ABSTRACT
We identified mutated genes in highly resistant subpopulations of methicillin-resistant Staphylococcus aureus (MRSA) that are most likely responsible for the historic failure of the β-lactam family of antibiotics as therapeutic agents against these important pathogens. Such subpopulations are produced during growth of most clinical MRSA strains, including the four historically early MRSA isolates studied here. Chromosomal DNA was prepared from the highly resistant cells along with DNA from the majority of cells (poorly resistant cells) followed by full genome sequencing. In the highly resistant cells, mutations were identified in 3 intergenic sequences and 27 genes representing a wide range of functional categories. A common feature of these mutations appears to be their capacity to induce high-level β-lactam resistance and increased amounts of the resistance protein PBP2A in the bacteria. The observations fit a recently described model in which the ultimate controlling factor of the phenotypic expression of β-lactam resistance in MRSA is a RelA-mediated stringent response.
IMPORTANCE
It has been well established that the level of antibiotic resistance (i.e., minimum concentration of a β-lactam antibiotic needed to inhibit growth) of a methicillin-resistant Staphylococcus aureus (MRSA) strain depends on the transcription and translation of the resistance protein PBP2A. Here we describe mutated loci in an additional novel set of genetic determinants that appear to be essential for the unusually high resistance levels typical of subpopulations of staphylococci that are produced with unique low frequency in most MRSA clinical isolates. We propose that mutations in these determinants can trigger induction of the stringent stress response which was recently shown to cause increased transcription/translation of the resistance protein PBP2A in parallel with the increased level of resistance.
INTRODUCTION
It is generally agreed that the appearance of methicillin-resistant Staphylococcus aureus (MRSA) strains among clinical isolates represents the single most serious blow to the chemotherapy of S. aureus infections, since the unique resistance mechanism carried by all MRSA strains provides protection against the single largest family of antibacterial agents—the β-lactam antibiotics (1). Since their first appearance in clinical specimens in 1960, this resistance mechanism has made its way into a large variety of S. aureus lineages and diverse clones of MRSA have spread throughout the globe to cause serious and often life-threatening infections both in hospitals and in the community (2–5).
Most MRSA strains carry an identical—acquired—genetic determinant mecA (6, 7) which is part of a mobile genetic element (staphylococcal cassette chromosome mec element [SCCmec]) (8) inserted into the S. aureus chromosome at a unique chromosomal site. mecA encodes a protein, PBP2A, a peptidoglycan transpeptidase with extremely low affinity for the entire large family of β-lactam antibiotics (9), and the presence of this protein plays a critical role in allowing MRSA strains to continue synthesis of peptidoglycan and bacterial growth in the presence of high concentrations of β-lactam antibiotics. A model for the mechanism of action of PBP2A on the molecular level has been proposed (10).
In contrast to the common molecular mechanism of resistance, individual MRSA clinical isolates differ widely in their susceptibility to β-lactam antibiotics with individual MRSA strains presenting methicillin MIC values as low as a few µg/ml up to several hundred µg/ml depending on the particular MRSA clone, and this variation in resistance level cannot be explained by transcriptional regulation of mecA through the activity of regulatory elements such as the mecl and mecR1 or blal and blaR1 genes (11, 12).
A detailed examination of the β-lactam susceptibility of cultures of MRSA strains presents an even more complex and intriguing picture. MRSA grown from single-cell inocula produce cultures that are highly heterogeneous with respect to their antibiotic susceptibility with most cells (more than 99%) showing only moderate- or low-level resistance often close to the MIC values of methicillin-susceptible isolates. On the other hand, the same cultures also contain bacteria with an extremely high level of resistance—in the MIC range of several hundred µg/ml—and the frequency of such highly resistant cells in a given culture (10−4 to 10−5) appears to be specific for the particular MRSA clone. This phenomenon has become known as the “heterogeneous” phenotype. It was first recognized and described in 1960, in the microbiological analysis of the historically first MRSA infection by Jevons who was surprised to recover two MRSA populations with widely different methicillin MIC values from a patient with an MRSA infection (13). The methicillin MIC of the majority of the bacteria was 2 µg/ml, but upon prolonged incubation of the specimen, more bacteria were recovered with a much higher antibiotic MIC value in the range of several hundred µg/ml.
Most contemporary clinical isolates of MRSA express β-lactam resistance in a similar heterogeneous fashion (14). Plotting the number of bacteria capable of forming colonies against the concentration of the antibiotic in the agar plates produce phenotypic profiles called population analysis profiles (PAPs), and the shape of the PAP is characteristic for the particular MRSA strain (15, 16). The PAP was subsequently shown to be a unique phenotypic marker of MRSA clones—highly reproducible in chronologically distinct isolates of the same MRSA lineage (17). The presence of highly methicillin-resistant cells in cultures of MRSA is of obvious relevance both for the detection of MRSA in clinical specimens and also for therapeutic options (18–20).
The stability of PAP for a given MRSA clone indicates that the heterogeneous composition of MRSA cultures is genetically controlled, i.e., the highly resistant subpopulations of bacteria must carry mutations in some genetic determinants that are “wild type” in the majority of less-resistant cells of the same clone.
Full genome sequences of a large number of MRSA strains are now available in the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). However, most of the sequenced strains show heterogeneous antibiotic resistance, and therefore, the sequence information available relates only to the genetic makeup of the “cell majorities” in each of these particular MRSA clones and provide no information about unique mutated genes that must be present in highly resistant subpopulations carried by each one of the sequenced MRSA strains.
The purpose of this study was to identify mutated genes associated with the rare highly resistant bacteria that are produced with a low frequency (10−4 to 10−5) during growth of four historically early MRSA lineages belonging to the archaic clone of MRSA (17). The four strains selected for the study carry the mecA determinant on SCCmec type I cassettes that do not contain an active form of the mecI or mecR regulatory genes (12). The β-lactam antibiotic (methicillin or oxacillin) MIC values for the majority of bacteria in these four strains were in the range of 2 up to 12 µg/ml of oxacillin. When these four “parental” MRSA strains were plated on oxacillin-containing agar for PAPs, rare colonies (10−4 to 10−5) capable of growing on agar containing 100 µg/ml of the antibiotic were also detected. Cultures of such colonies—named “H*R”—(for homogeneous and high-level resistance)—produced highly and homogeneously resistant populations of bacteria with MIC values in the range of several hundred µg/ml.
Chromosomal DNA was prepared from the H*R cultures along with DNA prepared from the corresponding heteroresistant “parental” (majority) cells followed by full genome sequencing. Mutated genes unique to the particular H*R culture were identified by comparison to the status of the same gene in the “parental” culture.
Mutations in 27 genes and 3 intergenic sequences were identified in the highly resistant H*R derivatives recovered from the four heteroresistant “families” of MRSA. While the mutated genes represent a range of functional categories, we suggest that a common feature of these mutations may be their capacity to induce a stringent stress response in the bacteria. This proposal is consistent with recent evidence that identified a key role of the relA gene complex in defining the level of β-lactam resistance in laboratory models of MRSA strains (20, 21). As experimental evidence strongly suggested this, each of the four heteroresistant “parental” MRSA strains described in the present communication could be made to change their mode of expression of resistance from heterogeneous to homogeneous by the use of mupirocin, i.e., by experimentally inducing the stringent stress response in the bacteria (22–30).
RESULTS
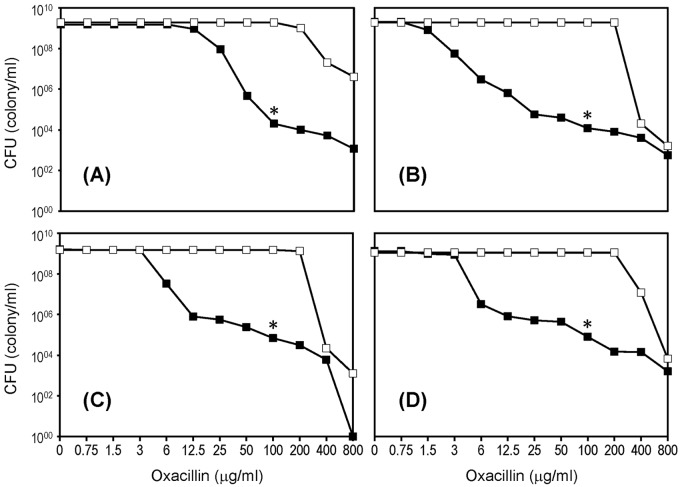
The majority of clinical MRSA isolates express β-lactam resistance in a heterogeneous fashion. As a first attempt to better understand the genetic basis of this phenomenon, we selected four genetically closely related and historically early MRSA strains which had very similar heteroresistant phenotypes, as indicated by the virtually superimposable population analysis profiles (PAPs) (Fig. 1). The four strains included UK13136, the historically first MRSA, isolated in 1960 in the United Kingdom (13) named in our study as the “parental” strain of family A. This strain has already been characterized by molecular techniques (31). The second MRSA strain, ST63/458, was also isolated in the United Kingdom in 1963 and was the “parental” strain of family B in our study. Both of these strains are of sequence type ST250 and carry an SCCmec type I. The two additional parental strains, E2125 (parental strain of family C) and strain E4278 (parental strain of family D), were both isolated in Denmark in 1964 and 1967, respectively, and both were ST247 carrying SCCmec type I. All four strains belonged to the “archaic” clone of MRSA (17).
FIG 1 .
Heterogeneous expression of β-lactam antibiotic resistance in four MRSA strains. (A to D) Population analysis profiles for strains UK13136 (A), UK63/458 (B), E2125 (C), and E4278 (D) are shown by the lines with solid squares. H*R isolates capable of growing in the presence of 100 µg/ml oxacillin were picked from the agar plates as indicated by an asterisk. Population analysis profiles of cultures of H*R isolates are shown by the lines with empty squares.
Figure 1 shows the rather similar PAPs of the four “parental” MRSA strains; in each strain, the majority of bacteria had relatively low oxacillin MIC values between 2 and 12 µg/ml. However, each culture also contained—with a low frequency of about 10−5—highly resistant (mutant) subpopulations of bacteria with an oxacillin MIC of ≥400 µg/ml.
Overnight cultures of the four parental strains were grown in tryptic soy broth (TSB), and 1-ml portions of the turbid overnight cultures were used to prepare the “parental” DNAs for sequencing. Aliquots of the rest of the overnight parental cultures were plated for population analysis, and 20 of the rare highly resistant colonies (named “H*R”) that appeared on the agar plates supplemented with 100 µg/ml oxacillin (see the asterisks in Fig. 1), were picked from the progeny of each of the four parental cultures. The H*R colonies were resuspended in TSB and restreaked on tryptic soy agar (TSA), and cultures of the 10 H*R colonies were grown in TSB, retested for oxacillin resistance by PAP, and used to prepare DNA for sequencing. Eventually, the DNA sequence of each H*R colony was compared to the sequence of the corresponding “parental” culture in order to identify the genes that were mutated in the H*R cultures.
On the basis of this comparison, mutations in 3 intergenic sequences and 27 genes were identified in the stably resistant H*R colonies recovered from the four heteroresistant MRSA strains (Table 1).
TABLE 1 .
Mutated loci in highly resistant (H*R) isolates identified in heteroresistant MRSA strains
| Mutation no. | Locus in S. aureus COL | Descriptiona | Nucleotide changeb | Amino acid change | Functional categoryc |
|---|---|---|---|---|---|
| 1 | SACOL0314 | Transcriptional regulator (rpiR family) | C349448T | Thr216Ile | b |
| 2 | SACOL0403 | Transcriptional antiterminator (bglG family) | C409253 Del | Frameshift after Ser461 | b |
| 3 | SACOL0434 | Hypothetical protein 1 | C440981T | Gln41Stop | f |
| 4 | SACOL0460 | IMP dehydrogenase (guaB) | C463203T | Arg310Cys | a |
| 5 | SACOL0461 | GMP synthase (guaA) | A463937 Del | Deletion after Ser27 | a |
| C464191T | Pro142Leu | ||||
| 6 | SACOL0495 | Hypothetical protein 2 | G497663T | Asp777Stop | f |
| 7 | SACOL0533 | Methionyl-tRNA synthetase (metS) | G542898T | Leu285Phe | c |
| 8 | SACOL0544 | Ribose-phosphate pyrophosphokinase (prsA) | C552272T | Pro291Leu | a |
| 9 | SACOL0554 | Hypoxanthine phosphoribosyltransferase (hpt) | G562925A | Met1Ile | a |
| 10 | SACOL0555 | Cell division protein (ftsH) | C565004A | Ala429Asp | e |
| 11 | SACOL0562 | Lysyl-tRNA synthetase (lysS) | G570441T | Arg11Leu | c |
| G570684A | Arg93His | ||||
| 12 | SACOL0574 | Glutamyl-tRNA synthetase (gltX) | G597328A | Glu439Lys | c |
| 13 | SACOL0576 | Cysteinyl-tRNA synthetase (cysS) | C599284G | His255Asp | c |
| 14 | SACOL0583 | Ribosomal protein L11 (rplK) | A603893T | Ile140Val | c |
| 15 | SACOL0588 | DNA-directed RNA polymerase, β-subunit (rpoB) | C608417T | Ala477Val | b |
| T610153A | Tyr1056Asn | ||||
| 16 | SACOL0589 | DNA-directed RNA polymerase, β′-subunit (rpoC) | G611391A | Arg239His | b |
| G611672T | Gly333Cys | ||||
| C611927A | Leu418Ile | ||||
| A612110T | Ile479Phe | ||||
| T612157A | Asp494Glu | ||||
| C612859A | Asn728Lys | ||||
| C612921A | Ala749Glu | ||||
| C613500T | Thr942Ile | ||||
| G613517T | Val948Leu | ||||
| 17 | SACOL0758 | 1-Phosphofructokinase (fruK) | G779646 Ins | Frameshift after Ala35 | d |
| 18 | Intergenic | Hypothetical protein 3/glucose-6-phosphate isomerase | C968358T | f | |
| 19 | SACOL0991 | Oligopeptide ABC transporter, permease (oppB) | G998492A | Asp265Asn | d |
| 20 | SACOL1689 | GTP pyrophosphokinase (relA2) | G1719144 Del | Frameshift after Met383 | a |
| C1719536T | Gln255Stop | ||||
| 21 | SACOL1710 | Valyl-tRNA synthetase (valS) | A1741737T | Asp177Val | c |
| 22 | Intergenic | Valyl-tRNA synthetase/DNA-3-methyladenine glycosylase | C1742564 Del | f | |
| 23 | SACOL1717 | Porphobilinogen deaminase (hemC) | C1747855T | Thr265Ile | d |
| 24 | SACOL1745 | Ribosome binding site of pyruvate kinase (pyk) | C1783697 Del | Deletion of RBS | d |
| 25 | SACOL2038 | tRNA N6-adenosine threonylcarbamoyltransferase (gcp) | G2098447A | Gly193Asp | c |
| 26 | SACOL2072 | DEAD box ATP-dependent RNA helicase (srmB) | G2137588A | Gly459Asp | b |
| 27 | SACOL2108 | Translation factor SUA5 (sua-5) | C2168647G | Pro104Arg | c |
| 28 | SACOL2117 | Fructose-bisphosphate aldolase (fbaA) | T2177986 Del | Frameshift after Lys167 | d |
| G2178061A | Gly143Arg | ||||
| C2178241T | His83Tyr | ||||
| 29 | SACOL2215 | Ribosomal protein S13/S18 (rpsM) | G2295683C | Ile308Leu | c |
| 30 | Intergenic | Hypothetical protein 4/hydroxymethylglutaryl-CoA reductase | G2617978A | f |
CoA, coenzyme A.
C349448T, C at position 349448 changed to T; C409253 Del, deletion of the C at position 409253; G779646 Ins, insertion of G at position 779646.
Functional categories a through g as defined in Table 2.
The mutated genes, their putative functions, and the nature of the nucleotide and amino acid change are listed in Table 1. The following six genes carried multiple mutations: fbaA (3 mutations), guaA (2 mutations), lysS (2 mutations), rpoB (2 mutations), rpoC (9 mutations), and relA2 (2 mutations).
These genes may be involved in (p)ppGpp-mediated stringent stress response: guaA, lysS, and relA2 are directly linked to the synthesis of (p)ppGpp which targets RNA polymerase, the product of rpoB and rpoC (32). The fbaA gene encoding fructose bisphosphate aldolase was reported to be downregulated in (p)ppGpp-mediated stringent stress response induced by serine hydroxamate (33). Of the 30 genetic loci carrying mutations, 27 are expected to alter function either by point mutations or by frameshifts. The remaining 3 determinants were in intergenic sequences with a change in a single nucleotide. Thus, mutations in 27 different genes would appear to be responsible for the increase in the resistance of H*R isolates to oxacillin either singly or through a concerted effect of all mutations.
Table 2 lists functional categories of the mutated genes identified in H*R isolates of the four MRSA families identified by capital letters A through D. Of the 27 mutated genes, 21 were in guanine metabolism (a), in transcription (b), in translation/ribosomal structure (c) and/or in transport (d). Interestingly, in most H*R isolates, mutation in a single gene was sufficient to produce the highly resistant phenotype (Table 3). Thirteen out of 17 genes listed in Table 3 are included in 3 functional categories: four (guaA, prsA, hpt, and relA2) in guanine metabolism; two (rpoB and rpoC) in transcription; and seven (metS, lysS, cysS, valS, gcp, sua-5, and rpsM) in translation/ribosome structure. Each of these mutations would be expected to trigger the stringent stress response and produce high and homogeneous resistance.
TABLE 2 .
Functional categories of mutations associated with highly resistant (H*R) isolates
| Functional category | No. of determinants | Mutation(s)a associated with H*R isolates in the following strain: |
|||
|---|---|---|---|---|---|
| UK13136 (family A) | ST63/458 (family B) | E2125 (family C) | E4278 (family D) | ||
| Guanine metabolism (category a) | 5 | guaA, guaB | guaA, relA2, prsA | relA2, hpt | |
| Transcription (category b) | 5 | bglG, rpoB, RNA helicase | rpoC | rpiR, rpoB, rpoC | |
| Translation/ribosomal structure (category c) | 9 | rpsM, lysS | rplK | gcp, cysS, valS | lysS, gltX, metS, sua-5 |
| Transport/metabolism (category d) | 5 | pyk, hemC | fbaA | oppB, fruK | |
| Cell division (category e) | 1 | ftsH | |||
| Unknown function (category f) | 5 | HP2, Intergenic | Intergenic | HP1, Intergenic | |
| Total | 30 | ||||
Genes potentially involved with induction of a stringent stress response are indicated in boldface print. The fbaA and oppB genes are downregulated by (p)ppGpp-mediated stringent stress response (33). HP stands for hypothetical protein.
TABLE 3 .
H*R isolates carrying mutations in a single gene
| Mutation no.a | Locus in S. aureus COL | Gene | H*R strain | Functional category |
|---|---|---|---|---|
| 5 | SACOL0461 | guaA | A3, BB9 | a |
| 7 | SACOL0533 | metS | DD8 | c |
| 8 | SACOL0544 | prsA | B5 | a |
| 9 | SACOL0554 | hpt | DD3 | a |
| 11 | SACOL0562 | lysS | AA9 | c |
| 13 | SACOL0576 | cysS | CC3 | c |
| 15 | SACOL0588 | rpoB | DD9 | b |
| 16 | SACOL0589 | rpoC | B4, B8, B9, BB2, BB3, DD6, DD7 | b |
| 17 | SACOL0758 | fruK | DD5 | d |
| 20 | SACOL1689 | relA2 | BB8, D3 | a |
| 21 | SACOL1710 | valS | C8 | c |
| 23 | SACOL1717 | hemC | AA2 | d |
| 24 | SACOL1745 | pyk | A5 | d |
| 25 | SACOL2038 | gcp | CC1 | c |
| 27 | SACOL2108 | sua-5 | DD1 | c |
| 28 | SACOL2117 | fbaA | B10, BB5, BB6 | d |
| 29 | SACOL2215 | rpsM | A2 | c |
Mutation numbers as in Table 1.
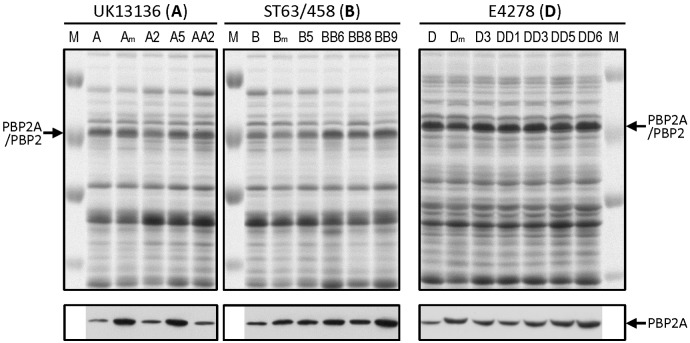
Twelve H*R isolates each carrying a single mutation (Table 3) were compared to their respective parental strains for the relative amounts of PBP2A. Cells were grown in the presence of 0.5 µg/ml oxacillin to induce the mecA gene, and membrane fractions were prepared for Western blotting. All H*R isolates showed at least 2-fold increase in PBP2A compared to their parental strains (Fig. 2) suggesting that each mutation resulted in the recruitment of increased amounts of PBP2A into the cell membranes.
FIG 2 .
Determination of PBP2A in membranes of H*R derivatives carrying single mutations. A group of H*R isolates carrying single mutations in a variety of genes associated with high-level oxacillin resistance (Table 3) were analyzed by SDS-PAGE and by testing the relative amounts of PBP2A by Western blotting with a monoclonal antibody prepared against PBP2A. Three isolates (A2, A5, and AA2) from family A, four isolates (B5, BB6, BB8, and BB9) from family B, and five isolates from family D (D3, DD1, DD3, DD5, and DD6) were included in the analysis. Lanes A, B, and D show the SDS-PAGE profiles and the Western blot analysis of PBP2A in these parental isolates. Lanes Am, Bm, and Dm contain parental samples in which the relative amounts of PBP2A were estimated in the presence of mupirocin. The single mutations carried by the H*R derivatives of family A were as follows: rpsM in lane A2, pyk in lane A5, and hemC in lane AA2. The single mutated genes analyzed in family B were prsA in lane B5, fbaA in lane BB6, relA2 in lane BB8, and guaA in lane BB9. The mutations analyzed in members of family D were relA2 in lane D3, sua5 in lane DD1, hpt in lane DD3, fruK in lane DD5, and rpoC in lane DD6. The M lanes contain molecular size markers (100, 70, 55, and 45 kDa).
DISCUSSION
Inspection of Table 1 through 3 and the figures indicates that a large number and different kinds of mutations can profoundly influence the phenotypic expression of oxacillin resistance in the four heteroresistant MRSA strains. Determinants include genes in cell division as well as genes associated with various aspects of intermediary metabolism. Such a diversity of genetic determinants is reminiscent of the large number of “auxiliary genes” (or fem genes) identified earlier as determinants essential for the optimal expression of high and homogeneous resistance in MRSA strains (34, 35). As a hypothesis to account for the polygenic nature of this phenomenon, it was proposed that the expression of antibiotic resistance involves a bacterial stress response (36).
This proposition, originally suggested to explain the multigenic nature of homogeneous oxacillin resistance, seems to also fit the mechanism of heterogeneous resistance analyzed in this communication. In a recent study, we described identification of the critical role that the S. aureus relA gene plays in the phenotypic expression of oxacillin resistance (20). RelA protein plays a central role in the control of biosynthetic activities through its catalytic product—ppGpp and pppGpp—that can interact with and regulate the ribosomal protein synthesis machinery.
The large number and functional diversity of the determinants that influence heterogeneous expression of resistance as described in this communication would fit the model in which a stress response—specifically, the stringent stress—is the ultimate controlling factor of the phenotypic expression of oxacillin resistance in MRSA (20, 21).
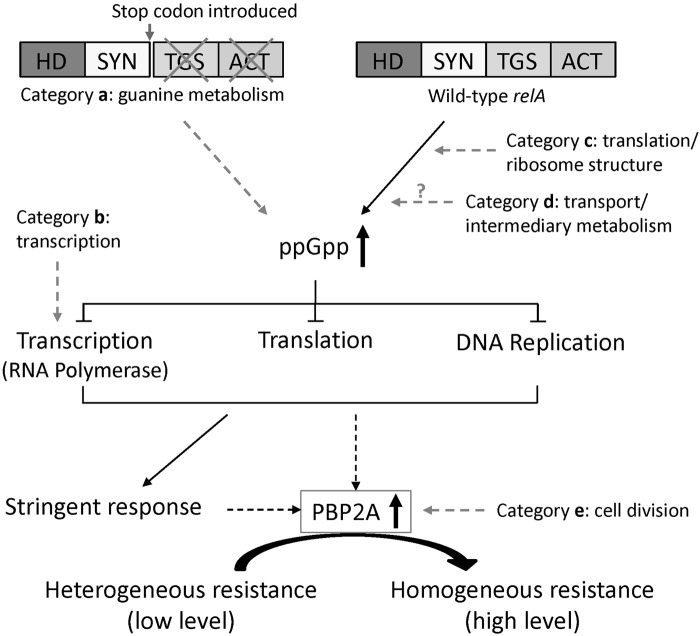
Figure 3 shows a modification of the relA model to indicate how diverse genetic determinants identified in the present communication could impact on the level of antibiotic resistance through specific interactions with the relA-controlled RNA polymerase system. In a recent communication (21), we began to test whether the physiological level of antibiotic resistance (i.e., the oxacillin MIC value) is paralleled by the cellular amounts of the mecA gene product PBP2A. In the model systems described in reference 21, increase in the MIC value was accompanied by a parallel increase in the cellular amounts of PBP2A, and increased amounts of PBP2A were also detected in the H*R derivatives described in this communication (Fig. 2).
FIG 3 .
Model for the triggering of the stringent stress response by mutations identified in the highly resistant (H*R) isolates. A schematic model for the postulated effect of H*R mutations on the relA-controlled stress response of S. aureus is shown. Functional categories of mutations are defined as in Table 2. HD, hydrolase; SYN, synthetase; TGS, a domain named after three enzymes that contain it [threonyl-tRNA synthetase (ThrRS), GTPase, and guanosine-3′,5′-bis(diphosphate) 3′-pyrophosphohydrolase (SpoT)]; ACT, the domain named after three proteins that carry it [aspartate kinase, chorismate mutase, and prephenate dehydrogenase (TyrA)].
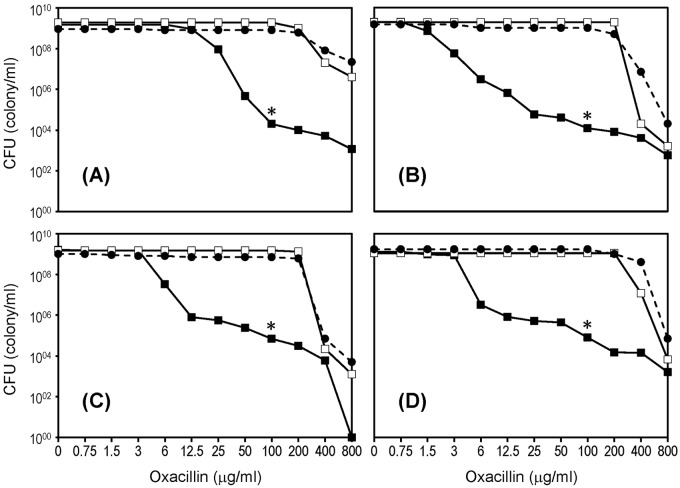
If the stringent stress response is the central controlling element of the level of oxacillin resistance in MRSA, then one would expect that artificial triggering of the stress response would convert heterogeneously resistant MRSA to highly and homogeneously resistant cultures. In an effort to test this, we repeated the population analysis of the four heteroresistant “parental” strains in the presence of sub-MICs of mupirocin, an agent capable of inducing stringent stress. Figure 4 demonstrates that each one of the four heteroresistant parental MRSA strains described in this communication would exhibit high and homogeneous resistance if the phenotype was assayed in the presence of sub-MICs of mupirocin added to the oxacillin-containing agar plates. Identical results were obtained when serine hydroxamate, an inhibitor of seryl-tRNA synthetase, was used instead of mupirocin. These observations may open up so-far untested avenues for the design of antibacterial agents that could influence resistance level of MRSA through a novel type of intervention.
FIG 4 .
Conversion of the heterogeneous population analysis profiles of four MRSA strains to high and homogeneous resistance by induction of the stringent stress response. Population analysis profiles (PAPs) of the four MRSA cultures (strains in Fig. 1A to D) (solid squares), PAPs of H*R derivatives (empty squares), and PAPs determined on agar plates on which the antibiotic was supplemented by sub-MICs (0.03 µg/ml) of mupirocin (solid circles). Asterisks indicate the concentration of oxacillin (100 µg/ml) at which the H*R colonies were picked.
MATERIALS AND METHODS
Aliquots (1 µl) of the four heteroresistant “parental” cultures of UK13136 (parental strain of family A), ST63/458 (parental strain of family B), E2125 (parental strain of family C), and E4278 (parental strain of family D) were inoculated into 5 ml of tryptic soy broth (TSB) and incubated at 37°C overnight with agitation. Portions (1 ml) of the overnight cultures were removed to prepare chromosomal DNAs representing the majority of cells (poorly resistant cells) of these cultures. The overnight cultures were diluted with TSB, and population analysis profiles (PAPs) were done on tryptic soy agar (TSA) plates containing increasing concentrations of oxacillin (Fig. 1). CFU were counted after 48-h incubation of the plates at 37°C. Twenty medium-size colonies capable of growing on TSA plates containing 100 µg/ml oxacillin were picked from the PAP plates of each of the four “parental” MRSA. These colonies were named “H*R” for homogeneous and high-level oxacillin resistance. H*R colonies were recovered from the plates with 1-µl loops and dispersed into Eppendorf tubes containing 200 µl of TSB. Portions (1 µl) from each Eppendorf tube were streaked onto a TSA plate which was incubated at 37°C for 48 h. The H*R isolates were next passaged three times onto fresh TSA plates, after which the isolates were retested for resistance level by Etest and population analysis. A total of 42 H*R isolates with high-level and homogeneous oxacillin resistance (oxacillin MIC of ≥400 µg/ml) were inoculated into 5 ml of TSB, incubated at 37°C with agitation overnight, and used to prepare H*R DNAs. The 42 H*R isolates included 10 colonies of UK13136, 12 of UK63/458, 10 of E2125, and 10 of E4278.
The antibiotic resistance profiles of the four heteroresistant strains were also determined by including sub-MICs of mupirocin in the antibiotic-containing plates used for population analysis (37, 38). Mupirocin is a known inducer of the stringent stress response in bacteria.
Genome sequencing.
Sequencing libraries were prepared according to previously published methods (39–41). Samples were run on an Illumina HiSeq 2000 sequencer operated according to the manufacturer’s instructions with 100 cycle paired-end runs. Data for the samples have been deposited in the European Nucleotide Archive (see below).
Detection of variations between H*R isolates and the “parental” strains.
The sequence of chromosomal DNAs isolated from the H*R colonies was compared to the DNA sequence of the corresponding “parental” strain in order to identify in the H*R isolates mutated loci that may be associated with the high-level oxacillin-resistant phenotype of these clones. This was done using three different approaches to call only high-confidence variants.
The first two approaches are based on de novo assemblers, which are capable of detecting variants (single nucleotide polymorphisms [SNPs]), insertions, and deletions while building the contigs. Both methods completely ignore reference genomes while calling variants between “parental” and H*R isolates. However, to make comparison between the methods easier, the results were mapped back to reference strain COL (GenBank accession number CP000046).
SGA v0.9.19 (42) commands “preprocess” and “index” were run using default settings. Variants were called using “graph-diff” with k-mer (−k) = 61 and min-discovery-count (−x) = 10. Cortex v1.0.5.15 (43) was run using the provided workflow pipeline using the joint variant discovery with k-mers between 31 and 63 (43).
The third approach is based on mapping. Each “parental” isolate and corresponding H*R isolates were mapped against strain COL using SMALT v.0.7.4 (http://www.sanger.ac.uk/resources/software/smalt). High-quality SNPs were called as described previously (44). Detection of indels was carried out using GATK (45). The obtained variant call format files (VCFs) were processed using an in-house script to remove variant calls due to the use of the reference genome COL in order to find only differences between the “parental” strain and H*R isolates. Variants found by all three methods were finally checked manually in order to carry on further analysis with high-quality variants.
Preparation of staphylococcal membrane proteins.
Membrane fractions were prepared from isolates belonging to families A, B, and D following the method described previously (21, 38) with slight modification. S. aureus strains were grown at 37°C in 200 ml of TSB in the presence of 0.5 µg/ml of oxacillin to induce transcription of the mecA gene. The strains analyzed included three H*R isolates from family A (A2, A5, and AA2), four H*R isolates from family B (B5, BB6, BB8, and BB9), and five H*R isolates from family D (D3, DD1, DD3, DD5, and DD6). Each analysis included the SDS-PAGE profiles of the corresponding parental strain: strain UK13136 for family A, strain ST63/458 for family B, and strain E4278 for family D. The relative amounts of PBP2A were determined in each of the isolates using Western blotting. The SDS-PAGE profiles and relative amounts of PBP2A were also compared for each of the parental strains with and without mupirocin (0.03 µg/ml) added to the growth medium. All cultures were harvested at an optical density at 620 nm (OD620) of 0.5, washed, and resuspended in 3 ml of 20 mM Tris-HCl (pH 7.6) containing 1× Halt protease inhibitor cocktail (Thermo Fisher Scientific, Inc.), 10 mM MgCl2, 100 µg/ml lysostaphin, 50 µg/ml lysozyme, 50 µg/ml DNase I, and 50 µg/ml RNase A. The cells were incubated at 37°C for 30 min and disrupted by sonication with pulse of 40% output for 5 min. The supernatants were transferred to fresh ultracentrifuge tubes after centrifugation at 7,000 × g for 20 min. Membrane fractions were collected by centrifugation at 100,000 × g for 1 h. The collected membranes were resuspended in 20 mM Tris-HCl (pH 7.6) and stored at −70°C. The concentration of total membrane proteins was determined by the bicinchoninic acid (BCA) assay.
Western blotting.
Western blotting with a monoclonal antibody prepared against PBP2A was used to determine PBP2A in membrane preparations as described previously with a few modifications (21, 38). The membrane proteins (50 µg for families A and B and 100 µg for family D) were loaded on the polyacrylamide gel (8% or 10% resolving gel; 4% stacking gel) for SDS-PAGE. The rabbit anti-PBP2A antibody was used as the primary antibody with dilution of 1:15,000, and the secondary horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (0.5 mg/ml; PerkinElmer) was diluted to 1:10,000. ChromPure human IgG Fc fragment (Millipore) was added to the blocking solution at a final concentration of 3 µg/ml in order to prevent the antibodies from nonspecific binding. Pierce enhanced chemiluminescence (ECL) 2 (Thermo Fisher Scientific, Inc.) substrate was used for visualization of PBP2A bands with X-ray film exposure.
Nucleotide sequence accession numbers.
Data for the genome sequencing samples have been deposited in the European Nucleotide Archive under the sample numbers ERS157365, ERS157381, ERS157396, ERS157409, and ERS157425 to ERS157449.
ACKNOWLEDGMENTS
This work was supported by a grant from the U.S. Public Health Service 2 RO1 AI457838-14 and by grant UL1 TR000043-07S1 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program awarded to A. Tomasz. S.D.B. is partly supported by the NIHR Cambridge BRC. This work was also supported by the Wellcome Trust Sanger Institute core grant 098051.
Footnotes
Citation Dordel J, Kim C, Chung M, Pardos de la Gándara M, Holden MTJ, Parkhill J, de Lencastre H, Bentley SD, Tomasz A. 2014. Novel determinants of antibiotic resistance: identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. mBio 5(2):e01000-13. doi:10.1128/mBio.01000-13.
REFERENCES
- 1. Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- 2. Fey PD, Saïd-Salim B, Rupp ME, Hinrichs SH, Boxrud DJ, Davis CC, Kreiswirth BN, Schlievert PM. 2003. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:196–203. 10.1128/AAC.47.1.196-203.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, Johnson SK, Vandenesch F, Fridkin S, O’Boyle C, Danila RN, Lynfield R. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976–2984. 10.1001/jama.290.22.2976 [DOI] [PubMed] [Google Scholar]
- 4. Salgado CD, Farr BM, Calfee DP. 2003. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin. Infect. Dis. 36:131–139 [DOI] [PubMed] [Google Scholar]
- 5. King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. 2006. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA300 clone as the predominant cause of skin and soft tissue infections. Ann. Intern. Med. 144:309–317. 10.7326/0003-4819-144-5-200603070-00005 [DOI] [PubMed] [Google Scholar]
- 6. Beck WD, Berger-Bächi B, Kayser FH. 1986. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J. Bacteriol. 165:373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inglis B, Matthews PR, Stewart PR. 1988. The expression in Staphylococcus aureus of cloned DNA encoding methicillin resistance. J. Gen. Microbiol. 134:1465–1469 [DOI] [PubMed] [Google Scholar]
- 8. Ito T, Katayama Y, Hiramatsu K. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hartman BJ, Tomasz A. 1984. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fuda C, Suvorov M, Vakulenko SB, Mobashery S. 2004. The basis for resistance to β-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 279:40802–40806. 10.1074/jbc.M403589200 [DOI] [PubMed] [Google Scholar]
- 11. Hackbarth CJ, Chambers HF. 1993. blaI and blaR1 regulate beta-lactamase and PBP2a production in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 37:1144–1149. 10.1128/AAC.37.5.1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. 1992. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 298:133–136. 10.1016/0014-5793(92)80039-J [DOI] [PubMed] [Google Scholar]
- 13. Jevons MP. 1961. Celbenin-resistant staphylococci. BMJ 1:124–125. 10.1136/bmj.1.5219.124 [DOI] [Google Scholar]
- 14. Chung M, Antignac A, Kim C, Tomasz A. 2008. Comparative study of the susceptibilities of major epidemic clones of methicillin-resistant Staphylococcus aureus to oxacillin and to the new broad-spectrum cephalosporin ceftobiprole. Antimicrob. Agents Chemother. 52:2709–2717. 10.1128/AAC.00266-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tomasz A, Nachman S, Leaf H. 1991. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 35:124–129. 10.1128/AAC.35.1.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Lencastre H, Figueiredo AM, Tomasz A. 1993. Genetic control of population structure in heterogeneous strains of methicillin resistant Staphylococcus aureus. Eur. J Microbiol. Infect. Dis. 12(Suppl 1):S13–S18. 10.1007/BF02389872 [DOI] [PubMed] [Google Scholar]
- 17. de Lencastre H, Chung M, Westh H. 2000. Archaic strains of methicillin-resistant Staphylococcus aureus: molecular and microbiological properties of isolates from the 1960s in Denmark. Microb. Drug Resist. 6:1–10. 10.1089/mdr.2000.6.1 [DOI] [PubMed] [Google Scholar]
- 18. Berger-Bächi B. 1994. Expression of resistance to methicillin. Trends Microbiol. 2:389–393. 10.1016/0966-842X(94)90617-3 [DOI] [PubMed] [Google Scholar]
- 19. Rohrer S, Maki H, Berger-Bächi B. 2003. What makes resistance to methicillin heterogeneous? J. Med. Microbiol. 52:605–607. 10.1099/jmm.0.05176-0 [DOI] [PubMed] [Google Scholar]
- 20. Mwangi MM, Kim C, Chung M, Tsai J, Vijayadamodar G, Benitez M, Jarvie TP, Du L, Tomasz A. 2013. Whole-genome sequencing reveals a link between β-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb. Drug Resist. 19:153–159. 10.1089/mdr.2013.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim C, Mwangi M, Chung M, Milheirco C, de Lencastre H, Tomasz A. 2013. The mechanism of heterogeneous beta-lactam resistance in MRSA: key role of the stringent stress response. PLoS One 8:e82814. 10.1371/journal.pone.0082814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hughes J, Mellows G. 1978. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Escherichia coli by pseudomonic acid. Biochem. J. 176:305–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sutherland R, Boon RJ, Griffin KE, Masters PJ, Slocombe B, White AR. 1985. Antibacterial activity of mupirocin (pseudomonic acid), a new antibiotic for topical use. Antimicrob. Agents Chemother. 27:495–498. 10.1128/AAC.27.4.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cassels R, Oliva B, Knowles D. 1995. Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in staphylococci. J. Bacteriol. 177:5161–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 188:6739–6756. 10.1128/JB.00609-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whitehead KE, Webber GM, England RR. 1998. Accumulation of ppGpp in Streptococcus pyogenes and Streptococcus rattus following amino acid starvation. FEMS Microbiol. Lett. 159:21–26. 10.1111/j.1574-6968.1998.tb12836.x [DOI] [PubMed] [Google Scholar]
- 27. Crosse AM, Greenway DL, England RR. 2000. Accumulation of ppGpp and ppGp in Staphylococcus aureus 8325-4 following nutrient starvation. Lett. Appl. Microbiol. 31:332–337. 10.1046/j.1472-765x.2000.00822.x [DOI] [PubMed] [Google Scholar]
- 28. Nascimento MM, Lemos JA, Abranches J, Lin VK, Burne RA. 2008. Role of RelA of Streptococcus mutans in global control of gene expression. J. Bacteriol. 190:28–36. 10.1128/JB.01395-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan X, Zhao C, Budin-Verneuil A, Hartke A, Rincé A, Gilmore MS, Auffray Y, Pichereau V. 2009. The (p)ppGpp synthetase RelA contributes to stress adaptation and virulence in Enterococcus faecalis V583. Microbiology 155:3226–3237. 10.1099/mic.0.026146-0 [DOI] [PubMed] [Google Scholar]
- 30. Geiger T, Francois P, Liebeke M, Fraunholz M, Goerke C, Krismer B, Schrenzel J, Lalk M, Wolz C. 2012. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 8:e1003016. 10.1371/journal.ppat.1003016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crisóstomo MI, Westh H, Tomasz A, Chung M, Oliveira DC, de Lencastre H. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. U. S. A. 98:9865–9870. 10.1073/pnas.161272898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Srivatsan A, Wang JD. 2008. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11:100–105. 10.1016/j.mib.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 33. Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. 2008. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 190:1084–1096. 10.1128/JB.01092-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berger-Bächi B, Strässle A, Gustafson JE, Kayser FH. 1992. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 36:1367–1373. 10.1128/AAC.36.7.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Lencastre H, Tomasz A. 1994. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2590–2598. 10.1128/AAC.38.11.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Lencastre H, Wu SW, Pinho MG, Ludovice AM, Filipe S, Gardete S, Sobral R, Gill S, Chung M, Tomasz A. 1999. Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb. Drug Resist. 5:163–175. 10.1089/mdr.1999.5.163 [DOI] [PubMed] [Google Scholar]
- 37. de Lencastre H, Sá Figueiredo AM, Urban C, Rahal J, Tomasz A. 1991. Multiple mechanisms of methicillin resistance and improved methods for detection in clinical isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 35:632–639. 10.1128/AAC.35.4.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim C, Milheiriço C, Gardete S, Holmes MA, Holden MT, de Lencastre H, Tomasz A. 2012. Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the beta-lactam-resistant phenotype. J. Biol. Chem. 287:36854–36863. 10.1074/jbc.M112.395962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. 2008. A large genome center’s improvements to the Illumina sequencing system. Nat. Methods 5:1005–1010. 10.1038/nmeth.1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quail MA, Swerdlow H, Turner DJ. 2009. Improved protocols for the Illumina genome analyzer sequencing system. Curr. Protoc. Hum. Genet. Chapter 18:Unit 18.12. 10.1002/0471142905.hg1802s62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Quail MA, Otto TD, Gu Y, Harris SR, Skelly TF, McQuillan JA, Swerdlow HP, Oyola SO. 2012. Optimal enzymes for amplifying sequencing libraries. Nat. Methods 9:10–11. 10.1038/nmeth.1814 [DOI] [PubMed] [Google Scholar]
- 42. Simpson JT, Durbin R. 2012. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 22:549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Iqbal Z, Turner I, McVean G. 2013. High-throughput microbial population genomics using the Cortex variation assembler. Bioinformatics 29:275–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. 10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43:491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]