ABSTRACT
While numerous viral microRNAs (miRNAs) expressed by DNA viruses, especially herpesvirus family members, have been reported, there have been very few reports of miRNAs derived from RNA viruses. Here we describe three miRNAs expressed by bovine foamy virus (BFV), a member of the spumavirus subfamily of retroviruses, in both BFV-infected cultured cells and BFV-infected cattle. All three viral miRNAs are initially expressed in the form of an ∼122-nucleotide (nt) pri-miRNA, encoded within the BFV long terminal repeat U3 region, that is subsequently cleaved to generate two pre-miRNAs that are then processed to yield three distinct, biologically active miRNAs. The BFV pri-miRNA is transcribed by RNA polymerase III, and the three resultant mature miRNAs were found to contribute a remarkable ∼70% of all miRNAs expressed in BFV-infected cells. These data document the second example of a retrovirus that is able to express viral miRNAs by using embedded proviral RNA polymerase III promoters.
IMPORTANCE Foamy viruses are a ubiquitous family of nonpathogenic retroviruses that have potential as gene therapy vectors in humans. Here we demonstrate that bovine foamy virus (BFV) expresses high levels of three viral microRNAs (miRNAs) in BFV-infected cells in culture and also in infected cattle. The BFV miRNAs are unusual in that they are initially transcribed by RNA polymerase III as a single, ∼122-nt pri-miRNA that is subsequently processed to release three fully functional miRNAs. The observation that BFV, a foamy virus, is able to express viral miRNAs in infected cells adds to emerging evidence that miRNA expression is a common, albeit clearly not universal, property of retroviruses and suggests that these miRNAs may exert a significant effect on viral replication in vivo.
INTRODUCTION
MicroRNAs (miRNAs) are a diverse class of ∼22-nucleotide (nt) regulatory RNAs that are expressed by all known multicellular eukaryotes (1). Cellular miRNAs are normally transcribed by RNA polymerase II (Pol II) to give rise to a long pri-miRNA that is cleaved in the nucleus by the microprocessor complex, consisting of the RNase III enzyme Drosha and the RNA-binding cofactor DGCR8, to release the ∼60-nt pre-miRNA intermediate, which contains an ∼22-bp imperfect stem with an ∼2-nt 3′ overhang (2). The pre-miRNA is then exported to the cytoplasm, where it is cleaved by a second RNase III enzyme, Dicer, which removes the terminal loop, leaving a second ∼2-nt 3′ overhang. One strand of the resultant miRNA duplex intermediate is then incorporated into the RNA-induced silencing complex (RISC), where it acts as a guide RNA to direct RISC to complementary mRNAs (2). The key to mRNA target recognition is complementarity to nucleotides 2 through 8 of the miRNA, the so-called seed sequence, and seed-mediated binding of RISC is sufficient to significantly inhibit mRNA function (1).
While cells express numerous miRNA species, several DNA viruses also express miRNAs that have been shown to play a role in immune evasion and can also regulate viral gene expression (3, 4). In particular, herpesviruses generally express multiple viral miRNAs, especially in chronically infected cells, and polyoma- and adenoviruses have also been shown to encode functional miRNAs. In contrast, analysis of numerous RNA viruses has so far failed to identify any viral miRNAs (5), with the exception of two retroviruses, i.e., bovine leukemia virus (BLV), which encodes five viral miRNAs (6, 7), and an avian leucosis virus, termed ALV-J, which was recently shown to express a single viral miRNA in infected cells (8). One reason that RNA viruses might have only rarely evolved the ability to express miRNAs as a way of downregulating the expression of host genes with antiviral potential is that the excision of a pre-miRNA from a pri-miRNA precursor, likely the viral genomic RNA in the case of RNA viruses, would result in the cleavage of that pri-miRNA by Drosha. This has the potential to result in a reduction in viral replication that would potentially exceed any enhancement caused by the viral miRNA, and this problem would become more extreme if several viral pre-miRNAs were excised from a single viral genome. Indeed, the ability of inserted pri-miRNA stem-loops to reduce viral titers in cis has been well documented in the case of retroviral vectors, although this reduction can largely be alleviated by knockdown of Drosha in the producer cells (9, 10).
As noted above, the retrovirus BLV does encode several viral miRNAs. However, in this case, each viral miRNA is initially transcribed by RNA polymerase III (Pol III) as a pre-miRNA that is directly exported to the cytoplasm for Dicer cleavage (6, 7). Importantly, while pre-miRNAs generally contain stems of ∼22 bp, pri-miRNA stems are ∼33 bp long, and Drosha cleavage is dependent on this longer stem length (11, 12). Therefore, BLV is able to express viral miRNAs, yet the BLV genomic RNA is not subject to cleavage by Drosha. In contrast, the single miRNA expressed by ALV-J has been reported to be excised by the canonical miRNA processing machinery (8), resulting in Drosha cleavage of some portion of the ALV-J genomic RNA and/or subgenomic envelope mRNA. Whether this results in a drop in the production of the ALV-J structural proteins is, however, currently unknown.
In this article, we present data demonstrating that bovine foamy virus (BFV), a member of the spumavirus subfamily of retroviruses (13), expresses three viral miRNAs at high levels in infected cells both in culture and in vivo. Interestingly, the BFV miRNAs, like the previously reported BLV miRNAs, are transcribed by Pol III and contain stems that are predicted to be too short for recognition by Drosha in the context of the BFV genomic RNA. These observations raise the possibility that a diverse range of retroviral species may have evolved the ability to express functional miRNAs in vivo.
MATERIALS AND METHODS
Molecular clones.
The complete BFV pri-miRNA sequence was amplified from DNA generated by random hexamer-primed reverse transcription (RT) of chronically BFV-infected total cellular RNA, using the forward primer 5′-GCCTAAGCTTGGGGTTCGGAGGATGGCTCA-3′ and the reverse primer 5′-ATCTGAATTCTCTAGTTATACATCTAAAAA-3′, and cloned into the HindIII and EcoRI sites of pGEM3Z. The EBV-BART-5 miRNA expression vector has been described previously (14). Luciferase-based reporter vectors were constructed by annealing target site oligonucleotides and ligating them into the XhoI and NotI sites in the 3′-untranslated region (3′ UTR) of the renilla luciferase gene (rluc) of psiCHECK2. The target sequences are complementary to miR-BF1-5p (5′-TTCGGAGGATGGCTCATCAAGC-3′), miR-BF1-3p (5′-TCCCTGAAGCCATATCCGAGGC-3′), miR-BF2-5p (5′-TCAGTAGAAAGACAGTACCTCGCC-3′), or miR-BF2-3p (5′-CAGGCGGTATGCTTTCTACTTTTT-3′).
Cell culture and virus propagation.
The BFV Riems isolate was initially propagated by cocultivation of infected and uninfected calf trachea cells (15). Subsequently, BFV was adapted to grow by cocultivation with Madin-Darby bovine kidney (MDBK) cells maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% fetal horse serum (Sigma). MDBK-derived BFV was subsequently serially passaged by cocultivation of BFV-infected and noninfected MDBK cells, and corresponding samples are designated chronically BFV-infected cells. In parallel, BFV, which is normally highly cell associated, was adapted to cell-free transmission by using MDBK cells in serial passages using cell-free culture supernatant rather than cell cocultivation. After 10 cell-free serial passages, BFV titers were ∼103 infectious particles/ml, and this stock was used to infect 106 MDBK cells (acute infection) at a multiplicity of infection of ∼0.01. BFV titers were determined using an MDBK-based indicator cell line containing the BFV long terminal repeat (LTR) promoter linked to an indicator gene, as previously described for primate foamy virus (16). The BFV LTR is activated by the BFV Tas transactivator upon infection, thus allowing the BFV titer to be determined readily. 293 and baby hamster kidney (BHK) cells were cultured in DMEM supplemented with 10% fetal bovine serum.
Peripheral blood leukocyte (PBL)-derived RNAs from experimentally BFV-infected calves.
Five male calves of the Holstein-Friesian breed (6 weeks old) that tested seronegative for BFV, BLV, and bovine viral diarrhea virus were used in this study. Animals were adapted to the housing and feeding conditions in the animal facilities of the National Veterinary Research Institute in Pulawy, Poland, before the start of the study. The study was approved by the local ethical commission and was performed in accordance with national regulations for animal experimentation.
Three animals were experimentally infected with the Polish BFV100 isolate (17), which is highly related to but distinct from the German BFV Riems isolate (15), by intravenous inoculation of 5 × 106 Cf2Th cells infected with BFV100. Two calves were inoculated with the same number of uninfected Cf2Th cells and served as negative controls, as described previously (18). Throughout the experiment, the control calves were kept isolated from the BFV-infected animals. Successful BFV infection in the three infected calves was confirmed by detection of BFV integrase gene DNA by nested PCR (19) and of BFV Gag-specific antibodies by enzyme-linked immunosorbent assay (ELISA) (18). Both control calves were negative in both assays, as expected. Six months after BFV infection, blood was collected from the jugular vein into tubes containing anticoagulant (EDTA-Na), hemolyzed with ice-cold H2O and 4% NaCl, and centrifuged. The PBLs were washed twice with phosphate-buffered saline (PBS), divided into aliquots of 5 × 106 cells, suspended in 0.75 ml of TRIzol (Invitrogen), and frozen at −70°C until RNA extraction. Extracted total RNA (10 ng) was then used for RT-PCR analysis as described below.
Deep sequencing of total small RNA.
Total RNA was isolated from BFV-infected MDBK cells by use of TRIzol. The RNA fraction containing ∼15- to 40-nt RNAs was isolated by polyacrylamide gel electrophoresis on 15% Tris-borate-EDTA (TBE)–urea gels (Bio-Rad), electroeluted from the excised gel slice by use of Gel Eluter (Hoefer), and processed using a TruSeq small RNA sample preparation kit (Illumina). Adapter-ligated small RNAs were reverse transcribed using SuperScript III (Life Technologies), amplified using GoTaq green PCR master mix (Promega) with the TruSeq 3′ indices, and sequenced on an Illumina HiSeq 2000 instrument. Initial reads were quality filtered with cassava 1.8.2.
RNA species of ≤200 nt were isolated from chronically BFV-infected MDBK cells by using a mirVana miRNA isolation kit (Ambion). To prepare the RNA for deep sequencing, the size-fractionated total RNA sample was first treated with calf intestinal phosphatase (CIP; New England BioLabs) in NEB buffer 3 at 37°C for 1 h and then precipitated using ethanol and sodium acetate (20). This treatment was designed to remove any 5′ triphosphates from the RNA. The RNA was then rephosphorylated with T4 polynucleotide kinase (T4 PNK; New England BioLabs) at 37°C for 1 h. RNAs of >25 nt were collected using Centri-Spin 40 columns (Princeton Separations) and sequenced with a TruSeq small RNA sample preparation kit as described above.
For the 50-bp single-end reads, adapter sequences were clipped, and reads of ≥15 nt were quality filtered using FASTX-toolkit v0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Read mates of ≥15 nt from the 50-bp paired-end sequencing were clipped and quality filtered using Trimmomatic v0.30 (21). Bowtie v.0.12.7 (22) was used to sequentially filter and assign reads to adapter sequences, the viral genome (accession no. JX307862.1), miRbase v.20 bovine miRNA hairpins (23), and Ensembl annotations of the UMD3.1 bovine noncoding RNAs and genome (24). While one mismatch was allowed for alignments to the viral and bovine RNA libraries, two mismatches were allowed when aligning to bovine genomic DNA.
Northern blotting.
Northern blotting was performed as previously described (25). Briefly, 25 μg of total RNA was fractionated in a 15% TBE-urea gel (Bio-Rad), transferred to a GeneScreen Plus hybridization transfer membrane (PerkinElmer), and UV cross-linked (Stratalinker; Stratagene). Membranes were prehybridized in ExpressHyb (Clontech) and then incubated at 37°C with 32P-end-labeled oligonucleotide probes complementary to miR-BF1-5p (5′-GCTTGATGAGCCATCCTCCGAA-3′), miR-BF1-3p (5′-GCCTCGGATATGGCTTCAGGGA-3′), miR-BF2-5p (5′-GGCGAGGTACTGTCTTTCTACTGA-3′), and miR-BF2-3p (5′-AGAAAGCATACCGCCTGACC-3′). Membranes were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 37°C and visualized by autoradiography.
Quantitative reverse transcription-PCR.
Custom TaqMan small RNA reverse transcription primers and PCR probes were ordered through Invitrogen. The target sequences were 5′-UUCGGAGGAUGGCUCAUCAAGC-3′ (for miR-BF1-5p) and 5′-UCAGUAGAAAGACAGUACCUCGCC-3′ (for miR-BF2-5p). Ten nanograms of total RNA was reverse transcribed on a programmable thermal controller (MJ Research) and amplified according to the manufacturer's protocol, using a StepOnePlus real-time PCR system (Applied Biosystems). Relative miRNA abundances in bovine cells were determined using the ΔΔCT method (26), with cellular U6 snRNA (assay ID 001973) as the reference.
To determine if BFV miRNA expression is Pol III dependent, 105 293 cells were plated in 24-well plates and treated with 50 μg/ml of α-amanitin (Sigma) the next day, as previously described (25). Briefly, 250 ng of each miRNA expression vector was transfected via calcium phosphate, with addition of α-amanitin or carrier at 2 h posttransfection. At 24 h posttransfection, cells were harvested with TRIzol (Ambion) and total RNA treated with RQ1 DNase (Promega) and then assayed as described above for miR-BF2-5p and EBV-BART-5-5p (assay ID 197237_mat), with RNU48 (assay ID 001006) as the reference. The mock sample was transfected with the parental expression vectors, lacking any miRNA sequence, and treated with carrier.
Luciferase indicator assays.
A total of 50 ng of each reporter plasmid was cotransfected with 500 ng of a pGEM3Z-based BFV-miRNA expression vector, using polyethylenimine (PEI), into 24-well plates containing 105 BHK cells per well, which had been plated the previous day. At 24 h posttransfection, lysates were harvested and luciferase activity measured using a dual-luciferase reporter assay system (Promega) on a TD-20/20 luminometer (Turner Designs). The ratio between the renilla luciferase (RLuc) and internal control firefly luciferase (FLuc) proteins was determined for each sample and normalized to the ratio for the parental psiCHECK2 vector.
RESULTS
Identification of three novel BFV microRNAs.
To identify potential BFV-encoded miRNAs, we performed deep sequencing of small RNAs present in newly BFV-infected MDBK cells as well as MDBK cells chronically infected with BFV. The chronically BFV-infected MDBK cells were derived by long-term cocultivation of BFV-infected and uninfected MDBK cells and were predicted to be essentially uniformly infected. In contrast, the acutely infected MDBK cells were generated by infection of MDBK cells at ∼0.01 infectious unit of BFV per cell, followed by cultivation for 72 h prior to RNA harvest. Therefore, only a small percentage, perhaps 1 to 5% of the MDBK cells, was likely to be infected at this time point. As shown in Fig. 1A, we detected three different small RNAs of BFV origin in acutely BFV-infected MDBK cells, one of which was recovered as two different isoforms differing by 1 nt at the 5′ end. These same BFV-derived small RNAs were also recovered from chronically BFV-infected cells, but, as expected, at far higher levels. The most highly expressed BFV small RNA, miR-BF2-5p, was found to contribute ∼52% of all small RNA reads in these chronically BFV-infected cells, while a second BFV-derived small RNA, miR-BF1-5p, contributed a further 19% of all small RNA reads (Fig. 1B). In contrast, the most highly expressed cellular miRNA, miR-21, comprised only ∼4% of all recovered small RNA reads. A third BFV-derived small RNA, miR-BF1-3p, contributed ∼1% of all recovered small RNA reads in the chronically infected cells (Fig. 1B), and no other small RNAs of BFV origin contributed significant numbers of sequence reads in either chronically or acutely infected cells (Fig. 1A).
FIG 1.
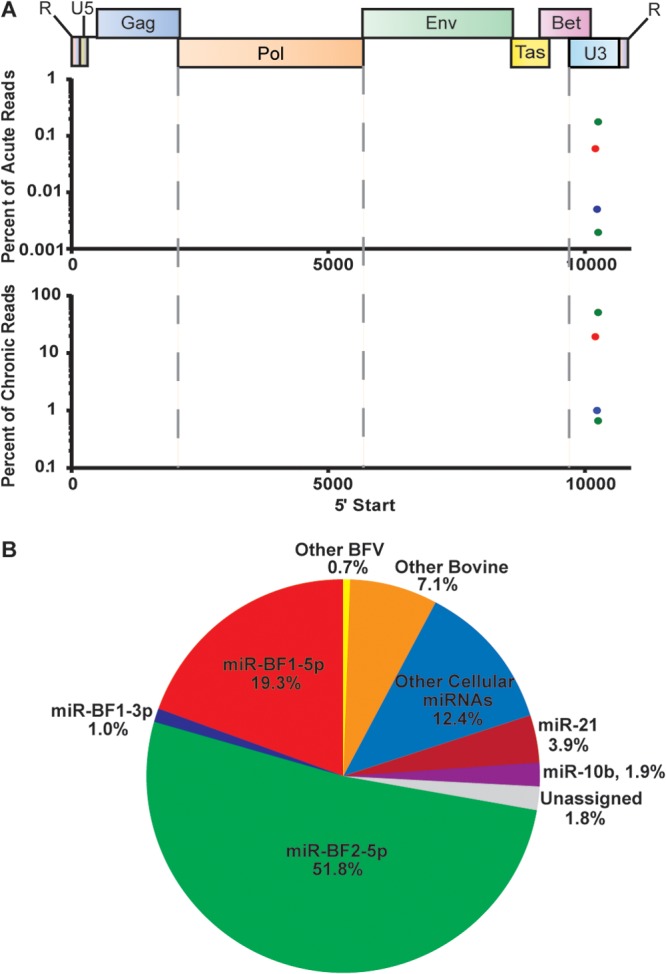
Alignment of small RNA reads to the BFV genome. (A) RNA reads that mapped to the viral genome are shown relative to their genomic origin (x axis). The number of reads is plotted as a percentage of the total number of small RNA deep-sequencing reads (y axis) from MDBK cells at 72 h postinfection (top) or from chronically infected MDBK cells (bottom). (B) Assignment of deep-sequencing reads in chronically BFV-infected MDBK cells.
Key characteristics of authentic viral miRNAs include the following: (i) they derive from a small number of discrete locations in the viral genome; (ii) they are close to the canonical miRNA size, which ranges from ∼20 nt to ∼25 nt; and (iii) they have a discrete 5′ end that results in a specific miRNA seed sequence (27). As shown in Fig. 1A, the BFV reads we obtained were indeed derived from one specific location in the BFV genome, located in the LTR U3 region, slightly 3′ to the end of the BFV bet gene. In the integrated BFV provirus, they would therefore be predicted to be present in two copies. Moreover, as shown in Fig. 2, the BFV reads we obtained all clustered at or near the predicted miRNA size of ∼22 nt. Finally, in Table 1, we provide a breakdown of the sequence reads derived from miR-BF1-5p, miR-BF1-3p, and miR-BF2-5p that contributed >2% of the reads obtained for that miRNA. As may be observed, essentially all the BFV reads obtained for each of these three viral miRNA species started with the same 5′ nucleotide. A small percentage (<1%) of the miR-BFV2-5p reads had the 5′ end displaced by one nucleotide from the more common 5′ start site. This subpopulation explains the additional miR-BF2-5p species shown in Fig. 1A. Therefore, these BFV-derived small RNA reads conform to the characteristics expected for authentic viral miRNAs. The miRNA sequence variation that was observed was almost entirely at the 3′ end, which plays a less important role than the seed sequence found near the miRNA 5′ end in promoting mRNA target recognition (1). In addition, we also saw some evidence for the addition of nontemplated bases at the viral miRNA 3′ ends, as has also been reported for some cellular miRNAs (28).
FIG 2.
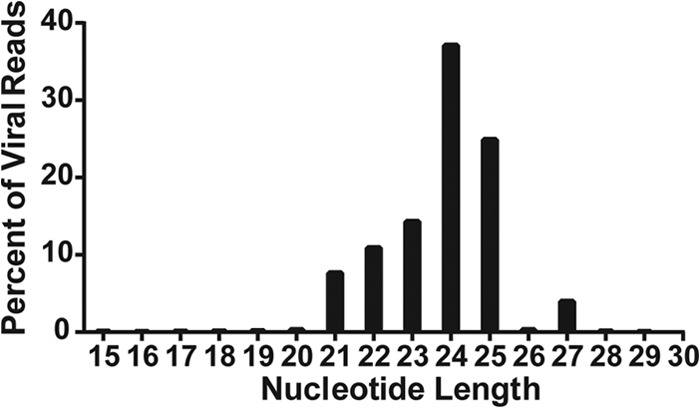
Small RNAs expressed by BFV cluster at the size expected for miRNAs. Reads from chronically BFV-infected MDBK cells mapping to the BFV genome for a given nucleotide length (x axis) are graphed as percentages of total reads aligning to the BFV genome (y axis).
TABLE 1.
Summary of BFV small RNAs recovered from chronically BFV-infected MDBK cellsa
| miRNA | Sequence | Length (nt) | Start position | End position | No. of reads | % of reads |
|---|---|---|---|---|---|---|
| miR-BF1-5p | UUCGGAGGAUGGCUCAUCAAGC | 22 | 11,190 | 11,211 | 878,086 | 32 |
| UUCGGAGGAUGGCUCAUCAAGCU | 23 | 11,190 | 11,212 | 836,116 | 30 | |
| UUCGGAGGAUGGCUCAUCAAG | 21 | 11,190 | 11,210 | 659,169 | 24 | |
| UUCGGAGGAUGGCUCAUCAAGCC | 23 | 11,190 | 11,212 | 127,154 | 5 | |
| miR-BF1-3p | UCCCUGAAGCCAUAUCCGAGGC | 22 | 11,224 | 11,245 | 86,127 | 58 |
| UCCCUGAAGCCAUAUCCGAGG | 21 | 11,224 | 11,244 | 28,224 | 19 | |
| UCCCUGAAGCCAUAUCCGAGGU | 22 | 11,224 | 11,245 | 6,142 | 4 | |
| UCCCUGAAGCCAUAUCCGAGGCU | 23 | 11,224 | 11,246 | 3,400 | 2 | |
| UCCCUGAAGCCAUAUCCGAGGCA | 23 | 11,224 | 11,246 | 3,185 | 2 | |
| miR-BF2-5p | UCAGUAGAAAGACAGUACCUCGCC | 24 | 11,246 | 11,269 | 3,514,215 | 47 |
| UCAGUAGAAAGACAGUACCUCGCCU | 25 | 11,246 | 11,270 | 2,052,969 | 28 | |
| UCAGUAGAAAGACAGUACCUCGCCC | 25 | 11,246 | 11,270 | 384,581 | 5 | |
| UCAGUAGAAAGACAGUACCUCGC | 23 | 11,246 | 11,268 | 369,154 | 5 | |
| UCAGUAGAAAGACAGUACCUCGCCUGU | 27 | 11,246 | 11,272 | 301,059 | 4 | |
| miR-BF2-3p | CCAGGCGGUAUGCUUUCUACU | 21 | 11,285 | 11,305 | 35 | 58 |
| CCAGGCGGUAUGCUUUCUACUU | 22 | 11,285 | 11,306 | 11 | 18 | |
| UCAGGCGGUAUGCUUUCUACU | 21 | 11,285 | 11,305 | 3 | 5 | |
| CCAGGCGGUAUGCUUUCUAC | 20 | 11,285 | 11,304 | 2 | 3 | |
| CCAGGCGGUAUGCUUUCUACUUUU | 24 | 11,285 | 11,308 | 2 | 3 |
RNA species that contributed ≥2% of the total reads for a given BFV miRNA are shown, with their positions relative to the reference BFV strain (accession no. JX307862.1) indicated by genome coordinates. Nontemplated nucleotides are indicated in bold.
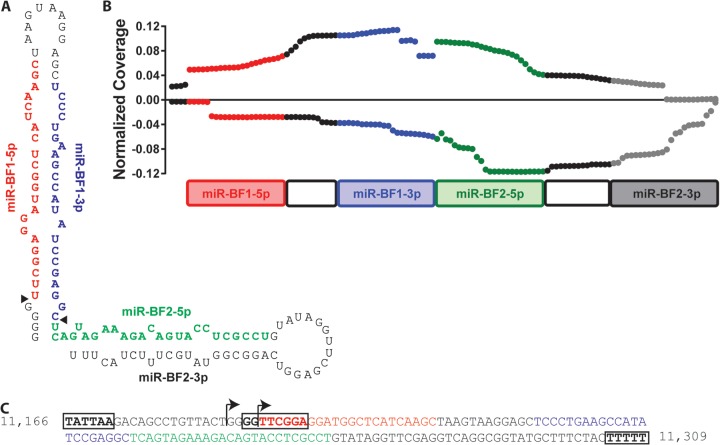
Mapping of the obtained reads onto the BFV genome showed that these small RNAs are closely adjacent to each other (Fig. 1A). In fact, miR-BF1-5p and miR-BF1-3p are separated by only 12 nt and show extensive complementarity, so they are predicted to fold into an RNA stem-loop structure (Fig. 3A). Moreover, miR-BF2-5p begins immediately after miR-BF1-3p ends and also has the potential to form a stem-loop structure (Fig. 3A). Further analysis of our deep-sequencing data indeed revealed a small number of reads that could represent the passenger strand for miR-BF2-5p, though this putative miR-BF2-3p RNA was present at a level ∼105 times lower than the highly expressed miR-BF2-5p (Table 1). Curiously, many of the reads obtained for miR-BF2-3p contained a nontemplated C residue at the 5′ end. This C is not present in any of the reported BFV sequences (15) and was not observed by sequencing of PCR products obtained from this region in infected MDBK cells, so its origin is unclear.
FIG 3.
Origin of BFV miRNAs. (A) Predicted sequence and RNA secondary structure of the BFV pri-miRNA. Black triangles indicate the sites of cleavage during BFV pri-miRNA processing to yield the two viral pre-miRNAs. (B) Paired-end deep sequencing of BFV sequences of ≥30 nt. The y axis shows the number of reads containing a given nucleotide divided by the total number of viral reads in the library. Values obtained with the forward sequencing primer are plotted on the positive y axis, with values obtained with the reverse primer plotted on the negative y axis. (C) Proviral DNA sequence with potential Pol III transcription factor binding sites and termination sequences boxed. Numbers represent the nucleotide locations within the reference BFV strain.
The ability of the small RNA reads of BFV origin to fold into two adjacent RNA stem-loop structures suggested that these might initially be transcribed as a single structured RNA containing two stem-loops (Fig. 3A). To address this possibility, we performed 50-bp paired-end deep sequencing of RNAs of <200 nt but >30 nt in length from chronically BFV-infected MDBK cells. As shown in Fig. 3B, we observed two major 5′ start sites, one coincident with the 5′ end of miR-BF1-5p and one located 4 nt 5′ to miR-BF1-5p. We also detected a third 5′ end, coincident with the 5′ end of miR-BF2-5p. At the 3′ end, we detected a major termination site after the third U of a run of 5 U residues and a minor termination site in a second U-rich sequence (5′-UUUCU-3′), located immediately 5′ to these 5 U residues (Fig. 3C). Runs of U residues, generally five or more, serve as terminators for Pol III, and a run of three Us at the end of a small RNA is characteristic of Pol III transcripts (29).
Interestingly, we observed that reads starting with a run of 4 G residues located 5′ to miR-BF1-5p invariably terminated at one of these two U stretches, while reads initiating at the first nucleotide of miR-BF1-5p generally terminated at the end of miR-BF1-3p. Runs initiating at the first nucleotide of miR-BF2-5p also ended at the distal U stretch. Based on this analysis, it appears that all three BFV small RNAs are initially transcribed as a single RNA that normally ends at the stretch of U residues at position ∼11,309 in the BFV genome and that folds into a structured pri-miRNA with two simple stem-loops, as shown in Fig. 3A. Because many full-length reads contained a 4-nt 5′ extension of 5′-GGGG-3′ (Fig. 3), while reads beginning at the first nucleotide of miR-BF1-5p generally ended at the last nucleotide of miR-BF1-3p, we favor the hypothesis that the BFV pri-miRNA is initially transcribed by Pol III, starting at the first G residue, at position 11,186, and is then processed by simultaneous cleavage after the 4-nt G run, at position 11,190, and between miR-BF1-3p and miR-BF2-5p, at position 11,245, to yield two canonical pre-miRNAs that are then exported to the cytoplasm and cleaved by Dicer to liberate the observed BFV miRNA species.
If this hypothesis is correct, then Northern analysis of RNAs derived from chronically BFV-infected MDBK cells should yield a single pri-miRNA, of ∼122 nt, recognized by probes specific for all four mature BFV miRNAs, and two pre-miRNAs, of 56 nt and 62 nt, recognized by both miR-BF1- and both miR-BF2-derived miRNAs, respectively. Indeed, as shown in Fig. 4, that is exactly what was observed. In particular, all four probes, including one specific for the miR-BF2-3p miRNA that was not recovered at significant levels as a mature miRNA (Table 1), recognized a single, ∼120-nt pri-miRNA. Moreover, the two miR-BF1 and two miR-BF2 miRNA pairs each recognized one of two distinct pre-miRNAs, with the latter, as expected, migrating slightly more slowly than the former. Finally, only three distinct mature miRNAs were recognized, although both miR-BF1-5p and, especially, miR-BF2-5p appeared to migrate as dimers, as expected based on the sequencing data (Table 1). Moreover, miR-BF2-5p appeared to be slightly larger than miR-BF1-5p or miR-BF1-3p, again as predicted by our sequencing data (Table 1).
FIG 4.
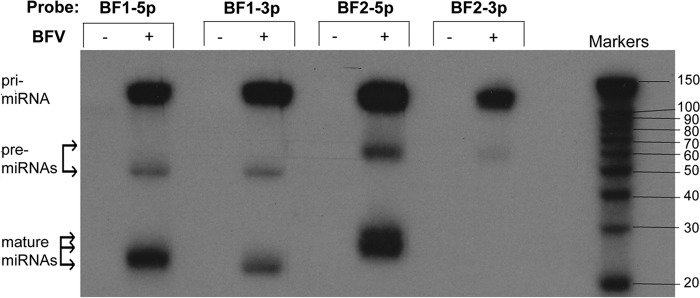
Northern blot analysis of RNAs from uninfected (−) and chronically BFV-infected (+) MDBK cells. The antisense oligonucleotide probes used for the indicated lanes are shown at the top. The predicted migration locations of the BFV pri-miRNA, the two pre-miRNAs, and the mature miRNAs are indicated.
BFV-derived microRNAs are transcribed by RNA polymerase III.
As noted above, the major putative BFV pri-miRNA species ends within a run of U residues and is only ∼122 nt long (Fig. 3A), and it is therefore predicted to be transcribed by cellular Pol III. Indeed, analysis of the BFV DNA sequence revealed the existence of a possible TATA box, located upstream of the pri-miRNA transcription start site, and a possible box B sequence (consensus sequence, 5′-GGTTCGANNCC-3′) that is closely adjacent to the pri-miRNA transcription start site (Fig. 3C). Canonical Pol III promoters often contain TATA boxes and/or box B sequences (30). However, we have not been able to identify a version of the box A sequence that is also found in many Pol III promoters.
To test whether the BFV miRNAs are indeed transcribed by RNA Pol III, we transfected 293 cells with a construct containing the entire BFV pri-miRNA sequence shown in Fig. 3C cloned into pGEM3Z, which does not contain any known eukaryotic Pol II or Pol III promoters. As a control, we also transfected 293 cells with an expression vector encoding the Epstein-Barr virus (EBV) miRNA miR-BART5, which is known to require transcription by Pol II (31). We then treated the transfected cells with α-amanitin, which is a specific inhibitor of Pol II-dependent transcription (6). At 24 h posttranscription, the cells were lysed, total RNA isolated, and expression of miR-BF2-5p and miR-BART5 measured by quantitative RT-PCR. Both viral miRNAs were readily detected in the untreated 293 cells (Fig. 5). However, in the α-amanitin-treated cells, miR-BART5 expression was almost entirely lost. In contrast, expression of miR-BF2-5p was still detected at levels comparable to those seen in the control cells. Therefore, these data confirm that the BFV miRNAs are indeed transcribed by cellular Pol III.
FIG 5.
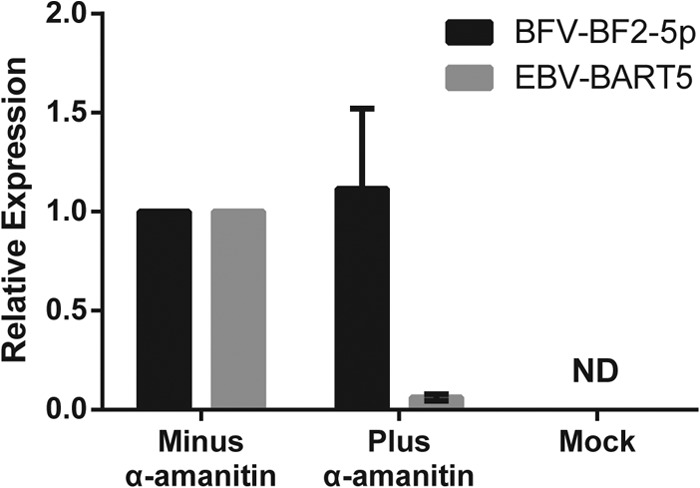
BFV miRNAs are transcribed by Pol III. Relative expression is shown for BFV miR-BF2-5p and EBV miR-BART5-5p in 293 cells transfected with the cognate miRNA expression vectors in the presence and absence of the Pol II inhibitor α-amanitin. Expression levels were determined by the ΔΔCT method (26), using RNU48 as the internal reference, with the carrier-treated cell value set at 1.0. “Mock” refers to 293 cells transfected with the parental vector lacking any miRNA sequence and treated with carrier. Averages for three independent experiments, with standard errors of the means, are shown. ND, no detectable amplification.
BFV microRNAs are functional.
While the BFV-derived small RNAs described above show all the characteristics of authentic miRNAs, we have not yet demonstrated that they are in fact able to repress the expression of an mRNA bearing complementary mRNA targets. To demonstrate that this is indeed the case, we utilized an indicator vector, psiCHECK2, which contains two independent luciferase genes, the firefly luciferase (fluc) and renilla luciferase (rluc) genes, under the control of two different promoters. We inserted a single artificial target sequence that was fully complementary to miR-BF1-5p, miR-BF1-3p, miR-BF2-5p, or miR-BF2-3p into the 3′ UTR of RLuc. We then cotransfected BHK cells with the pGEM3Z-based BFV pri-miRNA expression vector described above, along with one of the four BFV-miRNA indicator vectors or the parental psiCHECK2 vector. At 24 h postinfection, the cells were lysed and the levels of RLuc and the FLuc internal control determined. As shown in Fig. 6, we saw strong and specific downregulation of the RLuc gene present in the indicator plasmids specific for miR-BF1-5p, miR-BF1-3p, and miR-BF2-5p compared to either the psiCHECK2 negative control or the indicator specific for miR-BF2-3p, consistent with the very low expression of miR-BF2-3p determined by deep sequencing (Table 1) and Northern blotting (Fig. 4).
FIG 6.
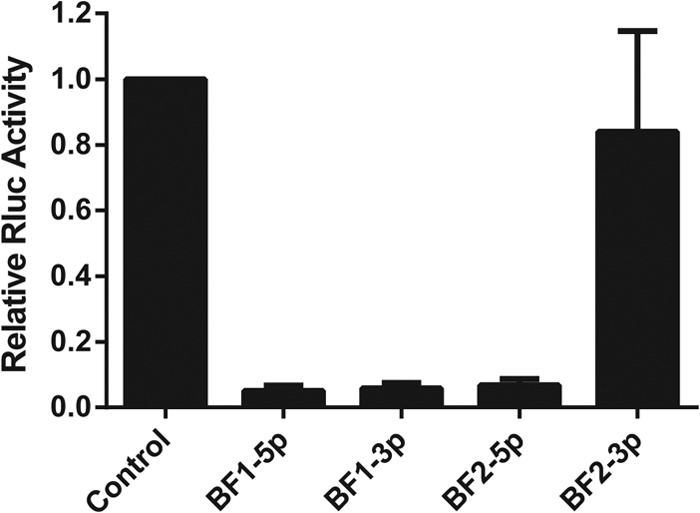
Functional analysis of BFV miRNAs. A single perfectly complementary target sequence for each BFV miRNA was inserted into the 3′ UTR of the RLuc indicator gene and cotransfected with a BFV pri-miRNA expression vector lacking any known exogenous Pol II or Pol III promoter. RLuc values are shown normalized to the FLuc internal control and to the level seen with the parental vector lacking an inserted miRNA binding site, which was set at 1.0. Averages for three independent experiments, with standard deviations, are indicated.
BFV microRNAs are expressed in vivo.
Our data demonstrate that BFV expresses three functional miRNAs in newly or chronically infected MDBK cell cultures. To test whether the same BFV miRNAs are also expressed in vivo, we used RT-PCR to test for the expression of miR-BF1-5p and miR-BF2-5p in PBLs drawn from three BFV-infected calves 6 months after experimental infection with BFV, as previously described (18). As controls, we used PBLs from two mock-infected control calves. Productive infection of the three BFV-infected calves, and lack of BFV infection of the two negative-control calves, was confirmed by nested PCR using primers specific for the BFV integrase gene (19) 1 month and 6 months after experimental infection and by detection of antibodies specific for the BFV Gag protein by ELISA exclusively in the three infected animals (18). As shown in Table 2, we did not detect any BFV miRNA expression in RNAs from uninfected animals, but we did detect miR-BF1-5p and miR-BF2-5p expression in the BFV-infected calves, at levels comparable to what was seen in newly BFV-infected MDBK cells, although we estimate that only ∼1% of the cells in the latter culture were actually infected by BFV at the time of RNA harvest. Therefore, we concluded that these BFV miRNAs are indeed expressed in infected cells in vivo.
TABLE 2.
BFV miRNAs are expressed in vivoa
| Sample | BFV infection | ΔΔCT value |
|
|---|---|---|---|
| BF1-5p | BF2-5p | ||
| Calf A PBLs | + | 1.00 | 1.00 |
| Calf B PBLs | + | 1.21 | 1.05 |
| Calf C PBLs | + | 2.02 | 1.97 |
| Calf D PBLs | − | 0.014 | ND |
| Calf E PBLs | − | 0.003 | ND |
| MDBK cells | − | 0.001 | ND |
| Acute | 2.68 | 10.7 | |
| Chronic | 3,880.00 | 10,390.00 | |
Relative expression of miR-BF1-5p and miR-BF2-5p in PBLs from three BFV-infected calves (+) (A, B, and C) and two uninfected calves (−) (D and E), as well as in uninfected, freshly infected, and chronically infected MDBK cells. Expression was calculated by the ΔΔCT quantitative PCR method (26), using the endogenous U6 snRNA as the reference, with the value for PBLs from calf A arbitrarily set to 1.0. ND, no detectable amplification.
DISCUSSION
Spumaviruses, more commonly referred to as foamy viruses, are an ancient group of generally nonpathogenic retroviruses that infect many mammalian species, with the notable exception of humans (13, 32). However, while no human foamy virus has been identified, there have been several reports documenting the zoonotic infection of individuals who came into contact with primates by a range of primate foamy virus (PFV) species (33, 34). Moreover, several PFV species replicate efficiently in human cells in culture, and retroviral vectors based on PFVs and other foamy viruses are under development for use in gene therapy or antigen delivery in humans and animals (35–37).
Among species that are susceptible to foamy viruses, infection is often common, and BFV has been detected in a considerable fraction of domesticated cattle in several different countries (18, 19). However, as is also the case for PFVs in either their natural hosts or zoonotically infected humans, there is little evidence that BFV exerts a significant deleterious effect in infected cows (13, 18).
Previous work has demonstrated that BLV, a deltaretrovirus that can induce B-cell lymphomas in cattle, expresses high levels of five viral miRNAs in BLV-transformed cells, and it has been proposed that these miRNAs can facilitate B-cell transformation (6, 7). Interestingly, these viral miRNAs were shown to be transcribed by Pol III, and it was predicted, based on computational analysis, that “a number of spumaviruses” might also encode Pol III-driven viral miRNAs (6). To test this hypothesis, we performed deep sequencing of small RNAs expressed in acutely or chronically BFV-infected cells, and our results indeed demonstrate that BFV expresses high levels of three distinct, fully functional viral miRNAs both in culture and in vivo. However, in contrast to BLV, which expresses each of its five viral miRNAs in the form of separate, ∼60-nt pre-miRNA stem-loop structures (6, 7), BFV encodes a pri-miRNA of ∼122 nt that must be processed to remove a 4-nt 5′ extension, and also cleaved between the two viral pre-miRNA species, to generate two distinct BFV pre-miRNAs (Fig. 3A and 4). At present, we do not know which nuclease(s) mediates this initial processing event. The finding, based on paired-end deep sequencing, that the BFV pri-miRNA precursor is apparently cleaved simultaneously on both sides of the miR-BF1 stem-loop, generating a pre-miRNA bearing a 2-nt 3′ overhang, may implicate the Drosha/DGCR8 microprocessor complex in this processing event, and this hypothesis is currently being tested. We note, however, that cleavage of cellular pri-miRNA stem-loops requires both an ∼33-bp stem and flanking single-stranded RNA sequences (11, 12), neither of which is present in the BFV pri-miRNA (Fig. 3A), so it is also possible that a different nuclease(s) is involved.
A comparison of the sequences of the BFV miRNAs defined in Table 1 shows that they are all fully conserved across the four reported BFV sequences, including sequences obtained for Chinese, American, and European BFV isolates (15). Indeed, the entire 122-nt BFV pri-miRNA sequence is fully conserved, with the exception of two nucleotides in the terminal loop of the miR-BF2 stem-loop that, based on previous work (31), are not predicted to affect miRNA processing or expression. Interestingly, comparison of the three highly expressed BFV miRNAs with known viral and cellular miRNAs revealed that the entire miR-BF1-5p seed sequence (nt 2 to 8) is conserved in miR-B5, which is expressed by bovine herpesvirus 1 (BHV-1) in infected cells (38). However, as the mRNA targets for miR-B5 are not currently known, this conservation is not currently informative as to miR-BF1-5p function.
An interesting aspect of the BFV miRNAs described in this report is that they all derive from a single locus in the U3 region of the BFV LTR, at a site located ∼50 nt 3′ to the end of the BFV bet gene (Fig. 1A). As such, they would be duplicated during reverse transcription, so the integrated BFV provirus would actually express these viral miRNAs from two independent locations. Perhaps more importantly, the Pol III transcription unit in the 5′ LTR would be located 5′ to both BFV Pol II transcription units, driven by the LTR promoter and the internal promoter located immediately 5′ to the viral tas gene, respectively (Fig. 1A). Therefore, this Pol III transcription unit would not be subject to transcriptional interference caused by these two viral Pol II-dependent promoters (39), and it is possible that the majority of BFV pri-miRNA transcription is actually derived from the Pol III transcription unit located in the 5′ LTR of BFV.
The observation that BLV, ALV-J, and BFV encode one or more viral miRNAs (6, 8), while HIV-1 and human T-cell leukemia virus type 1 (HTLV-1) do not (7, 27, 40), suggests either that miRNA expression has evolved independently in these three retroviral species or that HIV-1 and HTLV-1 (the latter, like BLV, is a deltaretrovirus) have lost the ability to express miRNAs. The former seems more likely, as BLV, ALV-J, and BFV express viral miRNAs that are initially transcribed by different RNA polymerases (Pol III for BLV and BFV and Pol II for ALV-J) and are subjected to different miRNA processing steps (Drosha independent for BLV and Drosha dependent for ALV-J). Regardless, it is now clearly of interest to examine whether other retroviral species, including other foamy viruses, encode miRNAs and whether, if they do, these exert any phenotypic effects in culture or in vivo. In this context, it is interesting that vectors based on a PFV isolate have been proposed as potential gene therapy vectors in humans (36, 41), and it appears to be important to determine whether PFV encodes any viral miRNAs and, if so, whether these are retained in these viral vectors. If this is the case, then it would be important to delete these miRNAs, or at least demonstrate that they do not exert any deleterious effects in transduced human cells. Similarly, while HIV-1 does not encode any miRNAs (27), so vectors based on this lentivirus do not face this concern, it would be important to examine any retrovirus, or indeed any virus, that has the potential for use as a gene therapy vector to confirm that these vectors do not encode any viral miRNAs.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grant R01-DA030086 and by Polish National Science Center grant NN308182438. A.W.W. received support from NIH grant T32-CA009111.
We thank Rebecca Skalsky for the miR-BART5 expression vector and Hal Bogerd for technical advice.
Footnotes
Published ahead of print 12 February 2014
REFERENCES
- 1.Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cullen BR. 2004. Transcription and processing of human microRNA precursors. Mol. Cell 16:861–865. 10.1016/j.molcel.2004.12.002 [DOI] [PubMed] [Google Scholar]
- 3.Grundhoff A, Sullivan CS. 2011. Virus-encoded microRNAs. Virology 411:325–343. 10.1016/j.virol.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen BR. 2013. MicroRNAs as mediators of viral evasion of the immune system. Nat. Immunol. 14:205–210. 10.1038/ni.2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, Doukas T, Paranjape S, Polacek C, dos Santos FB, Jalili R, Babrzadeh F, Gharizadeh B, Grimm D, Kay M, Koike S, Sarnow P, Ronaghi M, Ding SW, Harris E, Chow M, Diamond MS, Kirkegaard K, Glenn JS, Fire AZ. 2010. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 6:e1000764. 10.1371/journal.ppat.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kincaid RP, Burke JM, Sullivan CS. 2012. RNA virus microRNA that mimics a B-cell oncomiR. Proc. Natl. Acad. Sci. U. S. A. 109:3077–3082. 10.1073/pnas.1116107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosewick N, Momont M, Durkin K, Takeda H, Caiment F, Cleuter Y, Vernin C, Mortreux F, Wattel E, Burny A, Georges M, Van den Broeke A. 2013. Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma. Proc. Natl. Acad. Sci. U. S. A. 110:2306–2311. 10.1073/pnas.1213842110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Y, Smith LP, Nair V, Watson M. 2014. An avian retrovirus uses canonical expression and processing mechanisms to generate viral microRNA. J. Virol. 88:2–9. 10.1128/JVI.02921-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu YP, Vink MA, Westerink JT, Ramirez de Arellano E, Konstantinova P, Ter Brake O, Berkhout B. 2010. Titers of lentiviral vectors encoding shRNAs and miRNAs are reduced by different mechanisms that require distinct repair strategies. RNA 16:1328–1339. 10.1261/rna.1887910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandl A, Wittmann J, Jack HM. 2011. A facile method to increase titers of miRNA-encoding retroviruses by inhibition of the RNaseIII enzyme Drosha. Eur. J. Immunol. 41:549–551. 10.1002/eji.201040960 [DOI] [PubMed] [Google Scholar]
- 11.Zeng Y, Yi R, Cullen BR. 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24:138–148. 10.1038/sj.emboj.7600491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125:887–901. 10.1016/j.cell.2006.03.043 [DOI] [PubMed] [Google Scholar]
- 13.Kehl T, Tan J, Materniak M. 2013. Non-simian foamy viruses: molecular virology, tropism and prevalence and zoonotic/interspecies transmission. Viruses 5:2169–2209. 10.3390/v5092169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skalsky RL, Kang D, Linnstaedt SD, Cullen BR. 2014. Evolutionary conservation of primate lymphocryptovirus microRNA targets. J. Virol. 88:1617–1635. 10.1128/JVI.02071-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hechler T, Materniak M, Kehl T, Kuzmak J, Löchelt M. 2012. Complete genome sequences of two novel European clade bovine foamy viruses from Germany and Poland. J. Virol. 86:10905–10906. 10.1128/JVI.01875-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu SF, Linial ML. 1993. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J. Virol. 67:6618–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Materniak M, Bicka L, Kuzmak J. 2006. Isolation and partial characterization of bovine foamy virus from Polish cattle. Pol. J. Vet. Sci. 9:207–211 [PubMed] [Google Scholar]
- 18.Materniak M, Hechler T, Löchelt M, Kuzmak J. 2013. Similar patterns of infection with bovine foamy virus in experimentally inoculated calves and sheep. J. Virol. 87:3516–3525. 10.1128/JVI.02447-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romen F, Backes P, Materniak M, Sting R, Vahlenkamp TW, Riebe R, Pawlita M, Kuzmak J, Löchelt M. 2007. Serological detection systems for identification of cows shedding bovine foamy virus via milk. Virology 364:123–131. 10.1016/j.virol.2007.03.009 [DOI] [PubMed] [Google Scholar]
- 20.Umbach JL, Yen HL, Poon LL, Cullen BR. 2010. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio 1:e00204–10. 10.1128/mBio.00204-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 40:W622–W627. 10.1093/nar/gks540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozomara A, Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39:D152–D157. 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Garcia-Giron C, Gordon L, Hourlier T, Hunt S, Juettemann T, Kahari AK, Keenan S, Komorowska M, Kulesha E, Longden I, Maurel T, McLaren WM, Muffato M, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sheppard D, Sobral D, Taylor K, Thormann A, Trevanion S, White S, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Harrow J, Herrero J, Hubbard TJ, Johnson N, Kinsella R, Parker A, Spudich G, Yates A, Zadissa A, Searle SM. 2013. Ensembl 2013. Nucleic Acids Res. 41:D48–D55. 10.1093/nar/gks1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. 2010. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol. Cell 37:135–142. 10.1016/j.molcel.2009.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 27.Whisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, Stevenson M, Chen CH, Cullen BR. 2013. In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. mBio 4:e000193. 10.1128/mBio.00193-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman MA, Mani V, Hammond SM. 2011. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA 17:1795–1803. 10.1261/rna.2713611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arimbasseri AG, Rijal K, Maraia RJ. 2013. Transcription termination by the eukaryotic RNA polymerase III. Biochim. Biophys. Acta 1829:318–330. 10.1016/j.bbagrm.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dieci G, Conti A, Pagano A, Carnevali D. 2013. Identification of RNA polymerase III-transcribed genes in eukaryotic genomes. Biochim. Biophys. Acta 1829:296–305. 10.1016/j.bbagrm.2012.09.010 [DOI] [PubMed] [Google Scholar]
- 31.Cai X, Schäfer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. 10.1371/journal.ppat.0020023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meiering CD, Linial ML. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165–176. 10.1128/CMR.14.1.165-176.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, Prosser AT, Torimiro JN, Wright A, Mpoudi-Ngole E, McCutchan FE, Birx DL, Folks TM, Burke DS, Heneine W. 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363:932–937. 10.1016/S0140-6736(04)15787-5 [DOI] [PubMed] [Google Scholar]
- 34.Schweizer M, Falcone V, Gange J, Turek R, Neumann-Haefelin D. 1997. Simian foamy virus isolated from an accidentally infected human individual. J. Virol. 71:4821–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rethwilm A. 2007. Foamy virus vectors: an awaited alternative to gammaretro- and lentiviral vectors. Curr. Gene Ther. 7:261–271. 10.2174/156652307781369092 [DOI] [PubMed] [Google Scholar]
- 36.Erlwein O, McClure M. 2011. Gene delivery the key to gene therapy: the case for foamy viruses. Ther. Deliv. 2:681–684. 10.4155/tde.11.38 [DOI] [PubMed] [Google Scholar]
- 37.Schwantes A, Truyen U, Weikel J, Weiss C, Löchelt M. 2003. Application of chimeric feline foamy virus-based retroviral vectors for the induction of antiviral immunity in cats. J. Virol. 77:7830–7842. 10.1128/JVI.77.14.7830-7842.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glazov EA, Horwood PF, Assavalapsakul W, Kongsuwan K, Mitchell RW, Mitter N, Mahony TJ. 2010. Characterization of microRNAs encoded by the bovine herpesvirus 1 genome. J. Gen. Virol. 91:32–41. 10.1099/vir.0.014290-0 [DOI] [PubMed] [Google Scholar]
- 39.Cullen BR, Lomedico PT, Ju G. 1984. Transcriptional interference in avian retroviruses—implications for the promoter insertion model of leukaemogenesis. Nature 307:241–245. 10.1038/307241a0 [DOI] [PubMed] [Google Scholar]
- 40.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–276. 10.1038/nmeth746 [DOI] [PubMed] [Google Scholar]
- 41.Trobridge GD, Russell DW. 1998. Helper-free foamy virus vectors. Hum. Gene Ther. 9:2517–2525. 10.1089/hum.1998.9.17-2517 [DOI] [PMC free article] [PubMed] [Google Scholar]