ABSTRACT
Following human immunodeficiency virus type 1 (HIV-1) entry into the host cell, the viral capsid gradually disassembles in a process called uncoating. A proper rate of uncoating is important for reverse transcription of the HIV-1 genome. Host restriction factors such as TRIM5α and TRIMCyp bind retroviral capsids and cause premature disassembly, leading to blocks in reverse transcription. Other host factors, such as cyclophilin A, stabilize the HIV-1 capsid and are required for efficient infection in some cell types. Here, we show that a heat-labile factor greater than 100 kDa in the cytoplasm of cells from multiple vertebrate species slows the spontaneous disassembly of HIV-1 capsid-nucleocapsid (CA-NC) complexes in vitro. We identified the PDZ domain-containing protein 8 (PDZD8) as a critical component of the capsid-stabilizing activity in the cytoplasmic extracts. PDZD8 has been previously reported to bind the HIV-1 Gag polyprotein and to make a positive contribution to the efficiency of HIV-1 infection (M. S. Henning, S. G. Morham, S. P. Goff, and M. H. Naghavi, J. Virol. 84:8990–8995, 2010, doi:10.1128/JVI.00843–10). PDZD8 knockdown accelerated the disassembly of HIV-1 capsids in infected cells, resulting in decreased reverse transcription. The PDZD8 coiled-coil domain is sufficient for HIV-1 capsid binding, but other parts of the protein, including the PDZ domain, are apparently required for stabilizing the capsid and supporting HIV-1 infection. In summary, PDZD8 interacts with and stabilizes the HIV-1 capsid and thus represents a potentially targetable host cofactor for HIV-1 infection.
IMPORTANCE After human immunodeficiency virus type 1 (HIV-1) gains access to the interior of the target cell, host cell factors can influence virus infection in either a positive or negative way. HIV-1 depends upon certain host cell factors to assist processes that are required for virus replication. One example of such a host factor is PDZD8. This work shows that PDZD8 helps to stabilize the HIV-1 capsid, a huge complex of the viral RNA, enzymes, and protein. When PDZD8 is prevented from interacting with the HIV-1 capsid, the capsid becomes unstable and HIV-1 infection is inhibited. These results show that PDZD8 regulates the uncoating of the HIV-1 capsid. Interfering with the interaction of PDZD8 and capsid could prove to be a useful strategy for intervening in HIV-1 infection and transmission.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) RNA genome is contained within the viral capsid, a conical protein shell composed of ∼1,500 molecules of the capsid (CA) protein (1). The CA proteins of HIV-1 and other retroviruses oligomerize into hexamers by virtue of interactions among the N-terminal domains, and additional interactions involving the C-terminal and N-terminal domains promote interhexamer interactions that allow the capsid structure to be assembled (2, 3). Following virus entry into the host cell cytoplasm, the capsid disassembles and CA proteins gradually disassociate from the viral genome, a process called uncoating.
The process of retroviral uncoating is incompletely understood. Careful regulation of the rate of uncoating is critical to successful retroviral infection. Alterations in uncoating can affect reverse transcription or the nuclear import of viral cDNA. Some changes in CA, as well as in CA-binding small molecules, can alter CA-CA intermolecular interactions and lead to premature viral capsid uncoating; premature uncoating is associated with lower levels of reverse transcription and substantially decreased infectivity (4–6).
Over the past decade, considerable evidence has emerged indicating that the stability of the capsid is also dependent on components of the target cell cytoplasm. The cellular restriction factors TRIM5α and TRIMCyp have been shown to inhibit infection of multiple retroviruses by binding the capsid and inducing premature uncoating (7–9). Conversely, when CPSF6, which is normally located in the nucleus, is aberrantly localized in the cytoplasm, the uncoating of some retroviral capsids is slowed, resulting in decreased infection (10, 11). Some of the inhibitory effects on HIV-1 infection associated with knockdown of TNP03, a nuclear import factor, may be due to secondary effects on CPSF6 localization (12–14). Other cellular proteins affecting the kinetics of retroviral uncoating are positive cofactors of infection. Cyclophilin A, a CA-binding prolyl isomerase, stabilizes the HIV-1 capsid and is required for efficient infection in some cell types (15, 16). As-yet-unidentified host cell factors have been proposed to activate HIV-1 uncoating and induce reverse transcription in vitro (17). The mechanisms by which these cellular factors affect capsid stability and the relative contributions of different host factors to retroviral uncoating are under investigation.
Investigation of cellular cofactors of HIV-1 capsid stability and uncoating has proven particularly difficult due to the inherent instability of the viral cores in vitro, as well as the relatively small percentage of internalized CA protein associated with infectious cores during the early phase of retroviral infection (18). For this reason, we developed an assay to study host cell proteins that affect the stability of in vitro-assembled HIV-1 capsid-nucleocapsid (CA-NC) complexes. These multimeric assemblies of purified CA-NC protein form cylindrical tubes with a hexameric lattice similar to that of the viral capsid (19, 20). The inclusion of the uncleaved NC region of HIV-1 Gag permits oligonucleotide-mediated dimerization of the CA-NC protein, resulting in a capsid analog that is more stable than purified viral cores. Due to their capsid-like hexameric lattice and enhanced stability in various experimental conditions, in vitro-assembled CA-NC complexes have been used in both structural and protein-binding studies in place of the natural viral capsid (19, 21).
Here, we employed CA-NC complexes as the substrates in an assay designed to identify components of host cell cytoplasmic extracts that altered the disassembly of the capsid-like lattice. We found that cytosolic extracts from vertebrate cells, but not bacterial lysates, stabilize HIV-1 CA-NC complexes, allowing them to sediment more efficiently through a sucrose cushion. Fractionation of the cytoplasmic lysates and characterization of the biochemical properties of the capsid-stabilizing activity implicated a protein factor with a molecular mass greater than 100 kDa. Our analysis of candidate capsid-binding proteins identified the PDZ domain-containing 8 protein (PDZD8) as a critical component of the capsid-stabilizing activity in the cytoplasmic extracts.
Human PDZD8 has been previously identified as an HIV-1 cofactor, although its precise role in infection is unknown (22). PDZD8 coprecipitates with the HIV-1 Gag polyprotein, and knockdown of the PDZD8 protein inhibits HIV-1 infection (22). Overexpression of PDZD8 has been reported to enhance murine leukemia virus (MLV), simian immunodeficiency virus (SIV), and HIV-1 infection (22). The cellular function of PDZD8 is uncertain, although PDZD8 has been identified as a moesin-interacting factor as well as a potential regulator of microtubule stability (23).
We present evidence that PDZD8 acts to stabilize HIV-1 capsids. Knockdown of PDZD8 by small interfering RNA (siRNA) diminishes the ability of cytoplasmic lysates from human cells to stabilize HIV-1 CA-NC complexes in vitro. PDZD8 knockdown accelerates the disassembly of HIV-1 capsids in infected cells, resulting in a block to infection prior to reverse transcription. We examined the ability of PDZD8 variants to bind the HIV-1 CA-NC capsid complexes, identify the PDZD8 coiled coil as the capsid-binding domain, and show that capsid binding is not sufficient for stabilization of the CA-NC capsid complexes and enhancement of HIV-1 infectivity. The identification of PDZD8 as a capsid-stabilizing host factor represents a significant opportunity to advance our understanding of the retrovirus uncoating process.
MATERIALS AND METHODS
Cells, plasmids, and siRNA.
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 μg/ml streptomycin. Prior to transfection, 10,000 HeLa cells were seeded per well in a 24-well plate.
All of the studies reported herein were conducted with human PDZD8. FLAG-tagged wild-type PDZD8 and PDZD8 mutants were cloned in the pIRES2 DsRed2 vector (Clontech). PDZD8-expressing constructs were altered by QuikChange mutagenesis to include silent mutations that prevent targeting by the tested siRNA constructs. Twenty-four hours after seeding, cells were transfected with 0.2 μg plasmid DNA/well with Effectene transfection reagent (Qiagen), as described in the manufacturer's protocol.
For siRNA treatment, cells were transfected 24 h after the Effectene-mediated plasmid transfection with siRNA 21-mers siPDZD8#1 (Ambion; s42265) or siPDZD8#2 (Ambion; s42267). The siRNA was transfected at 20 pmol/well with RNAiMAX (Invitrogen), as described in the manufacturer's protocol.
Virus production and infectivity.
Retrovirus infectivity assays were carried out with single-round, vesicular stomatitis virus glycoprotein (VSVG)-pseudotyped viruses carrying luciferase reporter genes. The pseudotyped HIV-1, simian immunodeficiency virus (SIV), feline immunodeficiency virus (FIV), Moloney murine leukemia virus (Mo-MLV), and Rous sarcoma virus vectors expressing luciferase were produced in 293T cells by cotransfection with a VSVG expression vector, as previously described (24–30). HIV-1 was concentrated by centrifugation and analyzed by an exogenous 32P-reverse transcriptase (RT) assay (31). For infections, HeLa target cells were transfected with siRNAs and/or plasmids and then reseeded into 96-well plates at 6,000 cells/well 24 h prior to infection. Cells were incubated with pseudotyped viruses at various concentrations, as described in Results. After 4 h of virus-cell incubation, the virus-containing medium was removed and replaced with fresh medium. Forty-eight hours later, cells were lysed in 30 μl passive lysis buffer (Promega), and the levels of luciferase activity were assayed, as previously described (32). All infection assays were performed in triplicate.
Measurement of HIV-1 early reverse transcription products.
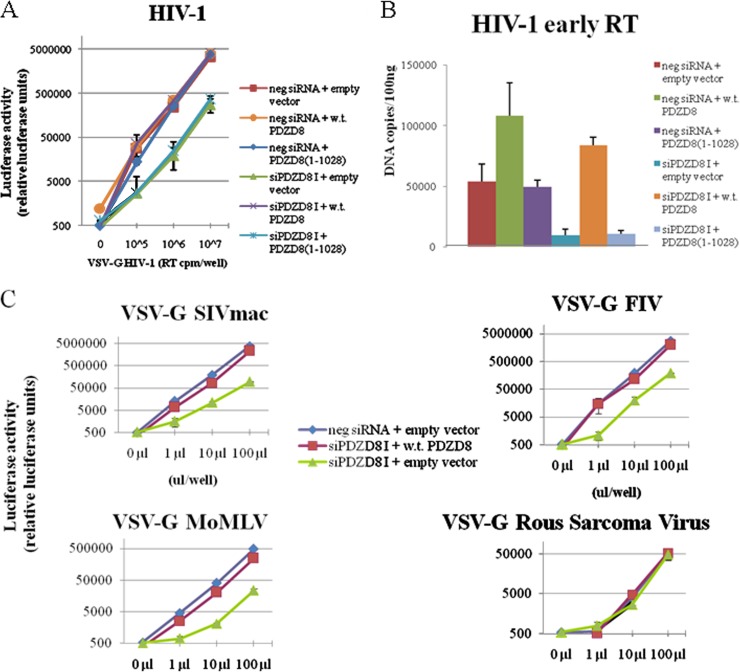
One day prior to infection, 4 × 105 transfected HeLa cells were seeded into 6-well plates in triplicate. VSVG-pseudotyped HIV-1 viruses were prepared as described above and were treated with 44 U/ml of Turbo DNase I (Ambion) at 37°C for 1 h. After 4 h of virus-cell incubation, cells were harvested and total DNA was extracted with the DNeasy kit (Qiagen). One hundred nanograms of total DNA was used as a template for quantitative real-time PCR, using previously described conditions (33). Duplicate measurements were performed for each of the triplicate samples.
Assay for stabilization of HIV-1 CA-NC complexes.
Purification of recombinant HIV-1 capsid-nucleocapsid (CA-NC) protein from Escherichia coli was carried out as previously described (19). High-molecular-mass HIV-1 CA-NC complexes were assembled using 300 μM CA-NC protein and 60 μM TG50 DNA oligonucleotide in a volume of 100 μl of 1× phosphate-buffered saline (PBS) and 500 mM NaCl (19). The reaction was allowed to proceed overnight at 4°C. Immediately before use, the assembled CA-NC complexes were spun at 10,000 × g for 1 min and resuspended in 1× PBS.
As the source of cellular lysate to assay for CA-NC stabilization, HeLa cells were transfected as described above. Forty-eight hours after transfection, 250,000 HeLa cells were resuspended in 200 μl 1× PBS and lysed by a 1-min treatment by a Kontes pestle. Cellular membranes and large complexes were then removed in a two-part centrifugation protocol. First, the lysed material was spun for 1 h at 15,000 × g at 4°C. Second, the supernatant from the first spin was loaded onto a 3-ml 55% (wt/vol) sucrose cushion and spun in a Beckman SW55Ti rotor at 115,000 × g for 2 h at 4°C. Following this centrifugation step, the supernatant above the sucrose layer was removed, quantitated by Coomassie (Bradford) protein assay (Pierce), and used in the CA-NC stabilization assay.
In the CA-NC stabilization assay, 10 μl of assembled CA-NC complexes was added to 250 μl of HeLa cell lysate (diluted to a protein concentration of 0.2 mg/ml). The CA-NC complexes and cell lysate were gently mixed at room temperature for 4 h (unless otherwise indicated). This mixture was then layered onto a 3.5-ml 70% sucrose cushion and spun at 50,000 × g for 20 min in an SW55Ti rotor at 4°C. Following centrifugation, 250-μl fractions (unless otherwise noted in Results) were removed from the centrifuge tube using a peristaltic pump. A final “pellet” fraction was created by resuspending any pelleted material in 250 μl of 1× PBS.
The CA-NC content of individual fractions was assayed by enzyme-linked immunosorbent assay (ELISA). Sucrose fractions were diluted 1:10 in BupHCarbonate-bicarbonate buffer (pH 9.4; Pierce). Fifty microliters of diluted fractions was then added to white flat-bottom 96-well plates (Nunc) for 1 h at room temperature. Plates were blocked with 20% FBS in 1× PBS for an additional 1 h. The plates were then treated with anti-p24 horseradish peroxidase (HRP)-conjugated antibody (AbCam) at 1 μg/ml in 1× PBS and 0.05% Tween for 1 h at room temperature. Plates were washed 2 times in blocking buffer and 3 times in 1× PBS and 0.05% Tween 20. HRP levels were then detected by SuperSignal Pico chemiluminescent substrate (Pierce), using a 96-well plate luminometer in accordance with the manufacturer's protocol. Known concentrations of CA-NC protein were included in the binding buffer in all plates to generate standard curves for quantitation. All fractions were quantitated in duplicate. The results reported are representative of data obtained from at least three independent experiments.
For some experiments, we used a CA-NC stabilization assay in which only the pellet fraction was measured. In this protocol, 1 μl of assembled CA-NC complexes was added to 250 μl of 0.5 mg/ml 293T cell lysate in 1× PBS and mixed for 1 h at room temperature. This mixture was then layered onto a 3.5-ml 70% sucrose cushion and spun at 110,000 × g for 60 min in an SW55Ti rotor at 4°C. The pellet fraction was then resuspended in 100 μl of 1× SDS running buffer and analyzed by SDS-PAGE gel electrophoresis and Coomassie blue staining.
PDZD8 binding to HIV-1 CA-NC complexes.
Binding of FLAG-tagged PDZD8 variants to HIV-1 CA-NC complexes was assessed with a protocol that uses the assembled CA-NC complexes and HeLa cell lysates described above. In the binding assay, 10 μl of assembled HIV-1 CA-NC complexes and HeLa cell lysates containing FLAG-tagged PDZD8 variants were gently mixed for 1 h at room temperature. The mixture was then layered onto a 3.5-ml 70% sucrose cushion and centrifuged at 110,000 × g for 2 h at 4°C in an SW55Ti rotor. The pellet fraction was resuspended in 100 μl of sodium dodecyl sulfate (SDS) sample buffer. This sample was then electrophoresed on a polyacrylamide gel, which was used for detection of CA-NC protein by Coomassie blue staining and FLAG-tagged protein by Western blotting, as previously described (21).
The fate-of-capsid assay.
Approximately 8 × 105 transfected HeLa cells/well were plated in 6-well plates. On the following day, the cells were incubated with VSVG-pseudotyped HIV-1 virus (5 × 105 RT counts) for 30 min at 4°C. Then, the cells were returned to a 37°C CO2 incubator until they were harvested 16 h later. The virus suspension was removed at the 4-h time point and replaced with fresh medium. The cells were washed three times with ice-cold PBS and detached by incubating with 1 ml of pronase (7 mg/ml in DMEM) for 5 min at 4°C. The cells were washed once in DMEM containing 10% FBS and twice in PBS. The washed cell pellet was resuspended in 250 μl hypotonic lysis buffer and placed on ice for 15 min. The cells were lysed by 1 min of treatment with a Kontes pestle. Cell debris was removed by centrifugation for 3 min at 2,000 × g. After centrifugation, 2 ml of lysate was layered onto a 7-ml 50% sucrose cushion (made in PBS) and centrifuged at 125,000 × g for 2 h at 4°C in a Beckman SW41 rotor. After centrifugation, 100 μl from the topmost part of the supernatant was collected and brought to a 1× concentration of SDS sample buffer. The pellet was resuspended in 100 μl of 1× SDS sample buffer. The samples were subjected to SDS-PAGE and Western blotting to detect the capsid proteins.
RESULTS
Stabilization of HIV-1 CA-NC complexes by cell lysates.
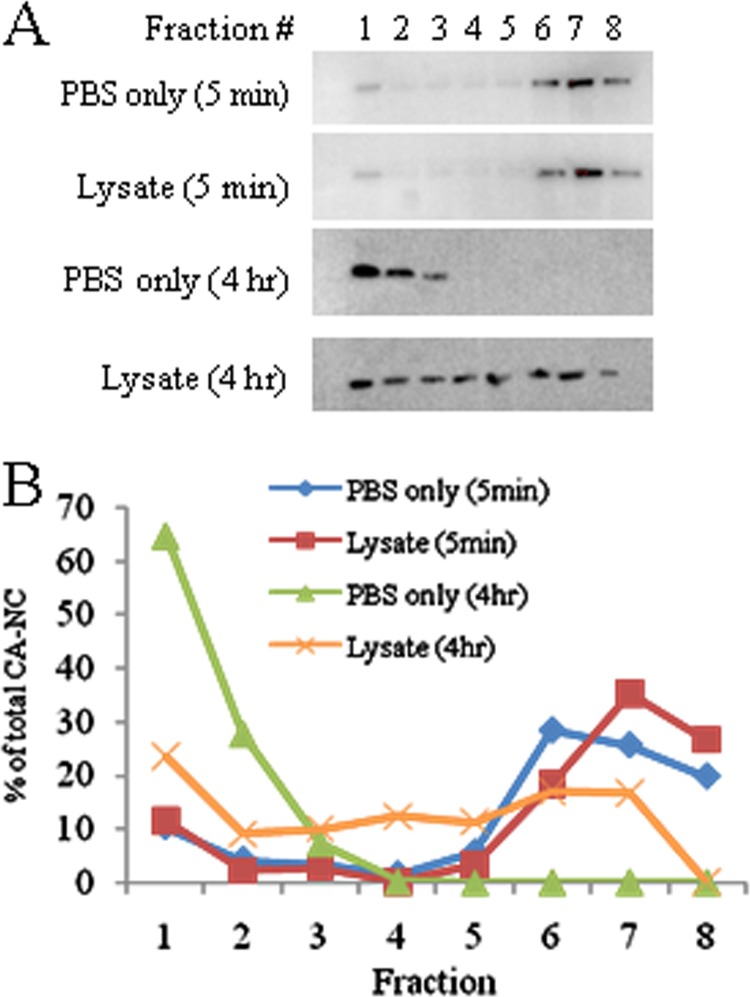
The binding of cellular proteins such as TRIM5α to HIV-1 capsid complexes has previously been studied by examining the cosedimentation of the host protein with CA-NC complexes assembled in vitro (7). During the course of these studies, we noted that a greater percentage of input CA-NC complexes passed through the high-density sucrose cushion following incubation with cytoplasmic lysates from a variety of vertebrate cells than that seen after incubation in PBS. To explore this finding further, we tracked the sedimentation of in vitro-assembled CA-NC complexes that were incubated with cell lysates or PBS. The CA-NC complexes were incubated at room temperature in PBS buffer alone or PBS buffer containing HeLa cell lysate for either 5 min or 4 h before being loaded onto a 3.5-ml 70% sucrose cushion. After centrifugation at 50,000 × g for 20 min, 500-μl fractions were taken, starting from the top (the supernatant was included in fraction 1, and any pelleted material was resuspended in fraction 8). The CA-NC protein was detected by Western blotting with anti-p24 antibodies (Fig. 1A). HIV-1 CA-NC complexes incubated for 5 min with the HeLa cell lysate or in PBS sedimented through the sucrose cushion at similar rates. In contrast, after a 4-h incubation, rapidly sedimenting CA-NC complexes were much more abundant after incubation in the cell lysate than in PBS. To quantify this effect, CA-NC levels within fractions taken from the sucrose cushion were determined by ELISA (Fig. 1B). For these experiments, smaller, 250-μl fractions of the sucrose cushion were collected, with the resuspended pellet kept separate (in fraction 8) from the supernatant fractions. These results confirmed that a 4-h incubation with HeLa cell lysates resulted in faster-sedimenting CA-NC complexes than a parallel incubation in PBS (Fig. 1B).
FIG 1.
Stabilization of HIV-1 CA-NC complexes by cell lysate. Ten microliters of the in vitro-assembled HIV-1 CA-NC complexes was gently mixed with 250 μl of HeLa cell lysate (0.2 mg/ml in 1× PBS) or 1× PBS buffer alone for 5 min or 4 h at 20°C. The mixture was then loaded onto a 3.5-ml 65% sucrose cushion and spun at 50,000 × g in a Beckman SW55Ti rotor for 20 min at 4°C. Following centrifugation, 500-μl fractions were taken from the top (250 μl supernatant included in fraction 1). The pellet was resuspended in 250 μl of 1× PBS and included in fraction 8. (A) Western blot of fractions with anti-p24-HRP-conjugated antibody (Abcam); (B) the CA-NC content of each fraction was determined by ELISA and normalized to the total CA-NC in the sample. This experiment was conducted three times, and the results from a typical experiment are shown.
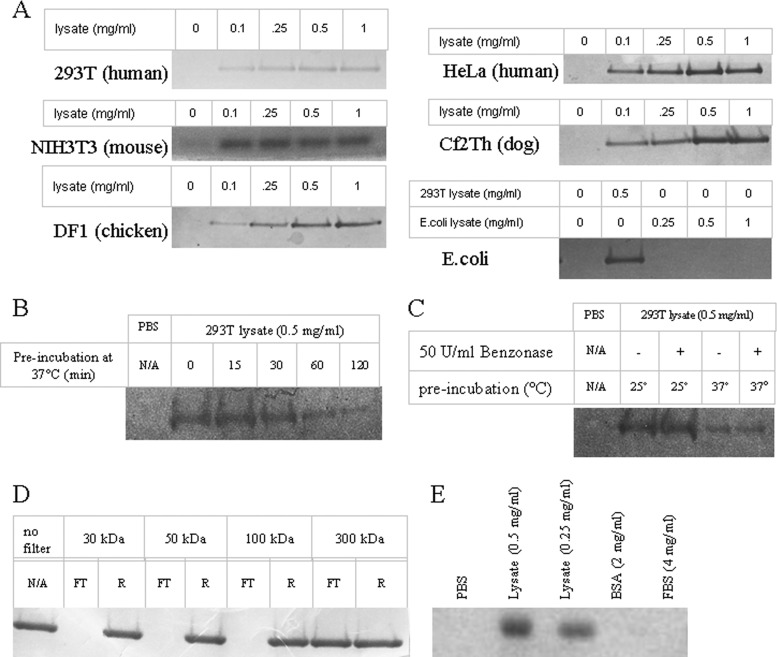
The CA-NC-stabilizing activity was detected in cells of several vertebrate species (human, mouse, dog, chicken) but not in E. coli (Fig. 2A). Analysis of the cell lysate suggested that the factor responsible for the stabilization of HIV-1 CA-NC complexes is heat labile, nuclease resistant, and >100 kDa (Fig. 2B to D). The stabilizing effect was protein content dependent but was not observed with equal protein concentrations of dialyzed bovine serum albumin (BSA), dialyzed FBS, or rehydrated milk in PBS (Fig. 2E). The loss of rapidly sedimenting CA-NC complexes over time is apparently irreversible; incubation of slowly sedimenting CA-NC complexes with cellular lysate did not restore a high sedimentation rate (data not shown). Together, this evidence suggests that specific components of cytoplasmic lysates from vertebrate cells slow the disassembly of HIV-1 CA-NC complexes in vitro. A recent publication reached a similar conclusion about the ability of human cell lysates to stabilize HIV-1 CA-NC cores (34).
FIG 2.
Characterization of the HIV-1 CA-NC complex-stabilizing factor in cell lysates. (A) Stabilization of CA-NC complexes by lysates from multiple cell types. Two microliters of the in vitro-assembled HIV-1 CA-NC complexes was gently mixed with 250 μl of 1× PBS buffer alone or 250 μl cell lysate at the indicated concentrations in 1× PBS for 1 h at 20°C. The mixture was then loaded onto a 3.5-ml 70% sucrose cushion and spun at 110,000 × g in a Beckman SW55Ti rotor for 60 min at 4°C. Following centrifugation, the pellet was resuspended in 100 μl 1× SDS loading buffer. Pellet fractions were analyzed by SDS-PAGE and stained with Coomassie blue. The SDS-PAGE gel regions containing CA-NC from the pellet fractions are shown. (B) Effect of preincubation at 37°C on the ability of a 293T cell lysate to stabilize HIV-1 CA-NC complexes. A 293T cell lysate at 0.5 mg/ml was incubated at 37°C for the indicated times. Following incubation, lysates were brought back to room temperature and used in a CA-NC stabilization assay as described for panel A. (C) Effect of Benzonase treatment on the HIV-1 CA-NC complex-stabilizing activity. Prior to use in the stabilization assay, 293T lysates were incubated at 20°C or 37°C with or without 50 U/ml of Benzonase nuclease. (D) Stabilization of HIV-1 CA-NC complexes by 293T cell lysate fractions. Following filtration of 0.5 mg/ml 293T cell lysate through filters of the indicated molecular mass cutoffs (30 kDa, 50 kDa, 100 kDa [Millipore]; 300 kDa [Sigma-Aldrich]), the retained (R) and flowthrough (FT) fractions were used in a CA-NC stabilization assay. These experiments were conducted three times, and the results from typical experiments are shown. (E) Comparison of CA-NC stabilization by a 293T lysate, purified bovine serum albumin (BSA), and filtered fetal bovine serum (FBS) at the indicated concentrations. Filtered 2 mg/ml milk solution also failed to stabilize CA-NC complexes (data not shown).
Contribution of PDZD8 to the in vitro stabilization of HIV-1 CA-NC complexes.
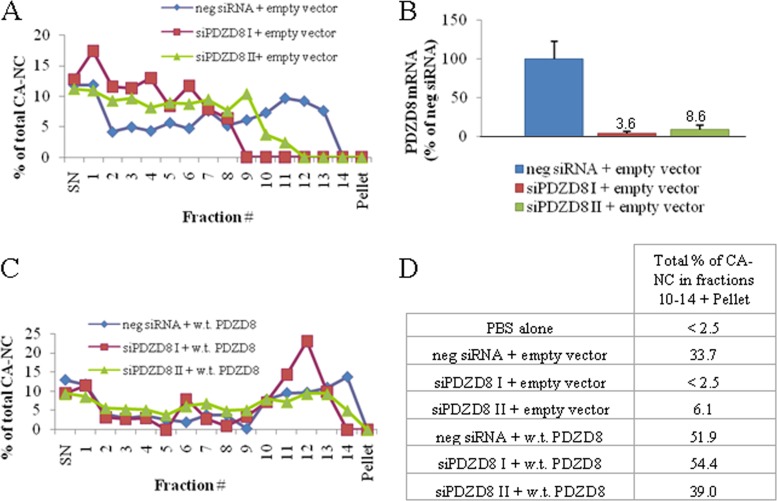
Human PDZD8, an approximately 160-kDa cytoplasmic protein of unknown cellular function, has been shown to bind HIV-1 Gag proteins and to enhance viral infectivity (22). Overexpression of PDZD8 in some cell types has been reported to enhance the production of HIV-1 reverse transcription products (22), indicating a potential role in early events in the retroviral life cycle. We hypothesized that PDZD8 might be a retroviral capsid-stabilizing factor and evaluated PDZD8 as a potential contributor to the in vitro stabilization of HIV-1 CA-NC complexes by cell lysates. Transfection with two different siRNAs targeting PDZD8 decreased the capacity of HeLa cell lysates to stabilize HIV-1 CA-NC complexes (Fig. 3A). A substantial loss of the fastest sedimenting CA-NC complexes was evident relative to the results obtained with lysate from cells treated with a negative-control siRNA. The loss of CA-NC-stabilizing ability was related to the degrees of PDZD8 mRNA knockdown produced by the two siRNAs (Fig. 3B). Add-back of PDZD8 by transfection with an siRNA-resistant plasmid expressing an N-terminally FLAG-tagged PDZD8 restored the CA-NC-stabilizing ability (Fig. 3C and D). The results suggest that PDZD8 is an important contributor to the in vitro HIV-1 CA-NC-stabilizing activity in cell lysates.
FIG 3.
PDZD8 contributes to the stabilization of HIV-1 CA-NC complexes in vitro. (A, C) HIV-1 CA-NC stabilization was assayed as described in the Fig. 1 legend, with the exception that 250-μl fractions were taken (including a supernatant fraction [SN] and a resuspended pellet fraction). HIV-1 CA-NC complexes and HeLa lysates were mixed for 4 h at room temperature before centrifugation over a 65% sucrose cushion. The HeLa cells that served as sources for the lysates were transfected with either the empty pIRES2 vector (A) or pIRES2 expressing FLAG-tagged, wild-type (w.t.) PDZD8 (C). The gene encoding wild-type PDZD8 was mutated to render it resistant to the siRNAs. After 24 h, cells were transfected a second time with either a negative-control noncoding siRNA or one of two PDZD8-targeting siRNAs (siPDZD8 I or siPDZD8 II). Cells were lysed 48 h after siRNA transfection, and the cell lysates were used in the HIV-1 CA-NC stabilization assay. Following the centrifugation of the lysate–CA-NC mixtures, the CA-NC content of individual fractions was determined by ELISA and normalized to the total CA-NC in each sample. (B) The efficiency of PDZD8 knockdown by the siRNA transfections described for panel A was assessed by quantitative RT-PCR. PDZD8 mRNA levels are reported as a percentage of that detected in cells transfected with the negative-control siRNA and empty pIRES2 vector. (D) The total percentage of CA-NC contained in fractions 10 to 14 plus the resuspended pellet fraction is shown for all samples. A result from a control stabilization assay in which 1× PBS alone was used in place of the HeLa lysate is also included. These experiments were conducted three times, and the results from a typical experiment are shown.
Contribution of PDZD8 to the stabilization of HIV-1 capsids in infected cells.
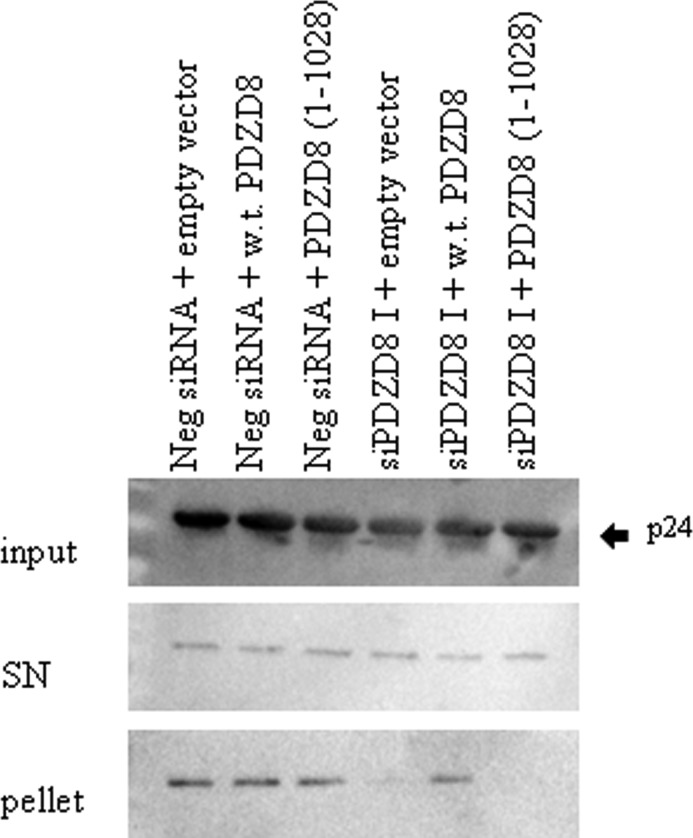
We hypothesized that PDZD8 plays a role in stabilizing the HIV-1 capsid that is introduced into the cytosol following virus entry into the host cell. To test this hypothesis, we utilized the previously described fate-of-capsid assay (7). Fate-of-capsid assays have been used to demonstrate increased rates of viral disassembly that are a consequence of viral restriction factors, small molecule inhibitors of infection, and destabilizing changes in the HIV-1 CA-NC protein (6, 7, 15). VSVG-pseudotyped HIV-1 virus was added to HeLa cells transfected with both the PDZD8-silencing and control siRNAs, as described above. Infected cells were lysed 16 h after infection, and particulate HIV-1 cores were isolated by centrifugation through a 50% sucrose cushion. Knockdown of PDZD8 by specific siRNA dramatically reduced the levels of pelletable core relative to the levels seen in cells transfected with the negative-control siRNA (Fig. 4). Add-back of FLAG-tagged wild-type PDZD8 by transfection with an siRNA-resistant expression plasmid restored the level of particulate cores that pelleted through the sucrose cushion. Notably, whereas overexpression of the putative hyperstabilizing restriction factor CPSF6 has been reported to increase the levels of particulate HIV-1 cores in this fate-of-capsid assay (12–14), expression of exogenous PDZD8 in the control siRNA-transfected cells did not substantially increase core recovery relative to empty vector-transfected controls. A PDZD8 mutant [PDZD8(1–1028)] that contains a C-terminal truncation that has been reported to result in a loss of HIV-1 Gag binding (22) did not restore the stability of the HIV-1 core in the HIV-1-infected cells in which endogenous PDZD8 expression had been knocked down; we show below that the PDZD8(1–1028) mutant lacks the ability to stabilize HIV-1 CA-NC complexes in vitro. These results indicate that wild-type PDZD8 contributes to the stability of the HIV-1 capsid in infected cells.
FIG 4.
Contribution of PDZD8 to stabilization of the HIV-1 capsid in infected cells. HeLa cells were transfected with the negative-control (Neg) siRNA or the PDZD8-targeting siPDZD8 I as well as the pIRES vector expressing FLAG-tagged wild-type (w.t.) PDZD8, the C-terminally truncated PDZD8(1–1028) mutant, or an empty vector. The genes encoding wild-type and truncated PDZD8 were mutated to resist the siPDZD8 I siRNA. The transfected HeLa cells were then incubated with VSVG-pseudotyped HIV-1. The fate-of-capsid assay was used to compare the rate of capsid uncoating. At 16 h after infection, cells were lysed in 250 μl hypotonic lysis buffer by a 1-min treatment with a Kontes pestle. Fifty microliters of each sample was saved as an input fraction. The remaining sample was loaded onto a 7-ml 50% sucrose cushion and spun at 125,000 × g for 2 h to separate soluble capsid protein in the supernatant (SN) from the particulate capsid in the pellet. The supernatant and resuspended pellet fractions were analyzed by SDS-PAGE and Western blotting using antibodies against p24. This experiment was conducted three times, and the results from a typical experiment are shown.
Contribution of PDZD8 to retrovirus infectivity.
Previous work has demonstrated a strong correlation between increased kinetics of HIV-1 core uncoating and decreases in viral infectivity (4, 7). Knockdown of PDZD8 by a specific siRNA resulted in an approximately 10-fold decrease in HIV-1 infectivity in HeLa cells (Fig. 5A). Transient transfection of an siRNA-resistant vector expressing wild-type PDZD8 restored infectivity to a level comparable to that seen in cells transfected with the negative-control siRNA. In contrast, expression of the PDZD8(1–1028) mutant with a C-terminal truncation did not rescue HIV-1 infectivity in HeLa cells in which PDZD8 expression was knocked down (Fig. 5A). These results suggest that wild-type PDZD8 contributes to HIV-1 infection, consistent with an earlier report (22).
FIG 5.
PDZD8 exerts positive effects on HIV-1 reverse transcription and infection of several retroviruses. (A) Knockdown of PDZD8 reduces HIV-1 infectivity, and infectivity is rescued by exogenous expression of siRNA-resistant PDZD8. HeLa cells were transfected with the pIRES2 vector expressing FLAG-tagged wild-type (w.t.) PDZD8, a C-terminal truncation mutant, PDZD8(1–1028), or an empty vector. These cells were also transfected with either a PDZD8-targeting siRNA, siPDZD8 I, or a negative-control (neg) siRNA. Forty-eight hours following siRNA transfection, cells were incubated with the indicated amount of VSVG-pseudotyped luciferase-expressing HIV-1 virus. Forty-eight hours after infection, cells were lysed and luciferase activity was assayed. The means and standard deviations from three independent experiments are reported. (B) Knockdown of PDZD8 inhibits HIV-1 reverse transcription. Cells transfected with PDZD8-targeting siRNA or control siRNA and siRNA-resistant PDZD8-expressing vectors were infected by VSVG-pseudotyped HIV-1, as described for panel A above. The cells were assayed by quantitative PCR to determine the levels of strong-stop cDNA. A heat-inactivated virus control yielded less than 5,000 DNA copies/ml (data not shown). The means and standard deviations from three independent experiments are reported. (C) HeLa cells transfected as described for panel A were incubated with the indicated concentrations of VSVG-pseudotyped luciferase-expressing simian immunodeficiency virus (SIVmac), feline immunodeficiency virus (FIV), Moloney murine leukemia virus (Mo-MLV), and Rous sarcoma virus for 4 h. Forty-eight hours following infection, luciferase activity was assayed. The means and standard deviations from three independent experiments are reported.
To determine the effect of PDZD8 knockdown on HIV-1 reverse transcription, we performed quantitative PCR to detect levels of strong-stop cDNA, an early reverse transcription product, at 4 h after infection. In PDZD8 knockdown cells, the level of HIV-1 strong-stop cDNA products was decreased (Fig. 5B). The production of strong-stop cDNA could be rescued by wild-type PDZD8 but not PDZD8(1–1028). Of note, in the negative-control siRNA-treated cells, the level of HIV-1 strong-stop cDNA was increased in the cells transfected with the plasmid expressing wild-type PDZD8 but not in cells expressing the PDZD8(1–1028) mutant. Although HIV-1 infectivity was not significantly increased in the cells overexpressing the wild-type PDZD8 (see above), this observation suggests that, for some aspects of early HIV-1 infection, endogenous PDZD8 levels may be rate limiting. The results support a model in which PDZD8 interaction with the HIV-1 capsid contributes to core stability and virus infectivity. Our findings also add to mounting evidence that premature capsid uncoating disrupts early HIV-1 reverse transcription (7, 35–37).
We tested the effect of PDZD8 knockdown and siRNA-resistant add-back of exogenous PDZD8 on the infectivity of a panel of additional VSVG-pseudotyped luciferase reporter retroviruses (Fig. 5C). PDZD8 knockdown decreased infection of cells by the pseudotyped SIVmac, Mo-MLV, and FIV. However, PDZD8 knockdown had no apparent effect on the infectivity of the pseudotyped Rous sarcoma virus vector. Apparently, PDZD8 contributes to the infectivity of a number of mammalian retroviruses.
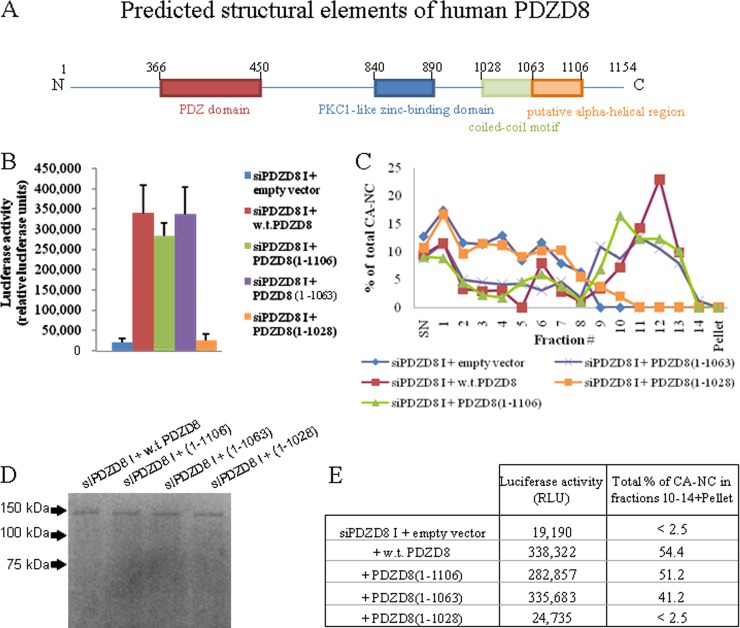
Functional contribution of the PDZD8 carboxyl terminus.
Although the structure of PDZD8 is unknown, putative domains in the protein have been identified by sequence similarity to established structures (Fig. 6A). PDZD8 was originally identified as a potential HIV-1 Gag-binding partner by a yeast two-hybrid screen. A fragment of PDZD8 comprised of amino acid residues 932 to 1110 bound a bait fragment of HIV-1 Gag (22). This PDZD8 fragment contains a putative coiled-coil domain (amino acid residues 1028 to 1060) as well as an additional region (residues 1063 to 1106) predicted to have significant helical character. To investigate the role of specific carboxy-terminal regions of PDZD8 in the activity of this protein, we created a series of FLAG-tagged PDZD8 mutants with carboxy-terminal truncations [PDZD8(1–1028), PDZD8(1–1063), and PDZD8(1–1106)]. HeLa cells transiently transfected with expression vectors encoding wild-type PDZD8 and these truncation mutants were used in infection assays and as a source of lysate for in vitro CA-NC stabilization assays (Fig. 6B and C). The truncated PDZD8 proteins were expressed at levels comparable to that of wild-type PDZD8 (Fig. 6D). PDZD8 residues from the carboxy terminus to residue 1063, including the putative alpha-helical region, were dispensable for rescue of HIV-1 infectivity and CA-NC stabilization. Further deletion of the putative coiled-coil motif (residues 1028 to 1060) eliminated the ability of exogenous PDZD8(1-1028) to rescue HIV-1 infectivity in HeLa cells in which the expression of endogenous PDZD8 was knocked down. The PDZD8(1-1028) mutant likewise did not stabilize HIV-1 CA-NC complexes in vitro. These results, summarized in Fig. 6E, support the importance of the coiled coil to PDZD8 function as an HIV-1 cofactor.
FIG 6.
Role of C-terminal sequences in PDZD8 function as an HIV-1 cofactor. (A) A schematic of the predicted structural elements of the human PDZD8 protein, with the amino acid residues noted. The domains in this figure are not drawn to scale. (B) HeLa cells transfected with the PDZD8-targeting siRNA siPDZD8 I were also transfected with pIRES vectors expressing N-terminally FLAG-tagged, wild-type (w.t.) PDZD8, PDZD8(1–1106), PDZD8(1–1063), PDZD8(1–1028), or an empty pIRES vector. The genes encoding the wt and truncated PDZD8 proteins were mutated to resist the siPDZD8 I siRNA. Cells were incubated with 1 × 106 cpm RT of VSVG-pseudotyped HIV-1-expressing luciferase. Luciferase activity was assessed at 48 h after infection. The means and standard deviations from three independent experiments are reported. (C) The HIV-1 CA-NC stabilization assay was performed with lysates from cells transfected as described for panel B. The CA-NC content of individual fractions was measured by ELISA and normalized to the total amount of CA-NC in the sample. This experiment was conducted three times, and the results from a typical experiment are shown. (D) Western blot of the cell lysates used for panel C, using an anti-FLAG antibody. (E) A chart comparing the fraction of total CA-NC located in fractions 10 to 14 plus the pellet in the HIV-1 CA-NC stabilization assay shown in panel C with the luciferase activity from the assay described for panel B.
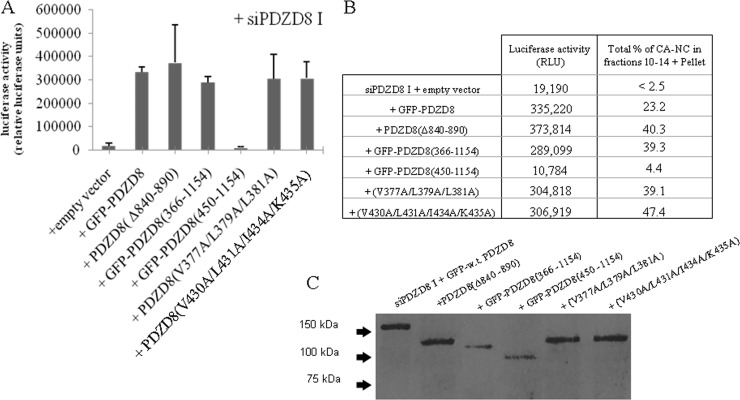
Role of the PDZ and PKC1-like domains in PDZD8 function.
Sequence analysis of PDZD8 indicates the presence of two additional conserved domains, a PDZ domain and a PKC1-like zinc-binding domain (Fig. 6A). To determine the importance of these regions to PDZD8 function, we progressively deleted N-terminal portions of the protein. However, we found that steady-state levels of PDZD8 N-terminal truncation mutants (both with and without N-terminal FLAG tags) were very low when transiently expressed. N-terminal fusion with green fluorescent protein (GFP) substantially increased expression to levels comparable to that of the full-length PDZD8 protein. Therefore, we constructed a series of fusion proteins in which an N-terminal FLAG-tagged GFP molecule is fused with either full-length PDZD8 or N-terminally truncated PDZD8. Additionally, we constructed a FLAG-tagged PDZD8 mutant with an internal deletion of the PKC domain (Δ840 to 890). Loss of the PDZD8 sequences N-terminal to the PDZ domain did not affect PDZD8 rescue of HIV-1 infection or in vitro stabilization of HIV-1 CA/NC complexes [see GFP-PDZD8(366–1154) in Fig. 7]. Further N-terminal truncation, which removed the PDZ domain [GFP-PDZD8(450–1154)], resulted in a loss of both CA-NC-stabilizing activity and the ability to rescue HIV-1 infection (Fig. 7). The PDZD8(Δ840–890) mutant, which lacks the PKC1-like domain, rescued HIV-1 infectivity in PDZD8 knockdown cells and CA-NC-stabilizing activity in PDZD8-depleted HeLa cell lysates (Fig. 7). Thus, the PKC1 domain is not apparently required for PDZD8 function with respect to HIV-1 infection. The PDZ domain, on the other hand, may contribute to the function of PDZD8 as an HIV-1 cofactor.
FIG 7.
Role of the PDZ and PKC1 domains in PDZD8 function as an HIV-1 cofactor. (A) HeLa cells were transfected with the PDZD8-targeting siRNA siPDZD8 I and either the empty pIRES vector or pIRES vectors expressing N-terminally FLAG-tagged, GFP-fused wild-type (w.t.) PDZD8, PDZD8(366–1154), and PDZD8(450–1154) or FLAG-tagged PDZD8(Δ840–890) with the PKC1-like domain deleted and full-length PDZD8 with alanine substitutions in the PDZ peptide-binding motif (V430A, L431A, I434A, K435A; V377A, L379A, L381A). The genes encoding the wild-type and variant PDZD8 proteins were mutated to resist the siPDZD8 I siRNA. Cells were incubated with 1 × 106 cpm RT of VSVG-pseudotyped HIV-1 expressing luciferase, and luciferase activity was assessed at 48 h following infection. (B) The HIV-1 CA-NC stabilization assay was performed with lysates from HeLa cells transfected as described for panel A. The CA-NC content of individual fractions was measured by ELISA and normalized to the total amount of CA-NC in the sample. The percentage of total CA-NC from each sample contained in fractions 10 to 14 plus the resuspended pellet fraction is reported. For comparison, the mean luciferase activities observed in the experiments described for panel A are listed as well. (C) Western blot of the cell lysates used for panel B with an anti-FLAG antibody. These experiments were conducted three times, and the results from a typical experiment are shown.
PDZ domains are common protein-protein interaction modules with a well-defined binding motif that most commonly binds the C-terminal peptides of target proteins (38). There are currently no known binding partners of the PDZD8 PDZ domain. To test the hypothesis that the protein-binding function of the PDZ domain is required for the capsid stabilization function of PDZD8, we used site-directed mutagenesis to alter the PDZD8 residues that are putatively located on both sides of the peptide-binding groove of the PDZ domain (39). These PDZD8 mutants rescued HIV-1 infectivity and in vitro CA-NC stabilization following PDZD8 knockdown with efficiencies comparable to those seen for wild-type PDZD8 (Fig. 7). Thus, the putative peptide-binding groove of PDZD8 can be substantially altered without affecting either PDZD8 stabilization of HIV-1 CA-NC capsids in vitro or rescue of HIV-1 infectivity.
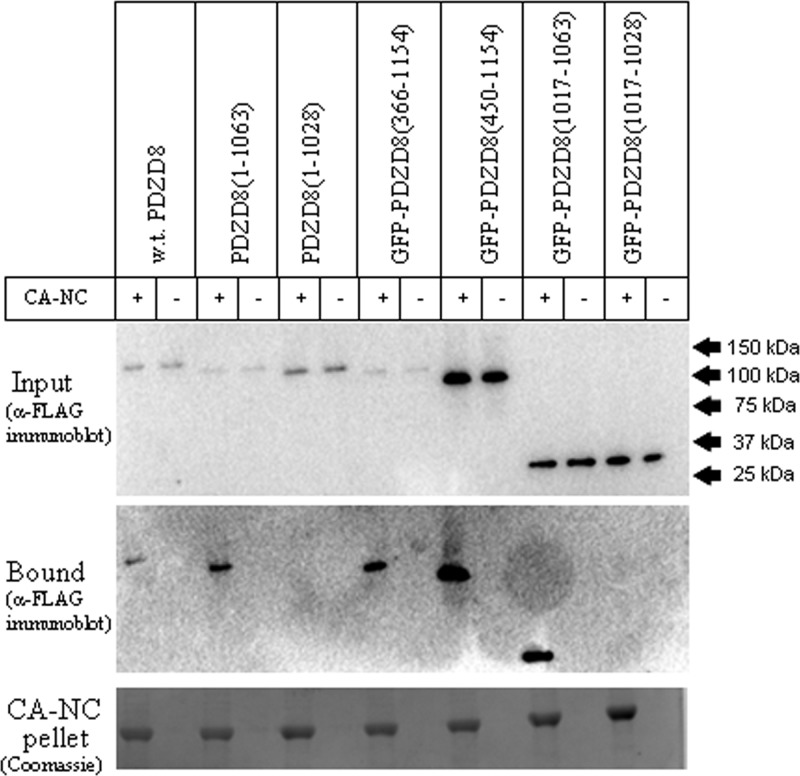
PDZD8 regions involved in HIV-1 capsid binding.
To investigate the contribution of PDZD8 domains to the interaction of PDZD8 with the HIV-1 core, we evaluated the ability of PDZD8 mutants to cosediment with HIV-1 CA-NC complexes. FLAG-tagged PDZD8 variants were expressed transiently in HeLa cells, and cell lysates were incubated with in vitro-assembled HIV-1 CA-NC complexes for 3 h. Binding of the PDZD8 variants to the CA-NC complexes was assessed after sedimentation through a 70% sucrose cushion and detection of the FLAG-tagged PDZD8 protein by Western blotting. Loss of the PDZ domain, which resulted in undetectable CA-NC-stabilizing ability (see above), did not affect the interaction with HIV-1 CA-NC complexes [see GFP-PDZD8(450–1154) in Fig. 8]. The putative coiled-coil motif of PDZD8 was both necessary and sufficient for HIV-1 CA-NC binding [see GFP-PDZD8(1017–1063) in Fig. 8]. These results are consistent with a previous report that deletion of a C-terminal fragment of PDZD8 (residues 1028 to 1154) abrogates the coimmunoprecipitation of PDZD8 by HIV-1 Gag/Pol (22). These results also indicate that HIV-1 capsid binding by PDZD8 is necessary but not sufficient for capsid stabilization and activity as an infection cofactor.
FIG 8.
The coiled-coil motif of PDZD8 is necessary and sufficient for binding to HIV-1 CA-NC complexes. Lysates derived from HeLa cells transfected with pIRES vectors expressing FLAG-tagged wild-type (w.t.) PDZD8 or the indicated PDZD8 variants were mixed with in vitro-assembled HIV-1 CA-NC complexes for 2 h at room temperature. As indicated, some truncations of PDZD8 were fused to an N-terminal GFP protein to enhance expression. Negative-control experiments lacking CA-NC complexes were performed in parallel. After mixing, 50 μl of the mixture was saved as an input fraction. The remaining mixture was loaded on a 3.5-ml 70% sucrose cushion and spun at 110,000 × g for 2 h. The supernatant and sucrose cushion were removed and the pellet fraction was resuspended in 100 μl 1× SDS sample buffer. Input and pellet fractions were analyzed by SDS-PAGE. The SDS-polyacrylamide gel was analyzed by staining with Coomassie blue to detect the CA-NC protein and Western blotted with an anti-FLAG antibody to detect bound PDZD8. The experiment was conducted three times, and the results from a typical experiment are shown.
DISCUSSION
Decreases in the stability of retroviral capsids have substantial negative impacts on viral infectivity, typically resulting in defective reverse transcription (4, 7). The retroviral capsid is a loose assemblage of ∼1,500 CA proteins and exists within the viral membrane in equilibrium with free CA monomers (40). Upon removal of the viral membrane, retroviral capsids are unstable in physiological buffers (4). These observations contrast with the results of fate-of-capsid assays in infected cells, where particulate retroviral capsids exhibit half-lives of 8 to 10 h (7). The existence of capsid-stabilizing factors in the cytoplasm of vertebrate host cells provides a solution to the contrasting behavior of retroviral capsids in vitro and in infected cells.
Host restriction factors that accelerate the uncoating of retroviral capsids have been extensively studied (41). The identity and mechanism of action of host factors that stabilize retroviral capsids and act as positive cofactors for infection are less well understood. Here, we describe an in vitro assay to measure the stabilization of HIV-1 capsid-like assemblies by host cell lysates. We used this assay to show that PDZD8, previously identified by Henning et al. (22) as a positively acting cofactor protein that promotes initiation of HIV-1 reverse transcription in cells, is a major capsid-stabilizing factor in cell lysates. By implicating PDZD8 in the stabilization of HIV-1 capsid-like assemblies in vitro and HIV-1 viral capsids in infected cells, we provide a mechanism for the observations of Henning et al. (22).
A very limited number of cellular proteins have been implicated in the regulation of retroviral capsid uncoating, and only one, cyclophilin A (CypA), is known to enhance infection by stabilizing the capsid (15). CypA represents an interesting reference for a consideration of the role of PDZD8 in the retroviral life cycle. Target cell PDZD8, like CypA, is apparently utilized by multiple diverse retroviruses. In the case of CypA, studies of archaic retroviruses indicate that retroviral CypA-CA interactions likely predate HIV-1 emergence by at least 12 million years, underscoring the utility of CypA as a capsid-stabilizing cofactor in the infection of multiple retroviruses (42). PDZD8 knockdown affected infection by several lentiviruses and a gammaretrovirus but not Rous sarcoma virus. Thus, although CypA and PDZD8 act as positive cofactors for diverse retroviruses, some retroviruses apparently do not utilize these host cofactors. It is of interest to understand whether the latter retroviruses employ other capsid-stabilizing strategies. The comparison to CypA also invites future work to consider the capsid stabilization effect of PDZD8 in a broad range of species and cellular contexts. Both CypA and PDZD8 are well conserved among vertebrate species. The effect of CypA-CA binding on HIV-1 infectivity is highly dependent on target cell type, potentially due to differing levels of expression of CypA or CypA-mediated modulation of the susceptibility of HIV-1 to coexpressed viral restriction factors (15, 43). Retroviral phenotypes that result from modulation of PDZD8 levels may be similarly context dependent. In CHME3 brain microglia and 293A kidney epithelial cells, overexpression of PDZD8 by transfection has been reported to increase HIV-1 infectivity and early reverse transcript production by approximately 10-fold (22). In HeLa cells, we observed only a small enhancement (<2-fold) of HIV-1 infectivity from a similar transfection. This difference is potentially related to differences in endogenous expression of PDZD8 among cell lines. Quantitation of PDZD8 protein levels is difficult due to the current unavailability of suitable antibodies for detection of endogenous PDZD8 protein. Alternatively, these discrepancies in infectivity phenotypes may reflect differences in other critical elements dictating capsid stability and reverse transcription.
We observed an excellent correlation between the PDZD8 mutant phenotypes in the in vitro HIV-1 CA-NC stabilization assay and the HIV-1 infectivity assay (Pearson's correlation coefficient = 0.90, P < 0.001). Deletion of the PDZD8 coiled-coil motif abrogated the stabilizing effect of PDZD8 on CA-NC assemblies in vitro and resulted in loss of the ability to support HIV-1 infection. Consistent with Henning et al.'s finding that the C-terminal region of PDZD8 is required for binding of the HIV-1 Gag protein (22), the coiled-coil domain appears to be necessary and sufficient for the interaction of PDZD8 with HIV-1 CA-NC complexes. The coiled-coil domain potentially plays a role in the multimerization of PDZD8, although this has yet to be demonstrated. Of note, multimerization of the capsid-destabilizing factor TRIM5α enhances binding to the viral capsid (44, 45). One model of restriction proposes that TRIM5α binding to the capsids of multiple unrelated retroviruses is achieved by recognition of conserved hexameric patterns on the assembled capsid surface by TRIM5α multimers (46). Future studies should address the natural oligomeric state of PDZD8 and its contribution to recognition of the retroviral capsid.
N-terminal deletion of the first 365 residues of PDZD8 exerted little impact on its function in stabilization of the HIV-1 capsid. However, a further N-terminal deletion that removed the PDZ domain resulted in an inactive PDZD8 protein, even though this mutant still bound the HIV-1 CA-NC complexes. Surprisingly, the peptide-binding motif of PDZD8, a common target for pathogenic viruses in other PDZ-containing proteins (47), does not appear to be required for capsid interaction, stabilization, or enhancement of infection. Additional studies will be required to determine the role of the PDZ domain in the function of PDZD8 as an HIV-1 cofactor.
The C-terminal region of PDZD8, which includes the coiled-coil motif, has also been implicated in the interaction of PDZD8 with the cytoskeletal regulation protein moesin (23). Moesin overexpression has pleiotropic effects on HIV-1 infection, including a negative postentry, prereverse transcription effect coincident with the timing of capsid uncoating (48–50). One explanation for moesin restriction of HIV-1 infection is that infectivity is diminished due to inhibition of PDZD8-capsid interaction as a result of the overexpressed moesin competing for access to PDZD8's C-terminal region.
Finally, the PKC1-like domain of PDZD8 appears to be completely dispensable for HIV-1 CA-NC-stabilizing activity and for the ability to act as a cofactor for HIV-1 infection.
Following retrovirus entry into the host cell, the uncoating of the viral capsid may be determined by the binding of stabilizing host factors, including PDZD8 and CypA, in competition with the binding of cellular restriction factors like TRIM5α and TRIMCyp. Additional inquiry into this critical stage of the viral life cycle may suggest interventional approaches, such as interruption of the PDZD8-capsid interaction, that could tip the balance toward host cell resistance.
ACKNOWLEDGMENTS
We thank Yvette McLaughlin and Elizabeth Carpelan for manuscript preparation.
We acknowledge the support of the National Institutes of Health (AI063987 and a Center for AIDS Research Award [AI06354]), the International AIDS Vaccine Initiative, and the late William F. McCarty-Cooper.
Footnotes
Published ahead of print 19 February 2014
REFERENCES
- 1.Ganser-Pornillos BK, Yeager M, Sundquist WI. 2008. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18:203–217. 10.1016/j.sbi.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganser-Pornillos BK, Cheng A, Yeager M. 2007. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell 131:70–79. 10.1016/j.cell.2007.08.018 [DOI] [PubMed] [Google Scholar]
- 3.Pornillos O, Ganser-Pornillos BK, Yeager M. 2011. Atomic-level modelling of the HIV capsid. Nature 469:424–427. 10.1038/nature09640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667–5677. 10.1128/JVI.76.11.5667-5677.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair WS, Pickford C, Irving SL, Brown DG, Anderson M, Bazin R, Cao J, Ciaramella G, Isaacson J, Jackson L, Hunt R, Kjerrstrom A, Nieman Ja, Patick AK, Perros M, Scott AD, Whitby K, Wu H, Butler SL. 2010. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 6:e1001220. 10.1371/journal.ppat.1001220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi J, Zhou J, Shah VB, Aiken C, Whitby K. 2011. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J. Virol. 85:542–549. 10.1128/JVI.01406-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U. S. A. 103:5514–5519. 10.1073/pnas.0509996103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sayah DM, Sokolskaja E, Berthoux L, Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573. 10.1038/nature02777 [DOI] [PubMed] [Google Scholar]
- 9.Diaz-Griffero F, Kar A, Lee M, Stremlau M, Poeschla E, Sodroski J. 2007. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology 369:400–410. 10.1016/j.virol.2007.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, Takemura T, Shelton K, Taniuchi I, Li Y, Sodroski J, Littman DR, Coffin JM, Hughes SH, Unutmaz D, Engelman A, KewalRamani VN. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233. 10.1016/j.chom.2010.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hori T, Takeuchi H, Saito H, Sakuma R, Inagaki Y, Yamaoka S. 2013. A carboxy-terminally truncated human CPSF6 lacking residues encoded by exon 6 inhibits HIV-1 cDNA synthesis and promotes capsid disassembly. J. Virol. 87:7726–7736. 10.1128/JVI.00124-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fricke T, Valle-Casuso JC, White TE, Brandariz-Nuñez A, Bosche WJ, Reszka N, Gorelick R, Diaz-Griffero F. 2013. The ability of TNPO3-depleted cells to inhibit HIV-1 infection requires CPSF6. Retrovirology 10:46. 10.1186/1742-4690-10-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Iaco A, Santoni F, Vannier A, Guipponi M, Antonarakis S, Luban J. 2013. TNPO3 protects HIV-1 replication from CPSF6-mediated capsid stabilization in the host cell cytoplasm. Retrovirology 10:20. 10.1186/1742-4690-10-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah VB, Shi J, Hout DR, Oztop I, Krishnan L, Ahn J, Shotwell MS, Engelman A, Aiken C. 2013. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J. Virol. 87:422–432. 10.1128/JVI.07177-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Kar AK, Sodroski J. 2009. Target cell type-dependent modulation of human immunodeficiency virus type 1 capsid disassembly by cyclophilin A. J. Virol. 83:10951–10962. 10.1128/JVI.00682-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatziioannou T, Perez-Caballero D. 2005. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 79:176–183. 10.1128/JVI.79.1.176-183.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auewarakul P, Wacharapornin P, Srichatrapimuk S, Chutipongtanate S, Puthavathana P. 2005. Uncoating of HIV-1 requires cellular activation. Virology 337:93–101. 10.1016/j.virol.2005.02.028 [DOI] [PubMed] [Google Scholar]
- 18.Aiken C. 2009. HIV protocols. Humana Press, Totowa, NJ [Google Scholar]
- 19.Ganser BK. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83. 10.1126/science.283.5398.80 [DOI] [PubMed] [Google Scholar]
- 20.Li S, Hill CP, Sundquist WI, Finch JT. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409–413. 10.1038/35030177 [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Li X, Stremlau M, Lee M, Sodroski J. 2006. Removal of arginine 332 allows human TRIM5alpha to bind human immunodeficiency virus capsids and to restrict infection. J. Virol. 80:6738–6744. 10.1128/JVI.00270-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henning MS, Morham SG, Goff SP, Naghavi MH. 2010. PDZD8 is a novel Gag-interacting factor that promotes retroviral infection. J. Virol. 84:8990–8995. 10.1128/JVI.00843-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henning MS, Stiedl P, Barry DS, McMahon R, Morham SG, Walsh D, Naghavi MH. 2011. PDZD8 is a novel moesin-interacting cytoskeletal regulatory protein that suppresses infection by herpes simplex virus type 1. Virology 415:114–121. 10.1016/j.virol.2011.04.006 [DOI] [PubMed] [Google Scholar]
- 24.Matreyek KA, Engelman A. 2011. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J. Virol. 85:7818–7827. 10.1128/JVI.00325-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. 2006. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology 353:396–409. 10.1016/j.virol.2006.05.035 [DOI] [PubMed] [Google Scholar]
- 26.Saenz DT, Barraza R, Loewen N, Teo W, Poeschla EM. 2012. Production and harvest of feline immunodeficiency virus-based lentiviral vector from cells grown in T75 tissue-culture flasks. Cold Spring Harb. Protoc. 2012:124–125. 10.1101/pdb.prot067553 [DOI] [PubMed] [Google Scholar]
- 27.Perron MJ, Stremlau M, Song B, Ulm W, Mulligan RC, Sodroski J. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. U. S. A. 101:11827–11832. 10.1073/pnas.0403364101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loewen N, Barraza R, Whitwam T, Saenz DT, Kemler I, Poeschla EM. 2003. FIV vectors. Methods Mol. Biol. 229:251–271. 10.1385/1-59259-393-3:251 [DOI] [PubMed] [Google Scholar]
- 29.Chen CM, Smith DM, Peters Ma, Samson ME, Zitz J, Tabin CJ, Cepko CL. 1999. Production and design of more effective avian replication-incompetent retroviral vectors. Dev. Biol. 214:370–384. 10.1006/dbio.1999.9432 [DOI] [PubMed] [Google Scholar]
- 30.Shun M-C, Daigle JE, Vandegraaff N, Engelman A. 2007. Wild-type levels of human immunodeficiency virus type 1 infectivity in the absence of cellular emerin protein. J. Virol. 81:166–172. 10.1128/JVI.01953-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willey RL, Smith DH, Lasky La, Theodore TS, Earl PL, Moss B, Capon DJ, Martin Ma. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pacheco B, Basmaciogullari S, Labonte Ja, Xiang S-H, Sodroski J. 2008. Adaptation of the human immunodeficiency virus type 1 envelope glycoproteins to new world monkey receptors. J. Virol. 82:346–357. 10.1128/JVI.01299-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dismuke D, Aiken C. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 80:3712–3720. 10.1128/JVI.80.8.3712-3720.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fricke T, Brandariz-Nuñez A, Wang X, Smith AB, Diaz-Griffero F. 2013. Human cytosolic extracts stabilize the HIV-1 core. J. Virol. 87:10587–10597. 10.1128/JVI.01705-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arfi V, Lienard J, Nguyen X-N, Berger G, Rigal D, Darlix J-L, Cimarelli A. 2009. Characterization of the behavior of functional viral genomes during the early steps of human immunodeficiency virus type 1 infection. J. Virol. 83:7524–7535. 10.1128/JVI.00429-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hulme AE, Perez O, Hope TJ. 2011. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. U. S. A. 108:9975–9980. 10.1073/pnas.1014522108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutluay SB, Perez-Caballero D, Bieniasz PD. 2013. Fates of retroviral core components during unrestricted and TRIM5-restricted infection. PLoS Path. 9:e1003214. 10.1371/journal.ppat.1003214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saras J, Heldin CH. 1996. PDZ domains bind carboxy-terminal sequences of target proteins. Trends Biochem. Sci. 21:455–458. 10.1016/S0968-0004(96)30044-3 [DOI] [PubMed] [Google Scholar]
- 39.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. 1996. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 85:1067–1076. 10.1016/S0092-8674(00)81307-0 [DOI] [PubMed] [Google Scholar]
- 40.Briggs JAG, Wilk T, Welker R, Kräusslich H-G, Fuller SD. 2003. Structural organization of authentic, mature HIV-1 virions and cores. Eur. Mol. Biol. Organ. J. 22:1707–1715. 10.1093/emboj/cdg143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malim MH, Bieniasz PD. 2012. HIV restriction factors and mechanisms of evasion. Cold Spring Harb. Perspect. Med. 2:a006940. 10.1101/cshperspect.a006940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldstone DC, Yap MW, Robertson LE, Haire LF, Taylor WR, Katzourakis A, Stoye JP, Taylor IA. 2010. Structural and functional analysis of prehistoric lentiviruses uncovers an ancient molecular interface. Cell Host Microbe 8:248–259. 10.1016/j.chom.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 43.Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138–1143. 10.1038/nm910 [DOI] [PubMed] [Google Scholar]
- 44.Javanbakht H, Yuan W, Yeung DF, Song B, Diaz-Griffero F, Li Y, Li X, Stremlau M, Sodroski J. 2006. Characterization of TRIM5alpha trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353:234–246. 10.1016/j.virol.2006.05.017 [DOI] [PubMed] [Google Scholar]
- 45.Li X, Sodroski J. 2008. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J. Virol. 82:11495–11502. 10.1128/JVI.01548-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. 2011. Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. U. S. A. 108:534–539. 10.1073/pnas.1013426108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Javier RT, Rice AP. 2011. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J. Virol. 85:11544–11556. 10.1128/JVI.05410-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naghavi MH, Valente S, Hatziioannou T, de Los Santos K, Wen Y, Mott C, Gundersen GG, Goff SP. 2007. Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells. EMBO J. 26:41–52. 10.1038/sj.emboj.7601475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubo Y, Yoshii H, Kamiyama H, Tominaga C, Tanaka Y, Sato H, Yamamoto N. 2008. Ezrin, radixin, and moesin (ERM) proteins function as pleiotropic regulators of human immunodeficiency virus type 1 infection. Virology 375:130–140. 10.1016/j.virol.2008.01.047 [DOI] [PubMed] [Google Scholar]
- 50.Barrero-Villar M, Cabrero JR, Gordón-Alonso M, Barroso-González J, Alvarez-Losada S, Muñoz-Fernández MA, Sánchez-Madrid F, Valenzuela-Fernández A. 2009. Moesin is required for HIV-1-induced CD4-CXCR4 interaction, F-actin redistribution, membrane fusion and viral infection in lymphocytes. J. Cell Sci. 122:103–113. 10.1242/jcs.035873 [DOI] [PubMed] [Google Scholar]