ABSTRACT
The major clonal lineages of the human pathogen Staphylococcus aureus produce cell wall-anchored anionic poly-ribitol-phosphate (RboP) wall teichoic acids (WTA) substituted with d-Alanine and N-acetyl-d-glucosamine. The phylogenetically isolated S. aureus ST395 lineage has recently been found to produce a unique poly-glycerol-phosphate (GroP) WTA glycosylated with N-acetyl-d-galactosamine (GalNAc). ST395 clones bear putative WTA biosynthesis genes on a novel genetic element probably acquired from coagulase-negative staphylococci (CoNS). We elucidated the ST395 WTA biosynthesis pathway and identified three novel WTA biosynthetic genes, including those encoding an α-O-GalNAc transferase TagN, a nucleotide sugar epimerase TagV probably required for generation of the activated sugar donor substrate for TagN, and an unusually short GroP WTA polymerase TagF. By using a panel of mutants derived from ST395, the GalNAc residues carried by GroP WTA were found to be required for infection by the ST395-specific bacteriophage Φ187 and to play a crucial role in horizontal gene transfer of S. aureus pathogenicity islands (SaPIs). Notably, ectopic expression of ST395 WTA biosynthesis genes rendered normal S. aureus susceptible to Φ187 and enabled Φ187-mediated SaPI transfer from ST395 to regular S. aureus. We provide evidence that exchange of WTA genes and their combination in variable, mosaic-like gene clusters have shaped the evolution of staphylococci and their capacities to undergo horizontal gene transfer events.
IMPORTANCE
The structural highly diverse wall teichoic acids (WTA) are cell wall-anchored glycopolymers produced by most Gram-positive bacteria. While most of the dominant Staphylococcus aureus lineages produce poly-ribitol-phosphate WTA, the recently described ST395 lineage produces a distinct poly-glycerol-phosphate WTA type resembling the WTA backbone of coagulase-negative staphylococci (CoNS). Here, we analyzed the ST395 WTA biosynthesis pathway and found new types of WTA biosynthesis genes along with an evolutionary link between ST395 and CoNS, from which the ST395 WTA genes probably originate. The elucidation of ST395 WTA biosynthesis will help to understand how Gram-positive bacteria produce highly variable WTA types and elucidate functional consequences of WTA variation.
INTRODUCTION
Interaction of bacterial pathogens with human hosts requires efficient mechanisms for colonization, infection, and evasion of human antimicrobial defense systems, such as the complement system. These processes depend on components of the bacterial cell envelope. As in most of the Gram-positive bacteria, the envelope of the opportunistic human pathogen Staphylococcus aureus is composed mainly of a thick peptidoglycan layer, surface proteins, and anionic polymers, represented by membrane-bound lipoteichoic acids (1, 2) and cell wall-anchored wall teichoic acids (WTA) (3, 4).
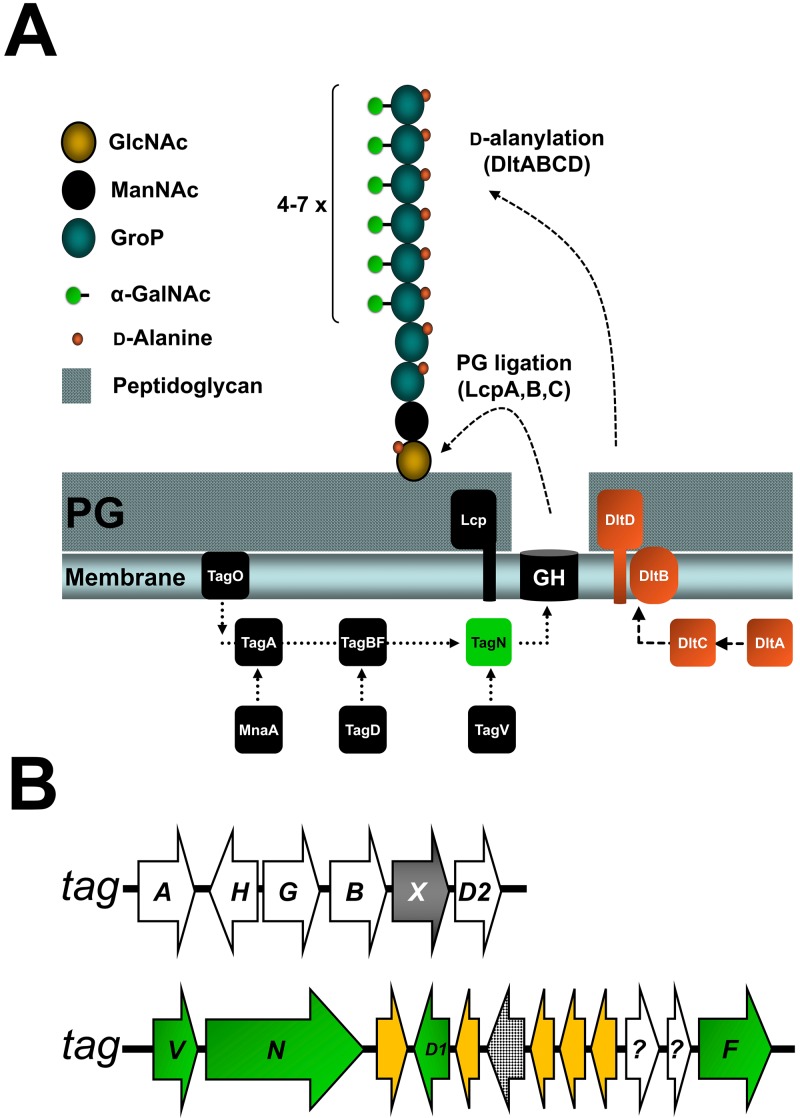
WTA structures have been shown to be highly variable among Gram-positive bacteria and are often species or even strain specific (5). Most of the S. aureus isolates produce poly-ribitol-phosphate (RboP) WTA with up to 40 repeating units substituted with d-alanine (d-Ala) and N-acetyl-d-glucosamine (GlcNAc). The WTA repeating units are covalently linked to the peptidoglycan component N-acetylmuramic acid via an anchor unit composed of GlcNAc-1-P (N-acetyl-d-glucosamine-1-phosphate) and N-acetyl-d-mannosamine followed by two glycerol-phosphate (GroP) units. In contrast to S. aureus, the WTA repeating units of most coagulase-negative staphylococci (CoNS), such as Staphylococcus epidermidis, are composed of GroP units substituted with variable sugar substituents (6). Recently, elucidation of the WTA structure of S. aureus strain PS187, a pneumonia isolate belonging to the ST395 lineage, has revealed a unique CoNS-like repeating unit composed of GroP units substituted with N-acetyl-d-galactosamine (GalNAc) (7) (Fig. 1A).
FIG 1 .
Proposed S. aureus PS187 WTA biosynthetic pathway. (A) Wall teichoic acid (WTA) biosynthesis in S. aureus PS187. Linkage unit synthesis (TagO, -A, -B, -D, MnaA), WTA polymerization (TagF, -D), glycosylation (TagN, -V), membrane transport (TagG, -H), d-alanylation (DltA, -B, -C, -D), and peptidoglycan ligation (LcpA, -B, -C) are indicated. The WTA repeating unit of PS187 is composed of GroP-GalNAc units. (B) WTA biosynthesis gene clusters found in PS187. Putative glycosyltransferases encoded by tagX (gray) and tagN (green), putative transposases (orange), istB-like protein (dotted); genes of unknown function (?) are indicated.
Extensive studies have revealed the complete WTA biosynthesis pathways of Bacillus subtilis and typical S. aureus during the past decades (8, 9). Major functional roles of S. aureus WTA encompass host-pathogen-associated interactions contributing to nasal colonization (10), binding to epithelial and endothelial cells (10, 11), activation of the human complement system (12–14), resistance to cationic antimicrobial peptides (15), fatty acids from human skin (16), and bacteriophages (phages) (17–20), along with basic cellular processes such as the positioning of penicillin-binding protein PBP4 and autolytic enzymes (21, 22). Remarkably, the recent elucidation of the S. aureus WTA glycosylation pathway encompassing the two unrelated WTA glycosyltransferases TarM and TarS has revealed that β-O-GlcNAc WTA modification is required for PBP2a-mediated β-lactam resistance (17) and enabled our recent studies on the role of WTA structure in phage-mediated interspecies and intergeneric horizontal gene transfer (HGT) among Gram-positive pathogens (7). Thus, WTA biosynthesis of S. aureus and potentially of other Gram-positive pathogens has become a very attractive drug target during the past couple years (23–27).
Not all S. aureus isolates carry the tarIJLF gene cluster for RboP WTA polymers and tarM and/or tarS glycosyltransferases (4). The unusual S. aureus strain PS187 does not bear homologs of these genes in its genome. Instead, PS187 harbors a unique genetic element replacing the tarIJLF cluster (7). Of note, this novel genetic element of unknown origin encodes several putative WTA biosynthesis genes, some of which are related to putative genes found in CoNS (7), but the roles of these genes in GroP WTA biosynthesis and glycosylation have remained unknown.
Here, we identified a unique S. aureus WTA α-O-GalNAc glycosyltransferase TagN in the ST395 lineage and show that α-O-GalNAc-modified WTA is the adsorption receptor for the ST395-specific bacteriophage Φ187. We also describe that a closely related TagN homolog found in Staphylococcus carnosus TM300 (TagN-Sc) can replace TagN, revealing an evolutionary connection between ST395 and CoNS. Finally, based on ectopic expression of ST395 WTA biosynthesis genes in classical RboP WTA-producing S. aureus, we identify the three enzymes, TagN, TagV, and TagF, required for GroP-GalNAc WTA biosynthesis in ST395 and provide insights into mechanisms enabling or interfering with Φ187-mediated exchange of pathogenicity islands (SaPIs) between ST395 and classical S. aureus.
RESULTS
The S. aureus ST395 lineage bears unique WTA biosynthesis genes.
We have recently sequenced the genome of ST395 isolate PS187 (7) and analyzed the genome for genes potentially involved in WTA biosynthesis. Similar to classical S. aureus strains, the ST395 prototype PS187 encodes the well-studied tagO gene and the tagAHGBXD gene cluster for WTA linkage unit biosynthesis and WTA translocation (Fig. 1B), which is in agreement with the notion that most Gram-positive bacteria have the same WTA linkage unit despite different polymer composition (5, 28). PS187 has been found to bear a novel genetic element with several transposon-related sequences replacing the tar cluster for poly-RboP WTA synthesis and glycosylation found in all sequenced non-ST395 S. aureus isolates (Fig. 1B).
The novel element was named S. aureus GroP WTA island (SaGroWI) because it turned out to be responsible for biosynthesis and glycosylation of GroP WTA (see below). Two of the proteins encoded by SaGroWI were related to previously characterized WTA biosynthetic enzymes over their entire length. One of these (TagF) was 24.06% and 24.1% identical to the GroP WTA polymerases of Bacillus subtilis 168 and of Staphylococcus epidermidis, respectively, but lacked an N-terminal portion of ca. 330 amino acids (see Fig. S1 in the supplemental material). The strongest similarity was found for the equally short product of a B. subtilis W23 gene (66% similarity), which has in vitro GroP polymerase activity but an uncertain role in WTA biosynthesis (29) (see Fig. S1). Nevertheless, the tagF homolog was the only putative GroP polymerase gene in the PS187 genome. SaGroWI encoded also a second homolog of TagD known to generate the CDP-glycerol substrate for the polymerase. However, the SaGroWI TagD was 26 amino acids shorter than that encoded in the tagAHGBXD gene cluster (see Fig. S2 in the supplemental material).
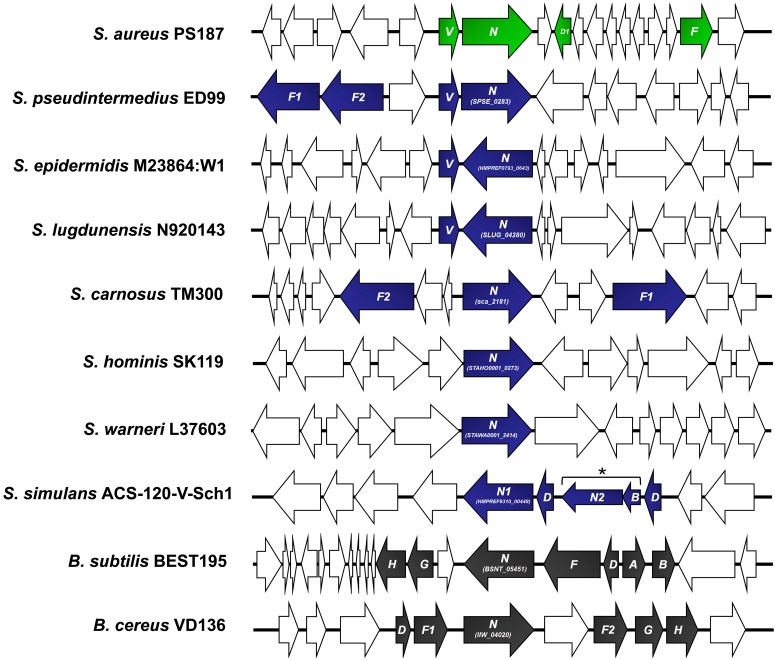
A third SaGroWI-encoded protein, TagN, has a domain related to the TagB superfamily at its N terminus and a C-terminal (WabH-like) glycosyltransferase domain previously not implicated in WTA biosynthesis. Because PS187 GroP WTA is glycosylated with α-linked GalNAc, it was tempting to speculate that TagN might be the GroP WTA GalNAc transferase. In line with this notion, all ST395 clones seem to contain tagN (7), and tagN homologs were also found in the genomes of GroP WTA-producing CoNS, such as S. carnosus and Staphylococcus simulans, and even in Bacillus cereus and Bacillus subtilis strain BEST 195 with an unusual set of WTA genes (Fig. 2; see also Fig. S3 in the supplemental material). Upstream of tagN, SaGroWI contains a potentially cotranscribed gene whose product shared similarity with NAD-dependent nucleotide sugar epimerases (Fig. 1B and 2). The protein was named TagV and was assumed to generate UDP-GalNAc, the putative donor substrate for GroP WTA glycosylation with GalNAc. tagV homologues were found in genomes of the GroP WTA-producing CoNS species S. epidermidis, Staphylococcus pseudintermedius, and Staphylococcus lugdunensis (Fig. 2). Thus, SaGroWI contains candidate genes for all steps required for GroP WTA polymer biosynthesis and its glycosylation with GalNAc.
FIG 2 .
CoNS share WTA biosynthesis genes with S. aureus ST395 clone PS187. Genetic organization of the tagV-tagN prototype cluster of S. aureus PS187 compared to CoNS, B. cereus VD136, and B. subtilis BEST195 bearing tagN homologs. Genes obviously involved in WTA biosynthesis (colored), genes with other functions (white), and tagN accession numbers are indicated. In S. simulans ACS-120-V-Sch-1, the asterisk (*) and brackets indicate a second copy of tagN (tagN2). (Note: the raw-sequence divides it into two open reading frames [ORFs], probably as a result of a sequencing error.)
TagN is required for GalNAc modification of GroP WTA in S. aureus PS187.
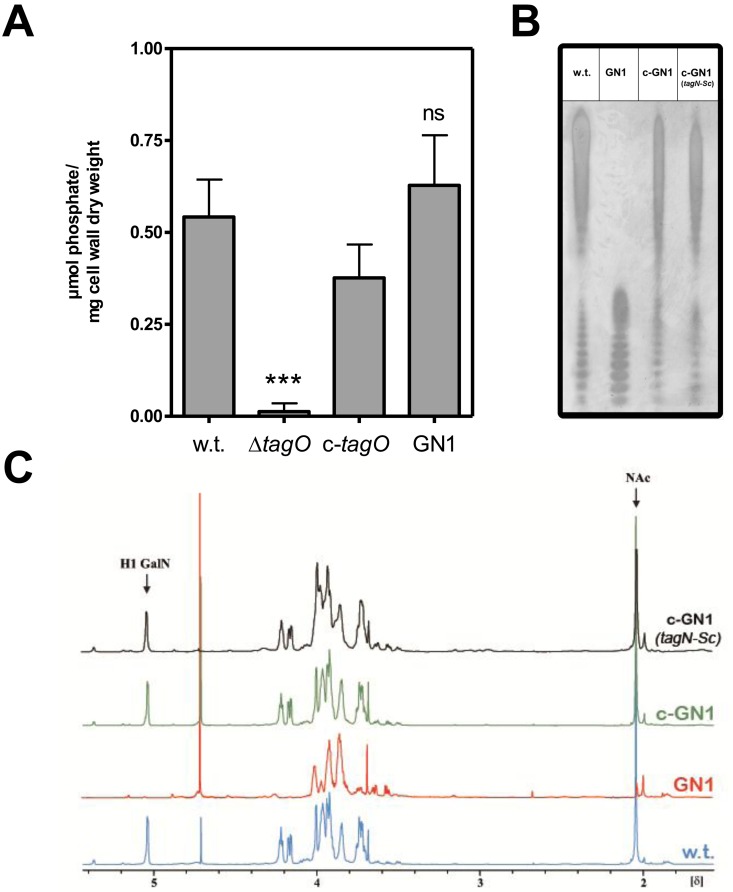
While inactivation of genes for WTA backbone synthesis is usually lethal because of the accumulation of lipid carrier-linked WTA precursors (30), the recently discovered genes for WTA glycosylation are dispensable for bacterial viability (17). In order to assess if TagN is indeed responsible for GroP WTA glycosylation, we set out to inactivate the entire gene in the genome of PS187, but none of our numerous attempts were successful, suggesting that TagN may have an essential role in GroP WTA biosynthesis. However, we easily succeeded with deletion of the C-terminal glycosyltransferase domain of tagN. WTA preparations from the resulting mutant GN1 and its parental strain contained similar amounts of phosphate, indicating that WTA amounts were the same in the two strains (Fig. 3A). When the samples were applied to polyacrylamide gel electrophoresis (PAGE), the WTA of GN1 migrated much faster than that of the parental strain (Fig. 3B). This pattern resembled the different electrophoretic mobilities of WTA from RboP WTA-producing S. aureus RN4220 and its GlcNAc-deficient mutant (14, 19), suggesting that GN1 may indeed lack the WTA GalNAc residues. When GN1 was complemented with a plasmid-encoded copy of tagN, WTA of the resulting strain c-GN1 migrated like that of the wild type (Fig. 3B).
FIG 3 .
Impact of two novel WTA glycosyltransferases, TagN and TagN-Sc, on electrophoretic mobility and composition of PS187 WTA. (A) Cell wall phosphate determination upon tagN disruption in the S. aureus PS187 panel compared to PS187 wild type (w.t.), PS187 ∆tagO strain lacking WTA, and its complemented strain (c-tagO). Statistically significant differences compared to the wild type calculated by the unpaired two-tailed Student t test are indicated: ns, not significant, P > 0.05; ***, P < 0.001. (B) WTA PAGE analysis of WTA preparations. Samples were resolved in polyacrylamide gels and visualized with alcian blue/silver staining. One representative experiment is shown. (C) 1H NMR spectrum of purified WTAs. Arrows indicate characteristic signals for GalNAc at δ 5.1 (anomeric proton signal) and δ 2.1 and for methyl group of NAc at δ 2.1. Wild type (w.t.), tagN mutant (GN1), tagN-complemented GN1 mutant (c-GN1), and tagN-Sc-complemented GN1 mutant (c-GN1 [tagN-Sc]) are indicated.
WTA preparations from GN1 were further purified by ion-exchange chromatography and analyzed by 1H nuclear magnetic resonance (1H-NMR) and heteronuclear 13C,1H single quantum correlation (HSQC) spectroscopy. Two characteristic signals of α-GalNAc (anomeric proton signal of α-GalN at δ 5.1 and NAc at δ 2.1) were present in the spectra of the wild type and the tagN-complemented mutant c-GN1 but absent in mutant GN1 (Fig. 3C; see also Fig. S4A and Table S1 in the supplemental material), demonstrating that GN1 WTA lacks GalNAc and requires tagN for WTA glycosylation.
To analyze if the TagN homologs of CoNS have similar functions as TagN, mutant GN1 was complemented with a tagN-like gene from S. carnosus (tagN-Sc). The resulting strain produced WTA that exhibited wild-type electrophoretic migration behavior (Fig. 3B) and contained GalNAc, as shown by NMR (Fig. 3C) and HSQC (see Fig. S4B and Table S1 in the supplemental material). Taken together, these data strongly suggest that tagN and its close homolog from S. carnosus encode GroP WTA α-O-GalNAc transferases.
WTA GalNAc modification is required for phage Φ187 infection of S. aureus ST395 and facilitates the HGT of SaPIs.
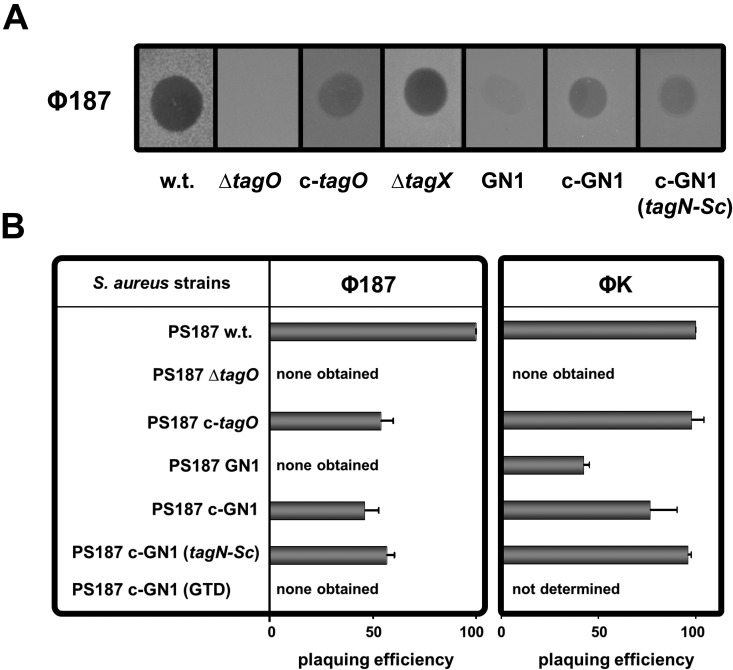
Most phages infecting RboP WTA-producing S. aureus, including transducing phages such as Φ11, require GlcNAc-modified WTA for efficient adsorption and infection (17–19). The ST395 lineage-specific Φ187 also requires WTA for adsorption and infection (7), but it has remained unknown if Φ187 has a preference for WTA with GalNAc residues. Disruption of tagN in GN1 resulted in resistance to Φ187 and in drastically reduced adsorption of Φ187, while complementation of GN1 with tagN restored both phage susceptibility and adsorption (Fig. 4A and B; see also Fig. S5A in the supplemental material), indicating that Φ187 is specific for host bacteria with GalNAc-modified WTA. Moreover, complementation of GN1 with the S. carnosus tagN homolog tagN-Sc restored Φ187 susceptibility, thereby confirming that TagN and TagN-Sc have similar functions (Fig. 4A and B). Of note, expression of only the C-terminal TagN glycosyltransferase domain along with an appropriate ribosomal binding site in GN1 did not restore Φ187 susceptibility, indicating that both the tagB-like and the glycosyltransferase domains of TagN are required for WTA glycosylation (Fig. 4B). tagX from the WTA gene cluster tagAH-GBXD found in RboP WTA-producing S. aureus strains encodes a putative glycosyltransferase of unknown function (17). Deletion of tagX in PS187 did not affect its susceptibility to Φ187, supporting the notion that it is probably not involved in WTA glycosylation (Fig. 4A).
FIG 4 .
Influence of tagN on phage Φ187 susceptibility. (A) Phage susceptibility assay for S. aureus PS187 strain panel. Φ187 lysates were spotted onto bacterial lawns and analyzed after overnight incubation for macroplaque formation. One representative experiment is shown. (B) Efficiency of plaquing (EOP) assay for S. aureus PS187 strain panel encompassing Φ187 and ΦK. Approximately 150 PFU were mixed with bacteria, incubated, mixed with soft agar, poured onto agar plates, and incubated overnight. PFUs were counted and EOP was indicated relative to the EOP on the parental wild-type strain, which was set as 100%. Values are given as means (n = 3 experiments) ± standard deviations (SD). Wild type (w.t.), tagO mutant (∆tagO), tagO-complemented ∆tagO mutant (c-tagO), tagX mutant (∆tagX), tagN mutant (GN1), tagN-complemented GN1 mutant (c-GN1 [tagN]), tagN-Sc-complemented GN1 mutant (c-GN1 [tagN-Sc]), and tagN glycosyltransferase domain-only-complemented GN1 mutant (c-GN1 [GTD]) are indicated.
The broad-host-range phage ΦK from the myovirus family has previously been shown to use the backbone of RboP-type WTA as a receptor for infection (18, 19). More recently, we have found that this phage is also virulent to strain PS187 (7). As shown in Fig. 4B, ΦK was unable to form plaques on the WTA-deficient ΔtagO mutant but could form plaques on GN1, albeit with reduced infection efficiency (and reduced adsorption; see Fig. S5B in the supplemental material) compared to the parental strain. This finding indicates that ΦK infection depends on the main WTA chain, while GalNAc residues on WTA are not essential but increase the plaquing efficiency (Fig. 4B).
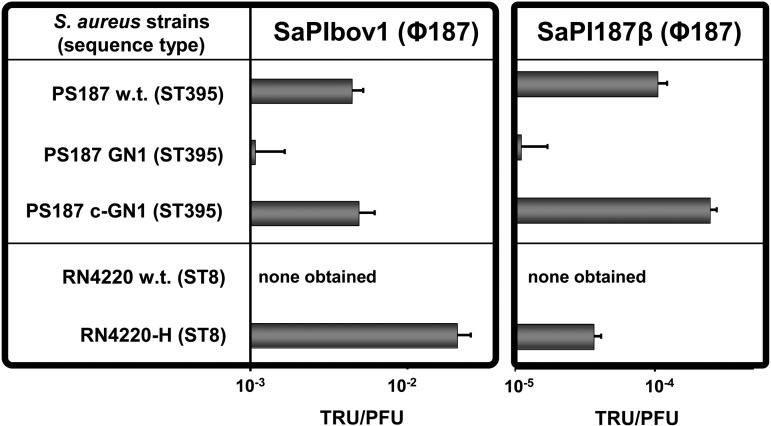
We have recently reported on WTA structure-dependent and Φ187-mediated transfer of mobile genetic elements (MGEs) such as SaPIs from S. aureus ST395 to CoNS and even to certain Listeria monocytogenes serotypes (7), which led to the conclusion that SaPI particles adopt the receptor requirements of cognate helper phages. Interestingly, although GN1 lacks GalNAc modification, SaPI transfer to GN1 via Φ187 occurred albeit at very low efficiencies (Fig. 5). These data suggest that Φ187-derived SaPI particles require GalNAc modification for efficient adsorption, although the naked GroP WTA backbone is sufficient to allow low-efficiency HGT. This property may explain why Φ187 can transfer SaPIs to various GroP WTA-producing CoNS, such as S. epidermidis, which glycosylate their WTA with sugars other than GalNAc (7).
FIG 5 .
The absence of a suitable Φ187 receptor prevents Φ187-mediated import of SaPIs to classical S. aureus. Phage Φ187-mediated transfer of SaPIbov1 or SaPI187β to the S. aureus PS187 strain panel, including GN1 lacking WTA GalNAc modification and S. aureus RN4220 wild type and RN4220-H. SaPI donor strains were VW1 (SaPIbov1) and VW7 (SaPI187β). Values represent the ratio of transduction units (TRU; transductants/ml phage lysate) to PFU (plaques/ml phage lysate on S. aureus PS187 w.t.) and are given as means (n = 3 experiments) ± SD. No TRU were observed in controls lacking phages or SaPI particles. PS187 and RN4220 wild type (w.t.), tagN mutant (GN1), tagN-complemented GN1 mutant (c-GN1 [tagN]) and RN4220-H expressing ST395 WTA are indicated.
Ectopic expression in S. aureus RN4220 reveals tagF, tagV, and tagN as the minimum set of genes required for GroP-GalNAc WTA biosynthesis.
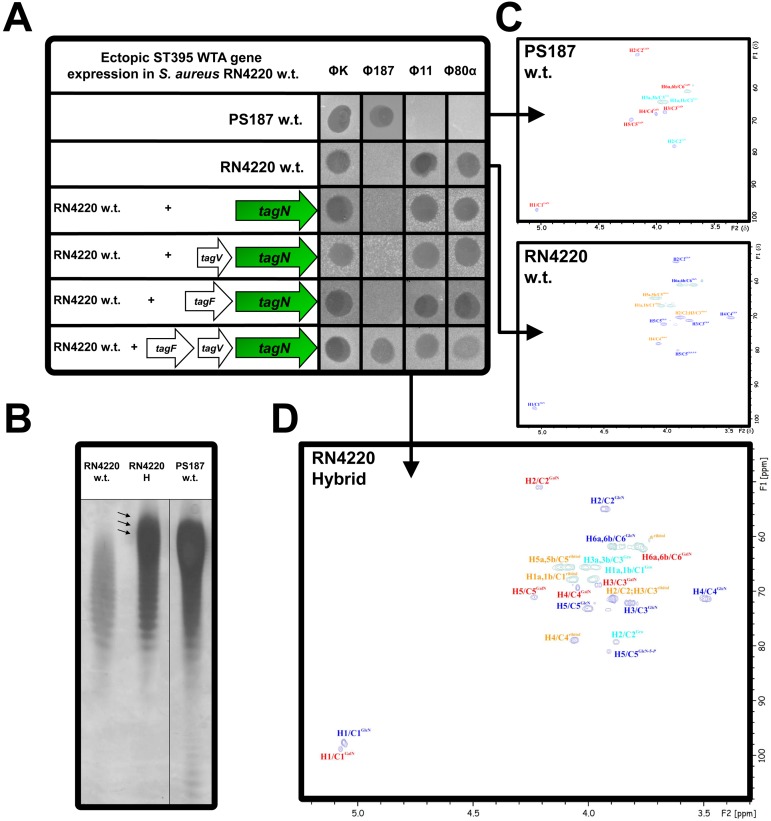
Expression of biosynthetic genes in a heterologous host has recently proved to be a successful strategy for defining the minimum set of genes required for WTA backbone biosynthesis and glycosylation (7). We cloned the genes tagF, tagV, and tagN, assumed to be responsible for biosynthesis of GalNAc-modified GroP WTA, in expression vector pRB474 (31) in a transcriptionally fused fashion and transferred the resulting plasmid to S. aureus RN4220, which produces RboP WTA. While RN4220 was resistant to Φ187, the resulting hybrid strain, RN4220-H, was susceptible and efficiently bound Φ187, suggesting that TagFVN are functional in the RN4220 background and are sufficient for production of the PS187-specific WTA polymer (Fig. 6A; see also Fig. S5C in the supplemental material). The hybrid strain RN4220-H retained susceptibility to RboP-GlcNAc-specific Φ11 and Φ80α, along with efficient adsorption, suggesting that both types of WTA are produced by this strain (Fig. 6A; see also Fig. S5D and E). PAGE analysis revealed that the most pronounced WTA species of RN4220 migrated faster than that of PS187 (Fig. 6B). Of note, RN4220-H WTA yielded a WTA band pattern migrating as slowly as that of PS187, which is in agreement with the assumption that RN4220-H produces also GalNAc-glycosylated GroP WTA (Fig. 6B). RN4220-H WTA was then analyzed by NMR, and spectra were compared to those derived from RN4220 and PS187 wild-type strains, which confirmed that RN4220-H indeed produced both types of WTA polymers, RboP-GlcNAc and GroP-GalNAc (Fig. 6C and D; see also Table S1 in the supplemental material).
FIG 6 .
Ectopic expression of ST395 WTA in S. aureus RN4220 leads to production of two types of WTA. (A) Phage susceptibility assay for S. aureus RN4220 strain panel encompassing engineered RN4220 strains expressing ST395 WTA biosynthesis genes tagN, tagV plus tagN, tagF plus tagN, and tagF in combination with tagV and tagN. Φ187, Φ11, Φ80α, and ΦK lysates were spotted onto bacterial lawns and analyzed after overnight incubation for macroplaque formation. One representative experiment is shown. (B) WTA PAGE analysis of WTA preparations of S. aureus PS187 wild type, RN4220 wild type, and RN4220 hybrid (RN4220-H). Samples were resolved in polyacrylamide gels and visualized with alcian blue/silver staining. Arrows indicate WTA alteration in RN4220-H compared to that in its parental strain. One representative experiment is shown. (C) Heteronuclear 13C,1H single quantum correlation (HSQC) spectrum from purified wild-type S. aureus RN4220 and PS187 WTA repeating units. (D) HSQC spectrum from purified S. aureus RN4220-H WTA expressing dual WTA. Arabic numerals label the proton/carbon atoms of the residues. Spectra were recorded at 700 MHz (at 600 MHz for RN4220 w.t.) and 300 K (PS187 w.t. and RN4220 w.t.) or 330 K (RN4220-H) relative to acetone (δH 2.225; δC 31.50). 1H and 13C chemical shifts are indicated in Table S1 in the supplemental material. Arrows indicate strain origin of WTA sample for HSQC spectra.
In order to elucidate if all three genes are required for biosynthesis of GroP-GalNAc WTA, they were expressed in different combinations in RN4220. When either tagF or tagV were omitted from the plasmid or tagN was expressed alone, the resulting RN4220 strain remained resistant to Φ187, indicating that tagF and tagV are indeed required in addition to tagN for biosynthesis of the unique ST395 WTA type (Fig. 6A). Thus, the genetic island SaGroWI in PS187 harbors all the genes required for GroP-GalNAc WTA polymer biosynthesis and glycosylation with GalNAc. Additional genes involved in initiation of WTA biogenesis, translocation across the cytoplasmic membrane, ligation to peptidoglycan, and d-alanylation are conserved in ST395 and RboP WTA-producing S. aureus and probably facilitate biosynthesis of both types of WTA.
Phage-mediated import of SaPIs from other Gram-positive bacteria to classical S. aureus-producing RboP WTA polymers has never been reported, and RN4220 is resistant to the acquisition of SaPIbov1 and SaPI187β from GroP WTA-producing S. aureus strain PS187, although RN4220 lacks restriction modification and clustered regular interspaced palindromic repeat (CRISPR) loci (7). However, expression of GroP-GalNAc WTA in RN4220 enabled uptake of SaPIs via Φ187, thereby confirming that similar WTA structures permit HGT of pathogenicity islands (Fig. 5).
DISCUSSION
The recent elucidation of the S. aureus RboP WTA pathway, including WTA glycosylation by TarM and TarS and WTA ligation to peptidoglycan by putative ligases, the LCP (LytR-CpsA-Psr) proteins (32–34), has completed the set of modules required for S. aureus RboP WTA biosynthesis and will help to understand functional roles of WTA and WTA glycosylation. Even though WTA structures of most of the S. aureus clones are identical, the ST395 lineage, including strain PS187, produces a unique WTA type, which is similar to those found in CoNS (7).
Based on functional characterization of defined mutants and by ectopic expression of predicted ST395 WTA genes, this study reveals three new WTA biosynthesis genes mediating GroP WTA biosynthesis in PS187, namely, tagV, tagN, and a tagF variant, all carried on a specific genetic element, SaGroWI. Although using the same substrate, CDP-glycerol, the PS187 TagF differs from characterized GroP WTA polymerases (35, 36) by the lack of a large N-terminal domain whose sequence varies largely between species and whose function is unknown (37). However, we provide evidence that even the short TagF is functional, which is in agreement with recent biochemical and structural data demonstrating that the C-terminal domain of S. epidermidis TagF is responsible for binding of the donor substrate CDP-glycerol and for polymerase activity (37). Our finding is also in agreement with the recently described in vitro GroP WTA polymerase activity of the short B. subtilis W23 TarF (29).
We describe a new type of WTA glycosyltransferase, TagN, which is unrelated to two recently identified WTA glycosyltransferases, TarM and TarS. The C-terminal domain of TagN, related to the GT1 family of glycosyltransferases, was required for GalNAc modification of PS187 WTA. It remains unclear why TagN encloses also an N-terminal domain that is weakly homologous to the TagB protein superfamily. The fact that we could not delete this part of the tagN gene suggests that it may have an additional role in WTA biosynthesis. Since several Staphylococcus and Bacillus genomes contain TagN homologs, the PS187 TagN appears like a prototype of a common GroP WTA biosynthetic module (see Fig. 2). Indeed, one of these homologs from S. carnosus TM300 (TagN-Sc) has an identical function.
The exact origin of WTA genes found on the SaGroWI element and when and how they may have jumped from CoNS to ST395 during evolution remains unknown. The fact that these genes are separated by transposase-related DNA fragments suggests that they have been combined by rather recent recombination events into a mosaic-like structure. WTA genes have been found on other MGEs, such as a copy of a WTA translocase gene on a multiresistance plasmid pSK1 (38), providing further evidence for the mobility of WTA biosynthetic genes. Of note, there is an evolutionary link between ST395 clones and CoNS since all three genes have their closest counterparts in different CoNS and at least some CoNS share the tagV-tagN cluster composition with ST395 clone PS187 (see Fig. 2). Although many CoNS produce GroP WTA and some of them even bear tagV and tagN homologs, neither S. epidermidis, S. lugdunensis, Staphylococcus warneri, nor Staphylococcus hominis have been found to glycosylate their WTA with GalNAc (6, 39). This discrepancy might result from strain-dependent differences in the presence or expression of the tagN homologs. Along this line, environmental conditions in the ecological niche might influence WTA gene expression levels and WTA glycosylation processes. However, while S. epidermidis and S. lugdunensis bear tagV and tagN next to each other but in opposite directions (Fig. 2), only S. pseudintermedius ED99 seems to share the operon-like organization found in PS187 (Fig. 2). Nevertheless, S. simulans has previously been found to glycosylate its GroP WTA with both GlcNAc and GalNAc (6), although tagN is not directly flanked by a tagV homolog in this species. Possibly, other UDP-4-glucose epimerases located at distant genome loci can replace tagV. In most of the CoNS species and even in B. subtilis strain BEST195, tagN and tagV are colocalized with other WTA biosynthesis genes in a cluster, which is a common feature of most WTA biosynthesis genes in Gram-positive bacteria. In contrast, tagN or tagV do not colocalize with other WTA biosynthetic genes in some CoNS (e.g., S. epidermidis), thereby resembling the isolation of the tarM locus in RboP-producing S. aureus (4) and suggesting that the staphylococcal WTA glycosylation genes are more variable than those for WTA backbone biosynthesis.
It remains unclear why ST395 clones produce an unusual WTA type. Early reports have suggested a canine origin of PS187-related strains, pointing to a specific niche of this lineage in dogs (40). The homology and the similar gene organization of tagV and tagN in S. pseudintermedius, which colonizes in particular dogs (41), further substantiates this hypothesis. Nevertheless, recent reports on ST395 from noses or infections of hospital patients in northern Germany and Poland (42, 43) indicate that ST395 clones are at least partly adapted to the human host. Since WTA has been found to be crucial for host-pathogen interaction (3), the discovery of tagN can help to unravel the role of WTA glycosyl residues in host-pathogen interaction and host specificity in the future.
MATERIALS AND METHODS
Bacterial strains and growth media.
All bacterial strains listed in Table S2 in the supplemental material were grown in basic medium (BM) (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% K2HPO4, 0.1% glucose) or Luria-Bertani (LB) broth supplemented with the appropriate antibiotics at a concentration of 10 µg/ml for chloramphenicol, 3 µg/ml for tetracycline, 2.5 µg/ml for erythromycin, or 100 µg/ml for ampicillin.
Molecular genetic methods.
For the construction of markerless ∆tagN or ∆tagX mutants in S. aureus PS187 by allelic replacement, the pKOR1 shuttle vector was used (44). Primers are listed in Table S3 in the supplemental material. Gene deletion was performed as described previously (44).
For the construction of tagN, tagVN, and tagN-Sc complementation vectors, PS187 wild-type tagN (and the tagVN operon) and S. carnosus TM300 wild-type tagN-Sc genes were amplified via PCR from PS187 wild-type or TM300 wild-type genomic DNA (for primers, see Table S3). The resulting PCR products were purified, digested, and cloned into the Escherichia coli/S. aureus shuttle vector pRB474 (31) at the PstI/BamHI (tagN, tagVN, and tagN-Sc) sites, resulting in plasmids pRB474-tagN, pRB474-tagVN, and pRB474-tagN-Sc. For cloning of the tagN glycosyltransferase domain (for primers, see Table S3), the PCR product was purified, digested, and cloned into pRB474 at the BamHI/EcoRI site, resulting in plasmid pRB474-tagN (GTD). For ectopic expression of ST395 WTA, all required genes, tagF, tagV, and tagN, were amplified from genomic DNA (PS187 wild type) using the corresponding primers P1, P2, P3, and P4 (see Table S3). The resulting PCR products (tagF and tagVN PCR products) were purified and cloned into pRB474 using the infusion cloning technique accordingly to the manufacturer’s instructions (Clontech), resulting in plasmid pRB474-FEpiN. The plasmid was isolated from E. coli TOP10 and used to transform RN4220 resulting in the dual WTA-producing strain RN4220 hybrid (RN4220-H). For the construction of the pRB474-tagFN plasmid, infusion cloning technique was used again (primers P1, P2, P4, and P5). Plasmid pRB474-tagO has been described recently (18).
Experiments with phages and SaPI particles.
Phage susceptibility was determined using the soft-agar spot assay described previously (19). A phage panel encompassing serogroup L phage Φ187 (45), two serogroup B phages, Φ11 (45) and Φ80α (46), and the broad-host-range phage ΦK (19) was used. Phages ΦK, Φ11, and Φ80α were propagated on S. aureus RN4220 wild type or on PS187 wild type (Φ187) as described previously (7).
Phage adsorption rates were determined as described previously (18). Briefly, the multiplicity of infection (MOI) was set to 0.005 (for Φ187), 0.0001 (for ΦK), 0.1 (for Φ11), or 0.05 (for Φ80α), and adsorption was calculated by determining PFU of the unbound phage in the supernatant and subtracting it from the total number of input PFU. Adsorption efficiency was indicated relative to the adsorption on the parental wild-type strain PS187 (for Φ187 and ΦK) or RN4220 wild type (for Φ11 and Φ80α), which was set to 100%.
Efficiency of plaquing (EOP) was determined by mixing 100 µl phage suspension (~150 PFU, titrated on PS187 wild type) with 100 µl of bacteria (~1.8 × 106 CFU) and incubating for 10 min at room temperature. After incubation, 4 ml soft agar was added, mixed well, poured onto BM agar plates supplemented with the appropriate antibiotics, and incubated overnight at 30°C. Single PFUs were counted, and EOP was indicated relative to the EOP of the parental wild-type strain, which was set as 100%.
Phage Φ187-mediated SaPI transfer was studied as described previously (7). Briefly, the recently described S. aureus PS187 SaPIbov1 and SaPI187β SaPI donor strains VW1 and VW7 bearing a resistance marker-labeled SaPIbov1 or SaPI187β (7) (see Table S2 in the supplemental material) were infected with phage Φ187 to generate lysates containing phage Φ187-derived SaPI particles. Subsequently ~8.0 × 107 bacteria were mixed with 100 µl of SaPI lysates SaPIbov1 (~1.7 × 106 PFU/ml) or SaPI187β (~1.1 × 108 PFU/ml), respectively, incubated for 15 min at 37°C, diluted, and plated on BM agar supplemented with the appropriate antibiotics. To exclude spontaneous uptake of nicked DNA, SaPI-containing lysates were also treated with DNase I in controls as described previously (7). Transductants were counted after overnight incubation at 37°C, and transduction efficiency was calculated. SaPI transfer was confirmed by molecular typing of resistance markers.
WTA extraction and purification.
WTA was isolated as described previously (7). Briefly, overnight cultures were washed with ammonium acetate buffer (20 mM, pH 4.6) and mechanically disrupted either in a cell disrupter (Euler; for large-scale isolation) or in buffer together with glass beads (Sigma; acid washed, 150 to 212 µm) using a FastPrep-24 MP apparatus (Biomedicals Europe) (50% beads, 50% cell paste, 10 cycles, 6 m/s). The resulting lysates were incubated at 37°C overnight in the presence of DNase and RNase. Subsequently, sodium dodecyl sulfate (SDS) was added to a final concentration of 2% followed by ultrasonication for 15 min using a Branson Sonifier 250 apparatus (pulse duration of 0.9 s, output control of 3). Cell walls were washed several times to remove SDS. A 5% trichloroacetic acid treatment for 4 h at 65°C was used to release WTA from cell walls. Peptidoglycan debris was separated via centrifugation (10 min, 14,000 × g). Determination of inorganic phosphate as described previously (10) was used for WTA quantification. WTA crude extracts were further purified and dialyzed as described before (7).
WTA polyacrylamide gel electrophoresis (WTA PAGE).
Dialyzed WTA samples were separated as described previously with minor modifications (19). Briefly, a 26% and 0.75-mm-thick resolving gel was used on which about 100 nmol phosphate was loaded per lane. Samples were electrophoresed at 4°C in a Bio-Rad Protean II Xi electrophoresis cell for 18 h using a current of 15 mA/gel to ensure that even the faster-migrating WTA species of GN1 is resolved from the wild-type WTA. The running buffer contained 0.1 M Tris base and 0.1 M Tricine at a pH of 8.2. The gel was developed using the alcian blue/silver staining method as described previously (47).
Protein sequence alignments.
Multiple protein sequence alignments were performed using ClustalW (48) and online resources (49).
General and analytical chemistry methods.
The amino sugar composition of isolated WTA components was determined as their amino alditols obtained after hydrolysis of the samples as described previously (7).
NMR spectroscopy.
NMR experiments were carried in D2O at 300 K or 330 K in the case of RN4220-H as described previously (7). Briefly all one-dimensional (1H and 13C) and two-dimensional homonuclear (COSY, TOCSY, and ROESY) and heteronuclear (HSQC-DEPT, HMBC, and 1H,31P-HMQC-TOCSY) experiments were recorded with a Bruker DRX Avance III 700-MHz spectrometer (operating frequencies of 700.75 MHz for 1H NMR, 176.2 MHz for 13C NMR, and 283.7 for 31P) using standard Bruker software for all samples but RN4220, whose NMR experiments were recorded with a Bruker DRX Avance 600-MHz spectrometer (operating frequencies of 600.03 MHz for 1H NMR, 150.89 MHz for 13C NMR, and 242.90 for 31P) using standard Bruker software. COSY, TOCSY, and ROESY were recorded using data sets (t1 by t2) of 4,096 by 512 points for PS187 (12, 16, and 16 scans were acquired for each t1 value in each case, respectively), GN1 (24, 32, and 32 scans were acquired for each t1 value in each case, respectively), c-GN1 (24, 32, and 48 scans were acquired for each t1 value in each case, respectively), c-GN1 (tagN-Sc) (4, 8, and 16 scans were acquired for each t1 value in each case, respectively), and RN4220-H (4, 8, and 8 scans were acquired for each t1 value in each case, respectively), while a data set (t1 by t2) of 2,048 by 512 was recorded for the sample RN4220 (16, 16, and 32 scans were acquired for each t1 value in each case, respectively). The TOCSY experiments were carried out in the phase-sensitive mode with mixing times of 100 ms for all samples. The 1H,13C correlations were measured in the 1H-detected mode via HSQC-DEPT with proton decoupling in the 13C domain acquired using data sets of 4,096 by 512 points for all samples except RN4220 (set of 2,048 by 512) and 56 (GN1), 40 (PS187 and c-GN1), 32 (RN4220 and c-GN1 [tagN-Sc]), and 10 scans (RN4220-H) for each t1 value. The HMBC spectra were acquired using data sets of 4,096 by 512 points for all but RN4220 (set of 2,048 by 512) and 96 (RN4220), 56 (GN1), 48 (RN4220-H), 40 (PS187 and c-GN1) and 32 scans (c-GN1 [tagN-Sc]) for each t1 value. The 1H,31P-HMQC-TOCSY experiment was recorded using a data set of 2,048 by 512 points (32 scans for each t1 value for c-GN1 [tagN-Sc], 16 for RN4220, PS187, and GN1, 12 for c-GN1, and 4 for RN4220-H, respectively), using a mixing time of 100 ms for all samples. No 1H,31P-HMQC-TOCSY experiment was recorded for the sample c-GN1 (tagN-Sc). Chemical shifts were reported relative to acetone (δH 2.225; δC 31.50).
SUPPLEMENTAL MATERIAL
Alignment of S. aureus. PS187 TagF with experimentally validated WTA polymerases. Protein sequence of PS187 TagF was compared and aligned with S. epidermidis ATCC 12228 TagF (NP_765503.1), B. subtilis 168 TagF (NP_391453.1), and B. subtilis W23 TarF (YP_003867853.1) using ClustalW. Accession number and sequence identities (*), strong similarity (:), weak similarity (.), and difference ( ) are indicated. Download
Protein sequence analysis of S. aureus PS187 TagD proteins. Protein sequences of the two PS187 TagD proteins (TagD1, encoded by SaGroWI; and TagD2, encoded by tagAHGBXD cluster) were compared and aligned by using ClustalW. Sequence identities (*), strong similarity (:), weak similarity (.), and difference ( ) are indicated. Download
TagN protein sequence alignments. Protein sequence of PS187 TagN was compared and aligned with TagN homologies of S. hominis SK119 (EEK11989), S. lugdunensis N920143 (YP_005759552), S. epidermidis M23864:W1 (EES41741), S. warneri L37603 (EEQ79684), S. carnosus TM300 (YP_002635271), S. pseudintermedius ED99 (YP_006014467), S. simulans ACS-120-V-Sch-1 (HMPREF9310_00448), B. subtilis BEST195 (YP_005562917), and B. cereus VD136 (EOP63090.1) using ClustalW. Protein accession numbers and sequence identities (*), strong similarity (:), weak similarity (.), and difference ( ) are indicated. Download
NMR analysis of WTA from the S. aureus PS187 GN1 mutant. (A) Heteronuclear 13C,1H single quantum correlation (HSQC) spectrum from the purified S. aureus PS187 GN1 WTA repeating unit reveals the GroP backbone lacks α-linked GalNAc. Arabic numerals label the proton/carbon atoms of the residues. Spectra were recorded at 700 MHz and 300 K relative to acetone (δH 2.225; δC 31.50). 1H and 13C chemical shifts are indicated in Table S1. (B) HSQC spectrum from purified S. aureus PS187 GN1 WTA complemented with tagN-Sc repeating unit. Arabic numerals label the proton/carbon atoms of the residues. Spectra were recorded at 700 MHz and 300 K relative to acetone (δH 2.225; δC 31.50). 1H and 13C chemical shifts are indicated in Table S1 in the supplemental material. Download
Correlation with WTA structure and Φ187 adsorption. (A) Φ187 adsorption to its host S. aureus PS187 requires α-linked GalNAc WTA modification. (B) Lack of WTA glycosylation results in reduced adsorption of ΦK. (C) RboP-GlcNAc WTA blocks Φ187 adsorption. (D, E) Adsorption rates of phages Φ11 and Φ80α to its host S. aureus RN4220, RN4220 WTA hybrid, and PS187. Values are given as means (n = 3 experiments) ± SD. Based on the unpaired two-tailed Student t test, statistically significant differences versus the phage Φ187 propagation or ΦK-susceptible strain PS187 w.t. or versus the Φ11 and Φ80α propagation strain RN4220 w.t. are indicated; ns, not significant, P > 0.05; *, P < 0.01 to 0.05; **, P < 0.001 to 0.01; ***, P < 0.001; ****, P < 0.0001. Wild-type (w.t.), GN1, tagN-complemented GN1 mutant (c-tagN), and RN4220 WTA hybrid (RN4220-H) strains are indicated. Download
1H and 13C chemical shifts of WTA from various S. aureus strains analyzed in this work. §NAc, 2.08/23.32; *NAc, 2.08/23.35; **NAc, 2.07/23.18; GlcN1, unsubstituted GlcN; GlcN2, GlcN-5-P; #NAc, 2.04/22.45; Terminal Gro1 CH2OH, 3.68/59.23; Gro1, nonglycosilated Gro.
Strains used in this study (50–52).
Primers used in this study.
ACKNOWLEDGMENTS
We thank Chunguang Liang and Thomas Dandekar for help with genome sequence analysis, Emir Kulauzovic for help with WTA isolation, Heiko Kässner for NMR recordings, and Sylvia Düpow for amino sugar analysis.
This work was supported by grants TRR34 to A.P. and SFB766 to A.P. and G.X. from the German Research Council, grants from the German Center for Infection Research (DZIF) to A.P. and G.X., and SkinStaph and Menage grants to A.P. from the German Ministry of Education and Research.
Footnotes
Citation Winstel V, Sanchez-Carballo P, Holst O, Xia G, Peschel A. 2014. Biosynthesis of the unique wall teichoic acid of Staphylococcus aureus lineage ST395. mBio 5(2):e00869-14. doi:10.1128/mBio.00869-14.
REFERENCES
- 1. Reichmann NT, Gründling A. 2011. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol. Lett. 319:97–105. 10.1111/j.1574-6968.2011.02260.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rahman O, Dover LG, Sutcliffe IC. 2009. Lipoteichoic acid biosynthesis: two steps forwards, one step sideways? Trends Microbiol. 17:219–225. 10.1016/j.tim.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 3. Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6:276–287. 10.1038/nrmicro1861 [DOI] [PubMed] [Google Scholar]
- 4. Winstel V, Xia G, Peschel A. 1 November 2013. Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int. J. Med. Microbiol. 10.1016/j.ijmm.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 5. Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686–723. 10.1128/MMBR.67.4.686-723.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Endl J, Seidl PH, Fiedler F, Schleifer KH. 1984. Determination of cell wall teichoic acid structure of staphylococci by rapid chemical and serological screening methods. Arch. Microbiol. 137:272–280. 10.1007/BF00414557 [DOI] [PubMed] [Google Scholar]
- 7. Winstel V, Liang C, Sanchez-Carballo P, Steglich M, Munar M, Bröker BM, Penadés JR, Nübel U, Holst O, Dandekar T, Peschel A, Xia G. 2013. Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat. Commun 4:2345. 10.1038/ncomms3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swoboda JG, Campbell J, Meredith TC, Walker S. 2010. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 11:35–45. 10.1002/cbic.200900557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xia G, Kohler T, Peschel A. 2010. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int. J. Med. Microbiol. 300:148–154. 10.1016/j.ijmm.2009.10.001 [DOI] [PubMed] [Google Scholar]
- 10. Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243–245. 10.1038/nm991 [DOI] [PubMed] [Google Scholar]
- 11. Weidenmaier C, Peschel A, Xiong YQ, Kristian SA, Dietz K, Yeaman MR, Bayer AS. 2005. Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J. Infect. Dis. 191:1771–1777. 10.1086/429692 [DOI] [PubMed] [Google Scholar]
- 12. Park KH, Kurokawa K, Zheng L, Jung DJ, Tateishi K, Jin JO, Ha NC, Kang HJ, Matsushita M, Kwak JY, Takahashi K, Lee BL. 2010. Human serum mannose-binding lectin senses wall teichoic acid glycopolymer of Staphylococcus aureus, which is restricted in infancy. J. Biol. Chem. 285:27167–27175. 10.1074/jbc.M110.141309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jung DJ, An JH, Kurokawa K, Jung YC, Kim MJ, Aoyagi Y, Matsushita M, Takahashi S, Lee HS, Takahashi K, Lee BL. 2012. Specific serum Ig recognizing staphylococcal wall teichoic acid induces complement-mediated opsonophagocytosis against Staphylococcus aureus. J. Immunol. 189:4951–4959. 10.4049/jimmunol.1201294 [DOI] [PubMed] [Google Scholar]
- 14. Kurokawa K, Jung DJ, An JH, Fuchs K, Jeon YJ, Kim NH, Li X, Tateishi K, Park JA, Xia G, Matsushita M, Takahashi K, Park HJ, Peschel A, Lee BL. 2013. Glycoepitopes of staphylococcal wall teichoic acid govern complement-mediated opsonophagocytosis via human serum antibody and mannose-binding lectin. J. Biol. Chem. 288:30956–30968. 10.1074/jbc.M113.509893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405–8410. 10.1074/jbc.274.13.8405 [DOI] [PubMed] [Google Scholar]
- 16. Kohler T, Weidenmaier C, Peschel A. 2009. Wall teichoic acid protects Staphylococcus aureus against antimicrobial fatty acids from human skin. J. Bacteriol. 191:4482–4484. 10.1128/JB.00221-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown S, Xia G, Luhachack LG, Campbell J, Meredith TC, Chen C, Winstel V, Gekeler C, Irazoqui JE, Peschel A, Walker S. 2012. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. U. S. A. 109:18909–18914. 10.1073/pnas.1209126109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xia G, Corrigan RM, Winstel V, Goerke C, Gründling A, Peschel A. 2011. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J. Bacteriol. 193:4006–4009. 10.1128/JB.01412-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia G, Maier L, Sanchez-Carballo P, Li M, Otto M, Holst O, Peschel A. 2010. Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J. Biol. Chem. 285:13405–13415. 10.1074/jbc.M109.096172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xia G, Wolz C. 2014. Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 21:593–601. 10.1016/j.meegid.2013.04.022 [DOI] [PubMed] [Google Scholar]
- 21. Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho MG, Filipe SR. 2010. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 107:18991–18996. 10.1073/pnas.1004304107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qamar A, Golemi-Kotra D. 2012. Dual roles of FmtA in Staphylococcus aureus cell wall biosynthesis and autolysis. Antimicrob. Agents Chemother. 56:3797–3805. 10.1128/AAC.00187-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Farha MA, Leung A, Sewell EW, D’Elia MA, Allison SE, Ejim L, Pereira PM, Pinho MG, Wright GD, Brown ED. 2013. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to beta-lactams. ACS Chem. Biol. 8:226–233. 10.1021/cb300413m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suzuki T, Swoboda JG, Campbell J, Walker S, Gilmore MS. 2011. In vitro antimicrobial activity of wall teichoic acid biosynthesis inhibitors against Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 55:767–774. 10.1128/AAC.00879-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H, Gill CJ, Lee SH, Mann P, Zuck P, Meredith TC, Murgolo N, She X, Kales S, Liang L, Liu J, Wu J, Santa Maria J, Su J, Pan J, Hailey J, McGuinness D, Tan CM, Flattery A, Walker S, Black T, Roemer T. 2013. Discovery of wall teichoic acid inhibitors as potential anti-MRSA beta-lactam combination agents. Chem. Biol. 20:272–284. 10.1016/j.chembiol.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Campbell J, Singh AK, Santa Maria JP, Jr, Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson BJ, Walker S. 2011. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6:106–116. 10.1021/cb100269f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Campbell J, Singh AK, Swoboda JG, Gilmore MS, Wilkinson BJ, Walker S. 2012. An antibiotic that inhibits a late step in wall teichoic acid biosynthesis induces the cell wall stress stimulon in Staphylococcus aureus. Antimicrob. Agents Chemother. 56:1810–1820. 10.1128/AAC.05938-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Araki Y, Ito E. 1989. Linkage units in cell walls of Gram-positive bacteria. Crit. Rev. Microbiol. 17:121–135. 10.3109/10408418909105745 [DOI] [PubMed] [Google Scholar]
- 29. Brown S, Meredith T, Swoboda J, Walker S. 2010. Staphylococcus aureus and Bacillus subtilis W23 make polyribitol wall teichoic acids using different enzymatic pathways. Chem. Biol. 17:1101–1110. 10.1016/j.chembiol.2010.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. D’Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA, Brown ED. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 188:4183–4189. 10.1128/JB.00197-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brückner R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187–192. 10.1016/0378-1119(92)90048-T [DOI] [PubMed] [Google Scholar]
- 32. Dengler V, Meier PS, Heusser R, Kupferschmied P, Fazekas J, Friebe S, Staufer SB, Majcherczyk PA, Moreillon P, Berger-Bächi B, McCallum N. 2012. Deletion of hypothetical wall teichoic acid ligases in Staphylococcus aureus activates the cell wall stress response. FEMS Microbiol. Lett. 333:109–120. 10.1111/j.1574-6968.2012.02603.x [DOI] [PubMed] [Google Scholar]
- 33. Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. 2011. A widespread family of bacterial cell wall assembly proteins. EMBO J. 30:4931–4941. 10.1038/emboj.2011.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chan YG, Frankel MB, Dengler V, Schneewind O, Missiakas D. 2013. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J. Bacteriol. 195:4650–4659. 10.1128/JB.00544-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pereira MP, Schertzer JW, D’Elia MA, Koteva KP, Hughes DW, Wright GD, Brown ED. 2008. The wall teichoic acid polymerase TagF efficiently synthesizes poly(glycerol phosphate) on the TagB product lipid III. Chembiochem 9:1385–1390. 10.1002/cbic.200800026 [DOI] [PubMed] [Google Scholar]
- 36. Schertzer JW, Brown ED. 2003. Purified, recombinant TagF protein from Bacillus subtilis 168 catalyzes the polymerization of glycerol phosphate onto a membrane acceptor in vitro. J. Biol. Chem. 278:18002–18007. 10.1074/jbc.M300706200 [DOI] [PubMed] [Google Scholar]
- 37. Lovering AL, Lin LY, Sewell EW, Spreter T, Brown ED, Strynadka NC. 2010. Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis. Nat. Struct. Mol. Biol. 17:582–589. 10.1038/nsmb.1819 [DOI] [PubMed] [Google Scholar]
- 38. Jensen SO, Apisiridej S, Kwong SM, Yang YH, Skurray RA, Firth N. 2010. Analysis of the prototypical Staphylococcus aureus multiresistance plasmid pSK1. Plasmid 64:135–142. 10.1016/j.plasmid.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 39. Freney J, Brun Y, Bes M, Meugnier H, Grimont F, Grimont PAD, Nerv C, Fleurette J. 1988. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int. J. Syst. Bacteriol. 38:168–172. 10.1099/00207713-38-2-168 [DOI] [Google Scholar]
- 40. Meyer W. 1967. A proposal for subdividing the species Staphylococcus aureus. Int. J. Syst. Evol. Microbiol. 17:387–389 [Google Scholar]
- 41. Bannoehr J, Guardabassi L. 2012. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet. Dermatol. 23:253–266, e51–e52. 10.1111/j.1365-3164.2012.01046.x [DOI] [PubMed] [Google Scholar]
- 42. Holtfreter S, Grumann D, Schmudde M, Nguyen HT, Eichler P, Strommenger B, Kopron K, Kolata J, Giedrys-Kalemba S, Steinmetz I, Witte W, Bröker BM. 2007. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 45:2669–2680. 10.1128/JCM.00204-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Piechowicz L, Garbacz K, Galiński J. 2008. Staphylococcus aureus of phage type 187 isolated from people occurred to be a genes carrier of enterotoxin C and toxic shock syndrome toxin-1 (TSST-1). Int. J. Hyg. Environ. Health 211:273–282. 10.1016/j.ijheh.2007.06.010 [DOI] [PubMed] [Google Scholar]
- 44. Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 45. Pantůcek R, Doskar J, Růzicková V, Kaspárek P, Orácová E, Kvardová V, Rosypal S. 2004. Identification of bacteriophage types and their carriage in Staphylococcus aureus. Arch. Virol. 149:1689–1703. 10.1007/s00705-004-0335-6 [DOI] [PubMed] [Google Scholar]
- 46. Christie GE, Matthews AM, King DG, Lane KD, Olivarez NP, Tallent SM, Gill SR, Novick RP. 2010. The complete genomes of Staphylococcus aureus bacteriophages 80 and 80α—implications for the specificity of SaPI mobilization. Virology 407:381–390. 10.1016/j.virol.2010.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pollack JH, Neuhaus FC. 1994. Changes in wall teichoic acid during the rod-sphere transition of Bacillus subtilis 168. J. Bacteriol. 176:7252–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Combet C, Blanchet C, Geourjon C, Deléage G. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147–150. 10.1016/S0968-0004(99)01540-6 [DOI] [PubMed] [Google Scholar]
- 50. Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- 51. Schleifer KH, Fischer U. 1982. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int. J. Syst. Bacteriol. 32:153–156. 10.1099/00207713-32-2-153 [DOI] [Google Scholar]
- 52. Asheshov EA, Jevons MP. 1963. The effect of heat on the ability of a host strain to support the growth of a Staphylococcus phage. J. Gen. Microbiol. 31:97–107. 10.1099/00221287-31-1-97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of S. aureus. PS187 TagF with experimentally validated WTA polymerases. Protein sequence of PS187 TagF was compared and aligned with S. epidermidis ATCC 12228 TagF (NP_765503.1), B. subtilis 168 TagF (NP_391453.1), and B. subtilis W23 TarF (YP_003867853.1) using ClustalW. Accession number and sequence identities (*), strong similarity (:), weak similarity (.), and difference ( ) are indicated. Download
Protein sequence analysis of S. aureus PS187 TagD proteins. Protein sequences of the two PS187 TagD proteins (TagD1, encoded by SaGroWI; and TagD2, encoded by tagAHGBXD cluster) were compared and aligned by using ClustalW. Sequence identities (*), strong similarity (:), weak similarity (.), and difference ( ) are indicated. Download
TagN protein sequence alignments. Protein sequence of PS187 TagN was compared and aligned with TagN homologies of S. hominis SK119 (EEK11989), S. lugdunensis N920143 (YP_005759552), S. epidermidis M23864:W1 (EES41741), S. warneri L37603 (EEQ79684), S. carnosus TM300 (YP_002635271), S. pseudintermedius ED99 (YP_006014467), S. simulans ACS-120-V-Sch-1 (HMPREF9310_00448), B. subtilis BEST195 (YP_005562917), and B. cereus VD136 (EOP63090.1) using ClustalW. Protein accession numbers and sequence identities (*), strong similarity (:), weak similarity (.), and difference ( ) are indicated. Download
NMR analysis of WTA from the S. aureus PS187 GN1 mutant. (A) Heteronuclear 13C,1H single quantum correlation (HSQC) spectrum from the purified S. aureus PS187 GN1 WTA repeating unit reveals the GroP backbone lacks α-linked GalNAc. Arabic numerals label the proton/carbon atoms of the residues. Spectra were recorded at 700 MHz and 300 K relative to acetone (δH 2.225; δC 31.50). 1H and 13C chemical shifts are indicated in Table S1. (B) HSQC spectrum from purified S. aureus PS187 GN1 WTA complemented with tagN-Sc repeating unit. Arabic numerals label the proton/carbon atoms of the residues. Spectra were recorded at 700 MHz and 300 K relative to acetone (δH 2.225; δC 31.50). 1H and 13C chemical shifts are indicated in Table S1 in the supplemental material. Download
Correlation with WTA structure and Φ187 adsorption. (A) Φ187 adsorption to its host S. aureus PS187 requires α-linked GalNAc WTA modification. (B) Lack of WTA glycosylation results in reduced adsorption of ΦK. (C) RboP-GlcNAc WTA blocks Φ187 adsorption. (D, E) Adsorption rates of phages Φ11 and Φ80α to its host S. aureus RN4220, RN4220 WTA hybrid, and PS187. Values are given as means (n = 3 experiments) ± SD. Based on the unpaired two-tailed Student t test, statistically significant differences versus the phage Φ187 propagation or ΦK-susceptible strain PS187 w.t. or versus the Φ11 and Φ80α propagation strain RN4220 w.t. are indicated; ns, not significant, P > 0.05; *, P < 0.01 to 0.05; **, P < 0.001 to 0.01; ***, P < 0.001; ****, P < 0.0001. Wild-type (w.t.), GN1, tagN-complemented GN1 mutant (c-tagN), and RN4220 WTA hybrid (RN4220-H) strains are indicated. Download
1H and 13C chemical shifts of WTA from various S. aureus strains analyzed in this work. §NAc, 2.08/23.32; *NAc, 2.08/23.35; **NAc, 2.07/23.18; GlcN1, unsubstituted GlcN; GlcN2, GlcN-5-P; #NAc, 2.04/22.45; Terminal Gro1 CH2OH, 3.68/59.23; Gro1, nonglycosilated Gro.
Strains used in this study (50–52).
Primers used in this study.