | 10.12 | 5.41–15.39 | Nov 1999 | Oct 1999-Dec 1999 |
| PA | |||||
| H7N1 | SRD06-SC-Skyline | 5.84 | 4.79–6.91 | Feb 1999 | Feb 1999-Mar 1999 |
| H7N1-LPAI | SRD06-SC-Skyline | 5.49 | 4.20–6.77 | Mar 1999 | Feb 1999-Mar 1999 |
| H7N1-HPAI | SRD06-SC-Skyline | 6.64 | 4.08–9.69 | Nov 1999 | Sep 1999-Dec 1999 |
| MA | |||||
| H7N1 | SRD06-SC-Skyline | 7.21 | 5.02–9.51 | Feb 1999 | Dec 1998-Mar1999 |
| H7N1-LPAI | SRD06-SC-Skyline | 4.87 | 2.77–7.26 | Feb 1999 | Dec 1998-Mar1999 |
| H7N1-HPAI | SRD06-SC-Skyline | 8.87 | 4.69–13.37 | Nov 1999 | Sep 1999-Dec 1999 |
| PB1 | |||||
| H7N1 | SRD06-SC-Skyline | 5.57 | 4.54–6.68 | Feb 1999 | Gen 1999-Mar1999 |
| H7N1-LPAI | SRD06-SC-Skyline | 5.70 | 4.31–7.15 | Feb 1999 | Jan 1999-Mar 1999 |
| H7N1-HPAI | SRD06-SC-Skyline | 5.48 | 3.60–7.39 | Oct 1999 | Aug 1999-Nov 1999 |
| PB2 | |||||
| H7N1 | SRD06-SC-Skyline | 6.81 | 5.54–8.04 | Feb 1999 | Dec 1998-Mar 1999 |
| H7N1-LPAI | SRD06-SC-Skyline | 6.69 | 5.12–8.23 | Jan 1999 | Dec 1998-Mar 1999 |
| H7N1-HPAI | SRD06-SC-Skyline | 7.22 | 5.0–9.64 | Oct1999 | Aug 1999-Nov 1999 |
| NP | |||||
| H7N1 | SRD06-SC-Skyline | 5.42 | 3.81–7.15 | Feb 1999 | Dec 1999-Mar1999 |
| H7N1-LPAI | SRD06-SC-Skyline | 5.11 | 3.00–7.39 | Feb 1999 | Dec 1999-Mar 1999 |
| H7N1-HPAI | SRD06-SC-Skyline | 4.91 | 2.61–7.73 | Nov 1999 | Oct 1999-Dec 1999 |
| NS | |||||
| H7N1 | SRD06-SC-Skyline | 7.72 | 5.76–9.91 | Jan 1999 | Oct 1998-Mar 1999 |
| H7N1-LPAI | SRD06-SC-Skyline | 7.19 | 4.85–9.66 | Jan 1999 | Nov 1998-Mar 1999 |
| H7N1-HPAI | SRD06-SC-Skyline | 6.44 | 2.90–10.31 | Jun 1999 | Jan 1999-Dec 1999 |
| CG | |||||
| H7N1 | SRD06-SC-Skyline | 6.67 | 5.97–7.34 | Mar 1999 | Feb 1999-Mar 1999 |
| H7N1-LPAI | SRD06-SC-Skyline | 6.82 | 5.81–7.87 | Feb 1999 | Feb 1999-Mar 1999 |
| H7N1-HPAI | SRD06-SC-Skyline | 6.41 | 5.25–7.62 | Oct 1999 | Sept 1999-Nov 1999 |
SC, strict molecular clock; Skyline, Bayesian skyline coalescent prior.
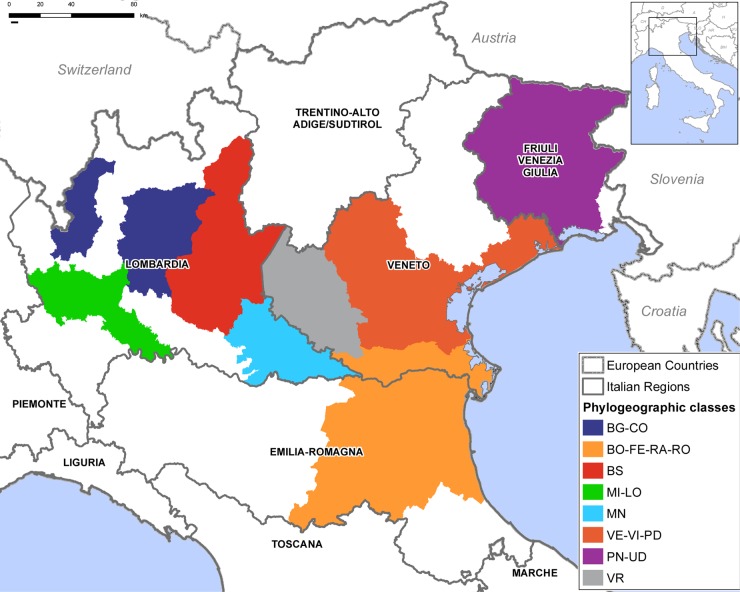
To infer the spatial spread of H7N1, we analyzed two data sets separately, one containing the HA gene from 139 viruses and another including the eight gene segments concatenated together for 103 isolates, after the removal of all gaps (sequence concatenation was appropriate, as we found no evidence for reassortment; see Results). Sequences of viruses sampled in regions for which fewer than three samples where available were excluded. We grouped the H7N1 sequences into eight discrete geographical locations, each representing one province or adjacent provinces such that each group had at least three sequences: VR (Verona), BS (Brescia), MN (Mantua), FE-RA-RO-BO (Ferrara, Ravenna, Rovigo, Bologna), VE-PD-VI (Venice, Padua, Vicenza), PN-UD (Pordenone and Udine), BG-CO (Bergamo and Como), and MI-LO (Milan and Lodi) (see Table S1 in the supplemental material) (Fig. 1). All these areas are located in northern Italy, and most are situated in the Veneto and Lombardia regions.
FIG 1.
Map of the discrete geographical locations used in the spatial analysis. Locations are colored gray for Verona; red for Brescia; light blue for Mantua; yellow for Ferrara, Ravenna, Rovigo, and Bologna; orange for Venice, Padua, and Vicenza; violet for Pordenone and Udine; blue for Bergamo and Como; and green for Milan and Lodi.
To determine the overall degree of geographical structure among the Italian H7N1 viruses, we employed the association index (AI) and parsimony score (PS) phylogeny-trait association statistics available in the Bayesian tip-association significance testing (BaTS) program (58). In this case, traits were defined as the eight geographical regions described above. This method uses the posterior distribution of trees obtained from the BEAST analysis and accounts for phylogenetic uncertainty in the data. The BaTS program also allowed us to assess the level of clustering in individual geographic locations using the monophyletic clade (MC) size statistic (58). In all cases, 1,000 random permutations of tip locations were undertaken to create a null distribution for each statistic. This statistic determines the association between sampling location and phylogeny by estimating the size of the largest cluster of sequences from each sampling location. Again, the LPAI and HPAI viruses were analyzed separately.
To infer phylogeographic histories in more detail, we utilized a Bayesian phylogeographic approach (3), employing the MCMC method implemented in BEAST. For this analysis, we utilized the date (exact day) of viral sampling, a Bayesian skyline prior, and the SRD06 model of nucleotide substitution, as described above. For the HA data set, a strict molecular clock was used, while for the concatenated genes a relaxed (uncorrelated lognormal) molecular clock model was preferred. Analyses were undertaken for the HA segment of the LPAI and HPAI viruses independently. The MCMC chain was run for 150 million iterations. All parameters reached convergence, as assessed using the Tracer v1.5 program (56). The initial 10% of the chain was removed as burn-in, and MCC trees were again summarized using TreeAnnotator v1.6.1. We also produced posterior summaries of the root and HPAI internal node geographical states as described by Lemey et al. (3).
Finally, to assess the role of poultry density in determining patterns of viral diffusion in northern Italy, we parameterized a discrete phylogeographic diffusion model using data derived from the Italian National Institute for Statistics (ISTAT) 5th Census of Agriculture (2000) (http://censagr.istat.it/). Specifically, we collected information on the poultry population size for three provinces in the Lombardia region (Mantua, Brescia, and Bergamo) and four in the Veneto region (Venice, Verona, Padua, and Vicenza) from which at least three sequences were available. From these data, we determined (i) the poultry population size of the province of origin, (ii) the poultry population size of the province of destination, and (iii) the product of the poultry population sizes from the province of origin and the province of destination, all of which were considered predictors in the phylogeographic analysis, in addition to geographic distance. Each of these predictors was incorporated into an asymmetric transition matrix that allows for separate directional rates between each pair of locations. We used a Bayes factor (BF) comparison (59) of the relative marginal likelihoods to select the most appropriate model for the data compared to a null model of equal migration rates.
Nucleotide sequence accession numbers.
All sequence data generated here have been submitted to GenBank and assigned accession numbers KF492991 to KF493575.
RESULTS
Phylogenetic analysis of LPAI and HPAI H7N1 viruses.
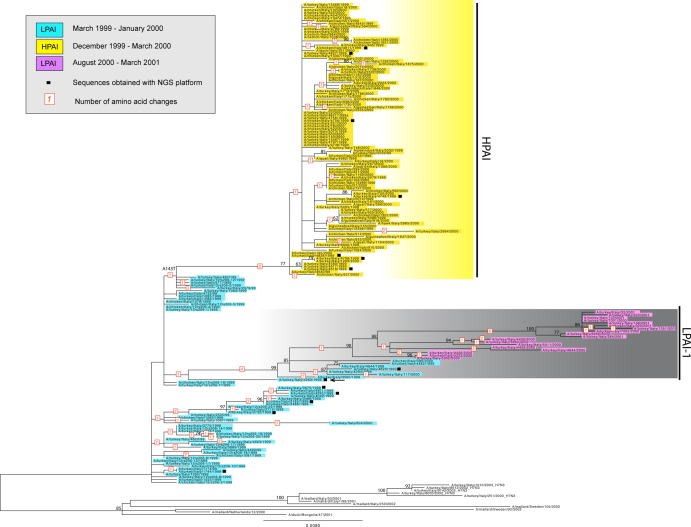
We inferred the evolutionary relationships of HA sequences of 144 avian influenza H7N1 viruses collected during the 1999-2001 Italian epidemic (96 sequences generated in this study and 48 obtained from GenBank). These data included 57 LPAI viruses sampled from March 1999 to January 2000, 69 HPAI isolates detected between December 1999 and April 2000, and 18 LPAI samples isolated from August 2000 to February 2001. As background, 443 Eurasian H7 sequences available in GenBank were included in the analysis. The topology of the HA phylogeny indicates that the LPAI and HPAI Italian H7N1 viruses clustered together within a monophyletic group that was distinct from all other European H7 viruses (see Fig. S1 in the supplemental material), suggesting a single introduction of the virus into Italy (Fig. 2). Within this clade, we identified two main phylogenetic clusters, one that includes all Italian H7N1 HPAI viruses (yellow shading) and another that contains all Italian H7N1 LPAI virus samples collected from both time periods: late September 1999 to January 2000 and August 2000 to February 2001 (i.e., before and after the HPAI outbreak). However, the two clusters are not monophyletic, as the HPAI viruses evolved from a subset of the LPAI viruses.
FIG 2.
ML tree of the HA gene segment of H7N1 avian influenza virus. Viruses are colored blue for LPAI viruses collected from March 1999 to January 2000, pink for LPAI viruses sampled between August 2000 and February 2001, and yellow for HPAI viruses (December 1999 to March 2000). The numbers at nodes represent bootstrap values (>70%), while branch lengths are scaled according to the numbers of nucleotide substitutions per site. The number highlighted in red represents the number of amino acid changes identified along the main branches of the tree. Sequences obtained using the NGS platform are marked with a black square. The closest relative of the LPAI-1 group is identified with a black arrow. The tree is midpoint rooted for clarity only.
To explore the genome-scale evolution of the viruses collected during this epidemic, ML trees were inferred for each of the eight gene segments (see Fig. S2 and S3 in the supplemental material) for a total of 109 H7N1 isolates: 63 HPAI viruses, 46 LPAI viruses sampled before (n = 28) and after (n = 18) the HPAI outbreak, and a subset of the most closely related sequences publicly available in GenBank. Phylogenies inferred for each of the seven other genome segments show a pattern identical to that observed in the HA tree: the H7N1 viruses represent a single viral introduction into Italy, and a distinct clade of Italian HPAI viruses (bootstrap values of >70%) has evolved from a subset of the precursor LPAI viruses. Importantly, there is no evidence for reassortment in the evolutionary history of all H7N1 viruses during this Italian epidemic. In addition to the HPAI viruses, a second genetically distinct cluster is present within the Italian H7N1 LPAI viruses, which we term LPAI-1 (shaded gray in Fig. 2). The LPAI-1 cluster is defined by high bootstrap values (>70%) and long branches in the HA, neuraminidase (NA), PB2, PA, and nonstructural (NS) phylogenies (Fig. 2; also see Fig. S2 and S3). This clade, which is characterized by a total of 16 amino acid signature mutations (Table 3), includes samples collected from the first wave of the LPAI epidemic (late September 1999 to January 2000) and from the second wave (August 2000 to February 2001), suggesting that this variant circulated undetected in Italy during the HPAI outbreak and reemerged in the commercial flocks almost 4 months after the eradication of the HPAI strain. The sequence A/chicken/Italy/3283/1999 (marked with a black arrow in the phylogenetic trees), isolated in early September 1999, is the most closely related to this cluster on the HA, NA, NS, PB2, and PB1 trees and possesses five out 16 amino acid substitutions specific to LPAI-1 isolates (Table 3).
TABLE 3.
List of the amino acid signature mutations of HPAI and LPAI-1 viruses
| Gene | HPAI virus |
LPAI-1 virus |
Comment | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Position | Amino acid |
Position | Amino acid |
||||||
| LPAI | LPAI-1 | HPAI | LPAI | LPAI-1 | HPAI | ||||
| PB2 | 398 | T | Ta | I | 527 | L | M | L | |
| PB1 | 154 | G | G | D | 211 | R | K | R | A/tk/It/3283/00 possesses the aa K |
| 216 | S | S | G | ||||||
| 745 | K | K | Eb | ||||||
| PA | 61 | I | I | T | 190 | S | P | S/Y | |
| 115 | N | N | K | ||||||
| 252 | E | E | Kc | 257 | I | V | I | ||
| HA | 130 | T | T | A | 86 | Q | Rh | Q | |
| 113 | E | G | E | ||||||
| 146 | A/S | A/S | Td | 143 | A/T | V | Ti | ||
| 228 | Ae | A | E | 148 | R | K | R | ||
| 454 | A | Af | T | 169 | A/T | Tj | A | A/tk/It/3283/00 possesses the aa T | |
| 554 | K | K | R | ||||||
| NP | 349 | T | T | A | |||||
| 376 | S | S | N | ||||||
| NA | 173 | R | R | K | 163 | V | M/L | V | A/tk/It/3283/00 possesses the aa L |
| 314 | I | L | I | ||||||
| 418 | M | I | Mk | ||||||
| M1 | 166 | V | V | A | |||||
| NS1 | 136 | V | V | I | 47 | G | C | G | |
| 139 | D | D | N | 85 | P | T | P | ||
| 225 | Rg | STOP | 221 | Y/Hl | STOP | Y | A/tk/It/3283/00 possesses a stop codon | ||
| NS2 | 64 | Tm | K | T | A/tk/It/3283/00 possesses the aa K | ||||
Except for 2 viruses which possess the amino acid (aa) I.
Except for 1 virus which possesses the aa G.
Except for 5 viruses which possess the aa E.
Except for 2 viruses which possess the aa A or S.
Except for 1 virus which possesses the aa E.
Except for 2 viruses which possess the aa T.
Except for 1 virus which possesses a stop codon at position 221.
Except for 2 viruses which possess the aa Q.
Except for 3 viruses which possess the aa A or K.
Except for 1 virus which possesses the aa I.
Except for 2 viruses which possess the aa I.
Except for 1 virus which possesses a stop codon.
Except for 1 virus which possesses the aa K.
Amino acid mutations of LPAI-1 and HPAI H7N1 viruses.
The evolution from the low-pathogenic to the highly pathogenic H7N1 viruses appears to have involved more than the multiple-basic-amino-acid insertion at the HA cleavage site. Across the viral genome, the HPAI isolates are characterized by 19 unique amino acid substitutions: five in the HA, one in the NA, one in the polymerase PB2, three in the polymerase PB1, three in the polymerase PA, two in the nucleoprotein (NP), one in the matrix protein 1 (M1), and three in the nonstructural protein 1 (NS1), including a nonsense mutation (Table 3). The latter mutation results in a C-terminal truncation of six amino acids that promotes the transport of NS1 into the nucleoli, where it may favor viral transcription and translation or may alter the cell cycle for viral needs (60). Of the 69 HPAI viruses analyzed here, 66 possess a substitution at position 143 of the HA gene (A143T) that introduces a potential additional glycosylation site at position 141, located close to the 130 loop of the receptor binding domain (61) and within the equivalent antigenic site A in H3 viruses (62). This mutation also was identified in an HPAI H7N7 virus isolated from a fatal human case in the Netherlands in 2003, where it was demonstrated to increase viral replication efficiency in MDCK cells and to alter viral attachment to human lung tissue (63). Of note, this additional glycosylation site was also found among the five low-pathogenicity precursor viruses that are the most closely related to the HPAI viruses on the HA tree that are likely to be their evolutionary precursors.
Similar to the HPAI viruses, the LPAI-1 isolates have a truncation in the NS1 protein and an additional glycosylation site (AGS) in the HA molecule. However, in contrast to the HPAI viruses, the truncation in the NS1 protein is of 10 rather than 6 amino acids in length (total length of 220 amino acids), and the potential AGS is located at position 169 (within the corresponding H3 and H5 antigenic site B of the HA molecule) instead of 143 (64, 65). The NS1 truncation may have the same effect as the six-amino-acid deletion observed in the HPAI viruses, while the new glycosylation site in the HA protein has been demonstrated to promote influenza virus replication in cell culture (66) and may also have an antigenic effect. Notably, the LPAI-1 viruses also possess an amino acid substitution at position 143 (A143V) of the receptor-binding domain of the HA. Unlike the HPAI strain, this mutation does not introduce any glycosylation site, but it could affect the receptor binding conformation. Of note, in the HA phylogeny, all of the viruses collected in south Verona and Padua from December 2000 to February 2001 following the introduction of the vaccination program (36) are separated from samples detected from the same area in the period immediately before (October 2000) by a long branch and at least six amino acid substitutions, two of them located in the H3-corresponding antigenic sites A (G151E) and B (G195V) (67).
Time-scaled evolutionary history of LPAI and HPAI H7N1 viruses.
Rates of nucleotide substitution and the times to the most recent common ancestor (tMRCAs) were estimated using a Bayesian coalescent approach (56) for each genome segment separately and for the eight concatenated segments. Evolutionary rates also were calculated for the entire H7N1 viral population and for the HPAI and LPAI viruses separately, although no significant difference in rates was observed between the LPAI and HPAI viruses for any of the genome segments (Table 2). For the entire viral population, nucleotide substitution rates were high for the genes encoding the surface glycoproteins, with a mean rate of 10.15 × 10−3 sub/site/year (95% HPD, 8.5 × 10−3 to 11.9 × 10−3) for the HA gene and 9.86 × 10−3 sub/site/year (95% HPD, 8.01 × 10−3 to 11.79 × 10−3) for the NA gene (Table 2). These rates are similar to those calculated for the HA and NA from viruses collected during the 2003 HPAI H7N7 outbreak in the Netherlands (6). The estimated rates for the other genome segments are lower, ranging from 5.42 × 10−3 sub/site/year (95% HPD, 3.81 × 10−3 to 7.15 × 10−3 sub/site/year) for the NP gene to 7.72 × 10−3 sub/site/year (95% HPD, 5.76 × 10−3 to 9.66 × 10−3) for the NS gene segment. Interestingly, the evolutionary rate for the branch leading from the low-pathogenic progenitor strain to the HPAI lineage was similar to the rest of the phylogeny, at 4.4 to 7.8 × 10−3 subs/site/year for the concatenated genome (95% HPD), with no evidence of a major rate elevation.
To explore the evolutionary origins of the H7N1 viruses in Italy, we estimated the tMRCA of (i) the entire H7N1 population, (ii) the HPAI viruses, and (iii) LPAI viruses for each genome segment and the eight concatenated segments. The tMRCAs for the entire Italian H7N1 viral population ranged from December 1998 to March 1999 (95% HPD value) for all of the gene segments (with the exception of NS), suggesting that the detection of the outbreak in March 1999 occurred no more than 3 months after the entry of the virus into Italy and possibly simultaneously with the appearance of the virus. Similarly, the tMRCAs for the HPAI viruses ranged from August 1999 to December 1999 (95% HPD), 0 to 5 months before the first detection of the HPAI strain, for all eight segments (Table 2). These dates overlap the period of circulation of the LPAI viruses and are consistent with the emergence of HPAI from the LPAI progenitor.
Selection pressures in the H7N1 genes.
An analysis of selection pressures on all virus genes revealed that the vast majority of codons were subject to purifying selection (mean dN/dS ratios ranged from 0.10 for PB2 to 0.83 for M2) (Table 4). Only the PB1-F2 protein, which is only 90 amino acids long, showed evidence of diversifying selection, although this is likely to be an artifact due to its encoding in an overlapping reading frame (dN/dS = 2.65) (Table 4) (68). Using the SLAC, FEL, IFEL, and MEME methods, we identified several sites (P < 0.05) in HA, NA, PB2, and NS2 with evidence of putative positive selection (Table 4). Interestingly, positions 143 and 146, adjacent to the receptor binding site in the HA gene, are highly polymorphic, which may affect virus interactions with the cell surface. Three (T, K, and A) and two (A and V) distinct amino acids were observed at position 143 in the HPAI and LPAI viruses, respectively. At position 146, three distinct amino acid substitutions (A, T, and S) were observed, with the majority of the HPAI and LPAI viruses retaining the amino acids T and A, respectively.
TABLE 4.
Amino acid sites under putative positive selection detected using different analytical models and mean dN/dS ratio for each gene
| Gene | Mean dN/dS | Result for positively selected sites |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SLAC | P value | FEL | P value | IFEL | P value | REL | Posterior probability | MEME | P value | ||
| HA | 0.37 | 143 | 0.04 | 77 | 0.03 | NA | NA | 146 | 0.05 | ||
| 468 | 0.04 | ||||||||||
| HA-HPAI | 0.41 | 264 | 0.04 | ||||||||
| 468 | 0.05 | ||||||||||
| HA-LPAI | 0.34 | 8 | 0.97 | ||||||||
| 86 | 0.87 | ||||||||||
| 143 | 0.97 | ||||||||||
| 146 | 0.97 | ||||||||||
| 169 | 0.97 | ||||||||||
| 195 | 0.97 | ||||||||||
| NA | 0.29 | NA | NA | 383 | 0.02 | ||||||
| PB2 | 0.10 | 195 | 0.07 | 195 | 0.07 | NA | NA | 195 | 0.0009 | ||
| 398 | 0.07 | 398 | 0.07 | 398 | 0.04 | ||||||
| 640 | 0.08 | 527 | 0.05 | ||||||||
| 702 | 0.04 | ||||||||||
| 752 | 0.02 | ||||||||||
| PB1 | 0.14 | NA | NA | ||||||||
| PA | 0.28 | NA | NA | ||||||||
| NP | 0.14 | NA | NA | ||||||||
| M1 | 0.29 | ||||||||||
| M2 | 0.83 | ||||||||||
| NS1 | 0.50 | ||||||||||
| NS2 | 0.31 | 46 | 0.99 | 46 | 0.04 | ||||||
| 49 | 0.99 | ||||||||||
| 68 | 0.99 | ||||||||||
| 116 | 0.99 | ||||||||||
| PB1-F2 | 2.65 | ||||||||||
Spatial movement of H7N1 in northern Italy.
To investigate the spatial diffusion of H7N1 avian influenza viruses within northern Italy, we inferred Bayesian phylogenies for the HA and the eight concatenated gene segments, considering eight discrete regions in northern Italy that are well sampled in our data (Fig. 1 and 3; also see Fig. S4 in the supplemental material). To quantitatively identify geographical structure in these data, we measured the extent to which isolates clustered by geographical location using a phylogenetic trait association test (see Table S2). Significant population subdivision was observed among the LPAI viruses in all locations (P < 0.01; see Table S2) except Mantua (P = 0.11; see Table S2). In contrast, HPAI viruses exhibited no statistically significant geographic structuring by province, indicative of greater spatial mixing (P > 0.13), as expected during a rapidly spreading outbreak.
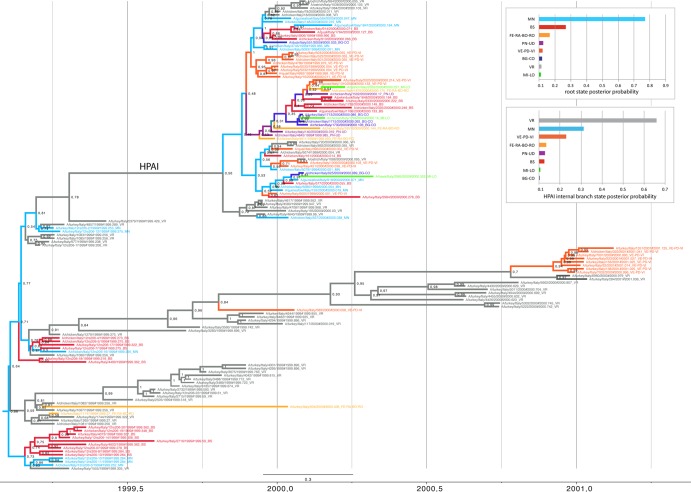
FIG 3.
MCC tree inferred for HA gene sequences of the Italian H7N1 viruses. Sequences are colored according to the province of origin. Light blue, Mantua; gray, Verona; red, Brescia; yellow, Ferrara-Ravenna-Bologna-Rovigo; orange, Venice-Padua-Vicenza; blue, Bergamo-Como; green, Milan-Lodi; violet, Pordenone-Udine. The numbers at branch points represent the probability of spatial states. The location state posterior probability distributions for the root and for the internal branch of the HPAI group are shown in the two graphs to the upper right of the phylogenetic tree.
The Bayesian MCC tree inferred for the HA segment (Fig. 3) identifies the Mantua and Verona provinces as the most important in the emergence and spatial dissemination of the H7N1 virus within northern Italy. Verona and Mantua have the highest root state posterior probabilities (0.56 and 0.67, respectively) (Fig. 3). These relatively low posterior probabilities, such that spatial origins are difficult to identify rigorously, are again suggestive of the rapid movement of viruses between geographical localities. The original LPAI H7N1 outbreak is most likely to have begun in Mantua (posterior probability, 0.67), with subsequent spread to Verona and Brescia and from Verona to the Padua and Rovigo provinces (Fig. 1 and 3), although the relatively low posterior probability values are indicative of rapid spread with little phylogenetic resolution. Although the HPAI virus appears to have originated first in Verona (posterior probability, 0.56), our phylogeographic analysis suggests that the poultry population in Verona has not played a major role in the spatial diffusion of this strain to other provinces. Instead, poultry from Mantua province most likely represented the key source for the spread of the HPAI virus to the six other provinces (Fig. 1 and 3). In addition, the second LPAI outbreak during August 2000 to February 2001 also is likely to have begun in Verona, with subsequent spread to Padua but not to other regions. Separate analyses of the LPAI and HPAI viruses gave results similar to those obtained from the complete data set (data not shown).
To quantitatively estimate the importance of poultry densities and geographical distances in the spatial dynamics of the virus, we encoded four potential predictors of viral dissemination between locations and fitted these models individually to the sequence data: (i) the poultry population in the province of origin, (ii) the poultry population in the province of destination, (iii) the product of the poultry population in the provinces of origin and destination, and (iv) the geographic distances between the different provinces. Bayes factor comparisons indicate that the spatial dissemination of the H7N1 viruses in Italian poultry are best described by the size of the poultry population in the region of destination (BF = 2.01 for the HA gene; BF = 3.99 for the complete genome; see Table S3 in the supplemental material), indicating viral migration from provinces of relatively low poultry population size (i.e., Mantua, Bergamo, and Venice) to provinces with large poultry populations (i.e., Verona) (see Table S3). Considering geographical distance as a predictor of viral movements also improved the marginal likelihood (BF = 1.41 for the HA gene; BF = 1.32 for the complete genome; see Table S3), indicating higher rates of viral dissemination among geographically closer provinces (see Table S3).
Intrahost genetic variation using NGS.
We used an ultradeep-sequencing approach to determine whether mutations associated with the HPAI phenotype were present at low levels in LPAI viral populations. Complete genomes were obtained for all 13 clinical samples included in the study, with a mean of 472,508 reads per sample (Table 1). The depth of coverage for the polymerase genes (PA, PB1, and PB2 segments) was lower (between approximately 1× and 1,000× per site) than that for the other five segments (between approximately 1,000 and 100,000× per site), indicating that there was a fragment length bias, where the longest fragments received less coverage (a coverage map is available for each sample in Fig. S5 and S6 in the supplemental material). We also obtained a smaller number of sequences (195,496) for sample 1744, but the coverage for the 5′ end of the PB1 segment was not sufficient to study population variation (Table 1; also see Fig. S5).
To assess the variation within each viral population and to identify potential HPAI progenitor viruses, the reads obtained from each sample were aligned to both the low-pathogenic and highly pathogenic influenza virus reference sequences. In particular, LPAI virus samples possessing genetic signatures typical of highly pathogenic viruses described previously (Table 3) and HPAI samples containing signature mutations of LPAI viruses were collated (Table 5). We identified one LPAI sample (4829-99), collected in Verona province immediately after the first identification of the HPAI virus, that contained a single read with an HA cleavage site associated with HPAI virus (Table 5). This sample contained a complete insertion of multiple basic amino acids at the HA cleavage site as well as a minority of variants (0.58%) with the mutation A166V in the M1 gene that is also typical of the HPAI virus (Table 5). Although at extremely low frequency, the occurrence of multiple HPAI-associated mutations suggests that it is unlikely to represent a sequencing artifact. However, none of the NGS-sequenced samples collected from birds infected with the low-pathogenicity virus possessed all of the amino acid signatures typical of the HPAI virus, and there was no evidence for the stepwise evolution of HPAI mutations at the cleavage site. A number of mutations were consistently found above the normal Illumina error level, such as A166V, found in six samples. Similar viral subpopulations were also present within some of the individuals. For example, samples 3675-99, 4295-99, and 3283-99 all have minority variants with mutations at site 745 of PB1, 349 of NP, and 166 of M1, with a 10-fold increase of variants with K at site 745 in sample 4295-99 collected in November 1999 relative to 3283-99 collected in September 1999. The HPAI samples also contained a number of low-frequency mutations typical of the LPAI viruses. Insertion of the multiple basic amino acids in the HA cleavage site was detected in all of the HPAI samples (Table 5).
TABLE 5.
Percentage of reads possessing mutations typical of the HPAI and LPAI virusesa
| Gene | Position | Amino acid |
% of reads with: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPAI signatures in LPAI samples |
LPAI signatures in HPAI samples |
||||||||||||||||
| HPAI | LPAI | 2732-99 | 3675-99 | 4295-99 | 3283-99 | 4829-99 | 1744-99 | 4618-99 | 4828-99 | 4827-99 | 4749-99 | 4708-99 | 4911-99 | 4756-99 | 4618-99 | ||
| PB2 | 398 | I | T | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PB1 | 154 | D | G | 0 | 0 | 0 | 0 | 0 | NAc | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PB1 | 216 | G | S | 0 | 0 | 0 | 0 | 0 | NAc | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PB1 | 745 | E | K | 0 | 0.34 | 2.8 | 0.23 | 0 | 0 | 0 | 1.12 | 0.68 | 0 | 0 | 0 | 0 | 0 |
| PA | 61 | T | I | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PA | 115 | K | N | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PA | 252 | K | E | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HA | 130 | A | T | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HA | 146 | T | A | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HA | 228 | E | A | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HA | 454 | T | A | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HA | 554 | R | K | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.14 | 0 | 0 | 0 | 0.87 | 0 |
| HA | 339 | Insb | 0 | 0 | 0 | 0 | 1 read | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| NP | 349 | A | T | 0.29 | 0.37 | 0.39 | 0.25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NP | 376 | N | S | 0 | 0.32 | 0 | 0 | 0 | 0 | 0.23 | 0.34 | 0.23 | 0.24 | 0.22 | 0.28 | 0.23 | 0.34 |
| NA | 173 | K | R | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.48 | 0 | 0.76 | 0 | 0 | 0 | 0.48 |
| M1 | 166 | A | V | 0.51 | 0.4 | 0.47 | 0.57 | 0.58 | 0.46 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NS1 | 136 | I | V | 0 | 0.29 | 0 | 0 | 0 | 0.20 | 0 | 0 | 0.17 | 0 | 0 | 0 | 0 | 0 |
| NS1 | 139 | N | D | 0 | 0 | 0 | 0 | 0 | 0.14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NS1 | 225 | NAc | R | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Reads were taken from the LPAI (2732-99, 3675-99, 4295-99, 3283-99, 4829-99, 1744-99) and HPAI (4618-99, 4828-99, 4827-99, 4749-99, 4708-99, 4911-99, 4756-99, 4618-99) samples after removal of mutations with frequencies below Illumina artifacts (P value < 0.05).
Insertion (Ins) refers to the presence of a complete insertion of basic amino acids (SRVR) at the HA cleavage site, which is typical of HPAI viruses. In this row, the results are expressed as the number of reads showing this insertion.
Minimum read depth to call variants (≤8) was not reached at these positions.
DISCUSSION
The continual reemergence of HPAI H7 avian influenza viruses presents a major concern for the poultry industry, as recently demonstrated by the 2012 H7N3 outbreak in Mexico and the 2013 Italian H7N7 outbreak (19, 69–71). However, efforts to control these outbreaks are undermined by a poor understanding of the genesis and evolution of HPAI viruses. Here, we employed newly developed deep-sequencing and Bayesian phylogenetic approaches to provide the most complete analysis of the evolution of high- and low-pathogenicity viruses from a single epidemic undertaken to date. We showed that all of the Italian LPAI and HPAI H7N1 isolates form a cluster that is distinct from all other Eurasian H7 isolates currently available (443 HA sequences), suggesting that the two epidemics resulted from a single viral introduction into Italy that occurred between December 1998 and March 1999. This inference is supported by the observation that the HPAI viruses have a common ancestor that dates from the period of circulation of the LPAI viruses (August-December 1999) and by the existence of minority genetic variants harboring signature mutations of the HP viruses in the ultradeep sequencing of the LP samples.
Although the original source of the Italian LPAI H7N1 outbreak has never been identified, some of the early isolates in our study lack the molecular signatures that frequently appear during the adaptation of viruses from aquatic bird hosts to domestic poultry (72, 73), including the stalk deletion in the NA protein and the additional glycosylation sites in the HA protein. Hence, it is possible that wild birds have been the source of this virus. However, the availability of background avian influenza virus sequences was too sparse to identify any closely related viruses in either domestic or wild birds.
Critically, the HPAI viruses form a distinct group within the Italian H7N1 viral population on the phylogenies inferred for all eight segments. This HPAI lineage rapidly diverged from the LPAI viruses, with 19 amino acid changes observed in all eight segments that were acquired over a time period of only 9 months. Despite their cocirculation in several Italian provinces, the phylogenies inferred for each segment show no evidence of reassortment between the HPAI and LPAI viruses, which continued to evolve as independent lineages. Among the low-pathogenicity AI populations, NGS technology identified minor variants with key mutations associated with the highly pathogenic viruses, including the insertion in the HA cleavage site. Although we employed a conservative approach to exclude sequencing errors, excluding PCR errors is more problematic. However, that identical mutations are identified in multiple samples (Table 5) reduces the likelihood that they are PCR errors.
These data confirm that the LPAI and HPAI epidemics shared a single common ancestor and suggest that some of the amino acid changes that characterize the HP virus cluster were already present with low frequency within several host viral populations from the beginning of the H7N1 epidemic, although the linkage between these amino acid sites could not be ascertained. Despite extensive sequencing, only complete insertions of the HA cleavage site were detected within the host viral populations of both the HP virus-infected farms and a single LP sample collected early during the HPAI epidemic. Previous studies have suggested that the insertion can be acquired through several possible evolutionary mechanisms, including (i) the duplication of purine triplets due to a transcription fault in the polymerase complex (14, 15); (ii) a stepwise accumulation of amino acid substitutions (14, 16); and (iii) nonhomologous recombination resulting in the insertion of a foreign nucleotide sequence (17, 18). The short length of the insertion (four amino acids) impedes efforts to distinguish between processes, as does the lack of deep-sequencing information from the index case of the HPAI epidemic wave. However, it was notable that our deep-sequencing data provided no evidence for the stepwise process of accumulation of mutations following polymerase complex errors. Hence, a disjunct evolutionary event, such as that caused by nonhomologous recombination, is a more likely causative mechanism for the generation of HPAI viruses.
Experimental studies also are required to determine if any of the mutations that were observed across the HPAI genome had important compensatory effects or contributed to the virulence of these viruses. Of note, some of these substitutions have been previously demonstrated to modulate avian influenza virus pathogenicity. In particular, the NS1 protein has a deletion of six amino acids at the C-terminal nucleolar localization signal (NoLS) (74), and two mutations (D139N and V136I) are located within or closely adjacent to the nuclear export signal (NES) (75). A recent study demonstrated that these mutations observed in the HPAI strain enhance cytoplasmic accumulation of NS1 and reduce beta interferon transcription, increasing viral pathogenicity (60). In addition, the C-terminal truncation promotes the transport of NS1 into the nucleoli, where it may favor viral transcription and translation or may alter the cell cycle to facilitate viral replication (60). Interestingly, some of the amino acid signatures identified in the Italian HPAI viruses have been previously described in other H7 HPAI strains, such as the mutation A146T in the HA protein, which was detected in some HPAI H7N3 viruses collected during a 2002 outbreak in Chile (17); the truncation at the NS1 protein, identified in most of the HPAI H7N3 viruses from Pakistan (31); and the additional glycosylation site at position 141 to 143 of the HA protein, found in both Pakistani (76, 77) and Dutch (6) isolates. The Dutch isolate is noteworthy because it has also been suggested to be under positive selection (6).
In September 1999, a few months before the identification of the HPAI variant, the LPAI-1 lineage evolved from the LPAI population, forming a distinct phylogenetic group in six of the eight phylogenies that is characterized by 16 amino acid changes. The closest relative of the LPAI-1 lineages appears to be the isolate A/chicken/Italy/3283/1999, which contains five of the 16 amino acid signatures of the LPAI-1 cluster. Isolate A/chicken/Italy/3283/1999 was detected only a few days prior to the first LPAI-1 virus (10 September 1999 and 27 September 1999, respectively), suggesting that these 11 mutations (PB2, L527M; PA, S190P, I257V; HA, Q86R, E113G, A143V, R148L; NA, V163M/L, M418I; NS1, G47C, P85T) were acquired over a short period of time. It is unclear whether this rapid accumulation of mutations reflects a strongly selectively driven event, as the virus responds to environmental changes, host switching, and/or control measures, or whether it was driven by a neutral population bottleneck and a subsequent founder effect. Although our Bayesian skyline analysis does not show any major reduction in population size (data not shown), the available epidemiological data do not provide sufficient resolution to trace the cause of this rapid evolution.
It is also notable that the time of the most recent common ancestor obtained for the HPAI variant dates to August 1999 to December 1999, which is coincident with the time scale of the emergence of the LPAI-1 group. Whether the parallel emergence of these lineages, which clearly differ in virulence, is due to common evolutionary or ecological pressure clearly merits further investigation. Our phylogeographic analysis shows that both lineages most likely emerged in Verona province, which has one of the highest densities of commercial poultry in Italy. Notably, these two viral lineages exhibited a degree of parallel evolution, including a glycosylation site in the HA protein and a truncation of the NS1 protein, suggesting that both viral populations have been subject to similar selection pressures despite their very different pathogenic effects. The presence of a potential additional glycosylation site in the HA gene and the deletion of the C terminus of the NS1 protein observed in the HPAI group have been previously demonstrated to confer replication and selective advantages on avian influenza viruses in the poultry hosts (31, 60, 78–80).
Our phylogeographic analysis also revealed striking spatial patterns of viral emergence and dissemination. The LPAI viruses were significantly more clustered by geographic region than the HPAI viruses. Although this could mean a slower spatial diffusion of LPAI, particularly in the provinces of Verona, Venice-Padua-Vicenza, and Brescia, the less frequent spatial clustering may also be related in part to the clinical manifestation of disease. In particular, LPAI may manifest with limited or unnoticeable signs of disease, such that it might be not promptly identified. Moreover, in contrast to HPAI isolates, LP strains were not subject to surveillance and control in 1999 to 2000 (81), and the notification of LP viruses was not mandatory at that time; therefore, cases of LPAI may have been unreported in some instances. In contrast, HPAI viruses exhibited little structuring by province, reflecting their rapid spread among all of the affected provinces, even those separated by large distances. Although the risk of infection has been found to be associated with proximity to an infected farm (39, 41, 44), the movement of contaminated people, trucks, equipment, and products can rapidly drive viral transmission between distant flocks during an HPAI outbreak (39, 42). Nevertheless, it is possible that we have missed the direct link between infected farms located in distant areas, as several outbreaks were identified only on the basis of results of serological, epidemiological, and clinical investigations (such that viruses and related genetic data were not available for these cases).
Although Verona province (located in the Veneto region) appears to be the most important in the initial emergence of the HPAI and LPAI-1 variants and the spread of LPAI viruses, the avian population in Mantua province (located in the Lombardia region) played the most important role in the spatial spread of the HPAI viruses to additional provinces. Indeed, limited dissemination of the virus toward other provinces was observed from Verona. These findings are in agreement with previous epidemiological studies (39, 40, 44) and possibly are explained by differences among the provinces in the control strategies employed at the time. In Veneto, a ban of restocking and a preemptive culling strategy were started 20 days earlier and on a greater number of farms than in Lombardia, reducing virus transmission within and outside the Veneto area. Hence, although the Mantua poultry population is markedly smaller than that of Verona and is not as likely to be the geographic origin of the HPAI virus, Mantua likely has acted as a bridge between the poultry population of Verona and the large poultry production of Brescia province, becoming essential for the westward spread of both the LPAI and HPAI viruses. This is supported in our phylogeographic analyses, which indicate that viral migration occurred from provinces of relatively lower poultry population size to those with the largest poultry populations.
Finally, although the precise role of the set of mutations identified in the H7 HPAI viruses needs to be elucidated, the identification of minority variants with HP key mutations in the LP samples that became prevalent in the HP viral population highlights the importance of implementing prompt and effective eradication measures to prevent the appearance of viruses with fitness advantages and unpredictable pathogenic properties. Furthermore, this study demonstrated the utility of deep sequencing to detect viral subpopulations carrying biologically relevant mutations. In particular, awareness of the existence of viral variants associated with differences in virulence, antiviral resistance, or pandemic potential may boost the laboratory community's responsiveness to influenza epidemics. Monitoring of such variants may be especially useful in the surveillance of low-pathogenic H7N9 avian influenza viruses in China, with the aim of predicting the emergence of variants resistant to antiviral drugs or with an increased capability of human-to-human transmission.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the European projects PREDEMICS (grant agreement no. 278433) and Epi-SEQ (research project supported under the 2nd Joint Call for Transnational Research Projects by EMIDA ERA-NET [FP7 project no. 219235]). E.C.H. is supported by an NHMRC Australia Fellowship. A.F. acknowledges the receipt of a fellowship from the OECD Cooperative Research Programme: Biological Resource Management for Sustainable Agricultural Systems in 2013.
We acknowledge Richard Orton (IBAHCM, University of Glasgow, Glasgow, United Kingdom) for his support with statistical analysis of data and Philippe Lemey (Rega Institute, Leuven, Belgium) for his useful suggestions in the application of phylogeographic analysis.
Footnotes
Published ahead of print 5 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03181-13.
REFERENCES
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615–619. 10.1038/nature06945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5:e1000520. 10.1371/journal.pcbi.1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fusaro A, Nelson MI, Joannis T, Bertolotti L, Monne I, Salviato A, Olaleye O, Shittu I, Sulaiman L, Lombin LH, Capua I, Holmes EC, Cattoli G. 2010. Evolutionary dynamics of multiple sublineages of H5N1 influenza viruses in Nigeria from 2006 to 2008. J. Virol. 84:3239–3247. 10.1128/JVI.02385-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattoli G, Fusaro A, Monne I, Coven F, Joannis T, El-Hamid HS, Hussein AA, Cornelius C, Amarin NM, Mancin M, Holmes EC, Capua I. 2011. Evidence for differing evolutionary dynamics of A/H5N1 viruses among countries applying or not applying avian influenza vaccination in poultry. Vaccine 29:9368–9375. 10.1016/j.vaccine.2011.09.127 [DOI] [PubMed] [Google Scholar]
- 6.Bataille A, van der Meer F, Stegeman A, Koch G. 2011. Evolutionary analysis of inter-farm transmission dynamics in a highly pathogenic avian influenza epidemic. PLoS Pathog. 7:e1002094. 10.1371/journal.ppat.1002094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baillie GJ, Galiano M, Agapow PM, Myers R, Chiam R, Gall A, Palser AL, Watson SJ, Hedge J, Underwood A, Platt S, McLean E, Pebody RG, Rambaut A, Green J, Daniels R, Pybus OG, Kellam P, Zambon M. 2012. Evolutionary dynamics of local pandemic H1N1/2009 influenza virus lineages revealed by whole-genome analysis. J. Virol. 86:11–18. 10.1128/JVI.05347-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes J, Allen RC, Baguelin M, Hampson K, Baillie GJ, Elton D, Newton JR, Kellam P, Wood JL, Holmes EC, Murcia PR. 2012. Transmission of equine influenza virus during an outbreak is characterized by frequent mixed infections and loose transmission bottlenecks. PLoS Pathog. 8:e1003081. 10.1371/journal.ppat.1003081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814–2822. 10.1128/JVI.79.5.2814-2822.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog. 9:e1003657. 10.1371/journal.ppat.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Röhm C, Horimoto T, Kawaoka Y, Süss J, Webster RG. 1995. Do hemagglutinin genes of highly pathogenic avian influenza viruses constitute unique phylogenetic lineages? Virology 209:664–670. 10.1006/viro.1995.1301 [DOI] [PubMed] [Google Scholar]
- 12.Banks J, Speidel EC, McCauley JW, Alexander DJ. 2000. Phylogenetic analysis of H7 haemagglutinin subtype influenza A viruses. Arch. Virol. 145:1047–1058. 10.1007/s007050050695 [DOI] [PubMed] [Google Scholar]
- 13.Banks J, Speidel ES, Moore E, Plowright L, Piccirillo A, Capua I, Cordioli P, Fioretti A, Alexander DJ. 2001. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch. Virol. 146:963–973. 10.1007/s007050170128 [DOI] [PubMed] [Google Scholar]
- 14.Horimoto T, Rivera E, Pearson J, Senne D, Krauss S, Kawaoka Y, Webster RG. 1995. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology 213:223–230. 10.1006/viro.1995.1562 [DOI] [PubMed] [Google Scholar]
- 15.Garcia M, Crawford JM, Latimer JW, Rivera-Cruz E, Perdue ML. 1996. Heterogeneity in the haemagglutinin gene and emergence of the highly pathogenic phenotype among recent H5N2 avian influenza viruses from Mexico. J. Gen. Virol. 77:1493–1504. 10.1099/0022-1317-77-7-1493 [DOI] [PubMed] [Google Scholar]
- 16.Spackman E, Senne DA, Davison S, Suarez DL. 2003. Sequence analysis of recent H7 influenza viruses associated with three different outbreaks in commercial poultry in the United States. J. Virol. 77:13399–13402. 10.1128/JVI.77.24.13399-13402.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suarez DL, Senne DA, Banks J, Brown IH, Essen SC, Lee CW, Manvell RJ, Mathieu-Benson C, Moreno V, Pedersen JC, Panigrahy B, Rojas H, Spackman E, Alexander DJ. 2004. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerging Infect. Dis. 10:693–699. 10.3201/eid1004.030396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasick J, Handel K, Robinson J, Copps J, Ridd D, Hills K, Kehler H, Cottam-Birt C, Neufeld J, Berhane Y, Czub S. 2005. Intersegmental recombination between the haemagglutinin and matrix genes was responsible for the emergence of a highly pathogenic H7N3 avian influenza virus in British Columbia. J. Gen. Virol. 86:727–731. 10.1099/vir.0.80478-0 [DOI] [PubMed] [Google Scholar]
- 19.Maurer-Stroh S, Lee RT, Gunalan V, Eisenhaber F. 2013. The highly pathogenic H7N3 avian influenza strain from July 2012 in Mexico acquired an extended cleavage site through recombination with host 28S rRNA. Virol. J. 10:139. 10.1186/1743-422X-10-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capua I, Alexander DJ. 2009. Avian influenza and Newcastle disease: a field and laboratory manual. Springer-Verlag, Milan, Italy [Google Scholar]
- 21.Hulse-Post DJ, Franks J, Boyd K, Salomon R, Hoffmann E, Yen HL, Webby RJ, Walker D, Nguyen TD, Webster RG. 2007. Molecular changes in the polymerase genes (PA and PB1) associated with high pathogenicity of H5N1 influenza virus in mallard ducks. J. Virol. 81:8515–8524. 10.1128/JVI.00435-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cauthen AN, Swayne DE, Sekellick MJ, Marcus PI, Suarez DL. 2007. Amelioration of influenza virus pathogenesis in chickens attributed to the enhanced interferon-inducing capacity of a virus with a truncated NS1 gene. J. Virol. 81:1838–1847. 10.1128/JVI.01667-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasilenko JL, Lee CW, Sarmento L, Spackman E, Kapczynski DR, Suarez DL. 2008. NP, PB1, and PB2 viral genes contribute to altered replication of H5N1 avian influenza viruses in chickens. J. Virol. 82:4544–4553. 10.1128/JVI.02642-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stech O, Veits J, Weber S, Deckers D, Schröer D, Vahlenkamp TW, Breithaupt A, Teifke J, Mettenleiter TC, Stech J. 2009. Acquisition of a polybasic hemagglutinin cleavage site by a low-pathogenic avian influenza virus is not sufficient for immediate transformation into a highly pathogenic strain. J. Virol. 83:5864–5868. 10.1128/JVI.02649-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abolnik C, Londt BZ, Manvell RJ, Shell W, Banks J, Gerdes GH, Akol G, Brown IH. 2009. Characterisation of a highly pathogenic influenza A virus of subtype H5N2 isolated from ostriches in South Africa in 2004. Influenza Other Respir. Viruses 3:63–68. 10.1111/j.1750-2659.2009.00074.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Londt BZ, Banks J, Alexander DJ. 2007. Highly pathogenic avian influenza viruses with low virulence for chickens in in vivo tests. Avian Pathol. 36:347–350. 10.1080/03079450701589134 [DOI] [PubMed] [Google Scholar]
- 27.Gaidet N, Cattoli G, Hammoumi S, Newman SH, Hagemeijer W, Takekawa JY, Cappelle J, Dodman T, Joannis T, Gil P, Monne I, Fusaro A, Capua I, Manu S, Micheloni P, Ottosson U, Mshelbwala JH, Lubroth J, Domenech J, Monicat F. 2008. Evidence of infection by H5N2 highly pathogenic avian influenza viruses in healthy wild waterfowl. PLoS Pathog. 4:e1000127. 10.1371/journal.ppat.1000127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam TT, Hon CC, Pybus OG, Kosakovsky Pond SL, Wong RT, Yip CW, Zeng F, Leung FC. 2008. Evolutionary and transmission dynamics of reassortant H5N1 influenza virus in Indonesia. PLoS Pathog. 4:e1000130. 10.1371/journal.ppat.1000130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vijaykrishna D, Bahl J, Riley S, Duan L, Zhang JX, Chen H, Peiris JS, Smith GJ, Guan Y. 2008. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 4:e1000161. 10.1371/journal.ppat.1000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cattoli G, Monne I, Fusaro A, Joannis TM, Lombin LH, Aly MM, Arafa AS, Sturm-Ramirez KM, Couacy-Hymann E, Awuni JA, Batawui KB, Awoume KA, Aplogan GL, Sow A, Ngangnou AC, El Nasri Hamza IM, Gamatié D, Dauphin G, Domenech JM, Capua I. 2009. Highly pathogenic avian influenza virus subtype H5N1 in Africa: a comprehensive phylogenetic analysis and molecular characterization of isolates. PLoS One 4:e4842. 10.1371/journal.pone.0004842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abbas MA, Spackman E, Swayne DE, Ahmed Z, Sarmento L, Siddique N, Naeem K, Hameed A, Rehmani S. 2010. Sequence and phylogenetic analysis of H7N3 avian influenza viruses isolated from poultry in Pakistan 1995-2004. Virol. J. 7:137. 10.1186/1743-422X-7-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Croville G, Soubies SM, Barbieri J, Klopp C, Mariette J, Bouchez O, Camus-Bouclainville C, Guérin JL. 2012. Field monitoring of avian influenza viruses: whole-genome sequencing and tracking of neuraminidase evolution using 454 pyrosequencing. J. Clin. Microbiol. 50:2881–2887. 10.1128/JCM.01142-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selleri M, Piralla A, Rozera G, Giombini E, Bartolini B, Abbate I, Campanini G, Rovida F, Dossena L, Capobianchi MR, Baldanti F. 2012. Detection of haemagglutinin D222 polymorphisms in influenza A(H1N1) pdm09-infected patients by ultradeep pyrosequencing. Clin. Microbiol. Infect. 10.1111/j.1469-0691.2012.03984.x [DOI] [PubMed] [Google Scholar]
- 34.Marangon S, Capua I, Rossi E, Ferrè N, Dalla Pozza M, Bonfanti L, Mannelli A. 2005. The control of avian influenza in area at risk: the Italian experience 1997–2005. In Koch G, Schrijver R. (ed), Avian influenza, prevention and control. Wageningen UR Frontis Series, Kluwer Academic Publisher, Wageningen, Germany [Google Scholar]
- 35.Capua I, Mutinelli F, Marangon S, Alexander DJ. 2000. H7N1 avian influenza in Italy (1999 to 2000) in intensively reared chickens and turkeys. Avian Pathol. 29:537–543. 10.1080/03079450020016779 [DOI] [PubMed] [Google Scholar]
- 36.Capua I, Marangon S, Cancellotti FM. 2003. The 1999-2000 avian influenza (H7N1) epidemic in Italy. Vet. Res. Commun. 27:123–127. 10.1023/B:VERC.0000014128.68876.31 [DOI] [PubMed] [Google Scholar]
- 37.Mulatti P, Bos MEH, Busani L, Nielen M, Marangon S. 2010. Evaluation of interventions and vaccination strategies for low pathogenicity avian influenza: spatial and space-time analyses and quantification of the spread of infection. Epidemiol. Infect. 138:813–824. 10.1017/S0950268809991038 [DOI] [PubMed] [Google Scholar]
- 38.Sartore S, Bonfanti L, Lorenzetto M, Cecchinato M, Marangon S. 2010. The effects of control measures on the economic burden associated with epidemics of avian influenza in Italy. Poultry Sci. 89:1115–1121. 10.3382/ps.2009-00556 [DOI] [PubMed] [Google Scholar]
- 39.Mannelli A, Ferrè N, Marangon S. 2006. Analysis of the 1999-2000 highly pathogenic avian influenza (H7N1) epidemic in the main poultry-production area in northern Italy. Prev. Vet. Med. 73:273–285. 10.1016/j.prevetmed.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 40.Mulatti P, Kitron U, Mannelli A, Ferré N, Marangon S. 2007. Spatial analysis of the 1999-2000 highly pathogenic avian influenza (H7N1) epidemic in northern Italy. Avian Dis. 51:421–424. 10.1637/7549-033106R.1 [DOI] [PubMed] [Google Scholar]
- 41.Busani L, Valsecchi MG, Rossi E, Toson M, Ferrè N, Pozza MD, Marangon S. 2009. Risk factors for highly pathogenic H7N1 avian influenza virus infection in poultry during the 1999-2000 epidemic in Italy. Vet. J. 181:171–177. 10.1016/j.tvjl.2008.02.013 [DOI] [PubMed] [Google Scholar]
- 42.Dorigatti I, Mulatti P, Rosà R, Pugliese A, Busani L. 2010. Modelling the spatial spread of H7N1 avian influenza virus among poultry farms in Italy. Epidemics 2:29–35. 10.1016/j.epidem.2010.01.002 [DOI] [PubMed] [Google Scholar]
- 43.Bos ME, Nielen M, Toson M, Comin A, Marangon S, Busani L. 2010. Within-flock transmission of H7N1 highly pathogenic avian influenza virus in turkeys during the Italian epidemic in 1999–2000. Prev. Vet. Med. 95:297–300. 10.1016/j.prevetmed.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 44.Mulatti P, Kitron U, Jacquez GM, Busani L, Mannelli A, Marangon S. 2010. Evaluation of the risk of neighbourhood infection of H7N1 highly pathogenic avian influenza in Italy using Q statistic. Prev. Vet. Med. 95:267–274. 10.1016/j.prevetmed.2010.04.005 [DOI] [PubMed] [Google Scholar]
- 45.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J. Virol. 83:10309–10313. 10.1128/JVI.01109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lunter G, Goodson M. 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21:936–939. 10.1101/gr.111120.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morelli MJ, Wright CF, Knowles NJ, Juleff N, Paton DJ, King DP, Haydon DT. 2013. Evolution of foot-and-mouth disease virus intra-sample sequence diversity during serial transmission in bovine hosts. Vet. Res. 44:12. 10.1186/1297-9716-44-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 51.Wilgenbusch JC, Swofford D. 2003. Inferring evolutionary trees with PAUP*. Curr. Protoc. Bioinformatics Chapter 6:Unit 6.4. 10.1002/0471250953.bi0604s00 [DOI] [PubMed] [Google Scholar]
- 52.Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- 53.Pond SL, Frost SD. 2005. A simple hierarchical approach to modeling distributions of substitution rates. Mol. Biol. Evol. 22:223–234. 10.1093/molbev/msi009 [DOI] [PubMed] [Google Scholar]
- 54.Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26:2455–2457. 10.1093/bioinformatics/btq429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8:e1002764. 10.1371/journal.pgen.1002764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185–1192. 10.1093/molbev/msi103 [DOI] [PubMed] [Google Scholar]
- 58.Parker J, Rambaut A, Pybus OG. 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect. Genet. Evol. 8:239–246. 10.1016/j.meegid.2007.08.001 [DOI] [PubMed] [Google Scholar]
- 59.Suchard MA, Weiss RE, Sinsheimer JS. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18:1001–1013. 10.1093/oxfordjournals.molbev.a003872 [DOI] [PubMed] [Google Scholar]
- 60.Keiner B, Maenz B, Wagner R, Cattoli G, Capua I, Klenk HD. 2010. Intracellular distribution of NS1 correlates with the infectivity and interferon antagonism of an avian influenza virus (H7N1). J. Virol. 84:11858–11865. 10.1128/JVI.01011-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569. 10.1146/annurev.biochem.69.1.531 [DOI] [PubMed] [Google Scholar]
- 62.Wiley DC, Wilson IA, Skehel JJ. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378. 10.1038/289373a0 [DOI] [PubMed] [Google Scholar]
- 63.de Wit E, Munster VJ, van Riel D, Beyer WE, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. 2010. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J. Virol. 84:1597–1606. 10.1128/JVI.01783-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaverin NV, Rudneva IA, Govorkova EA, Timofeeva TA, Shilov AA, Kochergin-Nikitsky KS, Krylov PS, Webster RG. 2007. Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies. J. Virol. 81:12911–12917. 10.1128/JVI.01522-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee MS, Chen MC, Liao YC, Hsiung CA. 2007. Identifying potential immunodominant positions and predicting antigenic variants of influenza A/H3N2 viruses. Vaccine 25:8133–8139. 10.1016/j.vaccine.2007.09.039 [DOI] [PubMed] [Google Scholar]
- 66.Wagner R, Wolff T, Herwig A, Pleschka S, Klenk HD. 2000. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: a study by reverse genetics. J. Virol. 74:6316–6323. 10.1128/JVI.74.14.6316-6323.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bush RM, Bender CA, Subbarao K, Cox NJ, Fitch WM. 1999. Predicting the evolution of human influenza A. Science 286:1921–1925. 10.1126/science.286.5446.1921 [DOI] [PubMed] [Google Scholar]
- 68.Holmes EC, Lipman DJ, Zamarin D, Yewdell JW. 2006. Comment on “Large-scale analysis of avian influenza isolates.” Science 313:1573. 10.1126/science.1131729 [DOI] [PubMed] [Google Scholar]
- 69.ProMED-mail. Accessed 26 October 2012 Avian influenza (54): Mexico (JA) high path H7N3, poultry. Archive no. 20120916.1295933. http://www.promedmail.org
- 70.Centers for Disease Control and Prevention (CDC). 2012. Notes from the field: highly pathogenic avian influenza A (H7N3) virus infection in two poultry workers–Jalisco, Mexico, July 2012. MMWR Morb. Mortal. Wkly. Rep. 61:726–727 [PubMed] [Google Scholar]
- 71.ProMED-mail. Accessed 21 August 2013 Avian influenza (83): Italy (Ferrara) poultry, HPAI, H7, OIE. Archive no. 20130820.1890944. http://www.promedmail.org
- 72.Giannecchini S, Clausi V, Di Trani L, Falcone E, Terregino C, Toffan A, Cilloni F, Matrosovich M, Gambaryan AS, Bovin NV, Delogu M, Capua I, Donatelli I, Azzi A. 2010. Molecular adaptation of an H7N3 wild duck influenza virus following experimental multiple passages in quail and turkey. Virology 408:167–173. 10.1016/j.virol.2010.09.011 [DOI] [PubMed] [Google Scholar]
- 73.Li J, zu Dohna H, Anchell NL, Adams SC, Dao NT, Xing Z, Cardona CJ. 2010. Adaptation and transmission of a duck-origin avian influenza virus in poultry species. Virus Res. 147:40–46. 10.1016/j.virusres.2009.10.002 [DOI] [PubMed] [Google Scholar]
- 74.Melén K, Kinnunen L, Fagerlund R, Ikonen N, Twu KY, Krug RM, Julkunen I. 2007. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J. Virol. 81:5995–6006. 10.1128/JVI.01714-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, Yamakita Y, Krug RM. 1998. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. U. S. A. 95:4864–4869. 10.1073/pnas.95.9.4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aamir UB, Naeem K, Ahmed Z, Obert CA, Franks J, Krauss S, Seiler P, Webster RG. 2009. Zoonotic potential of highly pathogenic avian H7N3 influenza viruses from Pakistan. Virology 390:212–220. 10.1016/j.virol.2009.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lebarbenchon C, Stallknecht DE. 2011. Host shifts and molecular evolution of H7 avian influenza virus hemagglutinin. Virol. J. 8:328. 10.1186/1743-422X-8-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suarez DL, Perdue ML. 1998. Multiple alignment comparison of the non-structural genes of influenza A viruses. Virus Res. 54:59–69. 10.1016/S0168-1702(98)00011-2 [DOI] [PubMed] [Google Scholar]
- 79.Suwannakhon N, Pookorn S, Sanguansermsri D, Chamnanpood C, Chamnanpood P, Wongvilairat R, Pongcharoen S, Niumsup PR, Kunthalert D, Sanguansermsri P. 2008. Genetic characterization of nonstructural genes of H5N1 avian influenza viruses isolated in Thailand in 2004-2005. Southeast Asian J. Trop. Med. Public Health 39:837–847 [PubMed] [Google Scholar]
- 80.Iqbal M, Essen SC, Xiao H, Brookes SM, Brown IH, McCauley JW. 2012. Selection of variant viruses during replication and transmission of H7N1 viruses in chickens and turkeys. Virology 433:282–295. 10.1016/j.virol.2012.08.001 [DOI] [PubMed] [Google Scholar]
- 81.CEC. 1992. Council Directive 92/40/EEC of 19 May 1992 introducing community measures for the control of avian influenza. Off. J. Eur. Comm. L167:1–15 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.