ABSTRACT
Human papillomavirus (HPV) causes a number of neoplastic diseases in humans. Here, we show a complex normal HPV community in a cohort of 103 healthy human subjects, by metagenomics analysis of the shotgun sequencing data generated from the NIH Human Microbiome Project. The overall HPV prevalence was 68.9% and was highest in the skin (61.3%), followed by the vagina (41.5%), mouth (30%), and gut (17.3%). Of the 109 HPV types as well as additional unclassified types detected, most were undetectable by the widely used commercial kits targeting the vaginal/cervical HPV types. These HPVs likely represent true HPV infections rather than transitory exposure because of strong organ tropism and persistence of the same HPV types in repeat samples. Coexistence of multiple HPV types was found in 48.1% of the HPV-positive samples. Networking between HPV types, cooccurrence or exclusion, was detected in vaginal and skin samples. Large contigs assembled from short HPV reads were obtained from several samples, confirming their genuine HPV origin. This first large-scale survey of HPV using a shotgun sequencing approach yielded a comprehensive map of HPV infections among different body sites of healthy human subjects.
IMPORTANCE This nonbiased survey indicates that the HPV community in healthy humans is much more complex than previously defined by widely used kits that are target selective for only a few high- and low-risk HPV types for cervical cancer. The importance of nononcogenic viruses in a mixed HPV infection could be for stimulating or inhibiting a coexisting oncogenic virus via viral interference or immune cross-reaction. Knowledge gained from this study will be helpful to guide the designing of epidemiological and clinical studies in the future to determine the impact of nononcogenic HPV types on the outcome of HPV infections.
INTRODUCTION
Human papillomaviruses (HPVs) are a group of double-stranded, nonenveloped, small DNA viruses that are widely prevalent among human populations. To date, 176 types of HPV isolated from different body sites have been identified and collected in two HPV databases (http://pave.niaid.nih.gov, http://www.hpvcenter.se) searched on 30 November 2013, and the number is growing (1–3).
HPV is a well-established cause of cervical cancers (4–8). It is well known that long-term persistence of HPV infection is a necessity for development of cervical cancers, and the majority of women with HPV infection show a “healthy” phenotype for most of their lifetime (9). HPV infection has been linked worldwide to several cancers outside the reproductive system, including cancers of the oropharynx, pharynx, larynx, tonsils, and so on (10–15). Around 30 HPV types, notably types 16 (i.e., HPV16) and 18, which showed a strong association with cervical cancers, were designated high- or low-risk types. Although more and more nonrisk types have been observed from various human samples by traditional HPV detection methods (16–18), our understanding of HPV prevalence has been limited by the narrowed spectrum of amplicon-based methods that were designed for detection of the few HPV types associated with cervical cancers.
As opposed to traditional methods, metagenomics analysis using whole-genome shotgun (WGS) sequencing is a nonselective approach that, in theory, allows for identification of all HPV sequences showing significant identities to the known HPV genomes in a sample. In recent years, this method has been used for HPV detection in some human samples and has resulted in the identification of several novel HPV types (19–24). Particularly, from condyloma samples negative for HPV using a traditional PCR method, abundant putative HPV sequences were found using metagenomics analysis (19). These studies suggest that HPV communities in different human body sites are much more complex than previously observed. So far, few studies have targeted HPV infections in healthy humans, and very little knowledge about the HPV infections in healthy populations is known. Thus, it is necessary to define the full picture of HPV (risk and nonrisk) infections in healthy human populations.
Benefiting from the development of next-generation sequencing technology, the NIH-sponsored Human Microbiome Project (HMP) has generated a large number of metagenomics shotgun sequence data sets from samples collected from various body sites of healthy humans (18). These sequences originate not only from human and microbial DNA but also from viruses and microbial phages (18). In this study, we take advantage of the published genomes of HPV prototypes to identify HPV reads from HMP sequencing data sets. These HPV reads were further assigned to specific types and assembled into large contigs. This nonselective analysis yielded a comprehensive map of the HPV community among different human body sites of healthy human subjects from a new angle.
MATERIALS AND METHODS
Sources of HPV and shotgun metagenomics sequences.
Of the 176 HPV types listed in the Papilloma Virus Episteme (PaVE) database (http://pave.niaid.nih.gov) and by the International HPV Reference Center (http://www.hpvcenter.se) (30 November 2013), 148 types (20 September 2012) had reference genomes and were downloaded to create a local HPV reference genome database (HPVrgdb) (1–3). Raw (unassembled) shotgun metagenomics sequences generated within HMP and their metadata were downloaded from MG-RAST (ftp://ftp.metagenomics.anl.gov/projects/385/), and the related metadata information was downloaded from NCBI (project name, Deep WGS Genome Sequencing of HMP Clinical Samples) (25). All HMP samples were from a healthy North American cohort, as previously described (26, 27). In particular, the vaginal samples were collected from healthy subjects without a diagnosis of condyloma or HPV within the 2 years prior to sampling.
Since paired-end sequencing used in HMP did not always yield usable pairs, only the forward read from each pair was selected for further analysis. This data set contains 1,606 sequencing files derived from 748 samples collected from 103 healthy subjects (see Table S1 in the supplemental material) (Fig. 1). The samples were collected from 4 organs, the skin, vagina, mouth, and gut. The skin samples were from 2 body sites, the anterior nares and the retroauricular crease; oral samples were from 9 body sites, the attached/keratinized gingiva, the buccal mucosa, the hard palate, the palatine tonsils, saliva subgingival plaque, supragingival plaque, the throat, and the tongue dorsum; vaginal samples were from 2 body sites, the posterior fornix and the mid-vagina; and gut samples were from the feces. Replicate sequence files of the same sample were separately analyzed and then combined into a single file for further analysis.
FIG 1.
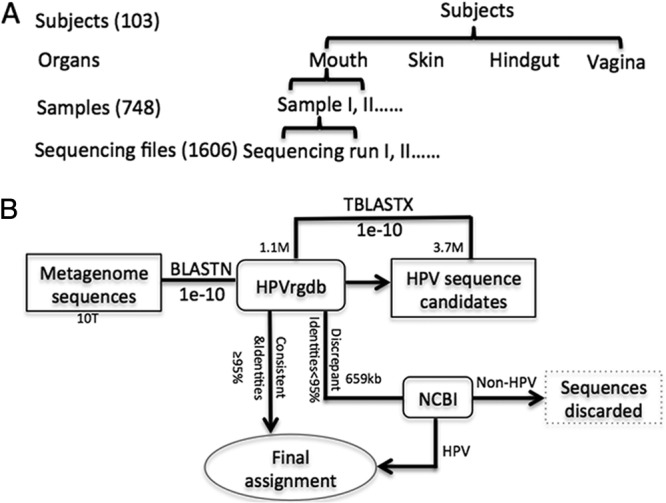
Bioinformatics method and samples. (A) Bioinformatics process. Initially, 1,606 sequence files from 748 samples collected from 103 healthy subjects were 10 terabytes (TB) in combined size after use of the quality filter and removal of human DNA sequences. The average read length was around 100 bp. All genomes of 148 prototypes of human papillomaviruses were downloaded from the Papilloma Virus Episteme (PaVE) database (http://pave.niaid.nih.gov) and the International HPV Reference Center (http://www.hpvcenter.se) (11 September 2012) to create a local HPV reference genomic database (HPVrgdb) of about 1.1 megabytes (MB) in size (1–3). BLASTN was employed to compare each shotgun read to the HPVrgdb, and those with an E value below 1E-10 were identified as HPV candidates; the combined size of these candidates was approximately 3.7 MB. The reads with more than 95% identities to their hits were assigned type numbers according to their hits. To confirm the assignment, all the HPV candidates were further compared by TBLASTX against the HPV database. Of the HPV candidates, 82% (3.04 MB) were confirmed to be HPV in origin with the same hits. The remaining HPV candidates, 0.66 MB in size, were further searched against NCBI GenBank (10 gigabytes [GB] in size). A read was assigned to a specific HPV type if its best hit (≤95% identities) was an HPV sequence with a definite type assignment, to an unclassified HPV type if its best hit was an HPV sequence without definite type assignment or it shared poor identities (<95%) with its best hit, or discarded if its best hit was to a non-HPV sequence. In total, 11 reads of the HPV candidates were discarded. (B) Hierarchical relationships of subjects, organs, samples, and sequencing files.
Bioinformatics approach for identification of HPV sequences.
We considered the approaches described in previous studies (19–22, 24) to identify HPV reads from the metagenomes, with modifications. We operated the analysis on the reads directly instead of using assembled contigs, because most reads in the HMP data sets could not be assembled into contigs. In brief, all metagenomics reads were compared against the HPVrgdb by BLASTN at a lenient threshold (E-value cutoff, ≤1E-10) (Fig. 1A). The hits were extracted as HPV candidate reads and then further examined by TBLASTX against the HPVrgdb to confirm their assignment. An HPV candidate read that hit the same HPV type sequence with high identity (≥95%) and full coverage by BLASTN was assigned to that HPV type; otherwise, an HPV candidate read was further searched in the NCBI GenBank. An HPV candidate read was assigned to HPV if its best hit was an HPV sequence or discarded if the best hit was a non-HPV sequence in NCBI GenBank. The HPV read identified was further assigned to a specific HPV type if it best matched with a sequence of genome of a known HPV type (≥95% identities and full coverage) or an unclassified HPV type if the best hit was to an HPV sequences without a type assignment or if the identities were low (<95%).
This method was validated using a mock-DNA data set, and the result showed that it had 99.3% sensitivity for recognizing known-type HPV sequences and 74.3% sensitivity for recognizing new/novel HPV sequences.
Assembly of HPV reads into contigs.
All reads with significant hits to HPV sequences were extracted from the data sets and then de novo assembled into contigs for each sample, respectively, using the software CLC Genomics Workbench (Qiagen) with the default parameters, and the contigs larger than 500 bp were kept for downstream analysis. All of these contigs were then compared against GenBank using NCBI BLASTN (MEGABLAST) with default parameters.
Definition of HPV positivity and prevalence.
A sample was designated HPV positive if one or more HPV sequences were detected in its sequence file(s). An organ was considered positive for HPV if an HPV sequence(s) was detected in at least one sample in the organ. A subject was considered positive for HPV if an HPV sequence(s) was detected in at least one sample from that subject. Subject prevalence was calculated as the number of subjects positive for HPV divided by the total number of subjects analyzed. Organ prevalence for each organ was calculated as the number of organs positive for HPV divided by the total number of organs analyzed.
Definition of relative abundance.
In order to keep the number of HPV reads comparable among samples, we computed the relative abundance of HPV for each sample by dividing the number of HPV reads in a sample by the total number of reads in the sample. The resultant fractions for all samples were normalized using the least common denominator. In this study, the final relative abundance was expressed as the number of HPV reads per 34 million total reads.
Construction of phylogenetic trees.
Available HPV genomes of 148 prototypes (20 September 2012) were downloaded from the Papilloma Virus Episteme (PaVE) database (http://pave.niaid.nih.gov) and from the International HPV Reference Center (http://www.hpvcenter.se). L1 genes of the prototype HPV genomes were predicted and used for phylogenetic tree building. Bootstrapped maximum-likelihood (ML) phylogenetic trees were constructed from a multiple sequence alignment using MEGA5.0 software. Bootstrap values were calculated from 1,000 resamplings of the multiple alignments (28). Figtree 1.4 software was used to visualize the tree (http://tree.bio.ed.ac.uk/software/).
RESULTS
A total of 1,606 metagenomics shotgun sequence files were available (see Tables S1 and S2 in the supplemental material) for 748 samples from 103 healthy human subjects in the HMP database. Based on similarity to the available genomic sequences of 148 classifiable HPV types, a total of 109 known HPV types and unclassified types were detected in these samples (Table 1) (see Table S1 in the supplemental material).
TABLE 1.
HPV detection in sequence files, samples, and subjects
| Detection in: | Parametera | No. detected in indicated organ |
Total | |||
|---|---|---|---|---|---|---|
| Gut | Mouth | Skin | Vagina | |||
| Sequence file | HPV+ | 26 | 50 | 131 | 56 | 263 |
| Total | 391 | 835 | 245 | 135 | 1,606 | |
| Sample | HPV+ | 19 | 38 | 72 | 25 | 154 |
| Total | 154 | 411 | 118 | 65 | 748 | |
| Subject | HPV+ | 17 | 27 | 46 | 17 | 71 |
| Total | 98 | 90 | 75 | 41 | 103 | |
HPV+, HPV-positive result. Total, total files, samples, or subjects tested.
HPV prevalence in different organs of healthy human subjects.
Of the 748 samples from 103 healthy human subjects, 65 were from the vagina of 41 subjects, 118 from the skin of 75 subjects, 411 from the mouth of 90 subjects, and 154 from the gut of 98 subjects (see Tables S1 and S2 in the supplemental material). HPV reads were detected in 154 samples (25 from the vagina, 72 from the skin, 38 from the mouth, and 19 from the gut). Overall, 71 subjects had an HPV read(s) detected in at least one sequence files, showing that the HPV prevalence in this healthy human cohort is approximately 68.9% (71/103). Of the 71 HPV-positive subjects, 42 (59.2%) had HPV detected in only one organ, 22 (31.0%) in two organs, 7 (9.9%) in three organs, and none in four organs (see Table S1 in the supplemental material) (Fig. 2). By organ, HPV prevalence was the highest in the skin (61.3%), followed by the vagina (41.5%), mouth (30%), and gut (17.3%) (Table 2).
FIG 2.
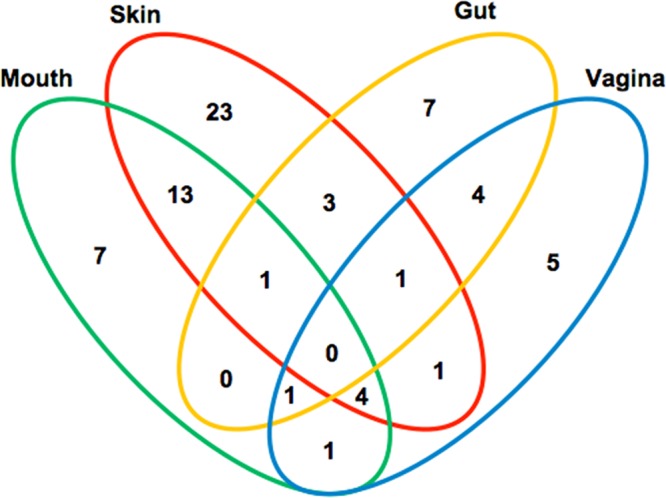
Detection of HPV DNA in single and multiple organs in healthy human subjects. The Venn diagram divides the 71 HPV-positive subjects into groups with single-, two-, and three-organ involvement, with specification to mouth, skin, gut, and vagina. The numbers show the number of subjects positive for HPV in a specific category.
TABLE 2.
HPV prevalence in human subjects by organ
| Organ | No. of subjects | No. of HPV+ subjectsa | HPV prevalence (%) |
|---|---|---|---|
| Vagina | 41 | 17 | 41.50 |
| Skin | 75 | 46 | 61.30 |
| Mouth | 90 | 27 | 30.00 |
| Gut | 98 | 17 | 17.30 |
| Total | 103 | 71 | 68.90 |
HPV+, HPV positive.
Distribution of HPV types in human organs.
The distribution patterns of the HPV types varied among the four human organs examined (Fig. 3). Each organ harbored specific HPV types and shared HPV types with others. Overall, the majority of the 109 HPV types (59.6%; 65 types) were detected in only one organ; 32.1% (35 types) were shared by two organs, 8.2% (9 types) by three organs, and none by all four organs. The skin samples harbored 80 HPV types, the highest among the four organs examined, accounting for 73.4% of the 109 HPV types detected. Forty HPV types were only found in skin samples, and the other 40 types were shared with the vagina (10 types), mouth (19 types), gut (2 types), mouth/vagina (6 types), and gut/vagina (3 types). The vaginal samples yielded 43 HPV types, including 20 HPV types specific to the vagina and 23 shared with the skin (10 types), mouth (3 types), gut (1 type), skin/gut (3 types), and mouth/skin (6 types). The oral samples generated 33 HPV types, of which only 5 types were organ specific while the large majority of types (75.8%; 25/33) were shared with the skin, fewer (27.2%; 9/33) with the vagina, and none with the gut. The gut had only six HPV types identified, the fewest among the 4 sites examined, of which all were shared with either the skin (5 types) or vagina (4 types) and none were organ specific.
FIG 3.
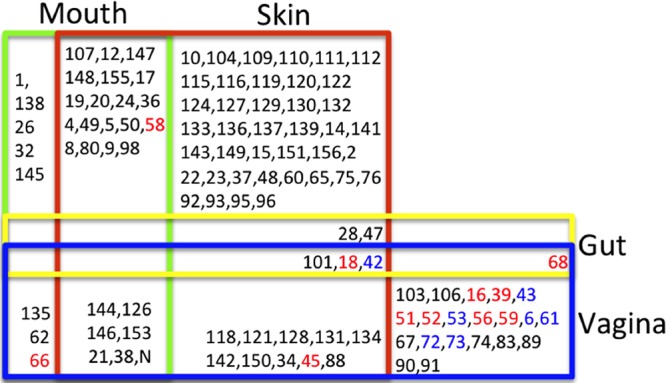
Distribution of HPV types in organs of healthy human subjects. The numbers represent the HPV types. High-risk types are marked in red and low risk types in blue. N, unclassified HPV type. The colored boxes represent different organs (vagina, blue; gut, yellow; skin, red; mouth, green). The numbers in a specific box mean that the corresponding HPV type was found in the organ(s). For example, “1” means that type 1 HPV was found only in the mouth, and “28” means that type 28 HPV was found in the skin, gut, and vagina simultaneously.
Dominant HPV types in human organs.
Although the large majority of known HPV types (73%; 109/148) were present in the samples from healthy human subjects, only a few HPV types dominated each organ in both relative abundance and prevalence.
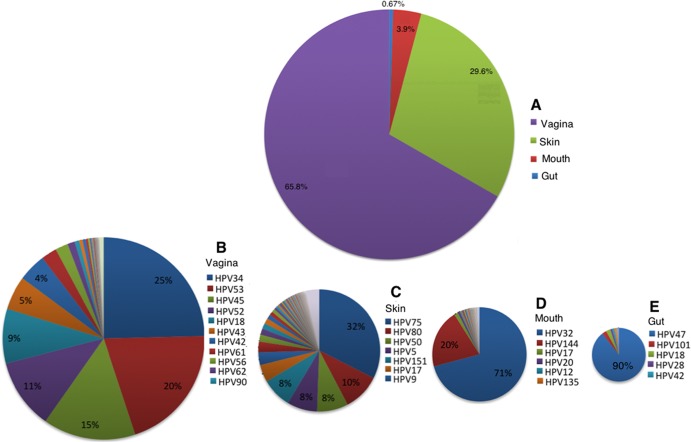
In total, 15,037 HPV reads were identified in this sequencing data set. Most of the HPV reads (10,034; 65.8%) were from vaginal samples, followed by the skin samples (4,512 reads; 29.6%), with a few from oral samples (596; 3.9%) and stool samples (106; 0.67%) (Fig. 4). The HPV types varied by organ in abundance (Fig. 4). In the vagina, of the 43 HPV types observed, the top four most abundant types (types 34, 53, 45, and 52) accounted for 71% of the reads from the vagina. In the skin, of the 86 HPV types identified, the top 7 most abundant types (types 75, 80, 50, 5, 151, 17, and 9) accounted for 74.5% of the reads, with type 75 alone accounting for 32.4% of the reads. In the mouth, of the 30 HPV types, the top two abundant types (types 32 and 144) accounted for 70.9% and 20.1% of the reads, respectively. In the stool, of the six HPV types, the most abundant type, type 47, accounted for 90% of the reads.
FIG 4.
Composition of HPV communities in healthy human subjects, by HPV type and organ. Distribution of HPV reads among four organs (A) and distribution of different HPV types in samples from the vagina (B), skin (C), mouth (D), and gut (E). Only the most abundant HPV types are shown.
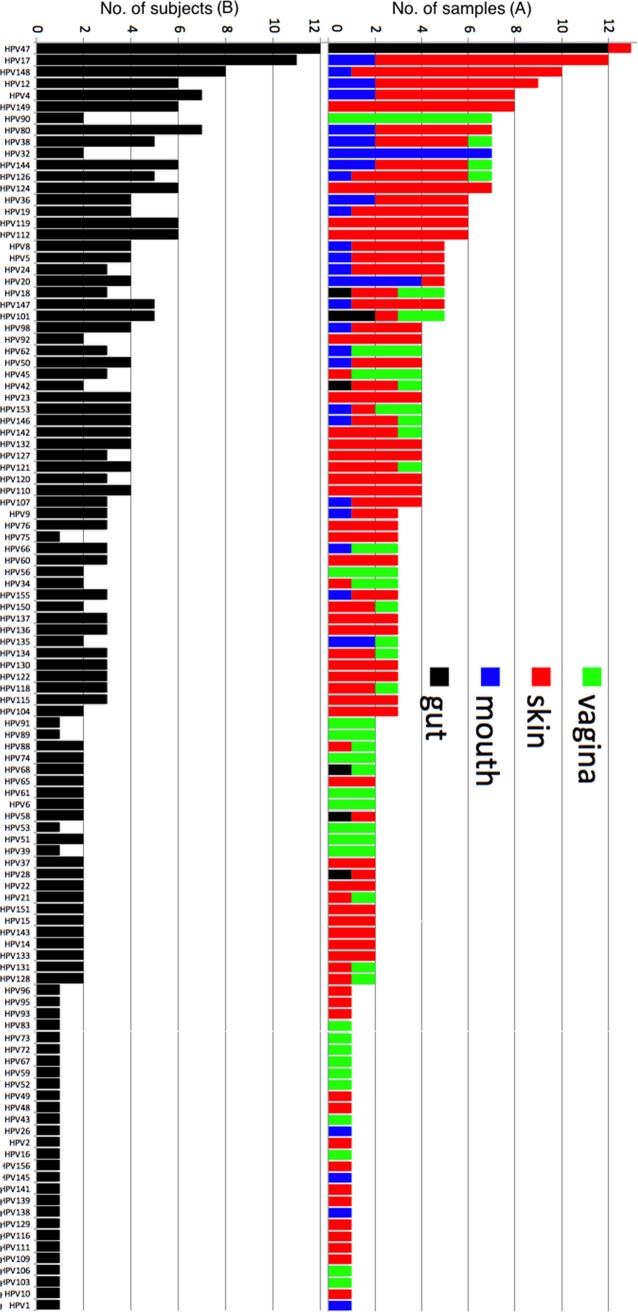
The HPV types also varied with organs in prevalence (Fig. 5). In the vagina, the most prevalent type was type 90, which was found in 7 of the 17 HPV-positive samples. Similarly, in the skin, the top three most prevalent types (types 17, 148, and 149) were present in 10, 9, and 8 of the 46 HPV-positive samples, respectively. In the mouth, the most prevalent type was type 32, which was detected in 7 of the 27 HPV-positive samples. In the gut, HPV type 47 was identified in 12 of the 17 HPV-positive samples.
FIG 5.
Prevalence of different HPV types detected in samples and subjects. (A) The y axis shows the number of samples in which a specific HPV type was found. The positive samples were further specified to organs by colors. (B) The y axis shows the number of human subjects for whom a specific HPV type was found.
In the HPV-positive samples, medians of the relative abundance of HPV reads were 4,123 per 34 million reads per sample from the vagina and 1,332 from the skin; the medians for the vagina and skin were significantly higher than (P ≪ 0.05) those for the mouth (median, 94) and gut (median, 3) (Fig. 6A) (Wilcoxon signed-rank test, R language). The samples with the highest relative abundance of HPV were the one with 37,053 in the vagina, the one with 34,079 in the skin, the one with 2,423 in the mouth, and the one with 14 in the gut.
FIG 6.
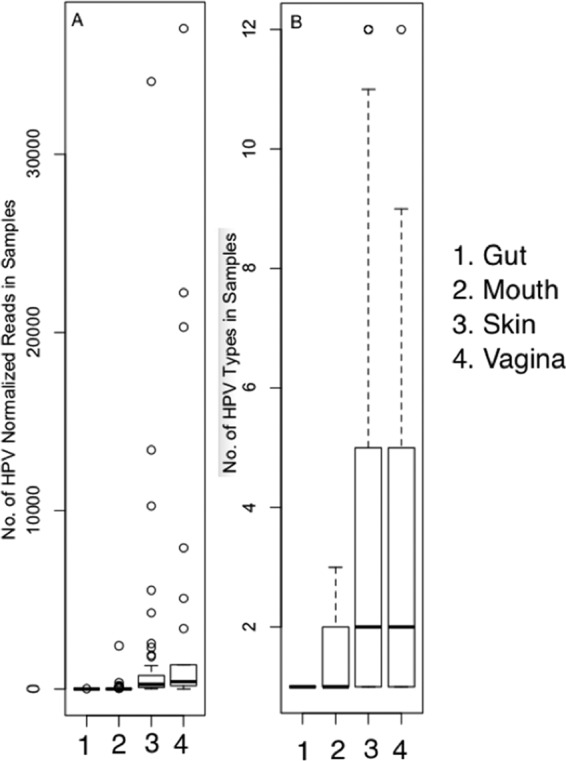
Relative abundance of HPV reads per sample and number of HPV types per sample in different organs. Relative abundance is expressed as the normalized number of HPV reads (per 34 million reads) in HPV-positive samples in the (1) gut, (2) mouth, (3) skin, and (4) vagina (A). Differences between organs were examined by a nonparametric Wilcoxon test of relative numbers of HPV reads between two organs: P4–3 = 0.127, P4–2 = 6.439E-08, P4–1 = 1.75E-07, P3–2 = 3.369E-12, P3–1 = 3.241E-15, and P2–1 = 5.72E-04. The number of different HPV types found in HPV-positive samples is expressed as an average number of HPV types found in samples from the (1) gut, (2) mouth, (3) skin, and (4) vagina (B). Median interquartile ranges and whiskers equivalent to 1.5 interquartile ranges are shown.
Coexistence of multiple HPV types in organs.
Two or more HPV types were detected in the majority of the 154 HPV-positive samples in the vagina (13/25 samples) and skin (51/72 samples) (Fig. 5 and 6B). In comparison, two or more HPV types were detected in 9 of the 38 HPV-positive samples of the mouth and 1 of the 19 HPV-positive gut samples. In particular, six skin samples (SRS058182, SRS020263, SRS019016, SRS019015, SRS016296, and SRS015051) and one vaginal sample (SRS048536) had more than 10 HPV types detected. Overall, more HPV types coexisted in skin samples and vaginal samples than in stool samples and oral samples.
Interactions between HPV types in organs.
We explored the set of normally occurring interactions (cooccurrence and coexclusion) between HPV types in the four organs. To overcome the bias stemming from sequencing depth and sampling, we normalized the number of reads corresponding to each HPV type by the total number of reads for each sample and expressed the normalized read counts as a relative percentage of all HPV reads. We used a metric of association appropriate for monitoring compositional data (29).
There was no evidence of interaction between HPV types within gut samples and within mouth samples (SparCC correlations, <0.2). Within skin samples, 5 HPV types among 82 types detected had evidence for weak interactions (SparCC correlations, >0.2) (Fig. 7A). Samples from the vagina had the largest number of interactions (SparCC correlations, >0.2) (Fig. 7B). Nineteen out of 43 HPV types detected in this organ had at least one interaction. Phylogenetically, the partners involved in these interactions were enriched in genus Alphapapillomavirus (17 out of 19 HPV types) compared to all HPV types detected in samples from the vagina (26 out of 43 HPV types). In addition, the interaction partners included all of three HPV types that were ubiquitous in all organs (types 18, 42, and 101). All but one of these interactions were cooccurrences, with the single coexclusion being between type 101 and untyped HPV. Oncogenic HPV type 18 had significant support (P < 0.01) for relatively strong cooccurrence interaction with HPV type 34 (SparCC correlation, 0.65) and HPV type 39 (SparCC correlation, 0.5).
FIG 7.
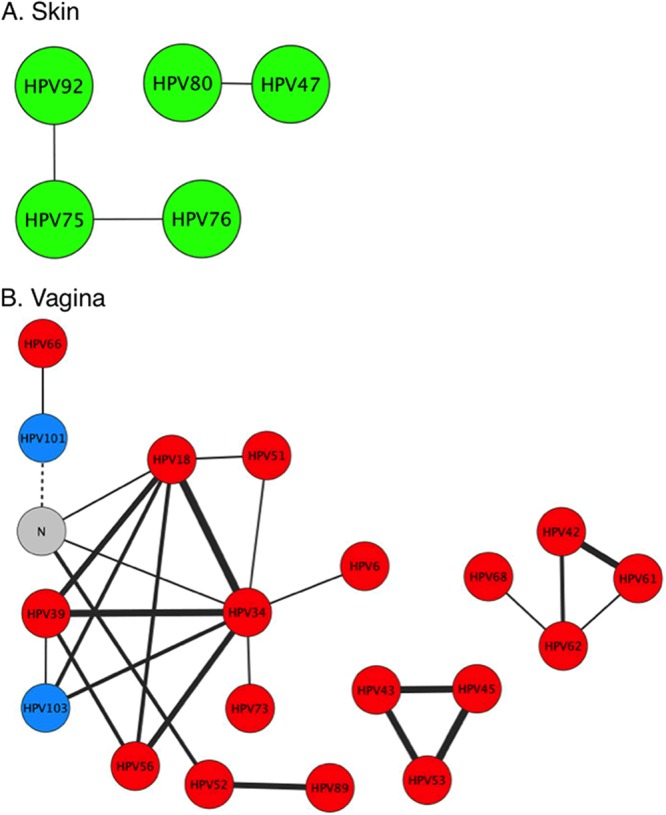
Cooccurrence/coavoidance networks of HPV types in the skin (A) and vagina (B). Nodes represent HPV types, colored by genus (red, Alphapapillomavirus; green, Betapapillomavirus; blue, Gammapapillomavirus) with edges showing interactions (SparCC correlations, ≥0.2; solid lines for cooccurrence, dashed lines for coavoidance) over all samples. Edge widths are proportional to the strength of the interaction. Only HPV types with at least one interaction in the corresponding organ are shown. N, unclassified HPV type.
Phylogenetic association of HPV types to human organ systems.
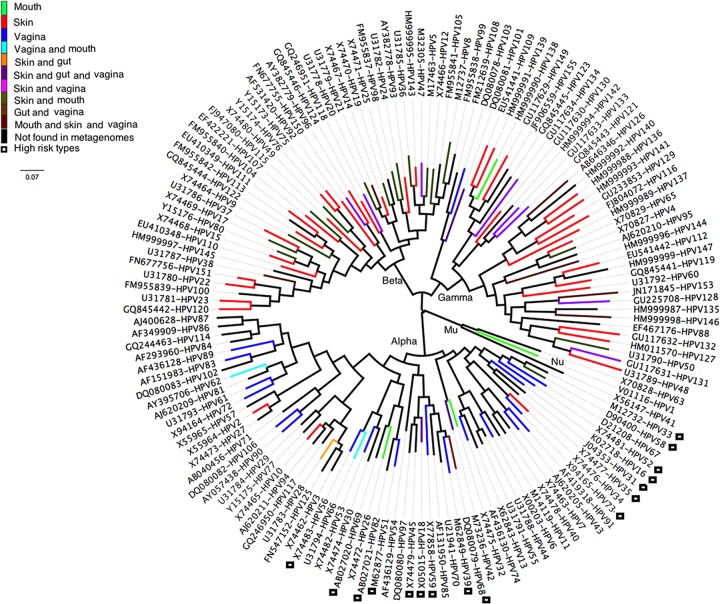
All known human papillomaviruses are phylogenetically grouped into five genera (Alpha-, Beta-, Gamma-, Mu-, and Nupapillomavirus) (30). Some studies suggested that HPV types from the genus Betapapillomavirus might play an important role in the development of squamous cell carcinoma of the skin (31–33). Based on the phylogenetic tree constructed with 148 known HPV prototypes (Fig. 8), most HPV types identified in the skin samples (64/80) were located in genera Betapapillomavirus and Gammapapillomavirus, while those from the vaginal samples (24/43) were in the genus Alphapapillomavirus (Fig. 8). The oral HPV types were dispersed in the genera Alpha-, Beta-, Gamma-, and Mupapillomavirus, but most (25/33) of the HPV types shared between oral and skin samples were found in the genus Betapapillomavirus. Only 6 types were found in gut samples, and all of them were shared with the skin or vagina, with 4 belonging to the Alphapapillomavirus group and 2 to the Betapapillomavirus group. Consistent with previous findings, it was shown that the 16 high-risk HPV types were concentrated in only two branches of the genus Alphapapillomavirus (Fig. 8) (34).
FIG 8.
Phylogenetic tree and the placement of the HPV types found in different organs. Phylogenetic tree based on L1 gene sequences of the prototypes of all known HPV types (148 types) with available genome sequences (20 September 2012) was constructed using MEGA5.0 and visualized using software Figtree 1.4.0. The colors on the branches of the tree show the organ where the HPV types were found in this study. High-risk HPV types are highlighted by black open boxes. Five HPV groups were marked as their respective genus (Alpha-, Beta-, Gamma-, Mu-, and Nupapillomavirus) based on previously described phylogenetic relationships (2, 30).
Presence of HPV confirmed by large assembled contigs.
The assembly generated 88 contigs larger than 500 bp, of which there were 38 larger than 1,000 bp, 9 were larger than 5,000 bp, and 6 had nearly full-length (>7,000-bp) HPV genomes. Searched against NCBI GenBank using BLASTN, all large contigs (>1,000 bp) have high identities, of between 96.7 and 99.9%, and full coverage (see Table S3 in the supplemental material) (Table 3) to HPV genomes of known types deposited in GenBank. Particularly, the 9 largest contigs showed ≥98.4% identities to known HPV genomes (see Table S3 in the supplemental material) (Table 2), of which the 7 from vaginal samples belong to types 52, 53, 34, 18, and 45, respectively, and the 2 from skin samples belong to type 75. The types observed based on contigs have abundant reads that have been assigned to the same types in the samples (see Tables S1 and S3 in the supplemental material). All of these cases confirm the HPV presence in the samples detected based on reads.
TABLE 3.
Large contigsa assembled from all reads of each HPV-positive sample and their best hits in GenBank
| Organ | Contig | Length (bp) | Best hitb and HPV type | Identity (%) | Coverage (%) |
|---|---|---|---|---|---|
| Vagina | SRR062364.hpv_contig_103 | 7,944 | HQ537734, HPV52 | 99.9 | 100.0 |
| Vagina | SRR062365.hpv_contig_1 | 7,940 | HQ537734, HPV52 | 99.9 | 100.0 |
| Vagina | SRR062073.hpv_contig_1 | 7,859 | EF546471, HPV53 | 100.0 | 100.0 |
| Vagina | SRR064532.hpv_contig_1 | 7,723 | X74476, HPV34 | 99.9 | 100.0 |
| Vagina | SRR064532.hpv_contig_2 | 7,446 | U89349, HPV18 | 99.7 | 100.0 |
| Skin | SRR062408.hpv_contig_1 | 7,143 | Y15173, HPV75 | 98.5 | 99.9 |
| Vagina | SRR062073.hpv_contig_3 | 6,581 | EF202156, HPV45 | 99.9 | 100.0 |
| Vagina | SRR060392.hpv_contig_4 | 5,318 | EF546473, HPV53 | 100.0 | 100.0 |
| Skin | SRR062412.hpv_contig_2 | 5,148 | Y15173, HPV75 | 98.4 | 100.0 |
Contigs of >5,000 bp are included.
The GenBank accession number for the best hit is given.
Unclassified HPV types.
Our results indicate that most of the HPVs in these samples, in particular the abundant viruses, belong to the known HPV types. Several recent papers have reported that new HPV types can be found frequently, especially using a high-throughput sequencing method (19–22, 24). Our analysis of the HMP data set also found some HPV sequences potentially belonging to new HPV types. Among the contigs larger than 500 bp, only one (570 bp) showed low identity (75.7%) to its best hit (type 156), suggesting that this contig originates from a novel HPV type (see Table S3 in the supplemental material). Of the 15,037 HPV reads identified, 232 had a best hit with unclassified HPV sequences from NCBI GenBank at a similarity range of between 85 and 100%, of which those with greater 95% identity were hits to unclassified HPV sequences. However, because of their low abundance, these reads could not be assembled into large contigs required for defining a novel HPV type for further analysis.
Because nearly all known HPV types were identified using HPV consensus primers, additional HPV genera with less similarity to the consensus sequences could have been missed by a simple similarity search. If additional HPV genera exist, it is possible that their nearest homologs would be animal papillomaviruses (PVs). We tested this possibility with a locally constructed searchable PV database containing all the reference genomes of HPV and animal PV strains from the papillomavirus database PaVE (http://pave.niaid.nih.gov). Searching against the local PV database by BLASTN with a cutoff E value of 1E-10 coupled with BLASTN and BLASTX against GenBank did not find any reads with consistent best hits to animal papillomaviruses. These results indicate that the likelihood of detecting animal PV in humans is very low. Lower stringency (cutoff of 1E-10 to 1E-03) could be a more productive way but failed because the large majority of hits were uncertain in identity due to a low level of similarity to known PVs in the local database but a higher level of similarity to non-PV sequences in GenBank. A new algorithm or method is needed to resolve the ambiguity in searching for new HPV genera.
Temporal stability of HPV infection in healthy human subjects.
In this HMP data set, as shown in Table 4, 13 subjects had a total of 34 repeat samples collected at multiple follow-up visits. The large majority of the repeat samples (13 out of 16) had at least one HPV type shared with the original samples. Recurrence of the same HPV types was observed for all repeat vaginal samples (5 repeats) and all repeat oral samples (3 repeats) but was less frequent for the skin (4 out of 6 repeats) and gut (1 out of 2 repeats).
TABLE 4.
HPV persistence in repeat samples in healthy human subjects
| Organ | Subject | Sample | Body site | HPV typea |
|---|---|---|---|---|
| Mouth | 159591683 | SRS013234 | Tongue dorsum | HPV20, HPV135, HPV147 |
| SRS024580 | Tongue dorsum | HPV20, HPV135, HPV17 | ||
| 604812005 | SRS043646 | Buccal mucosa | HPV32 | |
| SRS046623 | Buccal mucosa | HPV32, HPV107 | ||
| SRS045127 | Tongue dorsum | HPV32 | ||
| SRS055426 | Tongue dorsum | HPV32, HPV5 | ||
| Vagina | 638754422 | SRS022158 | Posterior fornix | HPV18, HPV34, HPV39, HPV56, HPV90, N |
| SRS048536 | Posterior fornix | HPV18, HPV34, HPV39, HPV56, HPV90, N, HPV103, HPV106, HPV51, HPV6, HPV73, HPV74 | ||
| 763577454 | SRS014465 | Vaginal introitus | HPV90 | |
| SRS015071 | Vaginal introitus | HPV90 | ||
| SRS014494 | Posterior fornix | HPV90, HPV45 | ||
| SRS015073 | Posterior fornix | HPV90 | ||
| 158883629 | SRS023850 | Posterior fornix | HPV91 | |
| SRS075419 | Posterior fornix | HPV91 | ||
| 763820215 | SRS014343 | Posterior fornix | HPV89, HPV131, HPV134, HPV52, N | |
| SRS054962 | Posterior fornix | HPV89, HPV118, N | ||
| Gut | 763678604 | SRS014235 | Stool | HPV47 |
| SRS019685 | Stool | HPV47 | ||
| 246515023 | SRS057717 | Stool | HPV68 | |
| SRS023346 | Stool | HPV101 | ||
| Skin | 763961826 | SRS019019 | Anterior nares | HPV107, HPV23 |
| SRS014682 | Anterior nares | HPV124, HPV14 | ||
| SRS019039 | Anterior nares | |||
| 763961826 | SRS019033 | Right retroauricular crease | HPV24, HPV75, HPV144, HPV92, HPV121, HPV130, N | |
| SRS019016 | Right retroauricular crease | HPV24, HPV75, HPV144, HPV92, HPV120, HPV104, HPV119, HPV147, HPV149, HPV153, HPV8, N | ||
| 764083206 | SRS015430 | Anterior nares | HPV148, HPV127, HPV149, HPV131, N | |
| SRS015450 | Anterior nares | HPV148, HPV127, HPV144, HPV132, HPV141, N | ||
| SRS019067 | Anterior nares | HPV148, HPV122, HPV149, HPV10, HPV115, HPV147, N | ||
| SRS019064 | Right retroauricular crease | HPV126, HPV14 | ||
| SRS019081 | Right retroauricular crease | HPV126 | ||
| 765094712 | SRS018585 | Anterior nares | HPV112, HPV132, HPV136, HPV148, N | |
| SRS051600 | Anterior nares | HPV119, HPV130, N | ||
| 159490532 | SRS017044 | Anterior nares | HPV12, HPV19, HPV28 | |
| SRS044474 | Anterior nares | HPV12, HPV19, HPV142, HPV17, HPV19, HPV88, N, HPV119, HPV134, HPV147 |
HPV types that were observed in multivisit samples are in boldface type. N, unclassified HPV type.
DISCUSSION
Our novel approach to survey for HPV sequences using shotgun metagenomics sequence data detected HPV DNA in 68.9% of healthy human subjects who do not have classical HPV-associated diseases. Among these subjects, the prevalence of HPV was found to be 17.3% in the gut, 30.0% in the mouth, 61.3% in the skin, and 41.5% in the vagina. This high prevalence could be attributed mostly to two factors. First, this study analyzed samples from multiple body sites. From each subject, up to 8 body sites in 3 or 4 organs were sampled. The cumulative prevalence increased from 33.3% (8/24) in subjects with one or two organs sampled to 79.8% (63/79) in subjects with 3 or 4 organs sampled. In comparison, most of the previous studies of healthy human subjects focused on only one body site, the vagina, and none on three or more sites. In previous studies, HPV prevalence in the vagina as determined using PCR-based assays ranges from 19.6% to 44.8% among healthy populations in North America (35), 53% on skin samples from Americans by using a multiplex serology method (31) and up to 77.8% of those from the Chinese by using an amplicon-based method (36), and on average 11% in the mouth (from 0% to 81%) (37–45). Second, the shotgun metagenomics sequencing method provides a broader coverage of HPV types than the amplicon-based PCR method. HPV genomes are far too heterogeneous to allow for design of a universal primer pair to amplify all 155 known HPV types. Because of this limitation, nearly all methods or HPV detection kits focus on a few HPV types that are known to cause cervical cancer in humans. With these kits, for example, based on PGMY09/11 primers, approximately 30 to 40 HPV types are detected in the vagina and cervix (46). Because it nonselectively sequences all DNA in a sample, in theory, shotgun sequencing is capable of detecting all 155 HPV types (47). In this study, although the vaginal samples were collected from healthy women without an HPV diagnosis within 2 years, in the vaginal samples, the shotgun method detected 43 HPV types, which include not only the already characterized high- and low-risk types but also many other types invisible to the current HPV detection method. Our study also suggests that the HPV prevalence in healthy human subjects could be higher if more body sites were sampled or if deeper shotgun sequencing were performed.
The significance of high HPV prevalence in healthy human subjects is unknown. Although the finding that the relative abundance of HPV was very low in a significant number of samples suggests that the HPV reads in these samples could be due to incidental contact with an environmental source that harbors HPV, our observations favor true HPV infection over incidental exposure. First, the HPV reads from the vagina and skin had their organ tropism, which would be unnecessary in the case of transitory exposure. For example, most HPV types identified in the skin samples belonged to the genera Betapapillomavirus and Gammapapillomavirus, while those from the vaginal samples to the genus Alphapapillomavirus (Fig. 8). Second, the marked temporal stability of the HPV community is consistent with colonization, as transitory exposure would yield a random profile. Thus, our finding of HPV in most healthy human subjects suggests that there is a form of subclinical HPV infection. The time factor may play a role in disease manifestation. Given that most of the 109 HPV types identified in our survey have previously been associated with various diseases, our finding of these HPV types in healthy human subjects could represent a preclinical form of HPV infection that had not yet developed to detectable disease. The preclinical and subclinical HPV infections might not develop into a clinical disease under the suppression of a competent immune system. For example, out of the 14 HPV types designated high-risk for cervical cancer by the IARC (48), 10 (types 16, 18, 39, 45, 51, 52, 56, 59, 66, and 68) were present in the vagina of nine women with no clinical diseases. Some high-risk HPV types were also detected in samples from the skin (types 18, 45, and 58), gut (types 18, 58, and 68), and mouth (type 66) in healthy subjects. Only three high-risk HPV types (types 31, 33, and 35) were not found in this group of healthy human subjects.
HPV causes nearly all cases of cervical cancer (49). Some of the known HPV types (http://pave.niaid.nih.gov) can be classified as “low risk” (wart causing) or “high risk” (cancer causing) for cervical cancers. The 14 high-risk HPV types cause 100% of cases of cervical cancers, 60 to 65% of cases of vaginal cancer, and 20 to 25% of cases of vulvar cancer (50). There are 12 low-risk HPV types that cause genital warts and are rarely associated with cervical cancers. Current HPV detection kits were designed to cover the HPV types present in the cervical region, with an emphasis on detecting high- and low-risk HPV types (51–53). Our findings indicate that the cervical HPV detection kits may not be appropriate for use in detection of HPV in other organs because of the difference in organ tropisms of HPV types as shown in the phylogenetic analysis and the difference in dominant HPV types between organs (Fig. 2, 3, and 8). For example, the vaginal HPV types are mainly in the genus Alphapapillomavirus, and the high-risk HPV types clustered together in only two small branches. Most of the skin types, however, belong to the genera Beta- and Gammapapillomavirus. The oral HPV types were dispersed in the genera Alpha-, Beta-, Gamma- and Mupapillomavirus. The oral HPV community is more similar to that of the skin than to that of the vagina. Of the 33 oral HPV types and 43 vaginal HPV types identified, only 9 were shared by these two organs, while 25 were shared by the mouth and skin. Also, the vagina and mouth are different in the most abundant HPV types (types 34, 53, 45, and 52 for the vagina and types 32 and 144 for the mouth) and in the most prevalent HPV types (types 90 for the vagina and type 32 for the mouth). The association between HPV and oral cancer has long been suspected but is controversial because HPV is detectable in only a small proportion of oral cancer cases by using the cervical HPV detection kits. Our findings indicate that the cervical detection kit could have missed the majority of HPV types in the mouth and that the underestimation of HPV prevalence in oral cavity cancer could be corrected by using a kit that has a broad coverage of HPV types or is specially designed based on new knowledge of HPV types in the mouth. The same could be true for other cancers outside the genital reproductive system, as suggested by a recent observation that Betapapillomavirus HPV infections might increase the risk of squamous cell carcinomas of the skin (32). The role of noncervical HPV types in cancers of other body sites needs further investigation with appropriate methods, and “high-risk” types could be redefined according to different organs.
It is known that most cervical-vaginal HPV infections in young females are temporary, lasting <1 year, with little long-term significance (54), and only a very small fraction of women infected with oncogenic HPV types eventually develop cervical cancer. However, it is unclear what factors determine the outcome of an HPV infection. Our metagenomics analysis revealed a complex normal HPV community in healthy human subjects that was only partially visible by the current HPV detection methods. Previous studies have reported coinfection with multiple HPV types in about 50% of HPV-positive cervical samples (55, 56), and coinfection of multiple HPV types is one of the factors that may influence the persistence of HPV infection (57). Similarly, we found that more than 50% of healthy subjects had coinfection with two to three HPV types in the vagina (13/25 samples) as well as in the skin (51/72 samples).
From an ecological perspective, nonrandom organization of HPV types is indicative of competitive or facilitative interactions among microbes. The cooccurrence patterns detected here based on viral relative abundances across many samples from the skin or vagina are unlike the biogeographical “cooccurrence” patterns of specific types within subjects from other studies (58). The mechanisms leading to any of the putative interactions we detected between HPV types in the skin or vagina may be quite diverse. Moreover, there is accumulating evidence for complex interactions between viruses and the immune environment in the genital tract (59, 60) and between microbial flora and viruses in the gut (61, 62). While our analysis provides a survey of putative interactions between HPV types in health, future studies of possible interactions with members of the bacterial flora and other members of the viral flora not assayed here are needed. Although the physiological role of the HPV community is yet to be defined, it can be hypothesized that some nononcogenic HPV types could reduce the cancer risk by eliminating oncogenic viral infection with cross immunity (63) and/or viral interference (56). In contrast, other HPV types might increase the cancer risk by maintaining a supportive cellular environment favoring a persistent infection in a coinfection with oncogenic HPV. Perturbation experiments with a high temporal resolution are required to define the nature of HPV infections in healthy human subjects.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants UH3CA140233 from the Human Microbiome Project of the NIH Roadmap Initiative and U01CA18237 from the National Cancer Institute and by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development.
Footnotes
Published ahead of print 12 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00093-14.
REFERENCES
- 1.Van Doorslaer K, Tan Q, Xirasagar S, Bandaru S, Gopalan V, Mohamoud Y, Huyen Y, McBride AA. 2013. The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res. 41:D571–D578. 10.1093/nar/gks984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Villiers EM. 2013. Cross-roads in the classification of papillomaviruses. Virology 445:2–10. 10.1016/j.virol.2013.04.023 [DOI] [PubMed] [Google Scholar]
- 3.Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79. 10.1016/j.virol.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee CA. 2012. Human papillomavirus (HPV) and cervical cancer. J. Insur. Med. 43:178–181 [PubMed] [Google Scholar]
- 5.Chelimo C, Wouldes TA, Cameron LD, Elwood JM. 2013. Risk factors for and prevention of human papillomaviruses (HPV), genital warts and cervical cancer. J. Infect. 66:207–217. 10.1016/j.jinf.2012.10.024 [DOI] [PubMed] [Google Scholar]
- 6.Bian ML, Cheng JY, Ma L, Cong X, Liu J, Chen Y, Chen X. 2013. Evaluation of the detection of 14 high-risk human papillomaviruses with HPV 16 and HPV 18 genotyping for cervical cancer screening. Exp. Ther. Med. 6:1332–1336. 10.3892/etm.2013.1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiMaio D, Liao JB. 2006. Human papillomaviruses and cervical cancer. Adv. Virus Res. 66:125–159. 10.1016/S0065-3527(06)66003-X [DOI] [PubMed] [Google Scholar]
- 8.Dell G, Gaston K. 2001. Human papillomaviruses and their role in cervical cancer. Cell. Mol. Life Sci. 58:1923–1942. 10.1007/PL00000827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sycuro LK, Xi LF, Hughes JP, Feng Q, Winer RL, Lee SK, O'Reilly S, Kiviat NB, Koutsky LA. 2008. Persistence of genital human papillomavirus infection in a long-term follow-up study of female university students. J. Infect. Dis. 198:971–978. 10.1086/591625 [DOI] [PubMed] [Google Scholar]
- 10.Gillison ML. 2008. Human papillomavirus-related diseases: oropharynx cancers and potential implications for adolescent HPV vaccination. J. Adolesc. Health 43:S52–S60. 10.1016/j.jadohealth.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isayeva T, Li Y, Maswahu D, Brandwein-Gensler M. 2012. Human papillomavirus in non-oropharyngeal head and neck cancers: a systematic literature review. Head Neck Pathol. 6(Suppl 1):S104–S120. 10.1007/s12105-012-0368-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkin DM. 2006. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 118:3030–3044. 10.1002/ijc.21731 [DOI] [PubMed] [Google Scholar]
- 13.Munoz N, Castellsague X, de Gonzalez AB, Gissmann L. 2006. Chapter 1: HPV in the etiology of human cancer. Vaccine 24(Suppl 3):S3/1–S3/10. 10.1016/j.vaccine.2005.01.101 [DOI] [PubMed] [Google Scholar]
- 14.Parkin DM, Bray F. 2006. Chapter 2: the burden of HPV-related cancers. Vaccine 24(Suppl 3):S3/11–S3/25. 10.1016/j.vaccine.2005.01.101 [DOI] [PubMed] [Google Scholar]
- 15.Tang KW, Alaei-Mahabadi B, Samuelsson T, Lindh M, Larsson E. 2013. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat. Commun. 4:2513. 10.1038/ncomms3513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clifford GM, Gallus S, Herrero R, Munoz N, Snijders PJ, Vaccarella S, Anh PT, Ferreccio C, Hieu NT, Matos E, Molano M, Rajkumar R, Ronco G, de Sanjose S, Shin HR, Sukvirach S, Thomas JO, Tunsakul S, Meijer CJ, Franceschi S, IARC HPV Prevalence Surveys Study Group. 2005. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis. Lancet 366:991–998. 10.1016/S0140-6736(05)67069-9 [DOI] [PubMed] [Google Scholar]
- 17.Antonsson A, Forslund O, Ekberg H, Sterner G, Hansson BG. 2000. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J. Virol. 74:11636–11641. 10.1128/JVI.74.24.11636-11641.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bottalico D, Chen Z, Kocjan BJ, Seme K, Poljak M, Burk RD. 2012. Characterization of human papillomavirus type 120: a novel betapapillomavirus with tropism for multiple anatomical niches. J. Gen. Virol. 93:1774–1779. 10.1099/vir.0.041897-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansson H, Bzhalava D, Ekstrom J, Hultin E, Dillner J, Forslund O. 2013. Metagenomic sequencing of “HPV-negative” condylomas detects novel putative HPV types. Virology 440:1–7. 10.1016/j.virol.2013.01.023 [DOI] [PubMed] [Google Scholar]
- 20.Mokili JL, Dutilh BE, Lim YW, Schneider BS, Taylor T, Haynes MR, Metzgar D, Myers CA, Blair PJ, Nosrat B, Wolfe ND, Rohwer F. 2013. Identification of a novel human papillomavirus by metagenomic analysis of samples from patients with febrile respiratory illness. PLoS One 8:e58404. 10.1371/journal.pone.0058404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocjan BJ, Steyer A, Sagadin M, Hosnjak L, Poljak M. 2013. Novel human papillomavirus type 174 from a cutaneous squamous cell carcinoma. Genome Announc. 1(4):e00445–13. 10.1128/genomeA.00445-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, Pariente K, Segondy M, Burguiere A, Manuguerra JC, Caro V, Eloit M. 2012. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One 7:e38499. 10.1371/journal.pone.0038499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Barry P, Yeh E, Glaser C, Schnurr D, Delwart E. 2009. Identification of a novel human gammapapillomavirus species. J. Gen. Virol. 90:2413–2417. 10.1099/vir.0.012344-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekstrom J, Bzhalava D, Svenback D, Forslund O, Dillner J. 2011. High throughput sequencing reveals diversity of Human Papillomaviruses in cutaneous lesions. Int. J. Cancer 129:2643–2650. 10.1002/ijc.26204 [DOI] [PubMed] [Google Scholar]
- 25.NIH HMP Working Group, Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. 2009. The NIH Human Microbiome Project. Genome Res. 19:2317–2323. 10.1101/gr.096651.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McInnes P, Cutting M. 29 July 2010. Manual of procedures for Human Microbiome Project. Core microbiome sampling protocol A, HMP protocol # 07-001. Version number: 12.0. http://hmpdacc.org/doc/HMP_MOP_Version12_0_072910.pdf
- 28.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman J, Alm EJ. 2012. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8:e1002687. 10.1371/journal.pcbi.1002687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. 2004. Classification of papillomaviruses. Virology 324:17–27. 10.1016/j.virol.2004.03.033 [DOI] [PubMed] [Google Scholar]
- 31.Karagas MR, Nelson HH, Sehr P, Waterboer T, Stukel TA, Andrew A, Green AC, Bavinck JN, Perry A, Spencer S, Rees JR, Mott LA, Pawlita M. 2006. Human papillomavirus infection and incidence of squamous cell and basal cell carcinomas of the skin. J. Natl. Cancer Inst. 98:389–395. 10.1093/jnci/djj092 [DOI] [PubMed] [Google Scholar]
- 32.Karagas MR, Waterboer T, Li Z, Nelson HH, Michael KM, Bavinck JN, Perry AE, Spencer SK, Daling J, Green AC, Pawlita M. 2010. Genus beta human papillomaviruses and incidence of basal cell and squamous cell carcinomas of skin: population based case-control study. BMJ 341:c2986. 10.1136/bmj.c2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovanda A, Kocjan BJ, Potocnik M, Poljak M. 2011. Characterization of a novel cutaneous human papillomavirus genotype HPV-125. PLoS One 6:e22414. 10.1371/journal.pone.0022414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention. 2012. Human papillomavirus: epidemiology and prevention of vaccine-preventable diseases, 12th ed. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/vaccines/pubs/pinkbook/hpv.html [Google Scholar]
- 35.Dunne EF, Unger ER, Sternberg M, McQuillan G, Swan DC, Patel SS, Markowitz LE. 2007. Prevalence of HPV infection among females in the United States. JAMA 297:813–819. 10.1001/jama.297.8.813 [DOI] [PubMed] [Google Scholar]
- 36.Li J, Pan Y, Xu Z, Wang Q, Hang D, Shen N, Liu M, Zhang C, Abliz A, Deng Q, Cai H, Ke Y. 2013. Improved detection of human papillomavirus harbored in healthy skin with FAP6085/64 primers. J. Virol. Methods 193:633–638. 10.1016/j.jviromet.2013.06.026 [DOI] [PubMed] [Google Scholar]
- 37.Sanchez-Vargas LO, Diaz-Hernandez C, Martinez-Martinez A. 2010. Detection of human papilloma virus (HPV) in oral mucosa of women with cervical lesions and their relation to oral sex practices. Infect. Agents Cancer 5:25. 10.1186/1750-9378-5-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreimer AR, Bhatia RK, Messeguer AL, Gonzalez P, Herrero R, Giuliano AR. 2010. Oral human papillomavirus in healthy individuals: a systematic review of the literature. Sex. Transm. Dis. 37:386–391. 10.1097/OLQ.0b013e3181c94a3b [DOI] [PubMed] [Google Scholar]
- 39.Esquenazi D, Bussoloti Filho I, Carvalho Mda G, Barros FS. 2010. The frequency of human papillomavirus findings in normal oral mucosa of healthy people by PCR. Braz. J. Otorhinolaryngol. 76:78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castellsague X, Drudis T, Canadas MP, Gonce A, Ros R, Perez JM, Quintana MJ, Munoz J, Albero G, de Sanjose S, Bosch FX. 2009. Human papillomavirus (HPV) infection in pregnant women and mother-to-child transmission of genital HPV genotypes: a prospective study in Spain. BMC Infect. Dis. 9:74. 10.1186/1471-2334-9-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rintala MA, Grenman SE, Puranen MH, Isolauri E, Ekblad U, Kero PO, Syrjanen SM. 2005. Transmission of high-risk human papillomavirus (HPV) between parents and infant: a prospective study of HPV in families in Finland. J. Clin. Microbiol. 43:376–381. 10.1128/JCM.43.1.376-381.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jay N, Moscicki AB. 2000. Human papillomavirus infections in women with HIV disease: prevalence, risk, and management. AIDS Read. 10:659–668 [PubMed] [Google Scholar]
- 43.Miller CS, White DK. 1996. Human papillomavirus expression in oral mucosa, premalignant conditions, and squamous cell carcinoma: a retrospective review of the literature. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 82:57–68. 10.1016/S1079-2104(96)80378-7 [DOI] [PubMed] [Google Scholar]
- 44.Kellokoski J, Syrjanen S, Yliskoski M, Syrjanen K. 1992. Dot blot hybridization in detection of human papillomavirus (HPV) infections in the oral cavity of women with genital HPV infections. Oral Microbiol. Immunol. 7:19–23. 10.1111/j.1399-302X.1992.tb00014.x [DOI] [PubMed] [Google Scholar]
- 45.Rautava J, Willberg J, Louvanto K, Wideman L, Syrjanen K, Grenman S, Syrjanen S. 2012. Prevalence, genotype distribution and persistence of human papillomavirus in oral mucosa of women: a six-year follow-up study. PLoS One 7:e42171. 10.1371/journal.pone.0042171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuessel Haws AL, He Q, Rady PL, Zhang L, Grady J, Hughes TK, Stisser K, Konig R, Tyring SK. 2004. Nested PCR with the PGMY09/11 and GP5(+)/6(+) primer sets improves detection of HPV DNA in cervical samples. J. Virol. Methods 122:87–93. 10.1016/j.jviromet.2004.08.007 [DOI] [PubMed] [Google Scholar]
- 47.Eisen JA. 2007. Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes. PLoS Biol. 5:e82. 10.1371/journal.pbio.0050082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaccarella S, Herrero R, Dai M, Snijders PJ, Meijer CJ, Thomas JO, Hoang Anh PT, Ferreccio C, Matos E, Posso H, de Sanjose S, Shin HR, Sukvirach S, Lazcano-Ponce E, Ronco G, Rajkumar R, Qiao YL, Munoz N, Franceschi S. 2006. Reproductive factors, oral contraceptive use, and human papillomavirus infection: pooled analysis of the IARC HPV prevalence surveys. Cancer Epidemiol. Biomarkers Prev. 15:2148–2153. 10.1158/1055-9965.EPI-06-0556 [DOI] [PubMed] [Google Scholar]
- 49.Mendez F, Munoz N, Posso H, Molano M, Moreno V, van den Brule AJ, Ronderos M, Meijer C, Munoz A. 2005. Cervical coinfection with human papillomavirus (HPV) types and possible implications for the prevention of cervical cancer by HPV vaccines. J. Infect. Dis. 192:1158–1165. 10.1086/444391 [DOI] [PubMed] [Google Scholar]
- 50.Burd EM. 2003. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 16:1–17. 10.1128/CMR.16.1.1-17.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kocken M, Uijterwaal MH, de Vries AL, Berkhof J, Ket JC, Helmerhorst TJ, Meijer CJ. 2012. High-risk human papillomavirus testing versus cytology in predicting post-treatment disease in women treated for high-grade cervical disease: a systematic review and meta-analysis. Gynecol. Oncol. 125:500–507. 10.1016/j.ygyno.2012.01.015 [DOI] [PubMed] [Google Scholar]
- 52.Nishino HT, Tambouret RH, Wilbur DC. 2011. Testing for human papillomavirus in cervical cancer screening: a review of indications and methodology. Cancer Cytopathol. 119:219–227. 10.1002/cncy.20161 [DOI] [PubMed] [Google Scholar]
- 53.Poljak M, Kocjan BJ. 2010. Commercially available assays for multiplex detection of alpha human papillomaviruses. Expert Rev. Anti Infect. Ther. 8:1139–1162. 10.1586/eri.10.104 [DOI] [PubMed] [Google Scholar]
- 54.Goldstein MA, Goodman A, del Carmen MG, Wilbur DC. 2009. Case records of the Massachusetts General Hospital. Case 10-2009—a 23-year-old woman with an abnormal Papanicolaou smear. N. Engl. J. Med. 360:1337–1344. 10.1056/NEJMcpc0810837 [DOI] [PubMed] [Google Scholar]
- 55.Goldman B, Rebolj M, Rygaard C, Preisler S, Ejegod DM, Lynge E, Bonde J. 2013. Patterns of cervical coinfection with multiple human papilloma virus types in a screening population in Denmark. Vaccine 31:1604–1609. 10.1016/j.vaccine.2012.12.084 [DOI] [PubMed] [Google Scholar]
- 56.Mejlhede N, Pedersen BV, Frisch M, Fomsgaard A. 2010. Multiple human papilloma virus types in cervical infections: competition or synergy? APMIS 118:346–352. 10.1111/j.1600-0463.2010.2602.x [DOI] [PubMed] [Google Scholar]
- 57.Asiaf A, Ahmad ST, Mohammad SO, Zargar MA. 14 October 2013. Review of the current knowledge on the epidemiology, pathogenesis, and prevention of human papillomavirus infection. Eur. J. Cancer Prev. 10.1097/CEJ.0b013e328364f273 [DOI] [PubMed] [Google Scholar]
- 58.Pons-Salort M, Letort V, Favre M, Heard I, Dervaux B, Opatowski L, Guillemot D. 2013. Exploring individual HPV coinfections is essential to predict HPV-vaccination impact on genotype distribution: a model-based approach. Vaccine 31:1238–1245. 10.1016/j.vaccine.2012.11.098 [DOI] [PubMed] [Google Scholar]
- 59.Auvert B, Marais D, Lissouba P, Zarca K, Ramjee G, Williamson AL. 2011. High-risk human papillomavirus is associated with HIV acquisition among South African female sex workers. Infect. Dis. Obstet. Gynecol. 2011:692012. 10.1155/2011/692012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marais DJ, Carrara H, Ramjee G, Kay P, Williamson AL. 2009. HIV-1 seroconversion promotes rapid changes in cervical human papillomavirus (HPV) prevalence and HPV-16 antibodies in female sex workers. J. Med. Virol. 81:203–210. 10.1002/jmv.21343 [DOI] [PubMed] [Google Scholar]
- 61.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334:249–252. 10.1126/science.1211057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. 2011. Successful transmission of a retrovirus depends on the commensal microbiota. Science 334:245–249. 10.1126/science.1210718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jenkins D. 2008. A review of cross-protection against oncogenic HPV by an HPV-16/18 AS04-adjuvanted cervical cancer vaccine: importance of virological and clinical endpoints and implications for mass vaccination in cervical cancer prevention. Gynecol. Oncol. 110:S18–S25. 10.1016/j.ygyno.2008.06.027 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.