ABSTRACT
Human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) strains differ in their capacity to replicate in macrophages, but mechanisms underlying these differences are not fully understood. Here, we identify a highly conserved N-linked glycosylation site (N173 in SIV, corresponding to N160 in HIV) in the V2 region of the SIV envelope glycoprotein (Env) as a novel determinant of macrophage tropism and characterize mechanisms underlying this phenotype. Loss of the N173 glycosylation site in the non-macrophage-tropic SIVmac239 by introducing an N173Q mutation enhanced viral replication and multinucleated giant cell formation upon infection of rhesus macrophages, while the addition of N173 to SIVmac251 had the opposite effect. The removal of N173 in SIVmac239 enhanced CD4-independent cell-to-cell transmission to CCR5-expressing cells. SIVmac239 with N173Q mediated CD4-independent cell-cell fusion but could not infect CD4-negative cells in single-round infections. Thus, CD4-independent phenotypes were detected only in the context of cell-to-cell contact. Similar results were obtained in SIVmac251 with and without N173. N173 decreased the neutralization sensitivity of SIVmac251 but had no effect on the neutralization sensitivity of SIVmac239. The N173Q mutation had no effect on SIVmac239 binding to CD4 in Biacore assays, coimmunoprecipitation assays, and enzyme-linked immunosorbent assays (ELISAs). These findings suggest that the loss of the N173 N-linked glycosylation site increases SIVmac239 replication in macrophages by enhancing CD4-independent cell-to-cell virus transmission through CCR5-mediated fusion. This mechanism may facilitate the escape of macrophage-tropic viruses from neutralizing antibodies while promoting spreading infection by these viruses in vivo.
IMPORTANCE In this study, we identify a genetic determinant in the viral envelope (N173) that increases replication and spreading infection of SIV strains in macrophages by enhancing cell-to-cell virus transmission. This effect is explained by a novel mechanism involving increased cell-to-cell fusion in the absence of CD4, the primary receptor that normally mediates virus entry. The same genetic determinant also affects the sensitivity of these viruses to inhibition by neutralizing antibodies. Most macrophage-tropic HIV/SIV strains are known to be neutralization sensitive. Together, these findings suggest that this efficient mode of virus transmission may facilitate the escape of macrophage-tropic viruses from neutralizing antibodies while promoting spreading infection by these viruses to cells expressing little or no CD4 in vivo.
INTRODUCTION
Human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) strains differ in their ability to infect and replicate in macrophages. Macrophage tropism is determined primarily by the envelope glycoprotein (Env), which forms trimers composed of three noncovalently bound heterodimers of gp120 and gp41 subunits. The gp120 exterior subunit interacts with CD4 and a coreceptor, mainly CCR5 (1, 2), and the gp41 transmembrane subunit mediates virus-cell fusion. Env determinants of both HIV and SIV macrophage tropism have been shown to confer reduced CD4 dependence, a phenotype frequently associated with enhanced neutralization sensitivity to soluble CD4 (sCD4) and antibodies targeting the CD4 and CCR5 binding sites (3–7). These findings suggest that macrophage-tropic Envs might adopt structures with more exposed CD4 and/or CCR5 binding sites, a phenotype that allows these Envs to overcome the restriction to virus entry imposed by low levels of CD4 expressed on macrophages by facilitating Env interactions with the receptors and thereby to enhance membrane fusion and entry in cells expressing low levels of CD4 (8–11).
As the only viral protein on the virion surface, HIV/SIV Env is the main target of humoral immune responses and therefore the main determinant of neutralization sensitivity. Mechanisms that these viruses evolve to prevent antibody recognition include high sequence variability and heavily glycosylated Env (12–15). The variable regions of Env, in particular the V1V2 region, can shield other conserved epitopes on Env, including the CD4 and CCR5 binding sites (16–19). Glycans cover 50% of the molecule and form a silent face protecting Env from the binding of neutralizing antibodies (12, 20). Recent findings suggest more complex roles of the V1V2 region and glycans in this region in determining Env structure and immunogenicity. Of particular interest are studies from the recent RV144 vaccine trial, which found that the presence of V2-specific antibodies was the only variable correlating with protection. These results suggest that the V2 region may be a key immunogenic epitope that plays an important role in modulating neutralization sensitivity (21, 22). Series of broadly neutralizing antibodies, including 2G12, PG9, PG16, PGT120s, and PGT130s, have been identified to recognize and target specific glycan patterns on HIV Env (23–28), suggesting that glycans are also susceptible to neutralizing antibody binding. Together, these findings show that immune selection pressures induce mutations in the V2 region, including changes at glycosylation sites, which in turn may alter Env structure.
Comparative studies of amino acid sequences, together with functional studies, identified viral determinants for macrophage tropism in the V1V2, V3, V4, C1, C2, and C3 regions of HIV/SIV Env (4, 11, 29–39). Of particular importance are determinants in the V1V2 region, which can strongly influence viral entry, replication, and cell-cell spread in macrophages (11, 30, 31, 36). Furthermore, changes in V1V2 sequences have been associated with the emergence of macrophage-tropic simian/human immunodeficiency virus (SHIV) in vivo (37, 38). Structural and mathematical modeling studies suggest that the V1V2 loop may interact with other regions of Env, including the V3 loop, which constitutes part of the coreceptor binding site, and thereby may modulate Env structure and interactions with CCR5 (40–43). However, relationships between changes in the V1V2 region that influence macrophage tropism and Env interactions with CD4/CCR5 are poorly understood.
In a previous study, we identified two N-linked glycosylation sites in the V2 and C5 regions of SIV Env that modulate macrophage tropism and enhance the neutralization resistance of SIVmac251 (P.-J. Yen, M. E. Mefford, J. A. Hoxie, K. C. Williams, R. C. Desrosiers, and D. Gabuzda, submitted for publication). The N-glycosylation site in V2, N173, is at a position analogous to HIV N160 (HxB2 numbering), a critical residue for PG9 binding (24) that is localized near the trimer apex in the recent HIV Env trimer crystal and cryo-electron microscopy (cryo-EM) structures (44, 45). The N-glycosylation site in C5, N481, is located near a region of the CD4 binding site. Here, we examined the functional roles of these N-glycosylation sites in macrophage tropism in SIVmac251 and SIVmac239 and the mechanisms by which they mediate effects on viral replication in macrophages.
MATERIALS AND METHODS
Recombinant SIV Envs and viruses.
N173 and N481 mutations were introduced into Env-expressing plasmids in pSIVΔgpv (46) by site-directed mutagenesis. The recombinant Envs were then subcloned into full-length SIVmac239 proviruses (293-FL, provided by Ronald Desrosiers) (47), which are used to transfect 293T cells for the production of replication-competent viruses. Pseudotyped viruses were generated by cotransfecting 293T cells with pSIVΔgpv and a SIV-based Env− luciferase vector (46). For generating SIVmac251 recombinant clones, T173N and S481N mutations were introduced by site-directed mutagenesis into SIVmac251BK28 (48). The gp120 and N-terminal gp41 (residues 1 to 213) regions of these plasmids were then subcloned into pSIVΔgpv and 293-FL (Yen et al., submitted). These SIVmac251 recombinant viruses express gp41 sequences from SIVmac239 and differ from the SIVmac239 sequence at only 4 positions (D633K, D637E, I697V, and V699T in the N-terminal region of gp41). Viruses used for infection were normalized by reverse transcriptase activity or the SIV p27 antigen concentration (enzyme-linked immunosorbent assay [ELISA] from Advanced Bioscience Laboratories, Inc., Kensington, MD).
Viral replication in peripheral blood mononuclear cells and monocyte-derived macrophages.
Peripheral blood mononuclear cells (PBMC) were isolated from rhesus macaque peripheral blood (New England Primate Research Center) by Histopaque (Sigma) density centrifugation and activated in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin (P/S), 20 U/ml interleukin-2 (IL-2), and 1 μg/ml phytohemagglutinin (PHA-P) for 3 days. Activated PBMC were then maintained in RPMI supplemented with 10% FBS, 1% P/S, and 20 U/ml IL-2 prior to infection with replication-competent viruses (10 ng p27) in duplicate wells in 96-well plates. At 3 h postinfection (p.i.), viruses were removed by washing cells three times with RPMI. To obtain monocyte-derived macrophages (MDM), PBMC were cultured in RPMI containing 15% FBS, 10% human serum type AB, 1% P/S, and 20 ng/ml macrophage colony-stimulating factor (M-CSF) for 5 days in 96-well plates. Nonadherent cells were then removed by washing three times with RPMI. Adherent cells were cultured in RPMI supplemented with 15% FBS, 5% human serum type AB, 1% P/S, and 20 ng/ml M-CSF for two additional days before infection. For infection, viruses (10 ng p27) were cultured with MDM for 24 h and then removed by washing once with RPMI. The culture supernatant was collected twice a week, and the p27 concentration in the supernatant was measured by ELISA (Advanced Bioscience Laboratories, Inc., Kensington, MD).
Cell-to-cell transmission.
Cf2 canine thymocyte donor cells were infected with vesicular stomatitis virus G protein (VSV-G)-pseudotyped SIV generated in 293T cells cotransfected with plasmids expressing VSV-G envelope and full-length replication-competent SIV proviruses. Two days after infection, donor cells were washed and mixed with target Cf2-luc reporter cells (49) at a 1:1 ratio, directly or in transwells (24-well plates). Cf2-luc target cells were transfected to express different levels of rhesus CD4 and CCR5, and cell surface receptor levels were quantified by flow cytometry. Viral transmission to target cells was quantified by measuring luciferase activity in cell lysates 2 days after coincubation. A cell-cell fusion assay was performed in a similar format by using Env-expressing instead of SIV-infected donor cells.
Env expression and cell-cell fusion assays.
Recombinant Envs were expressed in 293T cells transfected with pSIVΔgpv. At 2 days posttransfection, cells were lysed, and Env expression was examined by Western blotting. Env levels on virions were analyzed by Western blotting of virions, which were normalized by p27 levels. For cell-cell fusion assays, 293T cells cotransfected with pSIVΔgpv and pLTR-Tat, a Tat-expressing plasmid, were incubated with Cf2-luc cells transiently expressing rhesus CD4 and CCR5. Expression of the reporter luciferase gene in Cf2-luc cells is under the control of the HIV-1 long terminal repeat (LTR). The ratio of 293T cells to Cf2-luc cells was 1:10. Ten hours after coincubation, cells were lysed, and luciferase activity was quantified as an indication of cell-cell fusion. 293T cells transfected with green fluorescent protein (GFP) served as a negative control for background luciferase activity.
Affinofile, Cf2-luc, and TZM-BL cell single-round infection assays.
Affinofile cells (50) were seeded at 3 × 104 cells per well into 96-well plates 1 day prior to induction. Cells were induced with 0, 0.6, and 0.8 ng/ml doxycycline (to induce CD4) and 0, 0.25, and 0.5 μM ponasterone (to induce CCR5) in a 3-by-3 matrix format for 21 h at 37°C. CD4 and CCR5 expression was analyzed by flow cytometry and quantified with QuantiBRITE PE (R-phycoerythrin; BD Biosciences). The induced cells were infected with the indicated pseudotyped viruses in the presence of 40 μg/ml DEAE-dextran for 16 h. The viruses were then removed by replacing the medium, and luciferase activity was measured 48 h later as an indication of infection.
For Cf2-luc and TZM-BL cell infections, cells were infected with replication-competent viruses (10 ng p27) in the presence of 15 μg/ml of DEAE-dextran. At 2 days p.i., luciferase activity in cell lysates was measured. Cf2-luc cells were transfected with CD4 and CCR5 expression plasmids 24 h before infection.
Neutralization assays.
Viruses were preincubated with serial dilutions of heat-inactivated SIVmac251 antiserum (NIH AIDS Research and Reference Reagent Program), mouse ascites containing monoclonal antibodies (provided by James Hoxie) (51), or sCD4 (Immunodiagnostics) at 37°C for 1 h. After preincubation, TZM-BL cells (provided by Norman Letvin) were added with DEAE-dextran (final concentration, 15 μg/ml). Two days later, cells were lysed, and luciferase activity was measured.
Generation of soluble gp120.
To generate recombinant soluble gp120 (sgp120) proteins, 293F cells were transfected with a His-tagged sgp120-expressing plasmid by using 293 Fectin (Invitrogen). At 3 days posttransfection, the supernatant of transfected cells was harvested, filtered through a 0.45-μm filter, mixed with a 1/10 volume of equilibration buffer (500 mM NaCl, 500 mM NaH2PO4 [pH 7.9]) and Superflow Ni-nitrilotriacetic acid (NTA) beads (Qiagen), and rotated overnight at 4°C. The solution was then loaded onto a Poly-Prep chromatography column (Bio-Rad). The column was washed with wash buffer (300 mM NaCl, 50 mM NaH2PO4 [pH 7.9]). After washing, sgp120 was eluted stepwise with elution buffer (300 mM NaCl and 50 mM NaH2PO4 [pH 7.9] containing 10, 20, 50, 100, or 250 mM imidazole). The eluted sgp120 fractions were analyzed by SDS-PAGE. The 50 mM and 100 mM imidazole fractions were concentrated by Amicon Ultra-4 centrifugal filter units (30 kDa) and resuspended in phosphate-buffered saline (PBS).
Coimmunoprecipitation (co-IP).
sgp120 (2 μg) was mixed with human CD4-Ig (2 μg) and placed onto a rotator at room temperature for 1 h. Protein G Plus-agarose (Santa Cruz) was then added and incubated at room temperature for another hour. After incubation, the protein-agarose complex was washed three times with PBS, and binding of sgp120 to human CD4-Ig was analyzed by Western blotting.
Biacore analysis.
Kinetic analysis was performed on a Biacore 3000 optical biosensor (General Electric), as previously described (52), with the following modifications. sCD4 was immobilized onto flow cells 2, 3, and 4 on a CM5 sensor chip to surface densities of ∼700, ∼1,400, and ∼2,100 response units, respectively, Flow cell 1 was activated and deactivated and used as a control for nonspecific binding and refractive index changes. Different concentrations of sgp120 were injected over all flow cells at a flow rate of 50 μl/min for 1.2 min. Each concentration was injected in triplicates, the order of the injections was randomized, and dissociation was measured at the end of each injection for 10 min. The binding surfaces were regenerated after each injection by 2 sequential injections of 25 and 10 μl of 10 mM glycine (pH 2.5). All procedures were done at 25°C by using standard HBS (HEPES buffer saline; GE) as a running buffer. The response from the reference flow cell was subtracted from the responses from all active surfaces. The association and dissociation phase data of triplicate injections were fitted simultaneously with BIAevaluation (version 3.2) software using a 1:1 Langmuir model of binding.
Cell-based ELISA.
CD4 binding to Env trimers was examined by using a cell-based ELISA as previously described (53), in which we measured binding of human CD4-Ig and SIVmac251 antiserum to Env trimers expressed on the cell surface. Briefly, cells from the human osteosarcoma cell line (HOS cells) cultured in a 96-well plate were transfected with SIV Env-expressing plasmids. Three days later, the cells were incubated with human CD4-Ig or with SIVmac251 antiserum for 30 min at room temperature. The cells were then washed, and horseradish peroxidase (HRP)-conjugated secondary antibody was added. For samples incubated with human CD4-Ig, we used goat anti-human HRP-conjugated polyclonal antibody as the secondary antibody. For samples incubated with SIVmac251 antiserum, we used HRP-conjugated protein G. Binding was quantitated after adding Western Lightning reagents by measuring luminescence. Binding to SIVmac251 antiserum was used to normalize CD4-Ig binding relative to Env cell surface expression levels.
RESULTS
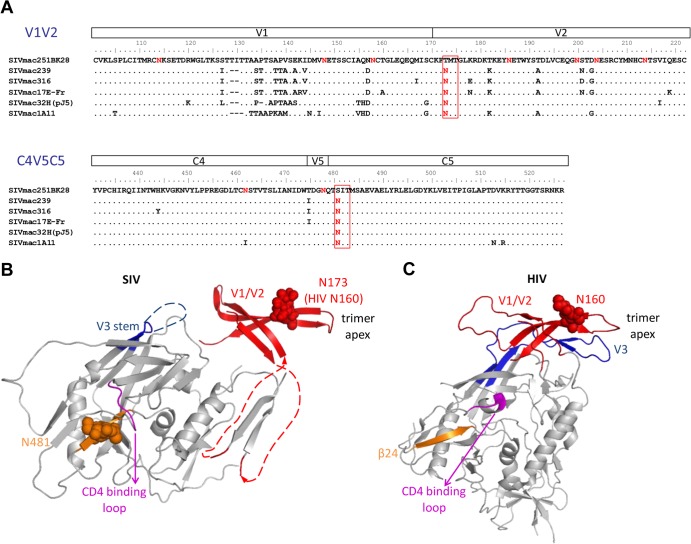
N173 and N481 N-linked glycosylation sites are highly conserved in SIV strains but are missing in SIVmac251BK28.
In a previous study, we identified two N-linked glycosylation sites in the V2 and C5 regions of gp120 that reduce macrophage tropism but enhance the neutralization resistance of SIVmac251BK28 (Yen et al., submitted). These N-linked glycosylation sites, N173 and N481, are present in other well-studied SIVmac clones and in sequences isolated from a SIVmac251 uncloned stock (47, 54–56) but are lost in SIVmac251BK28 (Fig. 1A). Furthermore, they are highly conserved in 7,119 SIV sequences in the Los Alamos HIV sequence database (99.79% and 99.83% for N173 and N481, respectively). The analogous glycosylation site of N173 in HIV, N160, is also highly conserved (91.75%) in 3,710 representative HIV sequences from all clades and in 14,630 of 16,318 (89.7%) sequences from clade B in the database. Crystal structures of gp120 show glycan moieties at SIV N481 and HIV N160, suggesting that these two glycosylation sites are indeed utilized, as predicted from the sequences (24, 41, 44). Modeling of the N173 and N481 glycans on these structures provides clues for their possible roles in Env function (Fig. 1B and C). N173 is located in the V2 region, which plays a role in modulating the formation and exposure of the CD4 binding site, interacting with V3 loop and modulating the CCR5 binding site, and Env trimer association (16–19, 40–43). Recent crystal and cryo-EM structures of a soluble cleaved HIV Env trimer show that V1/V2 is located at the top of the trimer and that the N173/N160 glycan is near the trimer apex (44, 45) (Fig. 1C). The V1/V2 and V3 loops are involved in intra- and interprotomer interactions at the trimer apex that are important for trimer association. These structures suggest that changes in the N173/N160 glycosylation site may affect V1/V2 and V3 interactions, which in turn may influence trimer structure and Env-CCR5 interactions, since V3 forms part of the CCR5 binding site. N-linked glycans in the V1/V2 region have also been identified as determinants of neutralization sensitivity, fusion activity, and CD4-independent infection (18, 57–64) and may alter the position of the V1/V2 loop (64). N481 is located in the outer domain of gp120, which is on the surface of the Env trimer, in close proximity to the CD4 binding loop. Therefore, glycosylation at this site might play a role in shielding gp120-neutralizing epitopes from antibody recognition and may also influence the structure of the CD4 binding site.
FIG 1.
N-linked glycosylation sites N173 and N481 are highly conserved among SIV strains but are missing in the SIVmac251BK28 clone. (A) Alignment of SIV gp120 sequences of the V1/V2 and the C4-V5-C5 regions shows that the N173 and N481 glycosylation sites are conserved in other well-studied SIV clones but are missing in SIVmac251BK28 (48). SIVmac239 and SIVmac32H (pJ5) are T-cell tropic (39, 95–97), whereas the other clones are macrophage tropic (6, 39, 48, 98–101). (B and C) N-linked glycosylation sites are shown in red, with N173 and N481 highlighted in red rectangles. The two N-linked glycosylation sites are mapped onto an unliganded SIV gp120 structure (41) (B) and an HIV gp120 structure from a soluble cleaved HIV Env trimer (44) (C), with the first sugar residues of the glycans and asparagine side chains shown as spheres. N173 and the V1/V2 region were substituted with a GAG linker in the original SIV gp120 structure. Here, we removed the GAG linker and modeled an HIV V1/V2 domain structure containing the analogous N160 glycan (red) (24), based on the orientation of V1/V2 in the HIV Env trimer structure. In the HIV Env trimer, V1/V2 forms a four-stranded β-sheet associated with V3 (blue) at the top of the Env trimer. The analogous N160 glycan is near the trimer apex. SIV N481 (orange) is at the β24 strand in the C5 region, in close proximity to the conserved GGDPE domain of the CD4 binding loop (magenta). The HIV Env protein shown here lacks the glycosylation site analogous to SIV N481. The β24 strand is shown in orange, as in the SIV gp120 structure.
Mutation of N173 in SIVmac239 enhances viral replication and syncytium formation in rhesus macrophages, while addition of N173 to SIVmac251 has the opposite effect.
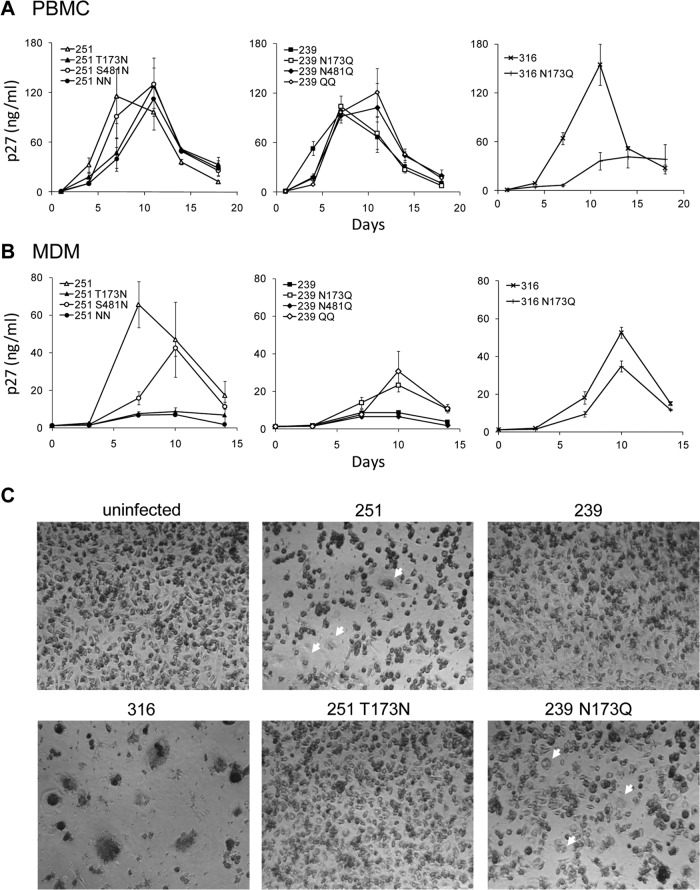
We previously showed that the introduction of both N173 and N481 decreased macrophage tropism of SIVmac251 (Yen et al., submitted). To dissect the roles of each N-glycosylation site, we introduced them individually into the SIVmac251 clone. We also generated N173Q and N481Q mutants of the T-cell-tropic clone SIVmac239 to test whether the removal of these glycosylation sites enhanced macrophage tropism. Mutations at either N-glycosylation site did not significantly influence SIVmac251 and SIVmac239 viral replication in rhesus PBMC (Fig. 2A). In contrast, the addition of N173 to SIVmac251 reduced viral replication in MDM, whereas the addition of N481 had only a modest effect, resulting in delayed replication and lower peak levels (Fig. 2B). The removal of N173 from SIVmac239 enhanced viral replication in MDM, while the removal of N481 had no significant effect. Infection with SIV lacking N173 (SIVmac251 and SIVmac239 N173Q) induced cytopathic effects and multinucleated giant cell (MNGC) formation in MDM (Fig. 2C). These results suggest that N173 was more important as a determinant of macrophage tropism and MNGC formation than N481. Effects of N173 on viral replication were also examined in the macrophage-tropic SIVmac316 clone. Although the introduction of N173Q into SIVmac316 reduced replication in rhesus PBMC (Fig. 2A), SIVmac316 N173Q replicated efficiently in rhesus macrophages but at slightly lower levels than did SIVmac316 (Fig. 2B). Thus, the effects of N173Q on SIV replication and macrophage tropism are strain dependent.
FIG 2.
SIVmac251 and SIVmac239 with N173 replicate well in rhesus macaque PBMC but poorly in rhesus macaque macrophages. (A) All SIVmac251 and SIVmac239 recombinant viruses replicated at high levels in rhesus PBMC, while SIVmac316 replication in these cells was reduced by the N173Q mutation. (B) Addition of N173 reduced SIVmac251 replication in macrophages, while mutation of N173 enhanced SIVmac239 replication in macrophages. SIVmac316 N173Q replicated efficiently in rhesus macrophages but at slightly lower levels than those of SIVmac316. (C) Infection of SIVmac251 and SIVmac239 N173Q induced MNGC (arrows) in MDM. Viruses used for infection were normalized by the p27 concentration (10 ng p27). Shown are means and standard deviations of samples from duplicate wells.
Mutation of N173 increases cell-to-cell transmission of SIV to CCR5+ cells expressing low levels of or no rhesus CD4.
SIV infection of rhesus macrophages with viruses lacking N173 (SIVmac251 and SIVmac239 N173Q) induced multinucleated giant cells (Fig. 2C), suggesting that these macrophage-tropic viruses mediate high levels of cell-cell fusion and may spread infection through cell-to-cell transmission. Previous studies showed that the ability of HIV/SIV to mediate fusion and viral entry with cells expressing low levels of CD4 is an important feature associated with enhanced tropism for macrophages, which express low levels of cell surface CD4 compared to CD4+ T cells (8–11). To determine whether the enhanced viral replication in rhesus macrophages is due to more efficient transmission between cells expressing little or no CD4, we designed and optimized a cell-to-cell transmission assay. First-round infection of donor Cf2 cells was normalized by using VSV-G-pseudotyped SIV. Cf2 target cells were transfected to express different levels of rhesus CD4/CCR5 (Fig. 3A). When target cells expressed high levels of CD4 and CCR5, such as TZM-BL cells expressing high levels of human CD4/CCR5 and Cf2-luc cells transfected to express high levels of rhesus CD4/CCR5, viruses with or without N173 were transmitted at similar high levels (Fig. 3B). In contrast, for target cells expressing rhesus CCR5 and low levels of or no rhesus CD4, the macrophage-tropic SIV viruses (SIVmac239 N173Q and SIVmac251) mediated higher levels of cell-to-cell transmission than the non-macrophage-tropic viruses (SIVmac239 and SIVmac251 T173N) (Fig. 3C). These differences in cell-cell transmission between SIVmac239 and SIVmac251 with versus without N173 were dependent on the levels of CD4. Both viruses showed significant effects of N173 at low levels of CD4. In contrast, the magnitude of the effect of N173Q in SIVmac239 was greatest in CD4− CCR5+ target cells, whereas differences between SIVmac251 with and SIVmac251 without N173 in these cells showed only a trend toward significance (P = 0.13). SIVmac239 N173Q mediated cell-to-cell transmission to CD4− CCR5+ target cells about 3-fold more efficiently than did parental SIVmac239. Viral transmission between donor and target cells was dependent on direct cell-cell contact, as it was abolished when donor and target cells were separated by transwells. In contrast, cell-free virus infection occurred efficiently when a large amount of cell-free virus (50 ng of SIV p27) was used for infections in a similar transwell assay format (data not shown). Cell-cell transmission was strictly CCR5 dependent, as these viruses were not transmitted to cells lacking both CD4 and CCR5. These results suggest that these macrophage-tropic viruses mediate efficient CD4-independent cell-to-cell transmission to CCR5-expressing cells and raise the possibility that the ability of viruses to mediate cell-to-cell transmission to target cells with low levels of or no CD4 is important for promoting viral replication and spreading infection in macrophages.
FIG 3.
Macrophage-tropic SIVmac239 N173Q mediates CD4-independent cell-cell transmission and enhanced cell-cell fusion with CCR5-expressing cells compared to non-macrophage-tropic SIVmac239. (A) Expression of rhesus CD4/CCR5 on Cf2-luc target cells used in both cell-cell transmission and cell-cell fusion assays. Percentages and mean fluorescent intensities (MFI) of CD4+ or CCR5+ cells are shown. Unstained cells and CD4-negative cells, included as negative controls, yielded signals similar to those of the isotype controls (data not shown). (B) VSV-G-pseudotyped SIVmac239 or SIVmac251 with or without N173 was transmitted at similar levels from infected Cf2 donor cells to TZM-BL target cells expressing human CD4/CCR5 or Cf2-luc target cells expressing high levels of rhesus CD4/CCR5. RLU, relative light units. (C and D) Mutation of N173 in SIVmac239 enhances CD4-independent cell-cell transmission (C) and Env-mediated fusion activity (D). Results are representative of 2 to 3 independent experiments. Error bars represent standard deviations for means of duplicate samples. An asterisk indicates a significant difference by Student's t test (P < 0.05).
Macrophage-tropic SIV Envs (SIVmac239 N173Q and SIVmac251) mediate CD4-independent fusion with CCR5-expressing target cells.
Next, we examined the relationship between cell-cell virus transmission and Env-mediated fusion. To address this question, we performed a cell-cell fusion assay in a format similar to that used for the above-described cell-to-cell transmission experiments but with Env-expressing instead of SIV-infected donor cells. Using this assay, we showed that cell-cell fusion activity showed patterns corresponding to the ability of viruses to mediate cell-to-cell transmission, with the macrophage-tropic Envs (SIVmac239 N173Q and SIVmac251) mediating CD4-independent fusion with CCR5-expressing target cells more efficiently than non-macrophage-tropic Envs (SIVmac239 and SIVmac251 T173N) (Fig. 3D). Similar to cell-to-cell transmission, CCR5 was required for cell-cell fusion, as fusion was not detected with target cells lacking both CD4 and CCR5. Together, these findings suggest that the loss of the N173 N-glycosylation site increases SIVmac239 macrophage tropism by enhancing Env-CCR5 interactions and CD4-independent cell-to-cell virus transmission during spreading infections in macrophages, which express very low levels of CD4.
Mutation of N173 enhances cell-cell fusion activity of SIVmac239 Env but has no significant effect on Env expression and processing.
Glycosylation plays an important role in the correct folding of HIV Env (65, 66). To examine whether N173 and N481 have effects on Env expression and processing, we examined the expression of the N173 and N481 mutants in 293T cells transfected with Env expression plasmids. Western blots showed similar expression levels for SIVmac239 and SIVmac251 recombinant Envs with and without the N-glycosylation sites (Fig. 4A). gp120 and gp160 protein bands for Envs with N173 and N481 migrated more slowly than those without N173 and N481, consistent with the addition of glycans at these sites (Fig. 4A).
FIG 4.
Mutation of N173 enhances SIV Env fusion activity but does not alter Env expression. (A) 293T cells transfected with Env expression plasmids were analyzed by Western blotting. N173 did not significantly affect Env expression. The shift in mobility of the gp120 and gp160 bands between the Envs with and those without N173/N481 glycosylation sites suggests the addition of glycans at these sites. (B) Env fusion activity was examined by a cell-cell fusion assay measuring fusion between 293T cells expressing SIV Envs and Cf2-luc cells expressing low or high levels of rhesus CD4 and CCR5. The loss of N173 enhanced cell-cell fusion mediated by SIVmac239 Env, whereas the addition of N173 reduced cell-cell fusion mediated by SIVmac251 Env. Results are representative of three independent experiments. Error bars represent standard deviations for means of duplicate samples. An asterisk indicates a significant difference versus the parental virus (P < 0.05 by Student's t test). A double asterisk indicates a significant difference versus the parental virus (P < 0.01).
We then examined the fusion activity of Envs expressing the N173 and N481 mutants, using a cell-cell fusion assay different from the one described above. In this alternative assay format, we used 293T cells as Env-expressing effector cells instead of Cf2 cells, the ratio of effector cells to target cells was 1:10 instead of 1:1, and the coincubation time was 10 h rather than 2 days. The N173 mutation induced a more significant difference in fusion activity under these conditions (compare Fig. 4B to 3D). Envs without N173 mediated higher levels of fusion than those with N173, with cells expressing either low or high levels of CD4. In contrast, the N481 mutation had only minor effects and did not significantly alter fusion activity when introduced into SIVmac251. Normalization of these fusion assay results to gp120 expression levels gave similar results, supporting the same conclusion (data not shown).
Viruses lacking N173 do not infect cells expressing low levels of or no CD4 more efficiently.
To examine whether changes in the N173 glycosylation site affect Env incorporation into virions, we examined viral proteins in virions by Western blotting. These experiments showed that Env levels on virions were similar between the parental and the N173 mutants (Fig. 5A), suggesting that N173 does not influence Env incorporation into virus particles. Macrophages express lower surface levels of CD4 than do CD4+ T cells (8–11). To test whether the enhanced replication of SIV clones in macrophages is due to increased infectivity on cells expressing low levels of CD4 or CCR5, we used Affinofile cells, an inducible cell line that can be simultaneously and independently induced to express different levels of human CD4 and CCR5 (50) (Fig. 5B). SIVmac239, SIVmac251, and the corresponding N173 mutants infected Affinofile cells with a similar pattern. The addition or removal of N173 did not alter the infection pattern of these viruses on Affinofile cells expressing different levels of CD4 and CCR5 (Fig. 5C). Unexpectedly, viruses expressing N173 showed higher infectivity than did those lacking N173 (Fig. 5C). Infection of Cf2-luc cells expressing rhesus CD4 and CCR5 showed similar results (data not shown). SIVmac239 and SIVmac251 without N173 did not infect cells with or without CD4, and the lack of N173 impaired the infectivity of these SIV clones in cell-free infections. Furthermore, the removal of N173 impaired the infectivity of SIVmac239 in TZM-BL cells, whereas the addition of N173 enhanced SIVmac251 infectivity in TZM-BL cells (data not shown). These results are in contrast to those of cell-cell transmission assays, in which viruses lacking N173 were transmitted to the same Cf2-luc target cells more efficiently than those with N173. In particular, these viruses mediated CD4-independent cell-cell transmission but could not infect CD4-negative cells in cell-free virus infections. Together, these findings suggest that CD4-independent infection required cell-cell contact.
FIG 5.
Mutation of N173 impairs single-round SIV infection of Affinofile cells. (A) Viral proteins in SIV virions were analyzed by Western blotting. (B) Affinofile cells were induced with doxycycline and ponasterone to express different levels of human CD4 and CCR5. Levels of expression were analyzed by flow cytometry and quantified by using QuantiBRITE. ABS, antibodies. (C) SIVs with Envs lacking N173 showed impaired infectivity compared to those with Envs with N173 and did not mediate CD4-independent infection in Affinofile cells.
The N173 mutation alters SIV neutralization sensitivity in a strain-dependent manner.
To probe structural changes induced by the N173 and N481 mutations, we next tested the neutralization sensitivity of the recombinant viruses to SIVmac251 antiserum and monoclonal antibodies targeting specific epitopes, including CD4 and CCR5 binding sites. Previous studies showed that the addition of both of these N-glycosylation sites enhanced the neutralization resistance of SIVmac251 (Yen et al., submitted). Here, we showed that this enhanced neutralization resistance was due to changes in N173 but not N481 (Table 1). N173 enhanced the neutralization resistance of SIVmac251 to SIVmac251 antiserum and monoclonal antibodies 5B11 and 7D3 (targeting CD4 and CCR5 binding sites, respectively) (51), resulting in >3-log differences in the 50% inhibitory concentrations (IC50s), whereas N481 did not alter neutralization sensitivity. In contrast to results for SIVmac251 clones, the removal of N173 and N481 did not alter the neutralization sensitivity of SIVmac239. SIVmac239 without N173 or N481 was still highly neutralization resistant, similar to parental SIVmac239. Thus, N173 enhanced the neutralization resistance of SIVmac251, while removal of N173 from SIVmac239 had no effect on neutralization resistance. These results suggest that N173 has strain-dependent effects on neutralization sensitivity of SIVmac251 and SIVmac239.
TABLE 1.
Neutralization sensitivity of SIV clones to SIVmac251 antiserum, monoclonal antibodies, and sCD4
The IC50 for the antiserum and monoclonal antibodies was calculated as the reciprocal dilution of antiserum or ascites required for achieving 50% inhibition of infection with no serum or ascites control, respectively. The IC50 for sCD4 is the concentration required for achieving 50% inhibition of infection with no sCD4 control.
The IC50 could not be achieved at 1:100 dilutions of antiserum or ascites containing the monoclonal antibodies.
Epitopes of 5B11 and 7D3 were mapped to CD4 and CCR5 binding sites, respectively (51).
N173 does not significantly affect SIV gp120-CD4 binding.
To investigate whether the enhanced macrophage tropism and cell-cell transmission of SIV lacking N173 was due to enhanced gp120 interactions with CD4, we first tested the neutralization sensitivity of these recombinant viruses to soluble CD4 (sCD4). Viruses with N173 were slightly more sensitive to sCD4 neutralization than their counterparts without N173 (Table 1). These results were unexpected, because we originally hypothesized that the removal of N173 would enhance gp120 interactions with CD4 and thereby increase fusion activity and the usage of low levels of CD4 on macrophages. However, sensitivity to sCD4 neutralization is not a direct indication of binding; other factors, such as a propensity to adopt CD4-induced conformational changes, could also be involved (53). To directly test interactions between the recombinant Envs and CD4, we purified His-tagged soluble gp120 (sgp120) from supernatants of transfected 293F cells (Fig. 6A) and examined binding to CD4 by co-IP (Fig. 6B) and Biacore (Fig. 6C) assays. For the co-IP assay, similar amounts of SIVmac239 and SIVmac239 N173Q sgp120 were pulled down by human CD4-Ig (Fig. 6B), suggesting similar binding to CD4 by both sgp120s. The binding kinetics and affinities between sgp120 and CD4 were analyzed by Biacore assays with human sCD4 immobilized on the chip surfaces at three different densities. Data from each surface were fit to a 1:1 binding model using BIAevaluation software to derive the kon, koff, and Kd (dissociation constant; Kd = Kon/Koff), the averages of which are shown in Fig. 6C. The results suggest that SIVmac239 and SIVmac239 N173Q gp120s bind to CD4 with similar kinetics and affinities. Finally, to examine the binding of Env trimers expressed on the cell surface to CD4, we used a cell-based ELISA as described previously (53, 67). CD4-Ig and SIVmac251 antiserum binding to cell surface Env trimers was measured, and CD4-Ig binding was normalized by SIVmac251 antiserum binding to control for Env cell surface expression levels. Consistent with results from the co-IP and Biacore assays, SIVmac239 and SIVmac239 N173Q Env trimers bound to CD4-Ig at similar levels (Fig. 6D). Thus, N173 does not appear to have significant effects on the binding of gp120 monomers or Env trimers to CD4.
FIG 6.
There are no significant differences between SIVmac239 and SIVmac239 N173Q sgp120 binding to human CD4. (A) Coomassie blue-stained SDS-PAGE gel of SIVmac239 and SIVmac239 N173Q sgp120 expressed in and purified from transfected 293F cells. (B) Co-IP of sgp120 with human CD4-Ig. (C) Kinetic analysis of the interactions between sgp120 and human sCD4. Direct binding was measured by Biacore assays. (D) The N173 mutation does not significantly affect CD4-Ig binding to Env trimers in a cell-based ELISA. Results shown are CD4-Ig binding after normalization by SIVmac251 antiserum binding.
DISCUSSION
In this study, we investigated functional roles of N173, a conserved N-linked glycosylation site in the V2 region of the SIV envelope glycoprotein, in macrophage tropism and neutralization sensitivity. The removal of N173 from SIVmac239 enhanced macrophage tropism and CD4-independent cell-to-cell transmission but had no significant effect on neutralization sensitivity. Similarly, the addition of N173 to SIVmac251 reduced viral replication in macrophages and decreased cell-to-cell transmission, but in contrast to SIVmac239, neutralization resistance was enhanced. Infection of macrophages by SIV lacking N173 was associated with the induction of MNGC formation, a phenotype that is likely to be explained by the increased fusion activity of Envs of these macrophage-tropic viruses when CD4 is at low levels or absent. These findings suggest that the loss of the N173 glycosylation site increases SIVmac239 replication in macrophages by enhancing CD4-independent cell-to-cell transmission through CCR5-mediated fusion. This mechanism may be important for promoting spreading infections by these viruses in tissues such as brain in vivo.
CD4-independent cell-to-cell transmission represents a novel mechanism to explain enhanced macrophage tropism. Consistent with our results, previous studies showed that infected macrophages can transmit HIV to T cells (68–70) and that Env determinants of macrophage tropism in the V1V2 region influence entry and spread infections in macrophages (11, 36). Cell-to-cell transmission is more efficient than cell-free virus infection (69, 71, 72) and has been shown to protect viruses from inhibition by neutralizing antibodies, antiretroviral drugs, and cellular restriction factors (72–77). This mechanism may be important to promote cell-to-cell spread and replication of macrophage-tropic SIVs in vivo, since these viruses are typically neutralization sensitive.
The CD4-independent SIV phenotypes were detected only in the context of cell-cell interactions but not cell-free virus infection. Although macrophage-tropic viruses mediated enhanced CD4-independent cell-cell fusion and cell-to-cell transmission, they were not able to infect cell lines in the absence of CD4 in single-round infection assays. This may be due to the transient and short-lived intermediate state of Env required for CD4-independent fusion. In SIV Envs lacking N173, the unliganded Env might have an increased propensity to sample a CD4-bound conformation that facilitates interactions with CCR5 and thereby allows CD4-independent fusion. This intermediate state may be short-lived before undergoing spontaneous and irreversible conformational changes and may not be sustained long enough to achieve cell-free virus infection. The rapid kinetics of cell-to-cell transmission compared to those of cell-free virus infection (minutes versus hours) may allow this transient short-lived form of Env to mediate CD4-independent cell-cell fusion. Within minutes of cell-cell contact, virus on the donor cell (78), and receptors on the target cell (79, 80) cluster to the junction of cell-cell contact. High local concentrations of CCR5 may facilitate CD4-independent infection in the context of cell-to-cell transmission. Previous studies showed that the addition of a glycan motif (D470N), which introduces an N-linked glycosylation site near the CD4-binding pocket of gp120, is associated with SIVmac251 CD4 independence and infection of CD4+ T-cell-depleted rhesus macaques (81, 82). Thus, SIV can evolve CD4 independence through different pathways.
To our knowledge, this is the first study to identify an N-linked glycosylation site as a determinant for cell-to-cell transmission of HIV/SIV. Results from fusion assays suggest that mutation of N173 enhances CCR5-mediated fusion activity, which in turn facilitates cell-to-cell transmission. Mutation of N173 may affect the structure and/or orientation of the V1V2 loop, which in turn may affect V3 loop and CCR5 binding site exposure and/or orientation, thereby increasing gp120-CCR5 interactions. Consistent with this model, the introduction of N173 enhanced neutralization resistance of SIVmac251 to 7D3, an antibody targeting the CCR5 binding site, a CD4-induced epitope. In a previous study, we showed that the addition of both N173 and N481 glycosylation sites enhances neutralization resistance of SIVmac251 to the V3-specific antibody 36D5 (Yen et al., submitted). This finding, along with similar phenotypes of SIVmac251 T173N and SIVmac251 NN (SIVmac251 containing both T173N and S481N mutations) observed in the present study, suggests that the addition of N173 may affect the position and/or exposure of V3. These models are also supported by recent structural studies showing that the V1/V2 loop is located at the trimer apex in association with the V3 loop and rotates upon CD4 binding to form an open conformation (44, 45, 83–90). The cryo-EM structure of an HIV Env trimer reveals a cluster of basic residues at the trimer apex (45). The removal of the N173 glycan may increase the exposure of these basic residues, which in turn may facilitate Env interactions with the negatively charged tyrosine-sulfated CCR5 N terminus and/or nonspecific attachment with cell membranes, thereby promoting fusion and cell-cell transmission. Further studies are required to examine whether these models are relevant for mechanisms by which changes at N173 affect cell-to-cell transmission.
The macrophage-tropic SIV clones (SIVmac251 and SIVmac239 N173Q) induced cell fusion and MNGC formation in macrophages and mediated higher levels of fusion in cell-cell fusion assays in cells expressing low levels of or no CD4 than their counterparts expressing N173. Our results suggest that these phenotypes are probably not due to enhanced gp120 interactions with CD4. In co-IP and Biacore assays, the removal of N173 had no effect on the binding of purified SIVmac239 gp120 monomers to CD4. Likewise, a cell-based ELISA showed no significant differences in CD4 binding between SIVmac239 and SIVmac239 N173Q Env trimers on the cell surface. In these binding assays, we used human sCD4 or CD4-Ig. Human CD4 and rhesus CD4 have similar amino acid sequences (>92% identical), and infectivity and cell-to-cell transmission showed similar patterns between cells expressing human CD4 and those expressing rhesus CD4 (Fig. 3B). Furthermore, overexpression of human or rhesus CD4 in macrophages enhances SIV infection and replication at comparable levels (8). Together, these results suggest that human CD4 is a reasonable surrogate for examining the usage of rhesus CD4. Together with results of assays testing CD4-independent cell-to-cell transmission and cell-cell fusion, these findings suggest that enhanced macrophage tropism of SIV lacking N173 may result from mechanisms other than enhanced Env-CD4 interactions, possibly enhanced Env-CCR5 interactions.
The effects of N173 on neutralization sensitivity and macrophage tropism appear to be strain dependent. Changes in N173 altered the neutralization sensitivity of SIVmac251 but not that of SIVmac239, suggesting differential effects on the exposure or conformation of neutralizing epitopes. Furthermore, although the introduction of N173 reduced macrophage tropism of SIVmac251, this N-glycosylation site is highly conserved among SIV strains and is present in other macrophage-tropic clones, including SIVmac316. SIVmac316 differs from SIVmac239 by 9 amino acids, with macrophage tropism being mapped to V67M, K176E, and G382R in gp120 and to K573T and a stop codon at position 767 in gp41 (39). Mutation of N173 in SIVmac316 did not enhance its high macrophage tropism (Fig. 2B). Several well-characterized macrophage-tropic HIV-1 clones, SF162, 89.6, R3A, TYBE, dBR02, dBR07, and aBL01, and 118 of 666 brain-derived HIV-1 clones (38%, with the majority coming from autopsied cases with advanced disease) in the HIV Brain Sequence Database (http://www.hivbrainseqdb.org/) lack N160. These findings are consistent with a model in which HIV/SIV macrophage tropism can be achieved through different pathways that can be influenced by specific determinants in Env in a strain-dependent manner. SIV macrophage tropism is often associated with CD4 independence, while macrophage-tropic HIV remains CD4 dependent. The roles of N160 in macrophage tropism and CD4 independence of HIV-1 and HIV-2 strains are an open question that merits further investigation.
The viruses examined in our study replicated at similar high levels in rhesus PBMC regardless of the presence or absence of N173. In contrast, viruses without N173 replicated more efficiently in macrophages but were impaired in single-round infection of Affinofile, Cf2-luc, and TZM-BL cells. Viruses without N173 may infect poorly in the first round of infection in PBMC and macrophages, as suggested by the single-round infectivity defect in these cell lines, while the second round of infection via cell-to-cell transmission is enhanced. N160/N173 is highly conserved among HIV and SIV strains, and a lack of this glycan may result in structural changes linked to the infectivity defect. Consistent with our results, the loss of N160 abrogates viral infection of HIV-1 FE in TZM-BL cell assays (91). This infectivity defect might be context dependent in HIV, since N160 mutations had only modest effects in HIV-1 YU-2 and Sc19-15 (91). An alternative explanation is that other cellular factors that facilitate infection by the viruses lacking N173 are present on primary macrophages but absent on these cell lines. Some HIV Envs interact with integrin α4β7, which may serve as an attachment factor and facilitate viral infection (92–94). α4β7, however, is unlikely to be involved in facilitating viral replication in the present study. Although primary rhesus macrophages expressed α4β7 after 7 days in culture with M-CSF and N173 is located 20 amino acids upstream of the putative α4β7 binding motif, anti-α4β7 antibody had no significant inhibitory effects on macrophage infection with viruses characterized in the present study (data not shown). Further studies are required to examine whether the infectivity defect in viruses lacking N173 is related to interactions with another cellular attachment factor, structural changes, or other mechanisms.
In summary, we identified an N-linked glycosylation site, N173 in the V2 region, as an important determinant of SIV macrophage tropism. Our results suggest that mutation of N173 increases SIVmac239 macrophage tropism by enhancing CD4-independent cell-to-cell transmission through CCR5-mediated fusion. This mechanism may facilitate the escape of macrophage-tropic viruses from neutralizing antibodies while promoting spreading infections by these viruses in vivo.
ACKNOWLEDGMENTS
We thank A. Engelman, R. Desrosiers, R. P. Johnson, D. Barouch, and B. Chen for helpful discussions and advice. We are also grateful to the NIH AIDS Research and Reference Reagent Program for providing SIVmac251 antiserum, J. Hoxie for providing 5B11 and 7D3 antibodies and SIVmac251BK28, B. Lee for providing Affinofile cells, and R. Desrosiers for providing SIV239-FL.
This work was supported by NIH grants MH83588 and MH97659 to D.G. Core facilities were supported by Harvard Medical School Center for AIDS Research (CFAR) and DFCI/Harvard Cancer Center grants. A.H. is the recipient of a Mathilde Krim fellowship in basic science from the American Foundation for AIDS Research.
Footnotes
Published ahead of print 19 February 2014
REFERENCES
- 1.Doms RW, Trono D. 2000. The plasma membrane as a combat zone in the HIV battlefield. Genes Dev. 14:2677–2688. 10.1101/gad.833300 [DOI] [PubMed] [Google Scholar]
- 2.Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888. 10.1126/science.280.5371.1884 [DOI] [PubMed] [Google Scholar]
- 3.Puffer BA, Altamura LA, Pierson TC, Doms RW. 2004. Determinants within gp120 and gp41 contribute to CD4 independence of SIV Envs. Virology 327:16–25. 10.1016/j.virol.2004.03.016 [DOI] [PubMed] [Google Scholar]
- 4.Musich T, Peters PJ, Duenas-Decamp MJ, Gonzalez-Perez MP, Robinson J, Zolla-Pazner S, Ball JK, Luzuriaga K, Clapham PR. 2011. A conserved determinant in the V1 loop of HIV-1 modulates the V3 loop to prime low CD4 use and macrophage infection. J. Virol. 85:2397–2405. 10.1128/JVI.02187-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, Kunstman K, Wolinsky SM, Gabuzda D. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. U. S. A. 103:15160–15165. 10.1073/pnas.0605513103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puffer BA, Pohlmann S, Edinger AL, Carlin D, Sanchez MD, Reitter J, Watry DD, Fox HS, Desrosiers RC, Doms RW. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76:2595–2605. 10.1128/JVI.76.6.2595-2605.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Means RE, Matthews T, Hoxie JA, Malim MH, Kodama T, Desrosiers RC. 2001. Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J. Virol. 75:3903–3915. 10.1128/JVI.75.8.3903-3915.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannert N, Schenten D, Craig S, Sodroski J. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J. Virol. 74:10984–10993. 10.1128/JVI.74.23.10984-10993.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, Moore JP, Wolinsky SM, Gabuzda D. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277–6292. 10.1128/JVI.76.12.6277-6292.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori K, Rosenzweig M, Desrosiers RC. 2000. Mechanisms for adaptation of simian immunodeficiency virus to replication in alveolar macrophages. J. Virol. 74:10852–10859. 10.1128/JVI.74.22.10852-10859.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter BL, Wehrly K, Swanstrom R, Platt E, Kabat D, Chesebro B. 2005. Role of low CD4 levels in the influence of human immunodeficiency virus type 1 envelope V1 and V2 regions on entry and spread in macrophages. J. Virol. 79:4828–4837. 10.1128/JVI.79.8.4828-4837.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pantophlet R, Burton DR. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24:739–769. 10.1146/annurev.immunol.24.021605.090557 [DOI] [PubMed] [Google Scholar]
- 13.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. 2012. A blueprint for HIV vaccine discovery. Cell Host Microbe 12:396–407. 10.1016/j.chom.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton DR, Stanfield RL, Wilson IA. 2005. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. U. S. A. 102:14943–14948. 10.1073/pnas.0505126102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlsson Hedestam GB, Fouchier RA, Phogat S, Burton DR, Sodroski J, Wyatt RT. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6:143–155. 10.1038/nrmicro1819 [DOI] [PubMed] [Google Scholar]
- 16.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson WE, Sanford H, Schwall L, Burton DR, Parren PW, Robinson JE, Desrosiers RC. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993–10003. 10.1128/JVI.77.18.9993-10003.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205–5215. 10.1128/JVI.78.10.5205-5215.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711. 10.1038/31514 [DOI] [PubMed] [Google Scholar]
- 21.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286. 10.1056/NEJMoa1113425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, Ahmed H, Juraska M, Chen L, Konopa P, Nariya S, Stoddard JN, Wong K, Zhao H, Deng W, Maust BS, Bose M, Howell S, Bates A, Lazzaro M, O'Sullivan A, Lei E, Bradfield A, Ibitamuno G, Assawadarachai V, O'Connell RJ, deSouza MS, Nitayaphan S, Rerks-Ngarm S, Robb ML, McLellan JS, Georgiev I, Kwong PD, Carlson JM, Michael NL, Schief WR, Gilbert PB, Mullins JI, Kim JH. 2012. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 490:417–420. 10.1038/nature11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol G Principal Investigators. Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. 10.1126/science.1178746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. 10.1038/nature10696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calarese DA, Lee HK, Huang CY, Best MD, Astronomo RD, Stanfield RL, Katinger H, Burton DR, Wong CH, Wilson IA. 2005. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. U. S. A. 102:13372–13377. 10.1073/pnas.0505763102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103. 10.1126/science.1213256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators. Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. 10.1038/nature10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. U. S. A. 110:4351–4356. 10.1073/pnas.1217537110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunfee RL, Thomas ER, Wang J, Kunstman K, Wolinsky SM, Gabuzda D. 2007. Loss of the N-linked glycosylation site at position 386 in the HIV envelope V4 region enhances macrophage tropism and is associated with dementia. Virology 367:222–234. 10.1016/j.virol.2007.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koito A, Harrowe G, Levy JA, Cheng-Mayer C. 1994. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J. Virol. 68:2253–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shioda T, Levy JA, Cheng-Mayer C. 1991. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature 349:167–169. 10.1038/349167a0 [DOI] [PubMed] [Google Scholar]
- 32.Stamatatos L, Wiskerchen M, Cheng-Mayer C. 1998. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV type 1 isolate on viral envelope structure, cell entry, and replication. AIDS Res. Hum. Retroviruses 14:1129–1139. 10.1089/aid.1998.14.1129 [DOI] [PubMed] [Google Scholar]
- 33.Dunfee RL, Thomas ER, Gabuzda D. 2009. Enhanced macrophage tropism of HIV in brain and lymphoid tissues is associated with sensitivity to the broadly neutralizing CD4 binding site antibody b12. Retrovirology 6:69. 10.1186/1742-4690-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown RJ, Peters PJ, Caron C, Gonzalez-Perez MP, Stones L, Ankghuambom C, Pondei K, McClure CP, Alemnji G, Taylor S, Sharp PM, Clapham PR, Ball JK. 2011. Intercompartmental recombination of HIV-1 contributes to env intrahost diversity and modulates viral tropism and sensitivity to entry inhibitors. J. Virol. 85:6024–6037. 10.1128/JVI.00131-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duncan CJ, Sattentau QJ. 2011. Viral determinants of HIV-1 macrophage tropism. Viruses 3:2255–2279. 10.3390/v3112255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toohey K, Wehrly K, Nishio J, Perryman S, Chesebro B. 1995. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology 213:70–79. 10.1006/viro.1995.1547 [DOI] [PubMed] [Google Scholar]
- 37.Imamichi H, Igarashi T, Imamichi T, Donau OK, Endo Y, Nishimura Y, Willey RL, Suffredini AF, Lane HC, Martin MA. 2002. Amino acid deletions are introduced into the V2 region of gp120 during independent pathogenic simian immunodeficiency virus/HIV chimeric virus (SHIV) infections of rhesus monkeys generating variants that are macrophage tropic. Proc. Natl. Acad. Sci. U. S. A. 99:13813–13818. 10.1073/pnas.212511599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Igarashi T, Imamichi H, Brown CR, Hirsch VM, Martin MA. 2003. The emergence and characterization of macrophage-tropic SIV/HIV chimeric viruses (SHIVs) present in CD4+ T cell-depleted rhesus monkeys. J. Leukoc. Biol. 74:772–780. 10.1189/jlb.0503196 [DOI] [PubMed] [Google Scholar]
- 39.Mori K, Ringler DJ, Kodama T, Desrosiers RC. 1992. Complex determinants of macrophage tropism in env of simian immunodeficiency virus. J. Virol. 66:2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961–1972. 10.1128/JVI.74.4.1961-1972.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841. 10.1038/nature03327 [DOI] [PubMed] [Google Scholar]
- 42.Rusert P, Krarup A, Magnus C, Brandenberg OF, Weber J, Ehlert AK, Regoes RR, Gunthard HF, Trkola A. 2011. Interaction of the gp120 V1V2 loop with a neighboring gp120 unit shields the HIV envelope trimer against cross-neutralizing antibodies. J. Exp. Med. 208:1419–1433. 10.1084/jem.20110196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu L, Cimbro R, Lusso P, Berger EA. 2011. Intraprotomer masking of third variable loop (V3) epitopes by the first and second variable loops (V1V2) within the native HIV-1 envelope glycoprotein trimer. Proc. Natl. Acad. Sci. U. S. A. 108:20148–20153. 10.1073/pnas.1104840108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. 10.1126/science.1245625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. 10.1126/science.1245627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcon L, Sodroski J. 1997. High degree of sensitivity of the simian immunodeficiency virus (SIVmac) envelope glycoprotein subunit association to amino acid changes in the glycoprotein 41 ectodomain. AIDS Res. Hum. Retroviruses 13:441–447. 10.1089/aid.1997.13.441 [DOI] [PubMed] [Google Scholar]
- 47.Bixby JG, Laur O, Johnson WE, Desrosiers RC. 2010. Diversity of envelope genes from an uncloned stock of SIVmac251. AIDS Res. Hum. Retroviruses 26:1115–1131. 10.1089/aid.2010.0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kornfeld H, Riedel N, Viglianti GA, Hirsch V, Mullins JI. 1987. Cloning of HTLV-4 and its relation to simian and human immunodeficiency viruses. Nature 326:610–613. 10.1038/326610a0 [DOI] [PubMed] [Google Scholar]
- 49.Etemad-Moghadam B, Sun Y, Nicholson EK, Fernandes M, Liou K, Gomila R, Lee J, Sodroski J. 2000. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J. Virol. 74:4433–4440. 10.1128/JVI.74.9.4433-4440.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnston SH, Lobritz MA, Nguyen S, Lassen K, Delair S, Posta F, Bryson YJ, Arts EJ, Chou T, Lee B. 2009. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J. Virol. 83:11016–11026. 10.1128/JVI.01242-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edinger AL, Ahuja M, Sung T, Baxter KC, Haggarty B, Doms RW, Hoxie JA. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J. Virol. 74:7922–7935. 10.1128/JVI.74.17.7922-7935.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herschhorn A, Oz-Gleenberg I, Hizi A. 2008. Quantitative analysis of the interactions between HIV-1 integrase and retroviral reverse transcriptases. Biochem. J. 412:163–170. 10.1042/BJ20071279 [DOI] [PubMed] [Google Scholar]
- 53.Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, Smith AB, III, Sodroski J. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 7:e1002101. 10.1371/journal.ppat.1002101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, Sun C, Chen Y, Yeh WW, Letvin NL, Mascola JR, Nabel GJ, Haynes BF, Bhattacharya T, Perelson AS, Korber BT, Hahn BH, Shaw GM. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134. 10.1084/jem.20082831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strickland SL, Gray RR, Lamers SL, Burdo TH, Huenink E, Nolan DJ, Nowlin B, Alvarez X, Midkiff CC, Goodenow MM, Williams K, Salemi M. 2011. Significant genetic heterogeneity of the SIVmac251 viral swarm derived from different sources. AIDS Res. Hum. Retroviruses 27:1327–1332. 10.1089/aid.2011.0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Del Prete GQ, Scarlotta M, Newman L, Reid C, Parodi LM, Roser JD, Oswald K, Marx PA, Miller CJ, Desrosiers RC, Barouch DH, Pal R, Piatak M, Jr, Chertova E, Giavedoni LD, O'Connor DH, Lifson JD, Keele BF. 2013. Comparative characterization of transfection- and infection-derived simian immunodeficiency virus challenge stocks for in vivo nonhuman primate studies. J. Virol. 87:4584–4595. 10.1128/JVI.03507-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogert RA, Lee MK, Ross W, Buckler-White A, Martin MA, Cho MW. 2001. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 gp120 affect coreceptor usage and cellular tropism. J. Virol. 75:5998–6006. 10.1128/JVI.75.13.5998-6006.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lue J, Hsu M, Yang D, Marx P, Chen Z, Cheng-Mayer C. 2002. Addition of a single gp120 glycan confers increased binding to dendritic cell-specific ICAM-3-grabbing nonintegrin and neutralization escape to human immunodeficiency virus type 1. J. Virol. 76:10299–10306. 10.1128/JVI.76.20.10299-10306.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reitter JN, Means RE, Desrosiers RC. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679–684. 10.1038/nm0698-679 [DOI] [PubMed] [Google Scholar]
- 60.Wolk T, Schreiber M. 2006. N-glycans in the gp120 V1/V2 domain of the HIV-1 strain NL4-3 are indispensable for viral infectivity and resistance against antibody neutralization. Med. Microbiol. Immunol. 195:165–172. 10.1007/s00430-006-0016-z [DOI] [PubMed] [Google Scholar]
- 61.Cole KS, Steckbeck JD, Rowles JL, Desrosiers RC, Montelaro RC. 2004. Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J. Virol. 78:1525–1539. 10.1128/JVI.78.3.1525-1539.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chackerian B, Rudensey LM, Overbaugh J. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719–7727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kolchinsky P, Kiprilov E, Sodroski J. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 75:2041–2050. 10.1128/JVI.75.5.2041-2050.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435–3443. 10.1128/JVI.75.7.3435-3443.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y, Luo L, Rasool N, Kang CY. 1993. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J. Virol. 67:584–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Land A, Braakman I. 2001. Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie 83:783–790. 10.1016/S0300-9084(01)01314-1 [DOI] [PubMed] [Google Scholar]
- 67.Haim H, Salas I, Sodroski J. 2013. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J. Virol. 87:1884–1889. 10.1128/JVI.02765-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Groot F, Welsch S, Sattentau QJ. 2008. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood 111:4660–4663. 10.1182/blood-2007-12-130070 [DOI] [PubMed] [Google Scholar]
- 69.Carr JM, Hocking H, Li P, Burrell CJ. 1999. Rapid and efficient cell-to-cell transmission of human immunodeficiency virus infection from monocyte-derived macrophages to peripheral blood lymphocytes. Virology 265:319–329. 10.1006/viro.1999.0047 [DOI] [PubMed] [Google Scholar]
- 70.Sharova N, Swingler C, Sharkey M, Stevenson M. 2005. Macrophages archive HIV-1 virions for dissemination in trans. EMBO J. 24:2481–2489. 10.1038/sj.emboj.7600707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sourisseau M, Sol-Foulon N, Porrot F, Blanchet F, Schwartz O. 2007. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J. Virol. 81:1000–1012. 10.1128/JVI.01629-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen P, Hubner W, Spinelli MA, Chen BK. 2007. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J. Virol. 81:12582–12595. 10.1128/JVI.00381-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abela IA, Berlinger L, Schanz M, Reynell L, Gunthard HF, Rusert P, Trkola A. 2012. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog. 8:e1002634. 10.1371/journal.ppat.1002634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sigal A, Kim JT, Balazs AB, Dekel E, Mayo A, Milo R, Baltimore D. 2011. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 477:95–98. 10.1038/nature10347 [DOI] [PubMed] [Google Scholar]
- 75.Vendrame D, Sourisseau M, Perrin V, Schwartz O, Mammano F. 2009. Partial inhibition of human immunodeficiency virus replication by type I interferons: impact of cell-to-cell viral transfer. J. Virol. 83:10527–10537. 10.1128/JVI.01235-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ganesh L, Leung K, Lore K, Levin R, Panet A, Schwartz O, Koup RA, Nabel GJ. 2004. Infection of specific dendritic cells by CCR5-tropic human immunodeficiency virus type 1 promotes cell-mediated transmission of virus resistant to broadly neutralizing antibodies. J. Virol. 78:11980–11987. 10.1128/JVI.78.21.11980-11987.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jolly C, Booth NJ, Neil SJ. 2010. Cell-cell spread of human immunodeficiency virus type 1 overcomes tetherin/BST-2-mediated restriction in T cells. J. Virol. 84:12185–12199. 10.1128/JVI.01447-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piguet V, Steinman RM. 2007. The interaction of HIV with dendritic cells: outcomes and pathways. Trends Immunol. 28:503–510. 10.1016/j.it.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cavrois M, Neidleman J, Greene WC. 2008. The Achilles heel of the Trojan horse model of HIV-1 trans-infection. PLoS Pathog. 4:e1000051. 10.1371/journal.ppat.1000051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Piguet V, Sattentau Q. 2004. Dangerous liaisons at the virological synapse. J. Clin. Invest. 114:605–610. 10.1172/JCI22812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Francella N, Gwyn SE, Yi Y, Li B, Xiao P, Elliott ST, Ortiz AM, Hoxie JA, Paiardini M, Silvestri G, Derdeyn CA, Collman RG. 2013. CD4+ T cells support production of simian immunodeficiency virus env antibodies that enforce CD4-dependent entry and shape tropism in vivo. J. Virol. 87:9719–9732. 10.1128/JVI.01254-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ortiz AM, Klatt NR, Li B, Yi Y, Tabb B, Hao XP, Sternberg L, Lawson B, Carnathan PM, Cramer EM, Engram JC, Little DM, Ryzhova E, Gonzalez-Scarano F, Paiardini M, Ansari AA, Ratcliffe S, Else JG, Brenchley JM, Collman RG, Estes JD, Derdeyn CA, Silvestri G. 2011. Depletion of CD4(+) T cells abrogates post-peak decline of viremia in SIV-infected rhesus macaques. J. Clin. Invest. 121:4433–4445. 10.1172/JCI46023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113. 10.1038/nature07159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.White TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, Bess JW, Lifson JD, Milne JL, Subramaniam S. 2011. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J. Virol. 85:12114–12123. 10.1128/JVI.05297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6:e1001249. 10.1371/journal.ppat.1001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8:e1002797. 10.1371/journal.ppat.1002797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci. U. S. A. 108:11440–11445. 10.1073/pnas.1101414108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meyerson JR, Tran EE, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S. 2013. Molecular structures of trimeric HIV-1 Env in complex with small antibody derivatives. Proc. Natl. Acad. Sci. U. S. A. 110:513–518. 10.1073/pnas.1214810110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. 2012. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat. Struct. Mol. Biol. 19:893–899. 10.1038/nsmb.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mao Y, Wang L, Gu C, Herschhorn A, Desormeaux A, Finzi A, Xiang SH, Sodroski JG. 2013. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc. Natl. Acad. Sci. U. S. A. 110:12438–12443. 10.1073/pnas.1307382110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang W, Nie J, Prochnow C, Truong C, Jia Z, Wang S, Chen XS, Wang Y. 2013. A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology 10:14. 10.1186/1742-4690-10-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. 2008. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat. Immunol. 9:301–309. 10.1038/ni1566 [DOI] [PubMed] [Google Scholar]
- 93.Cicala C, Martinelli E, McNally JP, Goode DJ, Gopaul R, Hiatt J, Jelicic K, Kottilil S, Macleod K, O'Shea A, Patel N, Van Ryk D, Wei D, Pascuccio M, Yi L, McKinnon L, Izulla P, Kimani J, Kaul R, Fauci AS, Arthos J. 2009. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc. Natl. Acad. Sci. U. S. A. 106:20877–20882. 10.1073/pnas.0911796106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, Salazar MG, Decker JM, Parrish EH, Berg A, Hopper J, Hora B, Kumar A, Mahlokozera T, Yuan S, Coleman C, Vermeulen M, Ding H, Ochsenbauer C, Tilton JC, Permar SR, Kappes JC, Betts MR, Busch MP, Gao F, Montefiori D, Haynes BF, Shaw GM, Hahn BH, Doms RW. 2012. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Pathog. 8:e1002686. 10.1371/journal.ppat.1002686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers RC. 1990. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science 248:1109–1112. 10.1126/science.2160735 [DOI] [PubMed] [Google Scholar]
- 96.Clarke S, Berry N, Ham C, Alden J, Almond N, Ferguson D. 2012. Neuropathology of wild-type and nef-attenuated T cell tropic simian immunodeficiency virus (SIVmac32H) and macrophage tropic neurovirulent SIVmac17E-Fr in cynomolgus macaques. J. Neurovirol. 18:100–112. 10.1007/s13365-012-0084-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rud EW, Cranage M, Yon J, Quirk J, Ogilvie L, Cook N, Webster S, Dennis M, Clarke BE. 1994. Molecular and biological characterization of simian immunodeficiency virus macaque strain 32H proviral clones containing nef size variants. J. Gen. Virol. 75(Part 3):529–543 [DOI] [PubMed] [Google Scholar]
- 98.Choi WS, Collignon C, Thiriart C, Burns DP, Stott EJ, Kent KA, Desrosiers RC. 1994. Effects of natural sequence variation on recognition by monoclonal antibodies neutralize simian immunodeficiency virus infectivity. J. Virol. 68:5395–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Naidu YM, Kestler HW, III, Li Y, Butler CV, Silva DP, Schmidt DK, Troup CD, Sehgal PK, Sonigo P, Daniel MD, Desrosiers RC. 1988. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J. Virol. 62:4691–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Luciw PA, Shaw KE, Unger RE, Planelles V, Stout MW, Lackner JE, Pratt-Lowe E, Leung NJ, Banapour B, Marthas ML. 1992. Genetic and biological comparisons of pathogenic and nonpathogenic molecular clones of simian immunodeficiency virus (SIVmac). AIDS Res. Hum. Retroviruses 8:395–402. 10.1089/aid.1992.8.395 [DOI] [PubMed] [Google Scholar]
- 101.Flaherty MT, Hauer DA, Mankowski JL, Zink MC, Clements JE. 1997. Molecular and biological characterization of a neurovirulent molecular clone of simian immunodeficiency virus. J. Virol. 71:5790–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]