FIG 4 .
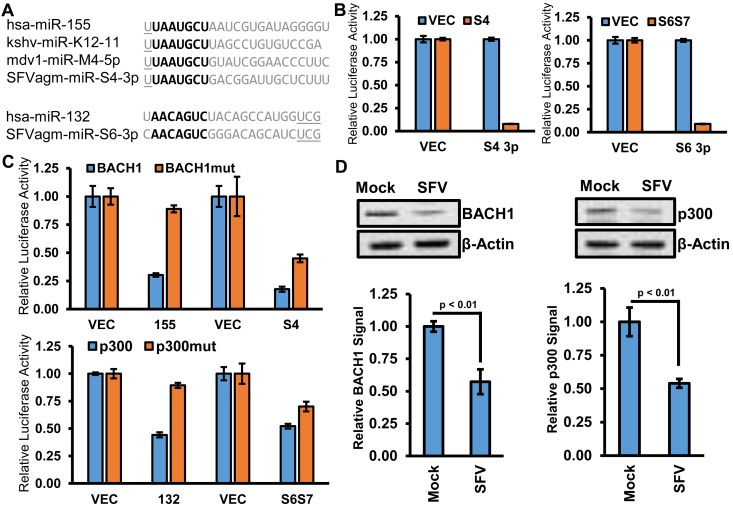
Some SFV-encoded miRNAs function as analogs of host miRNAs. (A) Sequence comparison of SFV miRNAs SFVagm-miR-S4-3p and SFVagm-miR-S6-3p to miRNAs hsa-miR-155 and hsa-miR-132, respectively. The miRNA seed sequence is indicated in bold. (B) RISC reporter assays for SFVagm miRNA candidates. HEK293T cells were cotransfected with either empty expression vector (VEC) or the indicated synthetic gene construct and both control firefly luciferase reporter and Renilla luciferase reporter plasmids with vector UTR (VEC) or two sites perfectly complementary to the indicated predicted miRNA arm (S4 3p or S6 3p). The mean relative luciferase activity ratios, normalized to the results for the control vector Renilla luciferase vector 3′ UTR, are graphed. Error bars represent the SD of three replicates. (C) BACH1 and p300 3′ UTR reporter assays were performed. HEK293T cells were cotransfected with either empty expression vector (VEC) or the indicated synthetic gene construct and the indicated 3′ UTR reporter. The pGL3-based BACH1 reporters and empty vector controls included cotransfection of pcDNA3.1dsLuc2CP control vector. The psiCheck2-based p300 reporters express both firefly and Renilla luciferase from the same plasmid. The BACH1mut and p300mut vectors contain mutations in seed regions complementary to miR-155 and miR-132, respectively (26, 27). The mean relative luciferase activity ratios, normalized to the results for the VEC control Renilla luciferase vector, are graphed. Error bars indicate SD. (D) Immunoblot analysis of protein extracted from HEK293 cells 72 h after infection with SFV or mock infection (Mock). A representative blot for each indicated target (BACH1 or p300) is provided, along with a blot for β-actin as a loading control. Graphs represent quantification of the means of three independent replicates relative to data for mock-infected cells. Error bars indicate SD, and P values were calculated with Student’s t test.