Abstract
Recent evidence identified multiple Henipavirus species in Africa distinct from those in Southeast Asia and Australia. The reported fusion glycoprotein (F) sequence of the African Gh-M74a strain (GhV-F) is likely incorrect: a single base pair deletion near the N terminus results in multiple aberrancies. Rectifying this by adding single nucleotide insertions results in a GhV-F that now possesses a signal peptide, is efficiently cell surface expressed, exhibits syncytium formation when coexpressed with GhV-G protein, and mediates pseudotyped viral particle entry.
TEXT
One recent global survey of ∼5,000 bat specimens identified 66 new species of paramyxoviruses—more species than were present in the GenBank database prior to the publication (1). This is worrisome, as paramyxoviruses are unusually susceptible to cross-species transfers, exhibiting one of the highest rates of host switching among RNA viruses (2). In particular, Drexler et al. reported >20 distinct viral clades within the genus Henipavirus, with Nipah (NiV), Hendra (HeV), and Cedar (CedPV) viruses from Southeast Asia and Australia representing only three of those clades (1, 3). Therefore, the phylogenetic diversity of henipaviruses (HNVs) is far greater than was previously thought. NiV and HeV cause lethal zoonoses in humans; infections can result in mortality rates exceeding 90% (4–6).
HNVs also exhibit a broader species tropism than other paramyxoviruses, in part due to the highly conserved ephrin-B2 and -B3 receptors that these viruses use (7). Although documented HNV spillover events are currently limited to Southeast Asia and Australia, the diversity of HNVs in endemic bat species (Eidolon helvum) throughout Africa (1, 8) raises the possibility of incipient spillover events. Indeed, a recent analysis of almost 500 human sera from Cameroon detected the significant presence of cross-neutralizing anti-NiV antibodies in groups considered to be at high risk for zoonotic exposure (e.g., bat bush meat butchers) (O. Pernet, B. S. Schneider, S. M. Beaty, M. LeBreton, T. E. Yun, A. Park, T. T. Zachariah, T. A. Bowden, P. Hitchens, C. M. R. Kitchen, P. Daszak, J. Mazet, A. N. Freiberg, N. D. Wolfe, B. Lee, submitted for publication; O. Pernet, S. M. Beaty, M. Lebreton, B. Schneider, N. Wolfe, B. Lee, presented at the Negative Strand Virus Meeting, Granada, Spain, 2013). The multiple clades of African HNVs were determined from sequence analysis of an L gene segment. However, the almost-full-length genomic sequence of one African HNV from Ghana (clone GH-M74a) was reported (GenBank accession number HQ660129.1); we term this virus GhV, and the fusion and attachment envelope glycoproteins of GhV are termed GhV-F′ and GhV-G, respectively. GhV-F′ and GhV-G share only 26 to 56% sequence identity with their NiV/HeV counterparts.
Functional properties of GhV envelope glycoproteins.
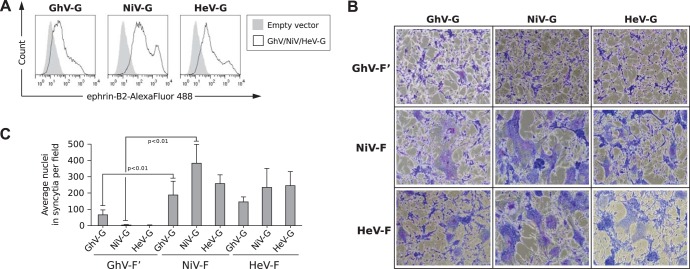
To examine the biological properties associated with GhV-F′ and GhV-G, we first codon optimized and expressed the GhV-F′ and -G genes from their predicted GenBank reference sequences (AFH96010.1 and AFH96011.1, respectively). Consistent with recently published data (9), cell surface GhV-G bound to soluble ephrin-B2 (Fig. 1A). Next, we examined the fusogenicity of GhV-F′ and GhV-G on human U87 glioblastoma cells, which express the HNV receptors ephrin-B2 and -B3 (10) and are highly permissive to HNV envelope (F/G)-mediated entry and syncytium formation (11). Curiously, while GhV-G cross-complemented NiV-F and HeV-F to form syncytia in permissive U87 cells (Fig. 1B), GhV-F′ exhibited little to no fusogenic activity when complemented with heterotypic NiV-G and HeV-G or even its homotypic GhV-G (Fig. 1B). Neither GhV-F′ (data not shown) nor GhV-G alone (see Fig. 5A) induced any syncytium formation. Thus, while GhV-G was fully functional for receptor binding and could trigger heterotypic HNV-F proteins, GhV-F′ appeared to be functionally compromised compared to NiV-F or HeV-F (Fig. 1C). For clarity, we designate this original GhV-F sequence GhV-F′.
FIG 1.
Functional properties of GhV envelope glycoproteins. (A) HNV-G proteins bind ephrin-B2. HEK293T cells were transfected with GhV-G, NiV-G, or HeV-G expression plasmids. Empty pcDNA3.1− vector was also transfected to serve as a negative control. At 27 h posttransfection, cells were processed for fluorescence-activated cell sorting (FACS) analysis using soluble ephrin-B2-(hu)Fc as the primary staining agent and Alexa Fluor 488-conjugated goat anti-human IgG (Fc) as the secondary antibody. (B) Fusogenicity of GhV-F′ and GhV-G proteins. Human U87 glioblastoma cells were transfected with plasmids encoding the indicated F (GhV-F′, NiV-F, and HeV-F) and G (GhV-G, NiV-G, and HeV-G) proteins at a 1:1 ratio, such that all homotypic and heterotypic combinations are represented. Cells were fixed and stained with Giemsa at 30 h posttransfection. Images were taken from 5 random fields for each condition. Representative images are shown. (C) Quantification of fusogenicity of GhV-F′ and GhV-G proteins. The images were quantified by manually counting the nuclei in syncytia in each field using ImageJ counting software. Results are presented as means and standard deviations (SD) from 5 fields. Significant differences were observed between GhV-F′ and NiV-F (P < 0.01; one-tailed Student t test).
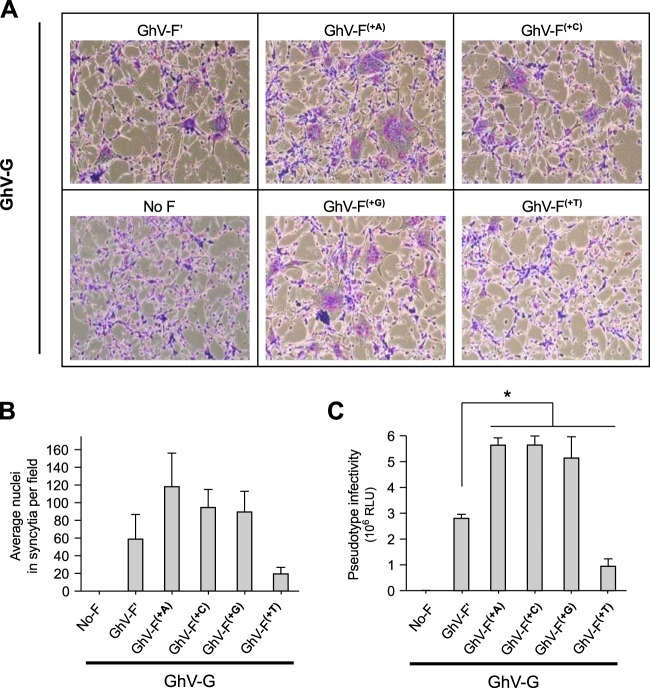
FIG 5.
Functionality of GhV-F(+1) variants in syncytium formation and virus infectivity assays. Human U87 glioblastoma cells were transfected in duplicate with plasmids encoding the GhV-F(+1) variants and GhV-G proteins at a 1:1 ratio. (A) Cells were fixed and stained with Giemsa at 30 h posttransfection. Representative images are shown. (B) Quantification of fusogenicity of GhV-F and GhV-G proteins. The images were quantified by manually counting nuclei in syncytia in each field using ImageJ software. Results are presented as means and SD from 10 fields per condition. (C) GhVpp infectivity. Purified GhVpp were produced as described for Fig. 4, and 5 μl was used to infect U87 cells. Infections were performed in triplicate. Infected cells were lysed and assayed for Renilla luciferase activity at 24 hpi. Activity was measured in relative luciferase units (RLU), and data are presented as means and SD. Statistical significance of differences between values for the GhV-F′ pseudotypes and the other groups was determined by a two-tailed Student t test followed by the Holm step-down procedure for multiple comparisons (*, P < 0.01).
The N terminus of GhV-F′ lacks a canonical signal peptide sequence.
Sequence alignment and homology modeling shows that GhV-F shares the major structural features associated with other HNV-F proteins, such as a cytoplasmic tail with the YXXL endocytic motif, HR1 and HR2 heptad repeat regions, a hydrophobic fusion peptide immediately following a basic cleavage site, and the conserved cysteines that are thought to link the F1 and F2 segments after protease cleavage (Fig. 2). However, the predicted N terminus of the GhV-F′ gene (GenBank no. AFH96010.1) contains ∼100 amino acids that are not conserved in the N termini of other HNV-F proteins (Fig. 2A and B), and bioinformatic analysis indicates the absence of any recognizable signal peptide motif (Fig. 3A and B; Table 1).
FIG 2.
Comparison of conserved features in NiV-F and GhV-F′. (Top) Comparison of conserved features across the full length of NiV-F and GhV-F′. The software PSIPRED was used to predict the secondary structure of the NiV-F and GhV-F′ protein sequences. A schematic of the two proteins highlights major features and conserved regions. The unconserved region of the N terminus in GhV-F′ consists of about 100 amino acids. (Bottom) Amino acid sequence of the full N-terminal region of GhV-F′. An extended version of Fig. 2A is shown, which includes the complete amino acid sequence of the unconserved N-terminal region of GhV-F′.
FIG 3.
A sequence correction in the N terminus of GhV-F′ creates a conserved signal peptide sequence. (A) Addition of one nucleotide in GhV-F′ creates a new start codon. Analysis of the protein and cDNA sequence in the unconserved region of GhV-F′ revealed an out-of-frame ATG (yellow box) close to an AT-rich region (red box). Insertion of one nucleotide (+N) at the indicated position (right after nucleotide 7,166 in the HQ660129.1 reference sequence) shifts the reading frame of F′, creates a new start codon from the upstream ATG (now position +1), and generates an amino acid insertion at position +4 (red X). (B) Signal peptide prediction for GhV-F′ and other HNV-F proteins. The first 70 amino acids of the indicated HNV-F proteins were bioinformatically analyzed for the presence of signal peptide sequences using four independent signal peptide prediction programs (Table 1). Results from the SignalP program are graphically presented, as described by Petersen et al. (13). SignalP is a neural network trained to predict signal peptides and provides three output scores for any query input sequence: the C score (raw cleavage site score [red]), S score (signal peptide score [green]), and Y score (combined cleavage site score [blue]). The C, S, and Y scores (y axis) for each amino acid (x axis) in the HNV-F query sequence are shown. In essence, the S score is high when the SignalP networks predict the sequence to be part of the signal peptide, while the C score is trained to recognize the SP cleavage site and is therefore highest at the first residue after the cleavage site. The Y score is a weighted score that helps identify the most likely cleavage site(s) when there are multiple C-score peaks. See http://www.cbs.dtu.dk/services/SignalP/output.php for details. (C) Alignment of HNV-F amino acid sequences. Insertion of a single nucleotide in GhV-F′ creates an isoleucine-/leucine-rich N terminus (dashed box) with a conserved signal peptide (green box). (D) Signal peptide prediction for the rectified GhV-F(+1) proteins GhV-F(+A), GhV-F(+C), GhV-F(+G), and GhV-F(+T). Signal peptide motifs were analyzed using SignalP software, as described for panel B.
TABLE 1.
Bioinformatic analysis of HNV-F proteins to determine the presence of a signal peptidea
| Protein | Location of SP detected with: |
|||
|---|---|---|---|---|
| SignalP | SPEPLip | PreDisi | Signal-BLAST | |
| GhV-F′ | Not found | Not found | 36b | Not found |
| GhV-F(+A) | 22–25 | 22 | 25 | 22 |
| GhV-F(+C) | 22–25 | 22 | 25 | 22 |
| GhV-F(+G) | 22–25 | 22 | 25 | 22 |
| GhV-F(+T) | 22–25 | 22 | 25 | 22 |
| NiV-FMalaysia | 24–28 | 24 | 26 | 23 |
| NiV-FBangladesh | 24–28 | Not found | 26 | 25 |
| HeV-F | 21–27 | 21 | 26 | 30 |
| CedPV-F | 23 | Not found | 22 | 21 |
The first 70 amino acids of the indicated HNV-F proteins were analyzed by four signal peptide prediction programs: SignalP, SPEPLip, PreDisi, and Signal-BLAST. Except for GhV-F′, at least three and often all four programs found signal peptide sequences at the N terminus of all HNV-F proteins examined, including the rectified GhV-F(+1). The predicted signal peptide cleavage sites (between 21 and 28) in all the HNV-F sequences examined are shown.
PreDisi gives a predicted cleavage site of 36 for GhV-F′, but the associated probability score that GhV-F′ has a signal peptide is zero.
Signal peptides (SPs) are usually located within the first 20 to 30 amino acids of the N termini of plasma membrane-associated or secreted proteins, and they are characterized by a preponderance of aliphatic and hydrophobic amino acids, such as leucines and isoleucines (12). Signal peptidases cleave SPs to generate the mature forms of the associated proteins. Although there is no canonical motif in SPs that predicts cleavage by signal peptidases, SP prediction programs can identify potential SP cleavage sites with high confidence via a combination of contextual cues (reference 13 and references therein). The median length of eukaryotic signal peptides is 22 amino acids (aa) (range, 11 to 55 aa) (12), and SPs were clearly present in the first ∼25 amino acids of the F proteins from NiVMalaysia, NiVBangladesh, HeV, and CedPV but not in GhV-F′ (Fig. 3B). Notably, four independent SP prediction programs failed to identify an SP within the first 70 aa of GhV-F′ (Table 1).
Closer inspection reveals an isoleucine-/leucine-rich region in GhV-F′ (105 to 115 aa) that aligns well with the predicted SPs in the other HNV-F proteins (Fig. 3C). Remarkably, the genomic sequence encoding residues 95 to 115 of GhV-F′ is 81% AT rich and includes an out-of-frame ATG start codon motif that is upstream of the isoleucine-/leucine-rich region. Poly(A/T) sequences are known to cause stuttering during PCR amplification (14), which can result in inadvertent insertions or deletions. Insertion of one nucleotide in silico, which resulted in an in-frame start codon from the nearby ATG, rectified GhV-F′ such that the N terminus of GhV-F(+1) aligned much better with the extant HNV-F proteins, both in terms of length and sequence similarity (Fig. 3A and C). Importantly, the rectified GhV-F(+1) was now predicted to have a bona fide SP (Fig. 3C; Table 1).
Expression and cleavage of rectified GhV-F(+1).
Depending on the identity of the inserted nucleotide, there are four possible versions of GhV-F(+1), each with a different amino acid at position +4 from the new initiator methionine (Fig. 3C). We generated all four versions of GhV-F(+1) and evaluated their expression and their F0 → F1+F2 cleavage levels, both in whole-cell lysates and in pseudotyped viral particles. Interestingly, while GhV-F′ is expressed in whole-cell lysate (Fig. 4A and B), the protein is not efficiently cleaved (Fig. 4C) or incorporated into pseudotyped virions (Fig. 4D and E). The minimal levels that are cleaved or incorporated suggest that an internal hydrophobic domain might be used inefficiently as a surrogate SP. The diminished incorporation of GhV-F′ into virions is consistent with inefficient trafficking through the secretory pathway due to the lack of any obvious SP sequence. GhV-F(+T) was poorly expressed in cell lysates (Fig. 4A and B) and on vesicular stomatitis virus (VSV)-ΔG-rLuc pseudotyped particles (Fig. 4D and E); however, cleavage was restored in the GhV-F(+A), GhV-F(+C), and GhV-F(+G) proteins (Fig. 4C and F), and all these proteins were more highly incorporated into pseudotyped virions than the fusion-compromised GhV-F′ (Fig. 4D and E).
FIG 4.
Expression of different GhV-F(+1) proteins in association with GhV-G and the resulting effect on pseudotyped virus formation and infectivity. Viral pseudotypes were generated on HEK293T cells transfected with plasmids encoding GhV-G and the GhV-F(+1) variants in a 1:1 ratio. Cells were simultaneously infected with a VSV-ΔG-rLuc virus complemented with VSV-G. At 24 h postinfection (hpi), pseudotype virus-containing media were clarified by low-speed centrifugation and then purified via ultracentrifugation through a 20% sucrose cushion. Purified GhV pseudotype particles (GhVpp) were resuspended in Opti-MEM, aliquoted, and stored at −80°C. (A and D) Expression of GhV-G and GhV-F′ or GhV-F(+1) variants in whole-cell lysates at 24 hpi (A) or in purified GhVpp (D). Primary anti-AU1 and anti-HA antibodies were used to detect AU1-tagged GhV-F and HA-tagged GhV-G, respectively. Fluorescent secondary antibodies were used for quantitative Western blotting, and the blots were imaged on a LI-COR Odyssey scanner. β-Tubulin is shown as a loading control. (B and E) The F0 and F1 bands from panels A and D were quantified using LI-COR Odyssey software, and expression levels for the GhV-F(+1) variants in whole-cell lysates (B) or GhVpp (E) were normalized to the expression of GhV-F′, which was set at 100%. (C and F) Cleavage efficiency of GhV-F(+1) variants in whole-cell lysates (C) or purified GhVpp (F) based on quantitative Western blot values. Data are presented as percent F cleavage, defined as [F1/(F0 + F1)] × 100. GhV-F(+T) was barely expressed in cell lysates and purified GhVpp, and thus, reporting cleavage efficiency values would be meaningless (asterisk). Results are representatives of 3 different experiments.
GhV-G levels of expression in cell lysates and incorporation into pseudotyped virions appeared to be inversely correlated with the expression levels of the GhV-F construct that was transfected in cis (Fig. 4A and D). The significance of this is unclear, as pseudotyped virions bearing GhV-F(+A), GhV-F(+C), and GhV-F(+G) were all more infectious than those bearing GhV-F′ or GhV-F(+T), despite the lower expression of GhV-G on the former (see Fig. 5C).
Functionality of rectified GhV-F(+1).
We evaluated the functionality of all four versions of GhV-F(+1) with a homotypic fusion assay. When coexpressed with GhV-G, GhV-F(+A) was the most fusogenic and generated the largest and most numerous syncytia. GhV-F(+C) and GhV-F(+G) were also fusogenic but less so than GhV-F(+A). GhV-F(+T) was the least fusogenic and appeared to be no better than GhV-F′ (Fig. 5A and B). These fusogenic phenotypes were reflected in VSV-based pseudotyped virus infection assays, where GhV-F(+T)/G and GhV-F′/G pseudotypes were also markedly less infectious than GhV-F(+A)/G, GhV-F(+C)/G, or GhV-F(+G)/G pseudotypes (Fig. 5C).
The functional defect of GhV-F′, but not its cause, was also recently recognized by Weis and colleagues (15). Our results implicate a single nucleotide deletion in the reference sequence for GhV-F′ as the cause of its dysfunction. Given the AT-rich context (Fig. 3A) of the putative deletion, and given that polymerase stuttering is most likely to result in an A or T deletion/insertion, we suggest that GhV-F(+A) be tentatively designated the working “wild-type” sequence for functional studies of this novel African henipavirus.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 AI069317 and a subproject grant from U54 AI065359 (PSWRCE) (to B.L.). S.B. was supported by UCLA Molecular Pathogenesis Training Grant T32-AI07323.
We thank members of the Lee lab for their critical comments on the manuscript.
Footnotes
Published ahead of print 12 February 2014
REFERENCES
- 1.Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Rasche A, Yordanov S, Seebens A, Oppong S, Adu Sarkodie Y, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stocker A, Carneiro AJ, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EK, Kruppa T, Franke CR, Kallies R, Yandoko ER, Herrler G, Reusken C, Hassanin A, Kruger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C. 2012. Bats host major mammalian paramyxoviruses. Nat. Commun. 3:796. 10.1038/ncomms1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kitchen A, Shackelton LA, Holmes EC. 2011. Family level phylogenies reveal modes of macroevolution in RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 108:238–243. 10.1073/pnas.1011090108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh GA, de Jong C, Barr JA, Tachedjian M, Smith C, Middleton D, Yu M, Todd S, Foord AJ, Haring V, Payne J, Robinson R, Broz I, Crameri G, Field HE, Wang LF. 2012. Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog. 8:e1002836. 10.1371/journal.ppat.1002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luby SP, Gurley ES, Hossain MJ. 2009. Transmission of human infection with Nipah virus. Clin. Infect. Dis. 49:1743–1748. 10.1086/647951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luby SP, Gurley ES. 2012. Epidemiology of henipavirus disease in humans. Curr. Top. Microbiol. Immunol. 359:25–40. 10.1007/82_2012_207 [DOI] [PubMed] [Google Scholar]
- 6.Marsh GA, Wang LF. 2012. Hendra and Nipah viruses: why are they so deadly? Curr. Opin. Virol. 2:242–247. 10.1016/j.coviro.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 7.Pernet O, Wang YE, Lee B. 2012. Henipavirus receptor usage and tropism. Curr. Top. Microbiol. Immunol. 359:59–78. 10.1007/82_2012_222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peel AJ, Sargan DR, Baker KS, Hayman DTS, Barr JA, Crameri G, Suu-Ire R, Broder CC, Lembo T, Wang L-F, Fooks AR, Rossiter SJ, Wood JLN, Cunningham AA. 2013. Continent-wide panmixia of an African fruit bat facilitates transmission of potentially zoonotic viruses. Nat. Commun. 4:2770. 10.1038/ncomms3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kruger N, Hoffmann M, Weis M, Drexler JF, Muller MA, Winter C, Corman VM, Gutzkow T, Drosten C, Maisner A, Herrler G. 2013. Surface glycoproteins of an African henipavirus induce syncytium formation in a cell line derived from an African fruit bat, Hypsignathus monstrosus. J. Virol. 87:13889–13891. 10.1128/JVI.02458-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakada M, Drake KL, Nakada S, Niska JA, Berens ME. 2006. Ephrin-B3 ligand promotes glioma invasion through activation of Rac1. Cancer Res. 66:8492–8500. 10.1158/0008-5472.CAN-05-4211 [DOI] [PubMed] [Google Scholar]
- 11.Palomares K, Vigant F, Van Handel B, Pernet O, Chikere K, Hong P, Sherman SP, Patterson M, An DS, Lowry WE, Mikkola HK, Morizono K, Pyle AD, Lee B. 2013. Nipah virus envelope-pseudotyped lentiviruses efficiently target ephrinB2-positive stem cell populations in vitro and bypass the liver sink when administered in vivo. J. Virol. 87:2094–2108. 10.1128/JVI.02032-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choo KH, Ranganathan S. 2008. Flanking signal and mature peptide residues influence signal peptide cleavage. BMC Bioinformatics 9(Suppl 12):S15. 10.1186/1471-2105-9-S12-S15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 14.Kieleczawa J. 2006. Fundamentals of sequencing of difficult templates—an overview. J. Biomol. Tech. 17:207–217 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2291785/ [PMC free article] [PubMed] [Google Scholar]
- 15.Weis M, Behner L, Hoffmann M, Kruger N, Herrler G, Drosten C, Drexler JF, Dietzel E, Maisner A. 2 December 2013. Characterization of African bat henipavirus GH-M74a glycoproteins. J. Gen. Virol. 10.1099/vir.0.060632-0 [DOI] [PubMed] [Google Scholar]