ABSTRACT
The success of future clinical trials with oncolytic viruses depends on the identification and the control of mechanisms that modulate their therapeutic efficacy. In particular, little is known about the role of autophagy in infection by attenuated measles virus of the Edmonston strain (MV-Edm). We investigated the interaction between autophagy, innate immune response, and oncolytic activity of MV-Edm, since the antiviral immune response is a known factor limiting virotherapies. We report that MV-Edm exploits selective autophagy to mitigate the innate immune response mediated by DDX58/RIG-I like receptors (RLRs) in non-small cell lung cancer (NSCLC) cells. Both RNA interference (RNAi) and overexpression approaches demonstrate that autophagy enhances viral replication and inhibits the production of type I interferons regulated by RLRs. We show that MV-Edm unexpectedly triggers SQSTM1/p62-mediated mitophagy, resulting in decreased mitochondrion-tethered mitochondrial antiviral signaling protein (MAVS) and subsequently weakening the innate immune response. These results unveil a novel infectious strategy based on the usurpation of mitophagy leading to mitigation of the innate immune response. This finding provides a rationale to modulate autophagy in oncolytic virotherapy.
IMPORTANCE In vitro studies, preclinical experiments in vivo, and clinical trials with humans all indicate that oncolytic viruses hold promise for cancer therapy. Measles virus of the Edmonston strain (MV-Edm), which is an attenuated virus derived from the common wild-type measles virus, is paradigmatic for therapeutic oncolytic viruses. MV-Edm replicates preferentially in and kills cancer cells. The efficiency of MV-Edm is limited by the immune response of the host against viruses. In our study, we revealed that MV-Edm usurps a homeostatic mechanism of intracellular degradation of mitochondria, coined mitophagy, to attenuate the innate immune response in cancer cells. This strategy might provide a replicative advantage for the virus against the development of antiviral immune responses by the host. These findings are important since they may not only indicate that inducers of autophagy could enhance the efficacy of oncolytic therapies but also provide clues for antiviral therapy by targeting SQSTM1/p62-mediated mitophagy.
INTRODUCTION
Understanding how a virus either escapes cell scrutiny or is sensed and incapacitated by the infected cell is of tremendous interest not only for antiviral strategies but also for virus-based anticancer or gene transfer therapies. Successful viral infection requires the virus to take advantage of the cellular machinery for replication, transcription, and translation of its own genome; however, hijacking a host cell requires sophisticated strategies to bypass cellular defenses (1).
Cells have evolved a variety of molecules that sense pathogen-associated molecular patterns (PAMPS) and mount innate immune responses (2). Antiviral responses are initiated through the detection of viral nucleic acids, lipids, or proteins by innate pattern recognition receptors (PRRs). Four members of the Toll-like receptor (TLR) family, TLR3, -7, -8, and -9, are located in endosomal membranes, in particular those of immune cells, whereas the DDX58/RIG-I like receptors (RLRs), comprising retinoic acid-inducible gene I (DDX58/RIG-I) and melanoma differentiation-associated protein 5 (IFIH1/MDA5), are located in the cytosol of immune and nonimmune cells. Upon activation, RLRs interact with mitochondrial antiviral signaling protein (MAVS/IPS-1) tethered to mitochondria. MAVS in turn relays signals to interferon regulatory factor 3 (IRF3), IRF7, and nuclear factor kappa B NFKB1/NF-κB1, leading to antiviral responses characterized by the production of type I interferons, such as alpha interferon or beta interferon 1 (IFNB1), and other innate immune response proteins, such as the chemokine C-X-C motif chemokine 10/interferon-induced protein 10 (CXCL10/IP-10) (2). This signaling pathway is then maintained by an amplification loop that increases RLRs and interferon expression, thus ensuring that a robust innate immune response is mounted (3–5).
Autophagy is a highly conserved homeostatic process that allows cells to recycle their components and to remove damaged organelles. This process begins with the formation of vesicles called autophagosomes that pack components targeted for degradation and is terminated upon Ras-associated protein RAB7 (RAB7A)-mediated fusion of the autophagosomes with lysosomes by the degradation of the cargo (6–8). Autophagy can induce bulk degradation or remove specific proteins or organelles upon recognition of autophagic receptors, such as sequestosome 1 (SQSTM1/p62), autophagy-related protein 32 (ATG32), or BCL2/adenovirus E1B (BNIP3) and BNIP3L/NIX (9–13). Autophagy plays a crucial role in the regulation of inflammation and constitutes an essential innate responsive mechanism against pathogens in primitive eukaryotic cells (14, 15). Bacteria and viruses can be degraded by autophagy after being engulfed by autophagosomes, a process called xenophagy (16–18). Viral peptides produced by autophagic digestion of viral particles are presented by major histocompatibility complex class II (MHC-II) and thus provide a link between autophagy and adaptive immunity (19, 20).
Despite some viruses being efficiently thwarted by autophagy, others have evolved countermeasures to abrogate the antiviral properties of autophagy. Several viral proteins inhibit key proteins involved in autophagy induction, such as Beclin 1 (BECN1), or block autophagic flux (8, 20). Contrary to intuition, recent studies indicate that viruses can actually benefit from autophagy, since it facilitates viral replication (21–23). Several underlying mechanisms are currently proposed to explain this effect. Some data suggest that membranes of autophagosomes form viroplasms, specific membranous intracellular compartments that facilitate the replication of some viruses (24, 25). Other novel findings indicate that autophagic proteins interfere with innate immune responses mediated by PRR upon infection by pathogens. This is the case of ATG5 and ATG12, two proteins essential for the formation of autophagosomes, which have been shown to inhibit innate immune responses by impeding the interaction of IFIH1 or DDX58 with MAVS (14, 26). In addition, mitophagy, a selective type of autophagy targeting mitochondria (27–29), reduces the innate response by preventing NOD-like receptor family pyrin-containing domain protein 3 (NLRP3) signaling and inflammation (30).
The attenuated measles virus of the Edmonston strain (MV-Edm) is an oncolytic single-stranded RNA virus that has entered clinical trials. Autophagy contributes to measles virus infection and replication both during interaction of MV-Edm with its entry receptor CD46 and later during production of viral protein C within the infected cell (22, 23, 31). Although recent studies show that the measles virus Edmonston strain employs autophagy for its replication (22, 23), how virus-induced autophagy counteracts innate immunity remains unclear. In this study, we demonstrated that autophagy and mitophagy contribute to replication of MV-Edm by interfering with the DDX58/RIG-I-like receptor-mediated innate immune responses.
MATERIALS AND METHODS
Cells, plasmids, and siRNA.
Human lung adenocarcinoma cell lines A549 (CCL-185), H1299 (CRL-5803), and Vero African green monkey kidney cells (CCL-81) were obtained from American Type Culture Collection (ATCC) (Manassas, VA). Cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 0.1 mM nonessential amino acids, 5% fetal bovine serum, and penicillin-streptomycin (all from Invitrogen). Expression plasmids were as follows: pBABEpuro-EGFP-LC3 (Addgene; 22405) was provided by Jayanta Debnath (University of California, San Francisco, CA), pCMV-myc-Atg5 and pCMV-myc-ATG7 (Addgene; 24922 and 24921) were provided by Toren Finkel (NHLBI, Bethesda, MD), pCI-neo was purchased from Promega (E1841), pCI-neo-hApg5-HA and pCI-neo-hApg5 (K130R)-HA (Addgene; 22948 and 22949) were provided by Noboru Mizushima (Tokyo Medical and Dental University, Tokyo, Japan). pCMV-myc was purchased from Beyotime (D2672). BEZ-235 was purchased from Selleck (S1009). Chloroquine (CQ) was purchased from Sigma (C6628). Small interfering RNA (siRNA) directed against SQSTM1 (p62) (NM_003900) was synthesized by GenePharma (sense, GGAGCACGGAGGGAAAAGAtt; antisense, UCUUUUCCCUCCGUGCUCCac). The siRNAs targeting ATG5 (Invitrogen; HSS114104), ATG7 (Invitrogen; HSS116182), BECN1 (Invitrogen; HSS112731), DDX58 (RIG-I) (Invitrogen; HSS177513), IFIH1 (MDA5) (Invitrogen, HSS127414), MAVS (IPS-1) (Invitrogen; HSS148538), SQSTM1 (p62) (Invitrogen; HSS121770), and RAB7 (Invitrogen; HSS187913) and negative-control siRNA (Invitrogen; 12935400) were all purchased from the Invitrogen Stealth RNAi collection.
Viruses.
Measles virus Edmonston vaccine lineage seed B (MV-Edm) and MV-Edm expressing a reporter gene luciferase (MV-Edm-luc, kindly provided by S. Russell, Mayo Clinic, Minnesota, USA) were propagated in Vero cells with a multiplicity of infection (MOI) of 0.02 in 2 ml Opti-MEM (Invitrogen; 31985-062) at 37°C for 3 h. The medium was changed to DMEM supplemented with 2% fetal calf serum (FCS), and cells were incubated at 37°C for 1 day before being transferred to 32°C for another day. Cells were harvested, and viral particles were released by two cycles of snap-freezing in liquid nitrogen and thawing in a 37°C water bath. Viral titers were determined by 50% endpoint dilution assays (50% tissue culture infective dose [TCID50]) on Vero cells.
Transfection.
One hundred nanomolar siRNA or 500 ng/ml expression plasmids coupled with Lipofectamine 2000 (Invitrogen; 11668-019) were used for transfection of A549 or H1299 cells on a 6- or 12-well plate according to the manufacturer's instructions.
Cell viability assay.
Cells were harvested by trypsin-EDTA and stained with trypan blue; viability was determined by a trypan blue exclusion assay.
Crystal violet staining.
Cells on 12-well plates were stained with a solution containing 0.4 mg/ml crystal violet with 5% formaldehyde in phosphate-buffered saline (PBS) and incubated for 30 min at room temperature. After washing twice with deionized water, the plates were air dried for scanning.
Western blot analysis.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer containing a protease inhibitor cocktail (Roche; 11873580001). The protein concentration was determined. Equal amounts of protein were separated by SDS-PAGE and electrophoretically transferred onto a polyvinylidene difluoride (PVDF) membrane (Roche; 03010040001). After blocking with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween 20, the membrane was incubated with specific primary antibodies, followed by incubation with appropriate horseradish peroxidase-conjugated secondary antibodies. Signals were detected using an enhanced chemiluminescence reagent (Millipore; WBKLS0500) and subjected to the Alpha Innotech Fluor Chem-FC2 imaging system. Antibodies used in this experiment were as follows: anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (Bioworld; MB001, 1:5,000 diluted), anti-microtubule-associated protein 1 light chain 3 beta (anti-MAP1LC3B/LC3) (Thermo Scientific, PAI-16930, 1:500 diluted), anti-SQSTM1/p62 (Abcam; ab109012, 1:5,000 diluted), anti-ATG5 (Cell Signaling Technology; 2630, 1:1,000 diluted), anti-DDX58/RIG-I (Cell Signaling Technology; 3743, 1:1,000 diluted), anti-IFIH1/MDA5 (Sigma-Aldrich; SAB3500356, 1:500 diluted), anti-MAVS/IPS-1 (Abcam; ab31334, 1:500 diluted), anti-phospho-IRF-3 (Cell Signaling Technology; 4947S, 1:1,000 diluted), anti-HSPD1/HSP60 (Epitomics; 1724-1, 1:10,000 diluted), and anti-COX4I1/COX4-1 (Abcam; ab16056, 1:500 diluted).
qPCR.
For quantitative reverse transcription (RT)-PCR (qPCR), total cellular RNA was extracted with TRIzol (Invitrogen; 15596-026), and 1 μg of RNA was reverse transcribed using the synthesis system (TaKaRa; DRR036A). qPCR was performed using the real-time PCR system (ABI 7300). Gene expression was calculated with the comparative threshold cycle (CT) method and normalized to the endogenous levels of GAPDH. Primer sequences used for qPCR are as follows: DDX58 (RIG-I), 5′-GCCATTACACTGTGCTTGGAGA-3′ and 5′-CCAGTTGCAATATCCTCCACCA-3′; IFIH1 (MDA-5), 5′-AGGAGTCAAAGCCCACCATCTG-3′ and 5′-ATTGGTGACGAGACCATAACGGATA-3′; MAVS (IPS-1), 5′-GCAATGTGGATGTTGTAGA G-3′ and 5′-CTGAAGGGT ATTGAAGAG-3′; ATG5, 5′-AAGCAACTCTGGATGGGATT-3′ and 5′-GCAGCCACAGGACGAAAC-3′; ATG7, 5′-ACCCAGAAGAAGCTGAACGA-3′ and 5′-AGACAGAGGGCAGGATAGCA-3′; BECN1, 5′-GGATGGATGTGGAGAAAGGCAAG-3′ and 5′-TGAGGACACCCAAGCAAGACC-3′; RAB7, 5′-TTACTTCGAGACCAGTGCCAAGGA-3′ and 5′-TGTCCAGTTTGATGGGTTCAGGGA-3′; SQSTM1 (p62), 5′-GAACTCCAGTCCCTACAGAT-3′ and 5′-CGATGTCATAGTTCTTGGTC-3′; CXCL10 (IP10), 5′-CTTCCAAGGATGGACCACACA-3′ and 5′-CCTTCCTACAGGAGTAGTAGCAG-3′; IFNB1, 5′-CTTGGATTCCTACAAAGAAGC-3′ and 5′-CATCTCATAGATGGTCAATGC-3′; IFI27, 5′-TGGCCAGGATTGCTACAGTTG-3′ and 5′-TATGGAGGACGAGGCGATTC-3′; OAS1, 5′-TCCAAGCTCAGTCAGCAGAA-3′ and 5′-TGTCAATGGCATGGTTGATT-3′; MV-Edm N-protein, 5′-ACATTAGCATCTGAACTCGGTATCAC-3′ and 5′-TTTTCGCTTTGATCACCGTGTA-3′; MV-Edm H-protein, 5′-GATGACAAGTTGCGAATGGAGA-3′ and 5′-GACAAGAC CCCGTATGAAGGAA-3′; GAPDH, 5′-CC ACCCATGGCAAATTCCATGGCA-3′ and 5′-TCTAGACGGCAGGTCAGGTCCACC-3′.
Luciferase assay.
The luciferase activity of cells infected by MV-Edm expressing luciferase (MV-Edm-Luc) was monitored by luminescence spectrometry after adding luciferin (Gold Biotechnology; 115144-35-9) at a concentration of 150 μg/ml according to the manufacturer's instructions.
ELISA.
Supernatants from treated or untreated cells were harvested, centrifuged, and stored at −80°C. Samples were then subjected to enzyme-linked immunosorbent assay (ELISA) for detection of CXCL10/IP10 (eBioscience; BMS284) and IFNB1/IFN-β (R&D Systems; 41410-1A) according to the manufacturer's protocol.
Confocal microscopy and immunofluorescence.
The pBABEpuro-EGFP-LC3/Map1lc3b (32) construct was transiently expressed in A549 cells 24 h prior to MV-Edm infection. For some experiments, cells were stained with MitoTracker red (Invitrogen, M7512) at a concentration of 100 nM for 20 min at 37°C in culture medium and then fixed with 4% paraformaldehyde. Cells were observed under a confocal microscope (Olympus), and images were obtained by using a digital camera with FV10-ASW software (Olympus) and analyzed using the software program ImageJ (National Institutes of Health, USA). Pearson's correlation coefficient was quantified using ImageJ software in 30 different cells per time point for each condition.
For some experiments, cells were fixed with 4% paraformaldehyde for 10 min and permeabilized in 0.1% PBS-Tween with 1% BSA–0.3 M glycine for 1 h. Cells were then incubated with anti-measles virus H protein (Santa Cruz; sc-57913, mouse monoclonal, 1:200 diluted) antibody overnight at 4°C, followed by incubation with Alexa Fluor 568-conjugated goat anti-mouse IgG (Invitrogen; A-11004, 1:1,000 dilution) for 1 h at room temperature. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen; D3571) before being subjected to confocal microscopy.
Flow cytometric analyses.
Cells were grown on 12-well plates with or without MV-Edm infection for 12, 24, and 48 h. MitoTracker green (Invitrogen; M7514) was then added for mitochondrion staining according to the manufacturer's instructions. The mean fluorescent intensity of cells was determined using a FACSCalibur instrument (Becton, Dickinson). Mitochondrial mass was quantified using mean fluorescence intensity averaged from 3 independent experiments. Uninfected and unstained cells were used to set maximum and minimum mean fluorescence intensities, respectively.
Electron microscopy.
A549 cells (5 × 104 cells/cm2) were seeded on sapphire discs (Brügger) in a 12-well plate and infected with MV-Edm at an MOI of 0.5 for 3 h. Cells were washed and incubated for an additional 9 h. Samples were frozen under high pressure, dehydrated, and chemically fixed. Ultrathin section were cut and stained with uranyl acetate and lead citrate. Cells were imaged using a Zeiss EM 10 transmission electron microscope using an acceleration voltage of 80 kV.
Cell fractionation.
Mitochondrial and cytoplasmic proteins were separated using a commercially available mitochondria/cytosol fractionation kit (Beyotime Institute of Biotechnology; C3601) according to the manufacturer's protocol. Briefly, cells were harvested and washed twice with ice-cold PBS. They were then incubated in 500 μl ice-cold mitochondrial lysis buffer on ice for 10 min. The cell suspension was then taken into a glass homogenizer and homogenized for 32 strokes using a tight pestle on ice. The homogenate was centrifuged at 800 × g for 10 min at 4°C to remove any unbroken cells. The supernatant was further centrifuged at 8,000 × g for 10 min at 4°C to obtain the mitochondrial fraction (pellet) and cytoplasmic proteins (supernatant). Samples of mitochondria were dissolved in lysis buffer, and proteins were subjected to immunoblotting.
Statistical analyses.
Student's t test was used for all statistical analyses.
RESULTS
MV-Edm-induced autophagy and autophagic flux enhance the oncolytic activity of the virus.
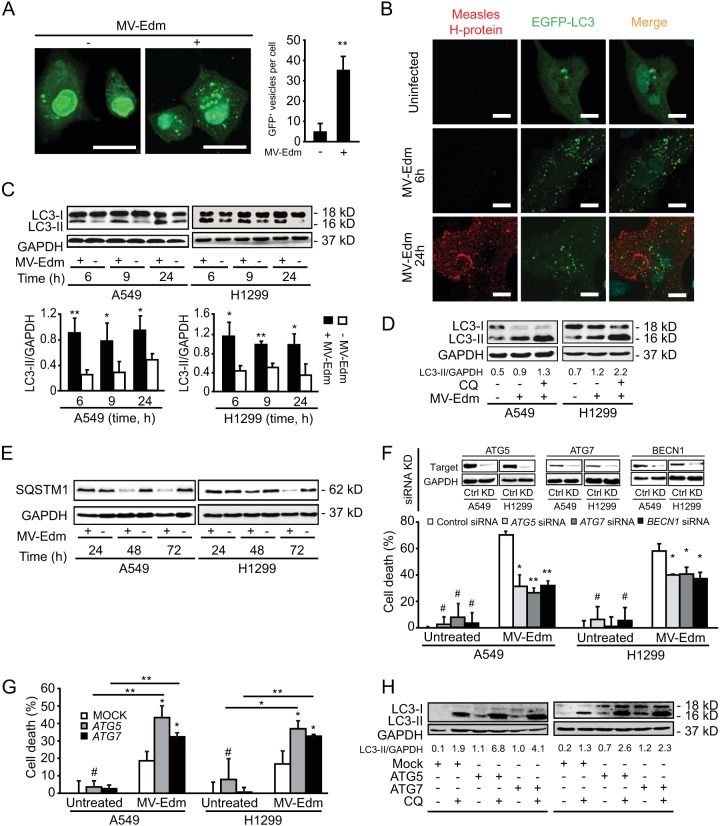
First, we investigated whether, in non-small cell lung carcinoma (NSCLC) cells, MV-Edm affected early stages of autophagy, i.e., autophagosome formation, and/or late stages of autophagy, which is referred to as autophagic flux. Enhanced green fluorescent protein (EGFP)–microtubule-associated protein 1 light chain 3 beta (MAP1LC3B/LC3) was overexpressed in NSCLC cells in order to follow the induction of autophagy in infected cells. Upon incubation with MV-Edm, we observed a significant change in the appearance of EGFP-MAP1LC3B from a homogenous to a punctuate distribution in the cytosol of the transfected cells (Fig. 1A). Autophagy was induced early upon infection (2 h) (Fig. 1A) and was sustained over 24 h while virus replicated (Fig. 1B). We also observed enhanced levels of lipidated MAP1LC3B/LC3 (LC3-II) in infected cells (Fig. 1C). Furthermore, lipidation levels of MAP1LC3B in the presence of chloroquine increased in infected cells when chloroquine abrogated the autophagic flux, thus arguing that autophagy is completed by lysosomal degradation of the cargo in infected cells (Fig. 1D). Along the same lines, we observed that SQSTM1 levels decreased between 24 and 48 h upon MV-Edm infection, which is consistent with degradation of SQSTM1 via a preserved autophagic flux (Fig. 1E). Taken together, these results indicate that MV-Edm triggers and sustains autophagy and autophagic flux both at early stages of viral infection in NSCLC cells and at later time points when viral replication continues. These observations are consistent with previous reports (23, 31).
FIG 1.
MV-Edm-induced autophagy and autophagic flux enhance oncolytic activity of the virus in NSCLC cells. (A) Translocation of EGFP-MAP1LC3B toward autophagosomes was followed by fluorescence microscopy in A549 cells transiently transfected with a plasmid encoding EGFP-MAP1LC3B grown for 2 h in the presence or absence of MV-Edm (MOI = 0.5). Bars represent 10 μm. The number of EGFP-MAP1LC3B-positive vesicles per cell was quantified by fluorescence microscopy. Bright punctuate structures are GFP+ vesicles, indicating autophagosomes. (B) A549 lung cancer cells were transiently transfected with a plasmid encoding EGFP-MAP1LC3B for 24 h followed by infection with MV-Edm (MOI = 0.5) for 6 and 24 h or were left uninfected and cultured in completed medium or in Dulbecco's PBS (DPBS) for 4 h. Cells were then stained for measles virus H protein. Aggregation of EGFP-MAP1LC3B at autophagosomes (green dots) and expression of measles virus H protein (red dots) were evaluated by fluorescence confocal microscopy. Scale bars represent 10 μm. (C) Levels of lipidated MAP1LC3B (LC3-II) were assessed by Western blotting in lysates obtained from A549 and H1299 lung cancer cells infected with MV-Edm at an MOI of 0.5 or left uninfected for 6, 9, and 24 h (upper panels). The LC3-II/GAPDH ratio was quantified by densitometric analysis (lower panels). (D) MAP1LC3B lipidation was analyzed and quantified in A549 and H1299 cells infected with MV-Edm and grown with or without chloroquine. (E) Degradation of SQSTM1 was monitored by immunoblotting of A549 and H1299 cells after infection by MV-Edm (MOI = 0.2) at 24, 48, and 72 h. (F and G) A549 and H1299 cells transfected with siRNA targeting ATG5, ATG7, or BECN1 or with nontargeting control siRNA (F) or with ATG5 or ATG7 expression plasmids or a mock plasmid (G) for 24 h were infected with MV-Edm (MOI = 0.2) for 48 h. Cell death was quantified using trypan blue staining. Knockdown efficiency for ATG5, ATG7, and BECN1 was monitored at the protein level by Western blotting (F, upper panels). One experiment representative of three (for F) or of two (for G) is shown. Results are means of triplicates. (H) Uninfected A549 and H1299 cells were transfected with plasmids encoding ATG5 or ATG7 or with a mock plasmid for 19 h. Cells were then grown in the presence or absence of chloroquine (CQ) (20 μM) for another 5 h. LC3-II was evaluated by Western blotting. The LC3-II/GAPDH ratio was quantified by densitometry. *, P < 0.05; **, P < 0.01.
The impact of autophagy on viral infection is diverse because autophagy can either potentiate or impede viral infection depending on the virus and possibly the cell type. To characterize the influence of autophagy on the oncolytic activity of MV-Edm in NSCLC cells, we modulated the activities of BECN1, ATG7, and ATG5, key proteins for initiation of preautophagosomal membrane formation in mammalians. Downregulating expression of these proteins decreased cell death induced by MV-Edm, indicating that autophagy contributes to virus-induced oncolytic activity (Fig. 1F). In line with this, enhanced autophagy upon overexpression of ATG5 or ATG7 (Fig. 1H) sensitized the A549 and H1299 cell lines for MV-Edm-induced cell death (Fig. 1G). Together, these results support the notion that autophagy contributes to MV-Edm-induced oncolysis in NSCLC.
MV-Edm-induced autophagy and autophagic flux facilitate replication and spread of MV-Edm.
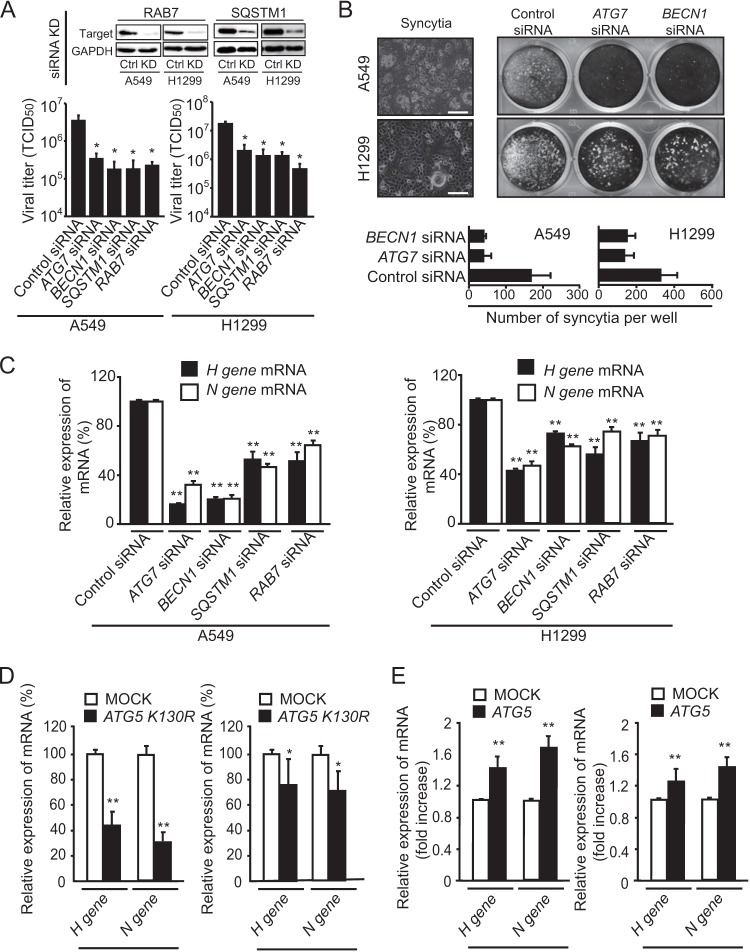
To elucidate how autophagy contributes to MV-Edm-induced oncolysis and spread, we quantified viral titers upon infection of cells downregulated for BECN1 and ATG7 (to inhibit formation of autophagosomes), SQSTM1 (to block cargo delivery to autophagosomes), and RAB7 (to abrogate fusion of autophagosomes with lysosomes) (Fig. 2A). All these approaches strongly decreased the titer of MV-Edm, indicating that both early and late stages of autophagy contribute to replication of MV-Edm. Knockdown of ATG7 or BECN1 reduced syncytium formation, the typical cytopathic effect of MV-Edm (Fig. 2B). Along the same lines, expression of mRNA encoding the viral proteins H and N was clearly decreased when ATG7, BECN1, SQSTM1, or RAB7 was knocked down (Fig. 2C). This was corroborated by decreased expression of viral H and N protein mRNA upon inhibition of autophagy by overexpressing dominant negative ATG5-K130R (ATG5 with a K130R substitution) (Fig. 2D). Conversely, overexpressing ATG5 promoted the expression of viral genes (Fig. 2E). These results support a proviral function of autophagy in MV-Edm-infected cells and are consistent with findings of previous studies performed in different cell lines (23).
FIG 2.
MV-Edm-induced autophagy and autophagic flux promote viral replication and spread in NSCLC cells. (A) Replication of MV-Edm was quantified in A549 and H1299 cells transfected with siRNA targeting ATG7, BECN1, SQSTM1, or RAB7 or with control siRNA, followed by MV-Edm infection at an MOI of 0.2 for 48 h. Viral particles were then harvested by two rounds of freezing-thawing cycles, and the viral titer was determined by calculating the TCID50 on Vero cells. Knockdown efficiency of RAB7 and SQSTM1 were monitored by Western blotting (upper panels). (B) Syncytium formation was observed by phase-contrast microscopy (left panel) and was further evaluated by crystal violet staining of A549 and H1299 cells transfected with siRNAs targeting ATG7 or BECN1 or with control siRNA and infected with MV-Edm (MOI = 0.2) for 48 h (right panel). Pictures representative of three independent experiments are shown. The mean number of syncytia is depicted (lower panel). (C to E) The expression of H and N viral structural genes was quantified by qRT-PCR in A549 and H1299 cells transfected with siRNA targeting ATG7, BECN1, SQSTM1, RAB7, or control siRNA (C) or with a plasmid expressing a mutant ATG5 gene encoding the K130R substitution (ATG5 K130R) (D) or functional ATG5 (E), followed by infection with MV-Edm (MOI = 0.2) for another 48 h. Results are mean of quadruplicates. *, P < 0.05; **, P < 0.01. Similar results were obtained in two independent experiments.
Antiviral innate immune response is impaired by MV-Edm-induced autophagy and autophagic flux.
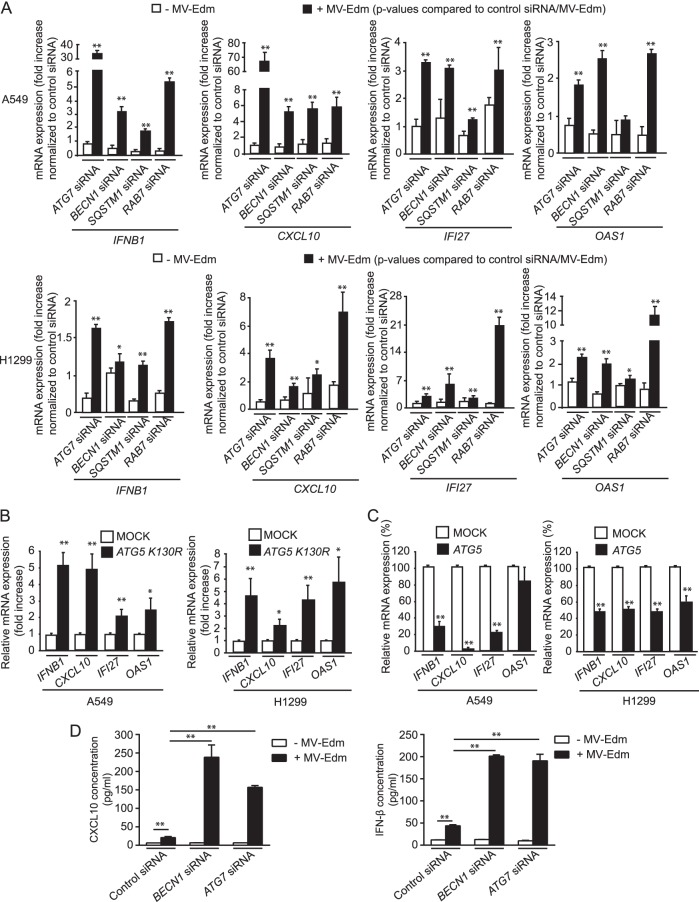
Cells can efficiently sense viruses and mount an innate antiviral response by producing cytokines that block viral replication, activate the immune system, or cause cell suicide. To evaluate the impact of autophagy on the elaboration of an innate immune response against MV-Edm infection, expression levels of the antiviral cytokines IFNB1, CXCL10 (a gamma interferon (IFN-γ)-inducible protein), IFI27, and 2′,5′-oligoadenylate synthetase 1 (OAS1) (which activates latent RNase L, causing viral RNA degradation) were determined by RT-PCR. As shown in Fig. 3A, impairment of autophagy mediated by RNAi against ATG7, BECN1, SQSTM1, or RAB7 in MV-Edm-infected cells enhanced expression of each of the cytokines assayed, except for IFNB1 in H1299 cells knocked down for BECN1. This effect was most pronounced with IFNB1 and CXCL10 in A549 cells, where mRNA levels increased 35- and 65-fold, respectively, when ATG7 was knocked down. In contrast, BECN1 downregulation in the same cell line resulted in only 3- and 4-fold elevation of IFNB1 and CXCL10 mRNA, respectively, suggesting that ATG7 and BECN1 are not equivalent in inducing proviral autophagy. Of note, completion of autophagy by fusion with lysosomes was also necessary for inhibition of the innate immune response, since knockdown of RAB7 significantly increased these cytokines (Fig. 3A). In line with these results, forced expression of ATG5-K130R led to a significant increase in these cytokines (Fig. 3B). In contrast, expression of IFNB1, CXCL10, IFI27, and OAS1 significantly decreased in cells overexpressing ATG5 (with the exception of OAS1 in A549 cells) (Fig. 3C). To assess whether enhanced gene expression of cytokines translated into protein production, we determined the protein levels of IFNB1 and CXCL10 in A549 cells where ATG7 or BECN1 was knocked down (Fig. 3D). CXCL10 and IFNB1 were barely detectable in uninfected cells with or without inhibition of autophagy. However, CXCL10 increased to approximately 20 pg/ml in cells transfected with a nontargeting siRNA and infected with MV-Edm and reached more than 237 or 157 pg/ml when BECN1 or ATG7 was knocked down, respectively. The IFNB1 concentration increased from approximately 44 pg/ml up to 201 or 195 pg/ml when BECN1 or ATG7, respectively, was downregulated (Fig. 3D). Taken together, these results show that autophagy and autophagic flux counteract the expression of crucial antiviral cytokines, which might enhance viral replication.
FIG 3.
MV-Edm-induced autophagy and autophagic flux impair antiviral immune responses. Gene expression of the antiviral cytokines IFNB1, CXCL10, OAS1, and IFI27 was quantified by qRT-PCR in A549 and H1299 cells transfected with siRNAs targeting ATG7, BECN1, SQSTM1, or RAB7 or with control siRNA (A), with an expression plasmid encoding the ATG5 mutant ATG5-K130R (B), or with functional ATG5 (C) and grown in the absence or presence of MV-Edm (MOI = 0.2) for 48 h. The fold increase in gene expression shown in panel A was normalized to levels for cells treated with control siRNA in the absence (open bars) or presence (filled bars) of MV-Edm. P values were obtained by comparison with results for cells transfected with control siRNA after MV-Edm infection. The percentage decrease in gene expression shown in panel B was compared to results for cells expressing ATG5-K130R and infected with MV-Edm. Means + SD for quadruplicates are shown. Similar results were obtained in two independent experiments. (D) CXCL10 and IFNB1 were quantified by ELISA in the supernatants of A549 cells transfected with siRNAs for ATG7, BECN1, or control siRNA followed by infection with MV-Edm for 48 h (filled bars) or left uninfected (open bars). Means + SD of two experiments are shown. *, P < 0.05; **, P < 0.01.
MV-Edm-induced autophagy and autophagic flux attenuate DDX58/IFIH1/MAVS signaling.
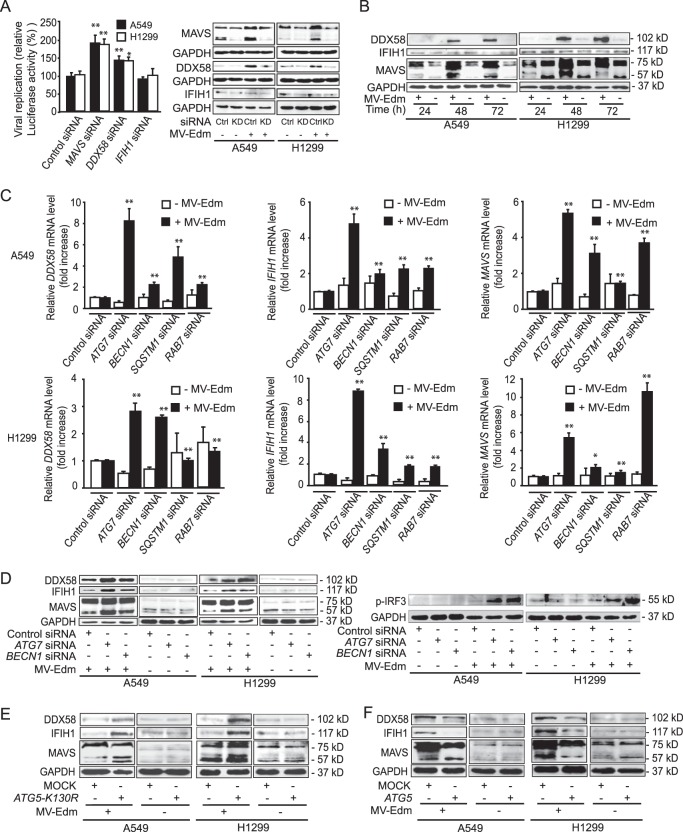
The DDX58/IFIH1/MAVS single-stranded RNA sensing and signaling pathway is central in the induction of antiviral responses, since it bridges early detection of viruses with expression of antiviral cytokines by the host. Having shown that MV-Edm-induced autophagy impairs expression of antiviral cytokines, we wanted to know whether this could be explained by MV-Edm-induced autophagy impairing the DDX58/IFIH1/MAVS signaling pathway. Indeed, replication of MV-Edm increased upon downregulation of MAVS or DDX58 (Fig. 4A). In addition, the amplification loop of RLR expression was activated following infection by MV-Edm, since expression of DDX58 and MAVS was enhanced at the protein level (Fig. 4B).
FIG 4.
MV-Edm-induced autophagy and autophagic flux mitigate DDX58/IFIH1/MAVS signaling. (A) Viral replication was quantified in A549 and H1299 cells transfected with siRNAs targeting DDX58, IFIH1, or MAVS or with nonspecific control siRNA after infection with MV-Edm-Luc (MOI = 0.2) for 24 h. Luciferase activity as a viral replication indicator was determined relative to that of MV-Edm-infected cells transfected with control siRNA. Results are compared to those for uninfected cells. Means + SD of quadruplicates are shown (left panel). Knockdown efficiency of MAVS, DDX58, and IFIH1 in NSCLC cells with or without MV-Edm infection was monitored by Western blot 48 h after siRNA treatment (right panel). Similar results were obtained in two independent experiments. *, P < 0.05; **, P < 0.01. (B) Expression of DDX58, IFIH1, and MAVS was determined by immunoblotting in A549 and H1299 cells infected with MV-Edm (MOI = 0.2) for 48 h. (C) Gene expression of DDX58, IFIH1, and MAVS was quantified by qRT-PCR in A549 and H1299 cells transfected with siRNAs targeting ATG7, BECN1, SQSTM1, or RAB7 or with nonspecific control siRNA and grown in the absence or presence of MV-Edm (MOI = 0.2) for 48 h. The fold increase in gene expression was normalized to results for cells treated with nonspecific control siRNA in the absence (open bars) or presence (filled bars) of MV-Edm. Means + SD of quadruplicates are shown. P values were obtained by comparison with results for cells transfected with control siRNA upon MV-Edm infection. *, P < 0.05; **, P < 0.01. Similar results were obtained in three independent experiments. (D) DDX58, IFIH1, and MAVS (left panel) and p-IRF3 (right panel) protein levels were evaluated by immunoblotting of cell lysates harvested from A549 and H1299 cells transfected with siRNAs targeting ATG7 or BECN1 or with a nonspecific control siRNA followed by MV-Edm infection (MOI = 0.2) for 48 h. A representative result from two independent experiments is shown. (E and F) A549 and H1299 cells were transiently transfected with a plasmid encoding ATG5-K130R (E) or ATG5 (F) and cultured in the presence or absence of MV-Edm (MOI = 0.2) for 48 h. Cell lysates were harvested, and DDX58, IFIH1, and MAVS protein levels were evaluated by immunoblotting. A representative result from two independent experiments is shown.
Unexpectedly, knockdown of BECN1 and ATG7 resulted in greater enhancement of DDX58, IFIH1, and MAVS expression at both the mRNA and protein levels in MV-Edm-infected cells (Fig. 4C and D). Similar results were obtained at the mRNA level when SQSTM1 or RAB7, which impairs recognition of autophagic cargos or autophagic flux, respectively, was silenced (Fig. 4C). Together this set of data indicates that autophagy represses the innate immune response mediated by RLRs in MV-Edm-infected cells. This was further confirmed by the dramatic increase in phospho-IRF3 observed after knockdown of BECN1 or ATG7 in infected cells (Fig. 4D). Along the same lines, protein levels of DDX58, IFIH1, and MAVS were increased by overexpression of ATG5-K130R in MV-Edm-infected cells, whereas the abundance of RLRs remained unchanged in noninfected cells (Fig. 4E). Conversely, stimulation of autophagy by overexpression of ATG5 reduced the abundance of RLRs in infected cells (Fig. 4F). Altogether, these results unmask that autophagy keeps innate immune responses in check in infected cells by mitigating the positive feedback loop of RLR signaling.
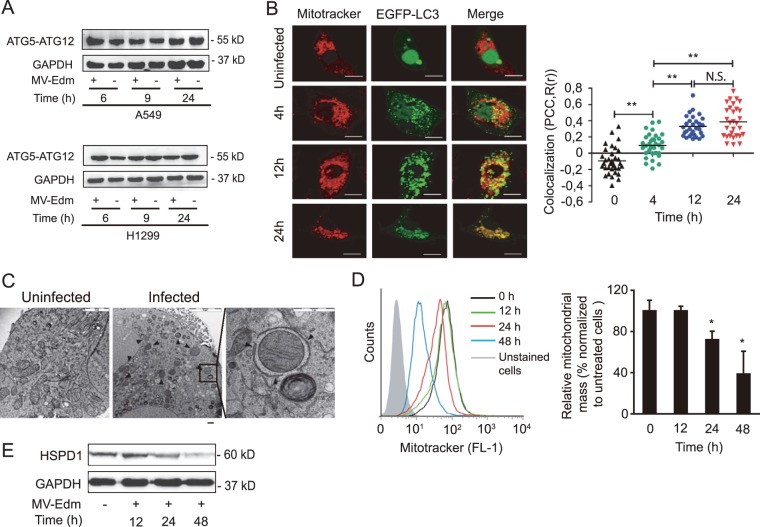
MV-Edm induces degradation of mitochondria by mitophagy.
Next, we investigated the specific mechanisms by which MV-Edm-induced autophagy interferes with the DDX58/IFIH1/MAVS pathway. ATG5-ATG12 heterodimers have recently been shown to sequester MAVS, resulting in negative regulation of genes containing MAVS-dependent promoters, which include RLR and MAVS genes themselves (26). We observed that MV-Edm does not affect the amount of ATG5-ATG12 complexes in NSCLC cells (Fig. 5A), suggesting that mitigation of RLR signaling occurs in the autophagic pathway downstream of ATG5-ATG12 complex formation.
FIG 5.
MV-Edm induces mitophagy leading to mitochondrial degradation. (A) Levels of ATG5-ATG12 conjugates were assessed by Western blotting in lysates obtained from A549 and H1299 lung cancer cells infected with MV-Edm at an MOI of 0.5 or left uninfected for 6, 9, and 24 h. A representative result from two independent experiments is shown. (B) Colocalization of autophagosomes and mitochondria was quantified in A549 cells transiently transfected with pBABEpuro-EGFP-Map1lc3b and infected by MV-Edm (MOI = 0.5) for 4, 12, and 24 h or left uninfected. Cells were then stained with MitoTracker stain and subjected to confocal microscopy (left panels). Scale bars = 10 μm. Colocalization (yellow dots) of mitochondria (red) with autophagosomes (green puncta) was quantified by calculating Pearson's correlation coefficient [PCC, R(r)] (right panels). Means are shown (n = 30 for each time point). **, P < 0.01; N.S, not significant. (C) Subcellular analysis of A549 cells infected without (left panel) or with (middle and right panels) MV-Edm (MOI = 1; 24 h) was performed by electron microscopy. Arrowheads depict double-layer structures that contain mitochondria in an MV-Edm-infected cell. Scale bar = 2 μm (left), 5 μm (middle), or 1 μm (right). (D) Mitochondrial mass was measured by cytometry in MitoTracker green-stained A549 cells 12, 24, and 48 h after infection with MV-Edm (MOI = 0.5). An overlay of histograms representative of 3 independent experiments (left panel), and quantification of mitochondrial mass as mean fluorescence intensity averaged from 3 independent experiments (right panel) are shown. (E) Mitochondrial HSPD1 protein level was determined by Western blotting in lysates obtained from A549 lung cancer cells infected with MV-Edm at an MOI of 0.5 for 12, 24, and 48 h or left uninfected. A representative result from two independent experiments is shown.
Since key proteins involved in the innate immune response, such as MAVS, are anchored to mitochondrial membranes, we hypothesized that MV-Edm controls innate immune response by mitophagy, a selective type of autophagy targeting mitochondria. To evaluate mitophagy, we analyzed the localization of mitochondria and of the autophagosomal marker EGFP-MAP1LC3B in MV-Edm-infected cells. Mitochondria and autophagosomes were found to colocalize 12 and 24 h after infection (Fig. 5B); in addition, double-layered structures characteristic of autophagosomes enveloped mitochondria 24 h after infection by MV-Edm, observed by electron microscopy (Fig. 5C). These observations indicate that MV-Edm induces mitophagy. In agreement with this assertion, colocalization of mitochondria with autophagosomes preceded a decrease in mitochondrial mass per cell after 24 h, as shown by cytometric analysis of MitoTracker green-stained cells (Fig. 5D) and by analysis of expression levels of the mitochondrial protein heat shock protein 60 (HSPD1/HSP60) (Fig. 5E). These observations further support the notion that mitochondria are degraded via mitophagy upon MV-Edm infection.
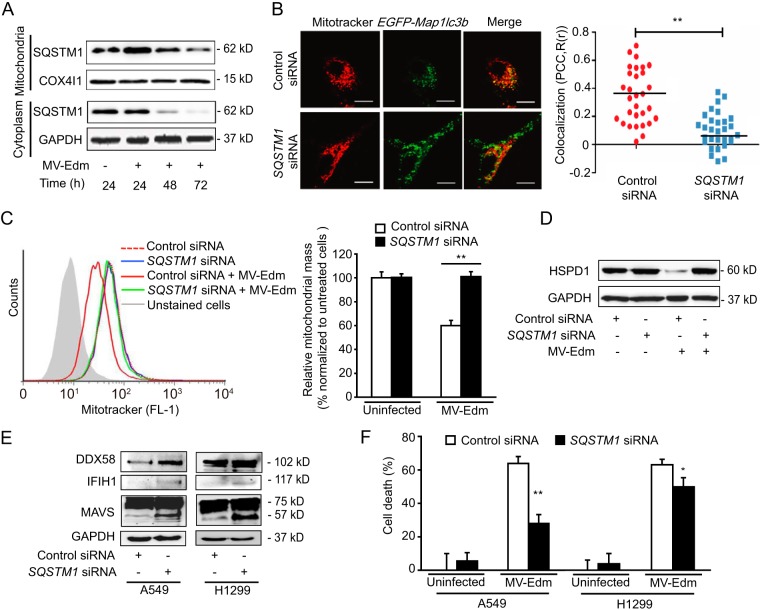
SQSTM1 impairs DDX58/IFIH1/MAVS signaling by mediating MV-Edm-induced mitophagy.
It has been reported that selective mitophagy occurs after recognition of altered mitochondria by autophagic receptors, such as SQSTM1, ATG32, or NIX. We postulated that these proteins might contribute to mitochondrial degradation in MV-Edm-induced cells. We found that SQSTM1 accumulated on mitochondria of infected cells following infection and mitophagy, eventually leading to reduction and exhaustion of SQSTM1 48 h and 72 h postinfection, respectively (Fig. 6A). Downregulation of SQSTM1 strongly reduced colocalization of mitochondria with autophagosomes (Fig. 6B) and resulted in preservation of the mitochondrial mass in MV-Edm-infected cells (Fig. 6C and D), confirming the role of SQSTM1 in targeting mitochondria toward autophagy. Together, these findings demonstrate that MV-Edm induces massive SQSTM1-mediated mitophagy that removes mitochondria from infected cells.
FIG 6.
SQSTM1 mediates MV-Edm-induced mitophagy, contributing to impaired DDX58/IFIH1/MAVS signaling. (A) Mitochondria were fractionated from A549 cells that were not infected or that were infected by MV-Edm at an MOI of 0.2 for 24, 48, and 72 h. Mitochondrion-associated and cytoplasmic SQSTM1 was detected by immunoblotting. COX4I1 and GAPDH were used as loading controls for mitochondrial and cytoplasmic samples, respectively. One representative blot from two independent experiments is shown. (B) Colocalization of autophagosomes and mitochondria was quantified in A549 cells transfected with SQSTM1 siRNA for 24 h followed by transient transfection with pEGFP-Map1lc3b for another 24 h. Cells were then infected with MV-Edm (MOI = 0.5) for 12 h and stained with MitoTracker red before being subjected to confocal microscopy (left panel). Scale bars = 10 μm. Colocalization (yellow dots) of mitochondria (red) with autophagosomes (green puncta) was quantified by calculating Pearson's correlation coefficient [PCC, R(r)] (right panel). Means are shown (n = 30 for each group). (C) Mitochondrial mass was measured by cytometry in MitoTracker green-stained A549 cells transfected with siRNA targeting SQSTM1 or nonspecific control siRNA and cultured in the presence or absence of MV-Edm (MOI = 0.5) for 48 h. An overlay of histograms representative of 3 independent experiments (left panel) and quantification of mitochondrial mass as mean fluorescence intensity averaged from 3 independent experiments (right panel) are shown. (D) The mitochondrial HSPD1 protein level was determined by Western blotting in lysates obtained from A549 lung cancer cells transfected with siRNA targeting SQSTM1 or nonspecific control siRNA and cultured in the presence or absence of MV-Edm (MOI = 0.5) for 48 h. A representative result from two independent experiments is shown. (E) Expression levels of DDX58, IFIH1, and MAVS were determined by immunoblotting in cell lysates from A549 and H1299 cells transfected with siRNA targeting SQSTM1 or with a nontargeting siRNA followed by infection with MV-Edm (MOI = 0.2) for 48 h. Similar results were obtained in two independent experiments. (F) Cell death was quantified by trypan blue exclusion in A549 and H1299 cells transfected with siRNA targeting SQSTM1 or with nontargeting control siRNA for 24 h and cultured in the presence or absence of MV-Edm (MOI = 0.2) for another 48 h. Means + SD for triplicates are shown; *, P < 0.05; **, P < 0.01. Similar results were obtained in three independent experiments.
To further assess the role of mitophagy in the regulation of innate immune response induced by MV-Edm, we abrogated mitochondrial clearance by knocking down SQSTM1 expression. This restored expression of DDX58, IFIH1, and MAVS (Fig. 6E). Consistent with the enhanced antiviral response upon SQSTM1 downregulation, cell death was significantly inhibited under these conditions (Fig. 6F). Taken together, these results converge to demonstrate that SQSTM1-mediated mitophagy contributes to mitigate the innate immune response upon MV-Edm infection, possibly by taking part in the degradation of mitochondrion-bound MAVS.
DISCUSSION
Autophagy is one of the primordial mechanisms of defense against pathogens (14, 15). However, autophagy also promotes infections, as shown here and in previous work (33, 34). MV-Edm triggers proviral autophagy and autophagic flux upon interaction with CD46, a receptor involved in the internalization of MV-Edm (22, 23, 31). Following entry of the virus into the cell, newly produced viral protein C sustains autophagy, which contributes directly to viral replication (23). As shown for other viruses, this could be related to assembly and maturation of viral particles into acidified autophagosomes (33, 34). Here we have shown that MV-Edm-induced mitophagy indirectly promotes viral replication and oncolytic activity of MV-Edm against NSCLC cells by mitigating the type I interferon innate immune response (Fig. 7). Overall, these results contribute to our understanding of how autophagy modulates the RLR signaling pathway and provide a rationale for optimizing virotherapy against cancer.
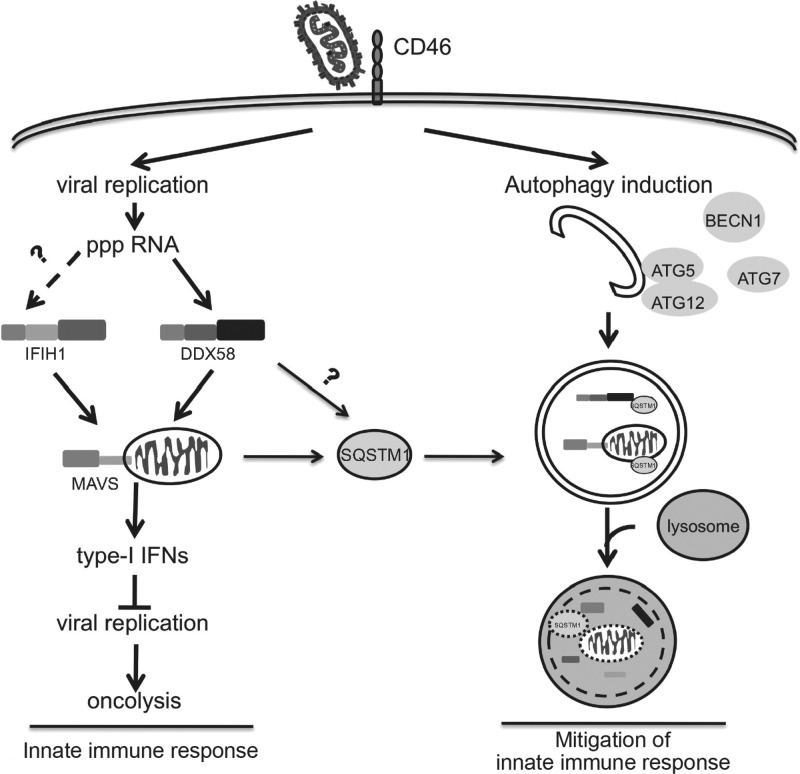
FIG 7.
MV-Edm subverts mitophagy to attenuate the innate immune response. In this model, MV-Edm induces an innate immune response via activation of DDX58/MAVS signaling while in parallel stimulating autophagy and mitophagy that degrade DDX58, IFIH1, and mitochondrion-anchored MAVS. SQSTM1 may play a crucial role in tipping the balance of anti- to proviral functions of autophagy, enhancing replication of MV-Edm, since it mediates mitophagy.
We have shown that MV-Edm subverts autophagy to mitigate the antiviral response mediated by RLRs and type I interferon-related cytokines at both the transcriptional and posttranscriptional levels. These results indicate that MV-Edm disrupts innate immune responses at early stages of the signaling pathways. A previous study has shown that complexes of ATG5 and ATG12, two key proteins involved in autophagy, can interact with MAVS at the level of mitochondria to abrogate early-stage innate immune signaling mediated by IFIH1 or DDX58 (26). However, we did not observe enhanced cellular levels of ATG5-ATG12 complexes after infection by MV-Edm in NSCLC cells. Instead, we found that completion of late stages of autophagy is required to attenuate RLR signaling, which argues that autophagy degrades components involved in this pathway. Previous reports demonstrate that autophagy contributes to MV-Edm replication (22, 23). Interestingly, we found that induction of autophagy in uninfected cells by overexpression of ATG5 or by treatment with a potent phosphatidylinositol 3-kinase inhibitor is insufficient to alter the expression levels of RLRs (data not shown). However, selective autophagy mediated by SQSTM1, a chaperone protein involved in specific degradation of ubiquitinated proteins by autophagy, is necessary to attenuate the expression of DDX58, IFIH1, or MAVS. Taken together, these observations suggest that autophagy plays multiple roles in MV-Edm infection via different types of autophagy. Bulk autophagy enhances viral replication as reported by others (23), while selective autophagy promotes viral replication by the attenuation of the innate immune response, as shown here.
The completion of late stages of autophagy is required to attenuate RLR signaling, which argues that autophagy degrades components involved in this pathway. Accordingly, we found that MV-Edm engages SQSTM1-mediated mitophagy, which also results in the degradation of mitochondrial tethered proteins that include MAVS. The trigger for mitophagy remains to be determined. It is possible that upon entry into the host cell, the virus induces mitochondrial damage or impairs mitochondrial dynamics, thereby targeting mitochondria toward autophagosomes (35). It is noteworthy that IRGM, which has been shown to promote proviral autophagy after infection by measles virus (22), is also implicated in the regulation of mitochondrial fission (36). Thus, it would be of interest to determine whether IRGM disrupts mitochondria dynamics and primes infected cells for mitophagy. Furthermore, measles virus protein C is involved in the inhibition of the IFN response (37, 38), and this protein has recently been shown to be essential for sustaining autophagy in infected cells (23). Whether protein C also contributes to mitophagy and whether it modulates IRGM activity remain to be determined.
We found that inhibition of autophagy decreases cell death in NSCLC cells, whereas recently published data demonstrate that apoptosis is enhanced upon inhibition of autophagy in HeLa cells infected by MV-Edm (23). Indeed, ongoing investigations in our lab also indicate that mitophagy inhibits apoptosis by removing damaged mitochondria before they release cytochrome c; however, mitophagy enhances overall oncolysis by nonapoptotic cell death (unpublished data). Thus, we postulate that autophagy-promoted oncolysis might be a consequence of excessive viral replication enhanced by mitigated innate immunity and/or by attenuated apoptosis.
SQSTM1-mediated mitophagy is transient following MV-Edm infection, since recruitment of SQSTM1 to mitochondria decreases over time. Our findings suggest two possible explanations for this observation. First, mitophagy might remove only a specific subset of mitochondria damaged by the virus, since mitochondrial mass is decreased roughly by half in infected cells. The second explanation is that the intracellular abundance of SQSTM1 and its degradation rate after autophagy and mitophagy induction define the time window where mitophagy occurs. In that case, attenuation of the innate immune response might be restricted to early stages of infection in cells with a low SQSTM1 reservoir, whereas high levels of SQSTM1 could allow anti-inflammatory mitophagy to be sustained for longer periods. A corollary is that variations of SQSTM1 abundance in different cells should influence oncolytic efficacy in a tissue-specific manner. Given that SQSTM1 is often upregulated in cancers displaying RAS dependency (39–41), this finding might contribute to explaining the targeted oncolytic activity of viruses against cancer.
In conclusion, this work unveils a novel strategy exploited by MV-Edm to mitigate the innate immune response of host cells. It has been shown that autophagy and mitophagy contribute to the defense against pathogens (36, 42) and attenuate inflammation to prevent organism damage under some circumstances (43). Our study demonstrates that MV-Edm usurps this homeostatic process in cancer cells to impair the antiviral immune response and to promote its replication. Finally, while our findings suggest that combination treatment with MV-Edm and drugs that stimulate autophagy should enhance oncolytic efficacy against cancer, they also underline that stratification according to SQSTM1 levels and appropriately timed modulation of autophagy might be needed to optimize mitophagy levels for viral replication.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81071860 and 81172143, to J.W., and 81301943, to A.J.), the Natural Science Foundation of Jiangsu Province of China (BK2010246, to J.W.), the Fundamental Research Funds for the Central Universities (1093021412 and 1112021402, to J.W.), the State Key Laboratory of Pharmaceutical Biotechnology (KF-GN-201410), and the Scientific Foundation of Graduate School of Nanjing University.
We thank Stephen J. Russell for providing modified oncolytic measles virus strains and Jie Dong for expert help with immunoblotting. We thank Meike Dahlhaus and the Zentrale Einrichtung Elektronenmikroskopie of Ulm University for electron microscopy and Paul Walther for expert help with analyzing the images.
We have no financial conflict of interest to declare.
Footnotes
Published ahead of print 26 February 2014
REFERENCES
- 1.Roy CR, Mocarski ES. 2007. Pathogen subversion of cell-intrinsic innate immunity. Nat. Immunol. 8:1179–1187. 10.1038/ni1528 [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988. 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- 3.Lou Y-J, Zhang Z-L, Pan X-R, Xu G-P, Jia P-M, Li D, Tong J-H. 2011. Intact JAK-STAT signaling pathway is a prerequisite for STAT1 to reinforce the expression of RIG-G gene. Exp. Cell Res. 317:513–520. 10.1016/j.yexcr.2010.10.025 [DOI] [PubMed] [Google Scholar]
- 4.Saito T, Hirai R, Loo Y-M, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U. S. A. 104:582–587. 10.1073/pnas.0606699104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall JC, Rosen A. 2010. Type I interferons: crucial participants in disease amplification in autoimmunity. Nat. Rev. Rheumatol. 6:40–49. 10.1038/nrrheum.2009.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pankiv S, Alemu EA, Brech A, Bruun J-A, Lamark T, Øvervatn A, Bjørkøy G, Johansen T. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188:253–269. 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen E-L. 2004. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 117:4837–4848. 10.1242/jcs.01370 [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873. 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- 9.Okamoto K, Kondo-Okamoto N, Ohsumi Y. 2009. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17:87–97. 10.1016/j.devcel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Ney PA. 2009. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 16:939–946. 10.1038/cdd.2009.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dötsch V, Ney PA, Dikic I. 2010. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51. 10.1038/embor.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. 2011. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One 6:e20975. 10.1371/journal.pone.0020975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. 2010. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12:119–131. 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- 14.Kudchodkar SB, Levine B. 2009. Viruses and autophagy. Rev. Med. Virol. 19:359–378. 10.1002/rmv.630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deretic V. 2012. Autophagy: an emerging immunological paradigm. J. Immunol. 189:15–20. 10.4049/jimmunol.1102108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine B. 2005. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120:159–162. 10.1016/S0092-8674(05)00043-7 [DOI] [PubMed] [Google Scholar]
- 17.Knodler LA, Celli J. 2011. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell. Microbiol. 13:1319–1327. 10.1111/j.1462-5822.2011.01632.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dötsch V, Bumann D, Dikic I. 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333:228–233. 10.1126/science.1205405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, Garcia E, Dinkins C, Leuba F, Wu L, Schwartz O, Deretic V, Piguet V. 2010. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 32:654–669. 10.1016/j.immuni.2010.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayward AP, Dinesh-Kumar SP. 2010. Special delivery for MHC II via autophagy. Immunity 32:587–590. 10.1016/j.immuni.2010.04.015 [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Rocha H, Gomez-Gutierrez JG, Garcia-Garcia A, Rao X-M, Chen L, McMasters KM, Zhou HS. 2011. Adenoviruses induce autophagy to promote virus replication and oncolysis. Virology 416:9–15. 10.1016/j.virol.2011.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grégoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, Diaz O, Deloire A, Azocar O, Baguet J, Le Breton M, Mangeot PE, Navratil V, Joubert P-E, Flacher M, Vidalain P-O, André P, Lotteau V, Biard-Piechaczyk M, Rabourdin-Combe C, Faure M. 2011. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 7:e1002422. 10.1371/journal.ppat.1002422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richetta C, Grégoire IP, Verlhac P, Azocar O, Baguet J, Flacher M, Tangy F, Rabourdin-Combe C, Faure M. 2013. Sustained autophagy contributes to measles virus infectivity. PLoS Pathog. 9:e1003599. 10.1371/journal.ppat.1003599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. 2006. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J. Virol. 80:6061–6071. 10.1128/JVI.02167-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suhy DA, Giddings TH, Kirkegaard K. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953–8965. 10.1128/JVI.74.19.8953-8965.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin K-Q, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. 2007. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. U. S. A. 104:14050–14055. 10.1073/pnas.0704014104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim I, Rodriguez-Enriquez S, Lemasters JJ. 2007. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462:245–253. 10.1016/j.abb.2007.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kissova I. 2004. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 279:39068–39074. 10.1074/jbc.M406960200 [DOI] [PubMed] [Google Scholar]
- 29.Wang K, Klionsky DJ. 2011. Mitochondria removal by autophagy. Autophagy 7:297–300. 10.4161/auto.7.3.14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou R, Yazdi AS, Menu P, Tschopp J. 2011. A role for mitochondria in NLRP3 inflammasome activation. Nature 469:221–225. 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
- 31.Joubert P-E, Meiffren G, Gregoire IP, Pontini G, Richetta C, Flacher M, Azocar O, Vidalain P-O, Vidal M, Lotteau V, Codogno P, Rabourdin-Combe C, Faure M. 2009. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe 6:354–366. 10.1016/j.chom.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 32.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mateo R, Nagamine CM, Spagnolo J, Mendez E, Rahe M, Gale M, Yuan J, Kirkegaard K. 2013. Inhibition of cellular autophagy deranges dengue virion maturation. J. Virol. 87:1312–1321. 10.1128/JVI.02177-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richards AL, Jackson WT. 2012. Intracellular vesicle acidification promotes maturation of infectious poliovirus particles. PLoS Pathog. 8:e1003046. 10.1371/journal.ppat.1003046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Twig G, Elorza A, Molina AJA, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446. 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, Komatsu M, Deretic V. 2010. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat. Cell Biol. 12:1154–1165. 10.1038/ncb2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaffer JA, Bellini WJ, Rota PA. 2003. The C protein of measles virus inhibits the type I interferon response. Virology 315:389–397. 10.1016/S0042-6822(03)00537-3 [DOI] [PubMed] [Google Scholar]
- 38.Nakatsu Y, Takeda M, Ohno S, Shirogane Y, Iwasaki M, Yanagi Y. 2008. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J. Virol. 82:8296–8306. 10.1128/JVI.00108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen H-Y, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137:1062-1075. 10.1016/j.cell.2009.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. 2008. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell 13:343–354. 10.1016/j.ccr.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 41.Thompson HGR, Harris JW, Wold BJ, Lin F, Brody JP. 2003. p62 overexpression in breast tumors and regulation by prostate-derived Ets factor in breast cancer cells. Oncogene 22:2322–2333. 10.1038/sj.onc.1206325 [DOI] [PubMed] [Google Scholar]
- 42.Orvedahl A, MacPherson S, Sumpter R, Tallóczy Z, Zou Z, Levine B. 2010. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7:115–127. 10.1016/j.chom.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. 2009. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. U. S. A. 106:2770–2775. 10.1073/pnas.0807694106 [DOI] [PMC free article] [PubMed] [Google Scholar]