Abstract
Human dipeptidyl peptidase 4 (hDPP4) was recently identified as the receptor for Middle East respiratory syndrome coronavirus (MERS-CoV) infection, suggesting that other mammalian DPP4 orthologs may also support infection. We demonstrate that mouse DPP4 cannot support MERS-CoV infection. However, employing mouse DPP4 as a scaffold, we identified two critical amino acids (A288L and T330R) that regulate species specificity in the mouse. This knowledge can support the rational design of a mouse-adapted MERS-CoV for rapid assessment of therapeutics.
TEXT
Middle East respiratory syndrome coronavirus (MERS-CoV) is a recently identified betacoronavirus that can infect the lower respiratory airway of humans, leading to acute respiratory distress syndrome (ARDS) with ∼43% mortality in hospitalized individuals (1). Disease symptoms associated with MERS-CoV are similar to those of severe acute respiratory syndrome coronavirus (SARS-CoV); however, MERS-CoV is phylogenetically more closely related to the bat coronaviruses HKU4 and HKU5 (2, 3). MERS-CoV also differs from SARS-CoV in terms of receptor usage, where MERS-CoV utilizes dipeptidyl peptidase 4 (DPP4) as an entry receptor (4). Given the importance of MERS-CoV as an emerging pathogen, there is a clear need for the development of new therapeutics, which requires the appropriate animal models. However, to date, nonhuman primates are the only reported animal model for MERS-CoV replication, while traditional small animal models, such as ferrets (5), hamsters (6), and mice (7), are nonpermissive. Given that species-specific differences in DPP4 may confound animal model development, it is important to identify determinants in DPP4 that govern MERS-CoV host range. Knowledge of DPP4 determinants may provide novel insights into interactions between DPP4 and MERS-CoV spike receptor binding domain (RBD), as well as support development of new small animal models.
To test whether mouse DPP4 (mDPP4) is capable of acting as an entry receptor for MERS-CoV, we compared mDPP4 with human DPP4 (hDPP4). An ectopic expression system was utilized to constitutively express mDPP4 and hDPP4 in human embryonic kidney 293T (HEK 293T) cells, which lack detectable expression of endogenous hDPP4 (data not shown). Human DPP4 and mouse DPP4 were expressed either as full-length proteins or as fusions to the Venus protein at the carboxy terminus. HEK 293T cells were transfected with 3 μg of the indicated DPP4 expression plasmid, and at ∼20 h posttransfection, cells were infected at a multiplicity of infection (MOI) of 5 with a recombinant MERS-CoV strain designed to express tomato red fluorescent protein (rMERS-CoV-red) (Fig. 1). The rMERS-CoV-red virus is derived from the EMC2012 substrain and was previously shown to infect and replicate in a manner similar to wild-type MERS virus (8). Transfection of the DPP4-Venus fusion constructs resulted in high transfection efficiency (nearly 100%) (Fig. 1A and B). Control HEK 293T cells were poorly permissive for MERS-CoV, while cells overexpressing hDPP4 were readily infected with rMERS-CoV-red virus (Fig. 1A), whereas mDPP4 overexpression did not support infection (Fig. 1B). Notably, the Venus protein does not interfere with the capacity of hDPP4 to allow viral infection (Fig. 1A and B). Despite the inability of mDPP4 to confer infection of rMERS-CoV-red, we could readily detect expression of the mDPP4 protein in 293T cells (Fig. 1C). Moreover, MERS-CoV S and N proteins were detected in cells expressing hDPP4 but not mDPP4 (Fig. 1D). These results indicate that the mDPP4 protein cannot support MERS-CoV infection, whereas hDPP4 readily promotes infection, as demonstrated previously (4).
FIG 1.
Mouse DPP4 (mDPP4) does not support MERS-CoV infection. HEK 293T cells were transfected with 3 μg of plasmid expressing human DPP4 (hDPP4) or hDPP4-Venus fusion (A) and mDPP4 or mDPP4-venus fusion (B). At ∼20 h posttransfection, cells were infected with rMERS-CoV-red virus at an MOI of 5. Venus fusion proteins were assessed by fluorescence microscopy at 48 h posttransfection. In independent experiments, infection with rMERS-CoV-red virus was assessed for red cells by fluorescence microscopy at ∼18 h postinfection. (C) Western blot analysis demonstrates overexpression of mDPP4 and hDPP4. Extracts were prepared at ∼48 h posttransfection using AV lysis buffer (3), and samples were heat inactivated for 60 min at 90°C for removal from a biosafety level 3 (BSL3) facility and resolved on an 8% SDS-PAGE gel. Blots were probed with primary goat-anti-DPP4 polyclonal antibody (R&D Systems) at 1:1,000 in 1× Tris-buffered saline–Tween (TBST) or goat anti-actin polyclonal antibody (Santa Cruz) and detected with a secondary rabbit anti-goat–horseradish peroxidase (HRP)-conjugated antibody (Sigma) at 1:10,000 in 1× TBST in 5% milk. (D) Western blot analysis of MERS-CoV S and N proteins. Lysates were collected at ∼18 h postinfection and treated as in panel C. Blots were probed with primary mouse polyclonal antiserum at 1:400, raised to S and N proteins as described previously (3), and detected with a secondary goat anti-mouse–HRP (GE Healthcare) at 1:10,000 in 1× TBST in 5% milk.
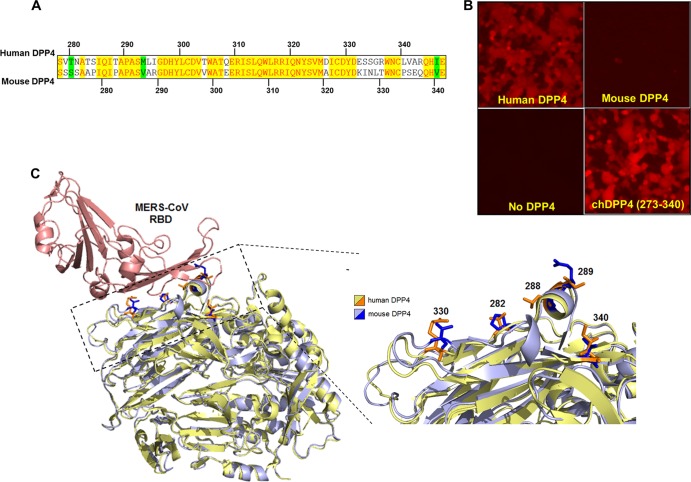
The recently published crystal structures of the MERS spike protein interacting with hDPP4 revealed a number of specific amino acids in blades IV and V of the β-propeller domain from hDPP4 that may facilitate an interaction with the spike protein receptor binding domain (RBD) (9, 10). Alignment of hDPP4 and mDPP4 sequences in this region exhibits amino acid variation that may account for differences in MERS-CoV susceptibility (Fig. 2A). We interrogated this region for the potential to confer infection onto mouse DPP4 using overlap extension PCR to replace amino acids 273 to 340 in mDPP4 with amino acids 279 to 346 of human DPP4 (Fig. 2A). Mouse DPP4 proteins bearing human DPP4 amino acids are referred to as “chimeric DPP4” (chDPP4) molecules, with the amino acid numbers relative to mDPP4 following in parentheses. Cells expressing chDPP4(273–340) were as susceptible to rMERS-CoV-red infection as cells expressing wild-type hDPP4 (Fig. 2B). These results clearly indicate that the hDPP4 region comprising amino acids 279 to 346 confers to mDPP4 the ability to support infection. In an effort to understand if mDPP4 is structurally distinct from hDPP4 in this region, we employed three-dimensional (3D) molecular visualization using PyMOL software. Overlaying mDPP4 on hDPP4 shows that the two proteins are highly similar in structure (Fig. 2C). This is especially important at the interface with the MERS spike RBD, where structural differences are anticipated based on distinct differences in susceptibility to MERS infection.
FIG 2.
Blades IV and V from the β-propeller of hDPP4 make mDPP4 permissible to MERS-CoV infection. (A) Vector NTI protein sequence alignment of human (top strand) and mouse (bottom strand) DPP4 molecules. Yellow highlighted regions indicate conserved amino acids, white regions signify amino acids that are functionally different (i.e., hydrophobic and hydrophilic), and green highlighting indicates amino acids that are different but functionally similar (i.e., the threonine and serine are both polar and uncharged). (B) HEK 293T cells were transfected with the indicated DPP4. At ∼20 h posttransfection, cells were infected with rMERS-CoV-red virus at an MOI of 5, and infection was assessed ∼18 h postinfection by fluorescence microscopy. (C) 3D molecular PyMOL software was employed to visualize the mDPP4 structure overlaid onto the hDPP4 structure. The hDPP4 structure was based upon the crystal structure resolved in context with the MERS S RBD (PDB code 4L72). MERS S protein is displayed in red, hDPP4 in yellow, and mDPP4 in blue. The mDPP4 sequence was threaded using the I-TASSER software (11). The expanded view depicts the DPP4 region at the interaction surface. Numbered and highlighted are the specific amino acids chosen for mutation in the mDPP4 protein.
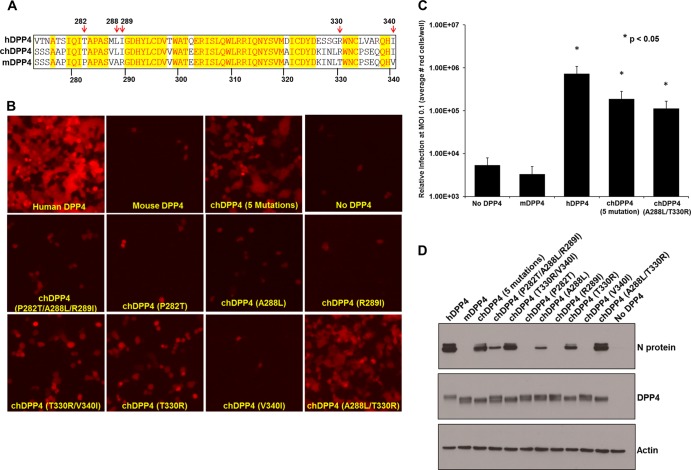
In the absence of obvious structural differences, we hypothesized that specific amino acids contribute to infection of cells that overexpress chDPP4(273–340). To confirm this, we initially mutated five functionally variant surface amino acids in this region of mDPP4 to the corresponding amino acid from hDPP4 using overlap extension PCR to generate various chDPP4 molecules (Fig. 3A). HEK 293T cells were transfected with the indicated chDPP4 constructs followed by infection with rMERS-CoV-red at an MOI of 1. A chDPP4 molecule containing all five mutations (P282T A288L R289I T330R V340I)—shown as “chDPP4 (5 mutations)” in Fig. 3—promoted highly efficient MERS-CoV infection (Fig. 3B), indicating that one or more of these mutations may be sufficient for infection. Previous structural studies of hDPP4 and the MERS RBD suggest that the interaction may require at least two distinct interactions localized to separate structural domains (10). Our T330R and V340I mutants are localized at or near one domain, while mutations comprising P282T, A288L, and R289I are located in the second domain. Accordingly, each group of mutations (T330R V340I and P282T A288L R289I) was independently introduced into mDPP4 and tested for its capacity to support infection of rMERS-CoV-red (Fig. 3B). Although both groups of mutations influence infection to different degrees, neither set recapitulated the levels of infection seen with all five mutations together (Fig. 3B). Further dissection of each group into single mutants revealed that A288L and T330R were partly responsible for the observed infection from each respective group (Fig. 3B). These results are substantiated by Western blot analysis demonstrating detection of the N protein in infected cells expressing either the A288L or T330R mutation but not in cells expressing chDPP4s with the P282T, R289I, or T330R mutation (Fig. 3D). Combination of these two mutations (chDPP4 A288L T330R) recapitulates the level of infection observed for chDPP4 containing all five mutations and approaches what is observed with hDPP4 (Fig. 3B). Quantitation of MERS-CoV-infected red cells (Fig. 3C) and Western blotting of infected cells (Fig. 3D) substantiate the permissibility of chDPP4 A288L T330R, exhibiting nearly a 1.5-log increase in infection compared to mDPP4 (Fig. 3C). Nevertheless, the mutants did not achieve the level of infection observed with hDPP4, indicating that additional amino acids that differ between mice and humans may also contribute to MERS-CoV infection. Regardless of the DPP4 mutation examined, all were efficiently expressed, and intriguingly, all chDPP4 mutants with the T330R mutation exhibit a shift from a doublet to a single lower-molecular-weight band, potentially indicating glycosylation at this site (Fig. 3D). Additionally, the possibility of a restriction factor in rodent cells could be eliminated since mouse and hamster cells ectopically expressing the indicated DPP4s were permissive to MERS-CoV-red infection, with the exception that mDPP4 does not support infection, as observed with HEK 293T cells (Fig. 4). Our results indicate that a successful infection requires a combination of at least two mutations (A288L and T330R), located at distinct structural domains on blades IV and V of the β-propeller of DPP4. These results are in agreement with previous crystal structure data from hDPP4 and the spike RBD, which suggest the hDPP4 equivalents (L294 and R336) are critical residues for binding and infection (9, 10). Our structure prediction model (Fig. 2B) indicates that A288 and L294 and T330 and R336 exhibit positional differences that may govern distinct functional interactions with the MERS-CoV spike protein RBD. Previous phylogenetic analyses indicate that mouse, bat, and ferret DPP4s are highly divergent from primate DPP4s (4, 5), despite the capacity of bat DPP4 to serve as a functional receptor for MERS-CoV infection (4). Moreover, swapping the hDPP4 region containing blades IV and V of the β-propeller into the ferret DPP4 resulted in a gain of infection, although the exact residues that mediate this function were not identified (5). Taken together, these data indicate that DPP4 proteins may be structurally conserved across mammalian species. Incorporation of the A288L and T330R modifications in the context of the mDPP4 will facilitate generation of a mouse model through the production of transgenics and can be used to adapt the MERS-CoV to use mDPP4.
FIG 3.
MERS-CoV infection is dependent upon specific amino acids in DPP4. (A) Vector NTI protein sequence alignment of hDPP4 (top strand) with chDPP4 (middle strand) and mDPP4 (bottom strand) indicating positions of introduced human mutations with red arrows. (B) HEK 293T cells were transfected with the indicated DPP4 molecule. At ∼20 h posttransfection, cells were infected with rMERS-CoV-red virus at MOI of 1, and infection was assessed ∼18 h postinfection by fluorescence microscopy. (C) In an independent experiment, cells overexpressing the indicated DPP4 constructs were infected with rMERS-CoV-red virus at MOI of 0.1, 0.01, and 0.001 on six-well plates. At ∼18 h postinfection, cells were scored at the following MOI: no DPP4 and mDPP4, 0.1; chDPP4 P282T A288L R289I T330R V340I [“chDPP4 (5 mutations)”] and chDPP4 A288L T330R, 0.01; and hDPP4, 0.001. Values were normalized to an MOI of 0.1 and expressed as relative infection at 0.1. Human DPP4, chDPP4 P282T A288L R289I T330R V340I, and chDPP4 A288L T330R showed a significant increase in infection over mDPP4 (*, P < 0.05, Student's t test). (D) Western blots demonstrating overexpression of hDPP4, mDPP4, and each chDPP4 molecule, N protein of infected cells, and β-actin as a loading control. Western blots were prepared and probed as described in Fig. 1C and D.
FIG 4.
Human and chimeric DPP4 molecules can support MERS-CoV infection in hamster and mouse cells. (A) Baby hamster kidney 21 (BHK-21) cells were electroporated with the indicated DPP4 molecules. At ∼20 h posttransfection, cells were seeded in 6-well plates and infected with rMERS-CoV-red at an MOI of 2 at 24 h posttransfection. (B) Mouse NIH 3T3 cells were transfected using Nucleofection (according to the Amaxa procedure) with the indicated DPP4 molecules. Cells were seeded into 12-well plates and infected with rMERS-CoV-red at an MOI of ∼4 at 24 h post-Nucleofection. All infections were assessed at 24 h postinfection by fluorescence microscopy.
ACKNOWLEDGMENTS
We thank members of the Heise and Baric laboratories for review of the manuscript. We also thank Blossom Damania for use of her Nucleofector.
This work was supported by grants NIH U19 AI107810 and NIH HHSN272201000019I-HHSN27200003-Task A57.
Footnotes
Published ahead of print 26 February 2014
REFERENCES
- 1.WHO MERS-CoV Research Group. 12 November 2013. State of knowledge and data gaps of Middle East respiratory syndrome coronavirus (MERS-CoV) in humans. PLoS Curr. 10.1371/currents.outbreaks.0bf719e352e7478f8ad85fa30127ddb8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham RL, Donaldson EF, Baric RS. 2013. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 11:836–848. 10.1038/nrmicro3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agnihothram S, Gopal R, Yount BL, Jr, Donaldson EF, Menachery VD, Graham RL, Scobey TD, Gralinski LE, Denison MR, Zambon M, Baric RS. 18 November 2013. Evaluation of serologic and antigenic relationships between Middle Eastern respiratory syndrome coronavirus and other coronaviruses to develop vaccine platforms for the rapid response to emerging coronaviruses. J. Infect. Dis. 10.1093/infdis/jit609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. 10.1038/nature12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raj VS, Smits SL, Provacia LB, van den Brand JM, Wiersma L, Ouwendijk WJ, Bestebroer TM, Spronken MI, van Amerongen G, Rottier PJ, Fouchier RA, Bosch BJ, Osterhaus AD, Haagmans BL. 2014. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4-mediated entry of the Middle East respiratory syndrome coronavirus. J. Virol. 88:1834–1838. 10.1128/JVI.02935-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Wit E, Prescott J, Baseler L, Bushmaker T, Thomas T, Lackemeyer MG, Martellaro C, Milne-Price S, Haddock E, Haagmans BL, Feldmann H, Munster VJ. 2013. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS One 8:e69127. 10.1371/journal.pone.0069127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman CM, Matthews KL, Goicochea L, Frieman MB. 2013. Wild type and innate immune deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J. Gen. Virol. 95:408–412. 10.1099/vir.0.060640-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. 2013. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 110:16157–16162. 10.1073/pnas.1311542110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF. 2013. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231. 10.1038/nature12328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen YH, Zhang L, Wang X. 2013. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 23:986–993. 10.1038/cr.2013.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]