Abstract
To monitor the kinetics of biological processes that take place within the minute time scale, simple and fast analytical methods are required. In this article, we present our discovery of an azide with an internal Cu(I)-chelating motif that enabled the development of the fastest protocol for Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC) to date, and its application toward following the dynamic process of glycan biosynthesis. We discovered that an electron-donating picolyl azide boosted the efficiency of the ligand-accelerated CuAAC 20–38-fold in living systems with no apparent toxicity. With a combination of this azide and BTTPS, a tris(triazolylmethyl)amine-based ligand for Cu(I), we were able to detect newly synthesized cell-surface glycans by flow cytometry using as low as 1 nM of a metabolic precursor. This supersensitive chemistry enabled us to monitor the dynamic glycan biosynthesis in mammalian cells and in early zebrafish embryogenesis. In live mammalian cells, we discovered that it takes approximately 30–45 min for a monosaccharide building block to be metabolized and incorporated into cell-surface glycoconjugates. In zebrafish embryos, the labeled glycans could be detected as early as the two-cell stage. To our knowledge, this was the first time that newly synthesized glycans were detected at the cleavage period (0.75–2 hpf) in an animal model using bioorthogonal chemistry.
Introduction
The glycome, the complete set of glycans produced by a cell, is a dynamic indicator of the cell’s physiology. Changes in the glycome reflect the changes in the cell’s developmental stages and transformation state of the cell.1 It has been heavily documented that aberrant glycosylation patterns, including both the under- and overexpression of naturally occurring glycans, as well as neoexpression of glycans normally restricted to embryonic tissues, are a hallmark of the tumor phenotype.2,3 The ability to visualize and monitor these changes in cells and in tissue samples would advance our understanding of the detailed roles of glycans in these diseases and provide new diagnostic tools for their treatment.
Monitoring the kinetics of glycan biosynthesis and recycling has been an attractive topic for glycobiologists over many years.4−7 Previous studies on this subject relied heavily on metabolic labeling with radiolabeled monosaccharides to follow the turnover of cell-surface glycoconjugates. This method is cumbersome and requires lengthy detection periods (1–2 days).7 To accurately monitor dynamic synthesis of glycoconjugates, more sensitive and efficient methods are required.
The nonradioactive detection of glycans has recently been enabled using a bioorthogonal chemical reporter strategy.8 Using this methodology, cells or organisms are first treated with a monosaccharide building block bearing a chemically reactive tag. The modified monosaccharide, when taken up by cells and metabolized, is incorporated into cell-surface glycoconjugates. The bioorthogonal chemical tag then allows covalent conjugation with fluorescent probes for visualization and analysis. The two most popular bioorthogonal reactions to date are the Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC)9,10 and strain-promoted copper-free click chemistry,11,12 the former being 10–100 times faster than the latter in aqueous solutions.13,14 CuAAC is a ligand accelerated process—ligands that stabilize the Cu(I) oxidation state in aqueous solutions can dramatically speed up this reaction.15−17 As discovered by Fokin and co-workers, the Cu(I)-catalyzed cycloaddition is initiated by the formation of a Cu(I) acetylide intermediate, which is then followed by the approach of a second Cu(I) to generate a dinuclear copper intermediate a (Scheme 1A).18 Based on this mechanistic rationale, organic azides bearing a Cu(I)-chelating motif could facilitate the coordination of the second Cu(I) species and further accelerate the CuAAC reaction. Indeed, pioneering work done by the Zhu19−22 and the Ting23 laboratories have showed that 2-(azidomethyl)pyridine derivatives could accelerate the CuAAC reaction 4–6-fold in the presence of a tris(triazolylmethyl)amine-based ligand compared to reactions using nonchelating azides in in vitro model systems. Ting et al. further showed that one of such 2-(azidomethyl)pyridine derivatives can be processed by an engineered Escherichia coli lipoic acid ligase for site-specific labeling of membrane proteins via CuAAC; they also showed that the same azide can be conjugated to Alexa Fluor 647 to label RNA molecules in fixed cells.23 To our knowledge, however, this chelating azide-assisted CuAAC has never been utilized to study other biological processes such as post-translational modification; the detection of post-translationally modified proteins in cellular systems represents one of the most exciting and powerful applications of bioorthogonal chemistry.
Scheme 1. Organic Azides with a Cu(I)-Chelating Motif Can Further Accelerate the Ligand-Assisted CuAAC.
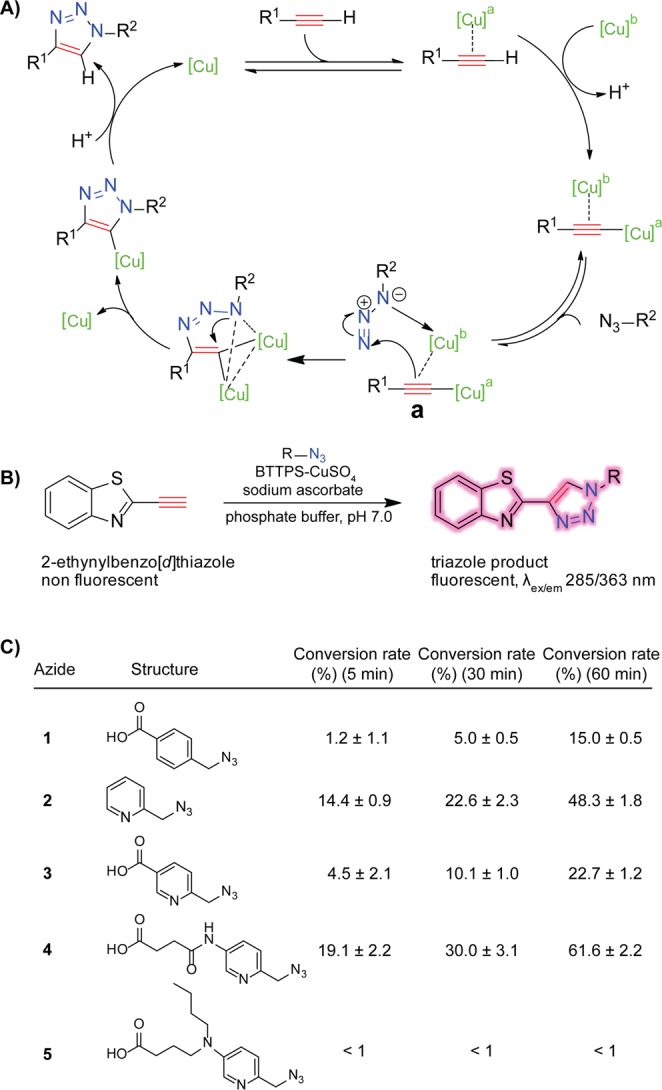
(A) Mechanistic rationale of CuAAC. (B) Fluorogenic assay for the qualitative measurement of the kinetics of CuAAC. Reaction conditions: potassium phosphate buffer (100 mM, pH 7.0), 2-ethynylbenzo[d]thiazole (50 μM), azide (25 μM), Cu-BTTPS (25 μM: 50 μM), sodium ascorbate (2.5 mM). (C) Triazole conversion at various time points.
In the work presented here, we examined a small library of azide probes bearing an internal chelating motif to identify the probe with the best kinetic behavior. By combining this new probe with the Cu(I)-stabilizing ligand 3-[4-({bis[(1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl]amino}methyl)-1H-1,2,3-triazol-1-yl]propyl hydrogen sulfate (BTTPS), and 3-(4-((bis((1-tert-butyl)-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl)-propan-1-ol (BTTP), the unsulfated version of BTTPS,24 we developed the fastest protocol of CuAAC to date. Using this protocol, we evaluated the detection limit of CuAAC semiquantitatively for labeling a single-alkyne containing protein in crude cell lysates and for labeling glycans on the cell surface for the first time. We discovered that this new protocol is at least 5-fold more sensitive than the one using a nonchelating azide in a Western blot-based detection; as low as 30 ng of protein could be detected in the new reaction system. Furthermore, a combination of the chelating azide and BTTPS afforded 20–38-fold enhancements in fluorescent labeling of alkyne-tagged glycans on the cell surface compared to that provided by using BTTPS alone. The new probe enabled us to monitor the dynamic glycan biosynthesis in mammalian cells and in zebrafish early embryogenesis. Our results revealed that cellular uptake and conversion of a monosaccharide building block into cell-surface glycoconjugates takes place within minutes.
Results and Discussion
Evaluating Rate Acceleration of the Chelating Azide-Assisted CuAAC Using a Fluorogenic Assay
Studies by the Zhu and the Ting laboratories showed that an azide with an internal chelating motif, e.g., picolyl azide 3, can accelerate CuAAC substantially. Inspired by these precedents, we synthesized five picolyl azide analogues and evaluated how electron-donating groups introduced to the pyridine ring impact the rate acceleration of the cycloaddition reaction. We monitored the progress of CuAAC using a fluorogenic alkyne—2-ethynylbenzo[d]thiazole, whose fluorescence is activated upon the cycloaddition reaction (Scheme 1B).25 The cycloaddition reaction was performed in aqueous solution in the presence of BTTPS.24 Our previous work showed that when coordinating with the in situ generated Cu(I), BTTPS and its analogue 2-[4-({bis[(1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl]amino}methyl)-1H-1,2,3-triazol-1-yl]acetic acid (BTTAA)26 provided the fastest and most biocompatible catalytic systems for CuAAC-mediated bioconjugation in live cells and living organisms. Under the reaction conditions specified in Scheme 1B, picolyl azide 4 exhibited the fastest kinetics and generated the desired triazole product with a 4-fold and nearly 3-fold higher yield during a 5- and 60-min reaction course, respectively, as compared to that of the azide 3 reported by Ting and co-workers (Scheme 1C).23 Interestingly, no rate acceleration was observed when azide 5 was used as the cycloaddition partner, presumably due to protonation of the tertiary amine under neutral pH conditions, converting it from an electron-donating group into an electron-withdrawing motif (Figure S2, Supporting Information).
Evaluating the Sensitivity of Chelating Azide-Assisted CuAAC in the Detection of a Single Alkyne-Tagged Protein in Cell Lysates
With the best performing azide identified, we first evaluated its activity toward the detection of a single alkyne-tagged protein in a complex protein mixture. We constructed an alkyne-tagged bovine serum albumin (BSA-alkyne), in which a single terminal alkyne group was introduced into cysteine34.27 The BSA-alkyne was mixed with the lysates of Jurkat cells, a human T lymphocyte cell line, in various weight ratios. The protein mixtures were then reacted with the conventional biotin azide probe 6 (Click Chemistry Tools, Cat. No. AZ104-25), or the new biotin azide probe 7, derivatized from the picolyl azide 4, in the presence of BTTP, and in situ generated Cu(I) ([BTTP]:[Cu] = 2:1), the catalytic system designed for the CuAAC-mediated bioconjugation in crude cell lysates for proteomics analysis.24 After a 1 h reaction, the protein mixtures were resolved by SDS polyacrylamide gel electrophoresis (SDS-PAGE) and probed with an anti-biotin horseradish peroxidase (HRP). As quantified by ImageJ, biotin azide 7 afforded 2-fold stronger signal than biotin azide 6, when 200 ng BSA-alkyne was presented in the cell lysates (Lane 3 vs Lane 2; Figure 1B). It was also capable of detecting low levels of dimeric BSA present in the protein mixtures, which was beyond the detection limit of the conventional biotin probe 6 (BSA dimerization is irreversible between pH 4.2 and 7.0 even after treatment with DTT).28 When only 50 ng of BSA-alkyne was included in the protein mixture, biotin azide probe 6 was not capable of detecting its presence, whereas biotin azide 7 still provided a robust signal (Lane 5 vs Lane 6; Figure 1B). As shown in Figure 1C, we discovered that biotin azide 7 was capable of detecting BSA-alkyne as low as 30 ng, whereas the detection limit of biotin azide probe 6 is approximately 150 ng (Figure S3, Supporting Information). To our knowledge, this study represents the first rigorous characterization of the detection limit of CuAAC in a complex protein mixture, paving the way for using this powerful chemistry to quantitatively analyze the dynamics of glycan biosynthesis.
Figure 1.
Comparison of the efficiency of biotin azide probes functionalized with and without a Cu(I)-chelating motif for labeling a BSA-alkyne in a mixture of cell lysates. (A) Chemical structures of biotin azide 6 and 7. (B) Comparison of the efficiency labeling the BSA-alkyne in cell lysates using biotin azide 6 and 7. (C) Evaluation of the detection limit of biotin azide 7 for labeling the BSA-alkyne in cell lysates.
Monitoring the Dynamic Glycan Synthesis in Live Cells
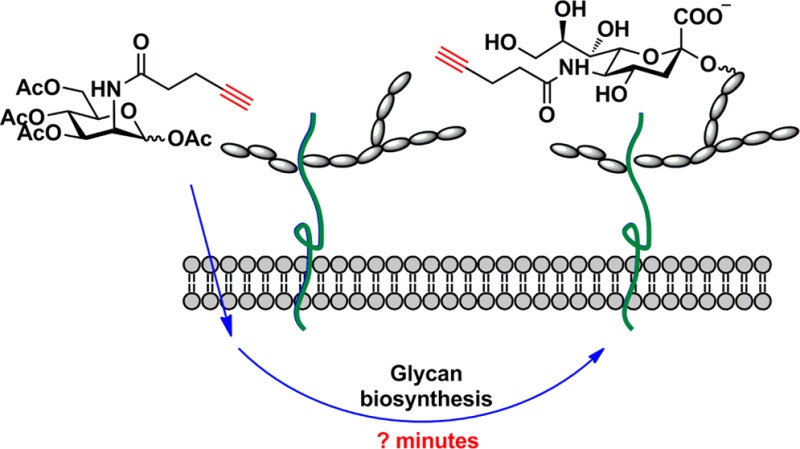
After the sensitivity of the new biotin probe was verified, we decided to explore its application toward monitoring dynamic glycan biosynthesis in cultured mammalian cells. The first question we chose to address was: how long does it take for a monosaccharide to be imported and incorporated into cell-surface glycoconjugates?
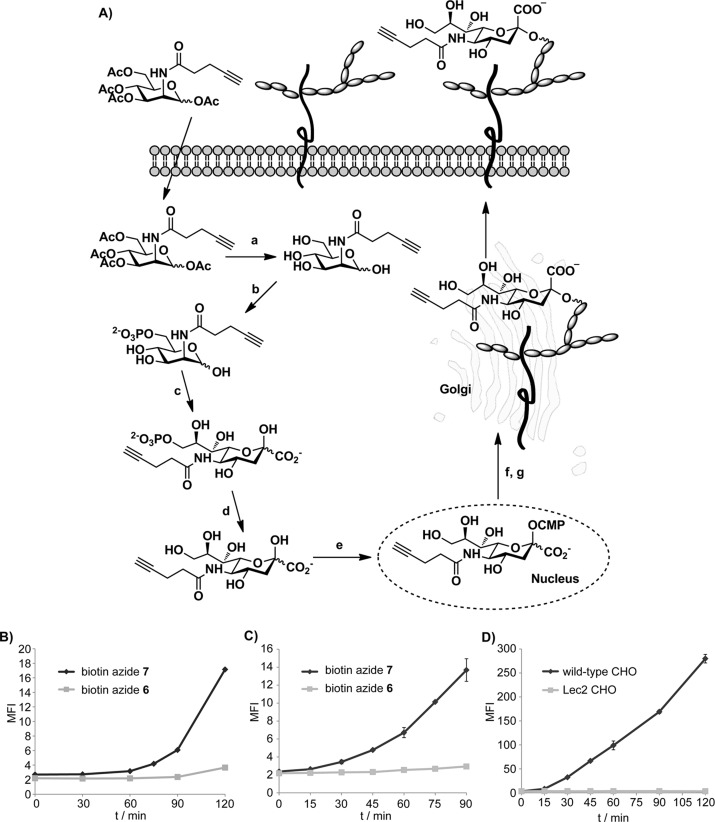
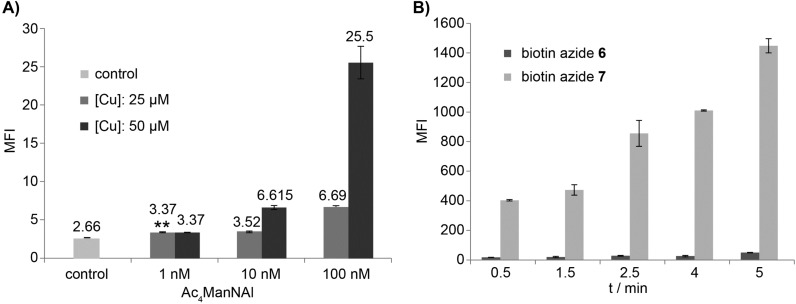
Toward this goal, we cultured Jurkat cells in media supplemented with peracetylated N-(4-pentynoyl) mannosamine (Ac4ManNAl), an alkyne-tagged metabolic precursor of sialic acid (Figure 2A).29−31 Peracetylation enhances the cellular uptake of the unnatural sugar.32,33 Once entering the cytoplasm, the ester groups in Ac4ManNAl are hydrolyzed by nonspecific esterases to release the free sugar that enters the sialic acid biosynthetic pathway, which allows for the metabolic incorporation of the alkyne tag into membrane sialylated glycoconjugates for their detection via CuAAC. A recent study from Lavis’s group showed that cytosolic esterases have high activity toward straight-chain esters and the hydrolyzed products of those esters can be detected within 30 min in different cell lines tested.34 However, it remained to be discovered how fast the unprotected unnatural precursor could be converted into the corresponding sialic acid analogue and integrated into cell-surface glycans. To answer this question, we first determined the sensitivity of the new biotin azide probe 7 in the detection of cell-surface alkyne-tagged sialic acids (SiaNAl) introduced by metabolic labeling (Figures S4–5, Supporting Information). We cultured Jurkat cells in media with various doses of Ac4ManNAl for 21 h before reacting with biotin-azide 7 in the presence of the BTTPS-Cu(I) catalyst ([BTTPS]:[Cu] = 5:1), the catalytic system designed for live cell bioconjugation. The treated cells were then probed with an Alexa Fluor 488-conjugated streptavidin (streptavidin-488) and analyzed by flow cytometry. As shown in Figure 3A, as low as 1 nM Ac4ManNAl could yield a detectable signal after a 21 h incubation, even at 25 μM Cu(I) concentration. In addition, similar experiments were performed to compare the efficiency of biotin-azide 7 with that of biotin-azide 6. In these experiments, cells were incubated with Ac4ManNAl (50 μM) for 72 h before reacting with 6 and 7 for 30 s to 5 min in the presence of the BTTPS-Cu(I) catalyst. We observed 20–38-fold enhancements in cell-associated fluorescent signal when biotin-azide 7 was used vs biotin-azide 6 (Figure 3B). Thus, the new biotin 7 is a significantly superior probe for the detection of alkyne-tagged glycans in live cells.
Figure 2.
Monitoring the kinetics of sialic acid biosynthesis in mammalian cell lines. (A) Sialic acid biosynthetic pathway. Metabolic conversion of Ac4ManNAl into a cell surface sialoside proceeds by the sequential action of nonspecific esterases (a), ManNAc 6-kinase (b), sialic acid 9-phosphate synthase (c), sialic acid 9-phosphatase (d), CMP-sialic acid synthetase (e), CMP-sialic acid Golgi transporter (f), and sialyltransferases (g). Monitoring the kinetics of sialic acid biosynthesis in Jurkat cells using (B) Ac4ManNAl (50 μM) and (C) SiaNAl (1.5 mM) as the metabolic precursor. (D) Monitoring the kinetics of sialic acid biosynthesis in CHO cells using SiaNAl (2 mM) as the metabolic precursor. Data shown in (D) were obtained using biotin azide 7.
Figure 3.
Sensitivity of biotin azide 7 for labeling alkyne-tagged glycans on the cell-surface of live cells. (A) Evaluation of the detection limit of biotin azide 7 for labeling the metabolically incorporated SiaNAl on the cell-surface of live Jurkat cells. Jurkat cells were incubated in RPMI medium containing 50 μM Ac4ManNAl for 21 h. The treated cells were reacted with biotin azide 7, probed by streptavidin-488, and the cell-associated MFI was quantified by flow cytometry. The two asterisks indicate a statistically significant difference from control group (p-value <0.01). (B) Comparison of the reactivity of biotin azides 6 and 7 in the labeling of metabolically incorporated SiaNAl on the cell-surface of live Jurkat cells. Jurkat cells were incubated in RPMI medium containing 50 μM Ac4ManNAl for 72 h. The treated cells were reacted with biotin azides 6 or 7 for 30 s to 5 min, probed by streptavidin-488, and the cell-associated MFI was quantified by flow cytometry.
To monitor the conversion of Ac4ManNAl into the corresponding sialic acid analogue and its integration into cell-surface glycans, we incubated Jurkat cells with 50 μM Ac4ManNAl. At various time intervals, we washed the cells, and reacted them with biotin azide probes 6 and 7 in the presence of the BTTPS-Cu(I) catalyst. After a 5-min reaction, we quenched the reaction with bathocuproine disulfonate (BCS), incubated the cells with streptavidin-488, and then quantified the mean fluorescence intensity (MFI) of the treated cells using flow cytometry analysis. Strikingly, with biotin azide 7 we were able to detect an increase in cell-associated fluorescence as early as 75 min after the cells were treated with Ac4ManNAl. By contrast, no increase in fluorescence could be detected until 45 min later when biotin azide 6 was used (Figure 2B). Because 30 min is estimated for a peracylated molecule to diffuse across the plasma membrane and its ester protecting groups to be hydrolyzed by nonspecific esterases, our results showed that it takes approximately another 45 min to convert the liberated ManNAl into the alkyne-bearing sialic acid and to incorporate it into cell surface glycans.
To confirm this result, we treated Jurkat cells with N-pentynoyl-neuraminic acid (SiaNAl) directly as the metabolic precursor to determine if similar or less time was required for the cells to generate an alkyne-dependent fluorescent signal. Previous studies showed that when sialic acid or its analogues were used directly as metabolic precursor, millimolar concentrations were required to achieve robust incorporation and metabolic remodeling of cell-surface sialylated glycoconjugates.35 Accordingly, we incubated Jurkat cells with 1.5 mM of SiaNAl. At various time intervals, we repeated the click labeling protocol and analyzed the MFI of the treated cells using flow cytometry. As shown in Figure 2C, the alkyne-dependent fluorescent signal could be detected at as early as 30 min after the cells were treated with the unnatural sugar, and the fluorescence intensity increased gradually over the time increments.
To validate that the observed increase in cell-associated fluorescence was indeed generated by the metabolically incorporated alkyne sugar, we repeated the same experiments using the Lec2 Chinese hamster ovary (CHO) cell mutant. The Lec2 mutant has a defect in its CMP-sialic acid Golgi transporter; thus, it does not produce any sialylated species on its cell surface.36 In control experiments, wild-type CHO cells with the normal sialic acid biosynthetic machinery were used. Analysis of the treated cells by flow cytometry revealed an increase in MFI for the wild-type CHO cells at 30 min after the cells were subjected to SiaNAl. By contrast, Lec2 mutants only showed background fluorescence even after a 2 h incubation (Figure 2D). Taken together, these results confirmed that the cell-associated fluorescence was produced by the metabolically incorporated alkyne-bearing SiaNAl.
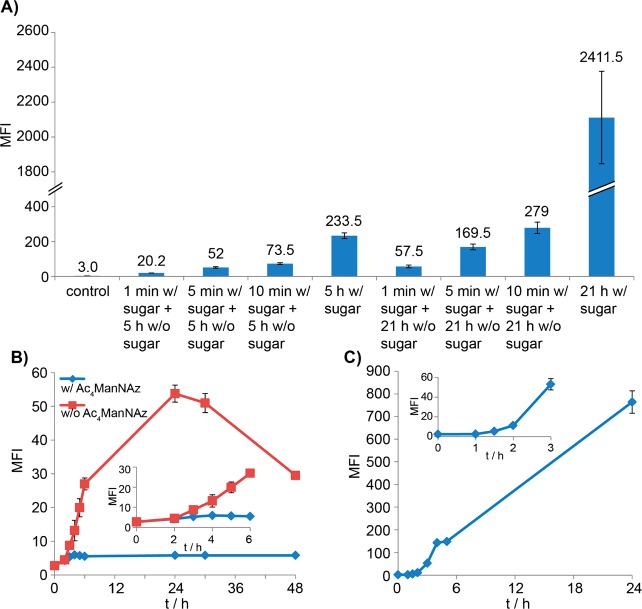
To determine the minimum amount of time required for cells to be subjected to a metabolic precursor to produce modified cell-surface glycans that could be detected by click chemistry, we performed a pulse-chase experiment, in which Jurkat cells were treated with Ac4ManNAl (50 μM) for 1, 5, or 10 min before cells were washed and transferred to the culture medium without the unnatural sugar. At 21 h post treatment with Ac4ManNAl, we performed the CuAAC-mediated biotin labeling and analyzed the MFI of the treated cells using flow cytometry. A significant increase in cell-associated fluorescence could be detected even for cells that were only subjected to Ac4ManNAl for 1 min (Figure 4A). Through analysis of this experimental group at various time intervals, we discovered that the alkyne-dependent fluorescent signal could be detected as early as 3 h and the fluorescent intensity reached the maximum the latest at 24 h post treatment (Figure 4B, red line). Next, we performed a pulse-chase experiment, in which Jurkat cells were pulse labeled with Ac4ManNAl for 1 min, then chased with the azide-containing ManNAc analogue, peraceylated N-azidoacetyl mannosamine (Ac4ManNAz) (50 μM). As shown in the blue line of Figure 4B, no significant increase in alkyne-dependent florescence could be detected as assayed by flow cytometry at various time points post the Ac4ManNAz treatment. Nevertheless, an azide-dependent fluorescence was observed as early as 90 min after the cells were switched to the Ac4ManNAz-supplemented media (Figure 4C). These findings suggest that the concentration of the endogenous sialic acid precursor is so low that it can be readily perturbed using exogenously supplemented analogs—even a 1 min treatment can produce a dramatic effect to remodel the cell-surface sialylated glycans.
Figure 4.
Monitoring the cellular uptake and metabolism of peracetylated ManNAc derivatives in Jurkat cells. (A) Monitoring the cellular uptake and metabolism of Ac4ManNAl in Jurkat cells by treating cells with 50 μM Ac4ManNAl for 1, 5, or 10 min, followed by growing the cells in Ac4ManNAl-free medium for 5 or 21 h. (B) Monitoring the cellular uptake and metabolism of peracetylated ManNAc derivatives in Jurkat cells by a pulse-chase labeling. Pulse, Ac4ManNAl (50 μM, 1 min); Chase, Ac4ManNAl-free medium with or without the addition of Ac4ManNAz. The red curve represents the alkyne-dependent MFI at various time intervals when chasing with an unnatural sugar-free medium. The blue curve represents the alkyne-dependent MFI at various time intervals when chasing with 50 μM Ac4ManNAz. (C) Monitoring the cellular uptake and metabolism of Ac4ManNAz in Jurkat cells that are treated with Ac4ManNAl (50 μM, 1 min) followed by Ac4ManNAz (50 μM) treatment. The curve shows azide-dependent fluorescence at various time intervals. Data shown in (A) and (B) were obtained using biotin azide 7.
Imaging Fucosylated Glycans in Zebrafish Early Embryogenesis
Finally, we monitored the kinetics of glycan biosynthesis in living organisms by using the zebrafish embryo as a model system, taking advantage of its transparency and external development. Previously, we demonstrated that microinjection of an alkyne-tagged GDP-fucose analogue, GDP-L-6-ethynylfucose (GDP-FucAl),37 into the yolk of zebrafish embryos at the one-cell stage allowed the nucleotide sugar to disperse into daughter cells and to be incorporated into cell-surface fucosylated glycoconjugates. Using the biocompatible CuAAC, the alkyne-tagged glycans could be detected as early as the blastula period (2.5 h post fertilization (hpf)).17 With the new, rate-accelerating azide in hand, we hypothesized that the newly synthesized glycans could be detected at earlier developmental stages.
Toward this end, we microinjected zebrafish embryos with GDP-FucAl at the one-cell stage. Following the synchronous cell divisions at the 2-, 4-, and 8-cell stages, which take place every 15–20 min and during which new membranes are delivered preferentially to the cleavage furrows, we fixed the embryos with paraformaldehyde—the fragile nature of early stage embryos prevented their direct treatment with the copper catalyst and extensive washing. We then reacted the treated embryos with azide 4 conjugated Alexa Fluor 488 (8) in the presence of the BTTPS-Cu(I) catalyst, which allowed us to visualize the newly synthesized glycans via fluorescence microscopy. Fluorescently labeled fucosides could be detected as early as the 2-cell stage with the most intense staining pattern observed at the newly formed membrane junctions (Figure 5A). When the embryos underwent further division to enter the 4-cell stage, the newly formed membrane junctions marked by the labeled fucosides emerged perpendicularly to the membrane formed at the 2-cell stage (Figure 5B). This distribution reflects the sites where new cell surface is secreted to promote cell cohesion during early embryogenesis.38 In a control experiment, we incubated embryos in medium supplemented with the AurB inhibitor ZM2 immediately after microinjection with GDP-FucAl. Consistent with a previous report, ZM2 caused a delay or a complete failure in furrow formation (Figure 5D).38 Upon labeling, no 488 fluorescence was observed on the cell-surface at 2 hpf; detectable fluorescence only accumulated in the cytoplasm as revealed by confocal microscopy analysis. To our knowledge, this was the first time that the newly synthesized glycans were detected at the cleavage period (0.75–2 hpf) using bioorthogonal chemistry. Our observation suggests that glycan addition likely contributes to the mechanisms allowing these otherwise nonadhesive cells to cohere following cell division.
Figure 5.
Monitoring the kinetics of fucoside biosynthesis in zebrafish embryos. Zebrafish embryos were microinjected with GDP-FucAl at the one-cell stage. At (A) 2-, (B) 4-, (C) 8-cell stages, and (D) 2 hpf of ZM2 treated embryos, embryos were fixed and reacted with Alexa Fluor 488 azide 8, and imaged using confocal microscopy. First row, side views of live embryos at time points equivalent to 2-, 4-, and 8-cell stages and 2 hpf of ZM2 treated embryos. Rows 2–4: animal pole views of fixed embryos labeled with 488. Nuclei were stained with Hoechst 33342.
Conclusion
In summary, by examining a small library of azides bearing an internal Cu(I)-chelating motif, we have identified a picolyl azide derivative that, when combined with the BTTPS-Cu(I) or BTTP-Cu(I) catalyst, provides the fastest reaction kinetics for a CuAAC to date. We discovered that this new protocol is at least 5-fold more sensitive than the one using the conventional, nonchelating azide and BTTP-Cu(I) in detecting a single-alkyne containing protein in crude cell lysates by Western blot. Therefore, the accelerating azide probe, as well as the new CuAAC protocol, represents an important contribution to the chemical biologist’s toolbox, which will permit enrichment of proteins with nanogram abundance for the analysis of their expression changes in a dynamic process or their ligand binding.
With this supersensitive reaction, we were able to follow the kinetics of sialic acid biosynthesis in cultured cells using as low as 1 nM metabolic precursor. We discovered that it takes approximately 30 and 45 min, respectively, for the alkyne-bearing SiaNAl and ManNAl to be converted into cell-surface sialylated glycoconjugates. Using zebrafish embryos as a vertebrate model, we discovered that the labeled fucosides could be observed as early as the 2-cell stage, and the newly synthesized glycans accumulate in the cell membrane junctions where new membranes are secreted. Currently, we are exploring if the optimized CuAAC protocol could be extended to track the early stages of glycan biosynthesis in the Golgi apparatus and to label glycoproteins in mice models.
Experimental Section
Chemical Synthesis
Detailed methods and characterization can be found in the Supporting Information.
Comparison of the Efficiency of Biotin Azide 7 and Biotin Azide 6 for Labeling BSA-Alkyne in a Mixture of Cell Lysates (Figure 1)
BSA-alkyne was synthesized by reacting BSA (100 μM) with acetylene-PEG4-maleimide (Click Chemistry Tools, Cat. No. TA104–25) (2 mM) in PBS overnight at 4 °C. Excess acetylene-PEG4-maleimide was removed by dialysis against PBS using a 10 kD molecular weight cutoff cellulose membrane.
Jurkat cell lysates were prepared by lysing 10 million cells in 200 μL lysis buffer containing 1% NP-40, 100 mM sodium phosphate (pH 7.5), 150 mM NaCl, and Roche protease inhibitor cocktail (EDTA free). Cell lysates were fast-frozen and thawed five times, and centrifuged at 15 000 g for 10 min at 4 °C. Supernatants were transferred to a new tube.
BSA-alkyne (200 ng, 50 ng, 40 ng, or 30 ng) was mixed with Jurkat cell lysate (20 μg). To the protein mixtures were added biotin azide 7 or biotin azide 6 (100 μM), a mixture of the BTTP-CuSO4 catalyst ([BTTP]:[CuSO4] = 2:1, [CuSO4] = 250 μM) and sodium ascorbate (2.5 mM) in a total volume of 20 μL. After incubating at 25 °C for 1 h, the reaction mixtures were resolved by SDS-PAGE. The samples were transferred to nitrocellulose, and incubated for 1 h at room temperature in blocking buffer (5% nonfat milk in TBST (Tris buffered saline with 0.1% Tween-20, pH 7.5)). The blocked membrane was incubated for 1 h at room temperature with an HRP–anti-biotin antibody (1:100 000 dilution) in blocking buffer, washed with 1× TBST (3×, 15 min/wash) and developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce). X-OMAT LS film (Kodak) was used to detect the chemiluminescence.
Detection of Cell Surface Glycans by Metabolic Labeling and Flow Cytometry Analysis (Figures 2, 3, and 4)
For the experiments of monitoring the sialic acid biosynthesis in live cells (Figure 2), Jurkat cells were seeded at 0.5 million/mL in untreated RPMI or RPMI medium containing 50 μM Ac4ManNAl or 1.5 mM SiaNAl for different time periods. CHO cells (wild-type and Lec2, seeded at 0.5 million/mL) were cultured in suspension in untreated α-Minimum Essential medium or medium containing 2 mM SiaNAl for different time periods. For the experiments monitoring the sensitivity of biotin azide 7 for labeling alkyne-tagged glycans in live cells (Figure 3A), Jurkat cells (seeded at 0.5 million/mL) were incubated in untreated RPMI or RPMI medium containing various concentrations of Ac4ManNAl for 21 h. For the CuAAC labeling efficiency evaluation experiment (Figure 3B), Jurkat cells were seeded at 0.15 million/mL and incubated for 3 days in untreated RPMI or RPMI medium containing 50 μM Ac4ManNAl. For the pulse-chase experiments of tracking alkyne-modified or azide-modified sialic acid expression on the surface of live cells (Figure 4), Jurkat cells (seeded at 0.5 million/mL) were incubated in 1 mL RPMI medium containing 50 μM Ac4ManNAl for various times. Then the cells were pelleted, washed 2× with RPMI medium, and resuspended in 2 mL untreated RPMI medium or RPMI medium containing 50 μM Ac4ManNAz, and allowed to grow for various times.
The metabolically labeled cells were harvested and washed 2× with labeling buffer (PBS, pH 7.4, 1% FBS), and were transferred into a 96-well round-bottom tissue culture plate (0.4 million cells in 90 μL/well) (Corning Inc.). To each well were added the following reagents in order: 50 μM biotin azide 7 or biotin azide 6 or biotin-PEG4-alkyne (Click Chemistry Tools, Cat. No. TA105–25), the BTTPS-CuSO4 complex ([BTTPS]:[CuSO4] = 5:1, [CuSO4] = 50 μM), and 2.5 mM sodium ascorbate. After reaction for 5 min (Figures 2, 3A, and 4) at room temperature, the reactions were quenched with 1 mM BCS. The cells were pelleted, washed 3× with 200 μL labeling buffer, and resuspended in the same buffer containing 1 μg/mL streptavidin-488 (Invitrogen). The plate was covered with aluminum foil and incubated at 4 °C for 30 min. The cells were then washed 3× with 200 μL labeling buffer and resuspended in 250 μL cold FACS buffer (Hank’s Balanced Salt Solution, pH 7.4, 1% FBS, 2 μg/mL 7-AAD, 0.2% NaN3) for flow cytometry analysis using an Eclipse iCyt flow cytometer. Similar results were obtained by using the BTTAA-Cu(I) catalyst (data not shown).
Metabolic Labeling of Fucosylated Glycans in Zebrafish Embryos by Microinjection with GDP-FucAl and Visualization via Confocal Microscopy (Figure 5)
Zebrafish embryos at the one-cell stage were collected, followed by microinjection of 1 nL of a 40 mM solution of GDP-FucAl and rhodamine-dextran (5% w/v) as a tracer in 0.2 M KCl. The embryos without microinjection were kept as a negative control group. The embryos were then cultured in E3 embryo medium at 28 °C. When embryos reached 2-cell, 4-cell, and 8-cell stages, they were transferred to a microcentrifuge tube containing 0.5 mL E3 embryo medium using a fire-polished glass Pasteur pipet. 0.5 mL of 8% paraformaldehyde (PFA) was then added to the tube immediately to fix the embryos overnight at 4 °C. In the ZM2 treated group, embryos were collected and treated with ZM2 solution as previously described.38
Before the CuAAC reactions, the fixed embryos were transferred to PBST and dechorionated by hand with forceps, followed by washing 4× with PBST. 92 μL PBST was added to each well of a 1% agarose-coated 96-well plate, followed by addition of Alexa Fluor 488 azide 8 (50 μM), BTTPS-CuSO4 complex ([BTTPS]:[CuSO4] = 2:1, [CuSO4] = 150 μM). Embryos were then transferred into these wells with less than five embryos per well. The solutions were gently shaken, and freshly prepared sodium ascorbate (5 mM) was added to initiate the CuAAC. After 15 min, the reaction was quenched with 1 mM BCS and diluted immediately with 100 μL PBST. The treated embryos were washed 4× with PBST, incubated in Hoechst 33342 solution for 10 min, followed by mounting on 35 mm glass bottom dishes (MatTek) in 3% methyl cellulose. All embryo images were acquired by Leica confocal microscopy SP5, and composite figures were prepared using ImageJ.
Acknowledgments
This work was supported partially by a DuPont Young Professor Award and the National Institutes of Health to P.W. (R01GM093282) and F.L.M. (R01GM089979).
Supporting Information Available
Synthesis details, experimental details, and supporting figures and illustrations. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
Hao Jiang and Tianqing Zheng contributed equally to this work.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Haltiwanger R. S.; Lowe J. B. (2004) Role of glycosylation in development. Annu. Rev. Biochem. 73, 491–537. [DOI] [PubMed] [Google Scholar]
- Hakomori S. (1989) Aberrant glycosylation in tumors and tumor-associated carbohydrate antigens. Adv. Cancer Res. 52, 257–331. [DOI] [PubMed] [Google Scholar]
- Dube D. H.; Bertozzi C. R. (2005) Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat. Rev. Drug Discovery 4, 477–88. [DOI] [PubMed] [Google Scholar]
- Fishman J. B.; Cook J. S. (1986) The sequential transfer of internalized, cell surface sialoglycoconjugates through the lysosomes and Golgi complex in HeLa cells. J. Biol. Chem. 261, 11896–905. [PubMed] [Google Scholar]
- Reichner J. S.; Whiteheart S. W.; Hart G. W. (1988) Intracellular trafficking of cell surface sialoglycoconjugates. J. Biol. Chem. 263, 16316–26. [PubMed] [Google Scholar]
- Huang K. M.; Snider M. D. (1993) Glycoprotein recycling to the galactosyltransferase compartment of the Golgi complex. J. Biol. Chem. 268, 9302–10. [PubMed] [Google Scholar]
- Pakula T. M.; Uusitalo J.; Saloheimo M.; Salonen K.; Aarts R. J.; Penttila M. (2000) Monitoring the kinetics of glycoprotein synthesis and secretion in the filamentous fungus Trichoderma reesei: cellobiohydrolase I (CBHI) as a model protein. Microbiology 146(Pt 1), 223–32. [DOI] [PubMed] [Google Scholar]
- Laughlin S. T.; Bertozzi C. R. (2009) Imaging the glycome. Proc. Natl. Acad. Sci. U.S.A. 106, 12–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. (2002) A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective ″ligation″ of azides and terminal alkynes. Angew. Chem., Int. Ed. 41, 2596–9. [DOI] [PubMed] [Google Scholar]
- Tornoe C. W.; Christensen C.; Meldal M. (2002) Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 67, 3057–64. [DOI] [PubMed] [Google Scholar]
- Baskin J. M.; Prescher J. A.; Laughlin S. T.; Agard N. J.; Chang P. V.; Miller I. A.; Lo A.; Codelli J. A.; Bertozzi C. R. (2007) Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. U.S.A. 104, 16793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning X.; Guo J.; Wolfert M. A.; Boons G. J. (2008) Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions. Angew. Chem., Int. Ed. 47, 2253–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett J. C.; Sletten E. M.; Bertozzi C. R. (2010) Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 132, 3688–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presolski S. I.; Hong V.; Cho S. H.; Finn M. G. (2010) Tailored ligand acceleration of the Cu-catalyzed azide-alkyne cycloaddition reaction: practical and mechanistic implications. J. Am. Chem. Soc. 132, 14570–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan T. R.; Hilgraf R.; Sharpless K. B.; Fokin V. V. (2004) Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org. Lett. 6, 2853–5. [DOI] [PubMed] [Google Scholar]
- Hong V.; Presolski S. I.; Ma C.; Finn M. G. (2009) Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew. Chem., Int. Ed. 48, 9879–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano del Amo D.; Wang W.; Jiang H.; Besanceney C.; Yan A.; Levy M.; Liu Y.; Marlow F. L.; Wu P. (2010) Biocompatible copper(I) catalysts for in vivo imaging of glycans. J. Am. Chem. Soc. 132, 16893–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrell B. T.; Malik J. A.; Fokin V. V. (2013) Direct evidence of a dinuclear copper intermediate in Cu(I)-catalyzed azide-alkyne cycloadditions. Science 340, 457–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherton W. S.; Michaels H. A.; Simmons J. T.; Clark R. J.; Dalal N. S.; Zhu L. (2009) Apparent copper(II)-accelerated azide–alkyne cycloaddition. Org. Lett. 11, 4954–7. [DOI] [PubMed] [Google Scholar]
- Kuang G. C.; Michaels H. A.; Simmons J. T.; Clark R. J.; Zhu L. (2010) Chelation-assisted, copper(II)-acetate-accelerated azide-alkyne cycloaddition. J. Org. Chem. 75, 6540–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang G. C.; Guha P. M.; Brotherton W. S.; Simmons J. T.; Stankee L. A.; Nguyen B. T.; Clark R. J.; Zhu L. (2011) Experimental investigation on the mechanism of chelation-assisted, copper(II) acetate-accelerated azide-alkyne cycloaddition. J. Am. Chem. Soc. 133, 13984–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels H. A.; Zhu L. (2011) Ligand-assisted, copper(II) acetate-accelerated azide-alkyne cycloaddition. Chem.—Asian J. 6, 2825–34. [DOI] [PubMed] [Google Scholar]
- Uttamapinant C.; Tangpeerachaikul A.; Grecian S.; Clarke S.; Singh U.; Slade P.; Gee K. R.; Ting A. Y. (2012) Fast, cell-compatible click chemistry with copper-chelating azides for biomolecular labeling. Angew. Chem., Int. Ed. 51, 5852–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Hong S.; Tran A.; Jiang H.; Triano R.; Liu Y.; Chen X.; Wu P. (2011) Sulfated ligands for the copper(I)-catalyzed azide-alkyne cycloaddition. Chem.—Asian J. 6, 2796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J.; Tung C. H. (2011) Development of benzothiazole ’click-on’ fluorogenic dyes. Bioorg. Med. Chem. Lett. 21, 320–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besanceney-Webler C.; Jiang H.; Zheng T.; Feng L.; Soriano Del Amo D.; Wang W.; Klivansky L. M.; Marlow F. L.; Liu Y.; Wu P. (2011) Increasing the efficacy of bioorthogonal click reactions for bioconjugation: a comparative study. Angew. Chem., Int. Ed. 50, 8051–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks A. J.; van Berkel S. S.; Hatzakis N. S.; Opsteen J. A.; van Delft F. L.; Cornelissen J. J.; Rowan A. E.; van Hest J. C.; Rutjes F. P.; Nolte R. J. (2005) Preparation of biohybrid amphiphiles via the copper catalysed Huisgen [3 + 2] dipolar cycloaddition reaction. Chem. Commun. 4172–4. [DOI] [PubMed] [Google Scholar]
- Brahma A.; Mandal C.; Bhattacharyya D. (2005) Characterization of a dimeric unfolding intermediate of bovine serum albumin under mildly acidic condition. Biochim. Biophys. Acta 1751, 159–69. [DOI] [PubMed] [Google Scholar]
- Hsu T. L.; Hanson S. R.; Kishikawa K.; Wang S. K.; Sawa M.; Wong C. H. (2007) Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc. Natl. Acad. Sci. U.S.A. 104, 2614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. V.; Chen X.; Smyrniotis C.; Xenakis A.; Hu T.; Bertozzi C. R.; Wu P. (2009) Metabolic labeling of sialic acids in living animals with alkynyl sugars. Angew. Chem., Int. Ed. 48, 4030–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J.; Meledeo M. A.; Wang Z.; Khanna H. S.; Paruchuri V. D. P.; Yarema K. J. (2009) Metabolic glycoengineering: sialic acid and beyond. Glycobiology 19, 1382–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A. K.; Fritz T. A.; Taylor W. H.; Esko J. D. (1995) Disaccharide uptake and priming in animal cells: inhibition of sialyl Lewis X by acetylated Gal beta 1-->4GlcNAc beta-O-naphthalenemethanol. Proc. Natl. Acad. Sci. U.S.A. 92, 3323–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. B.; Teng H.; Rhee J. K.; Lahar N.; Baskaran G.; Yarema K. J. (2004) Characterization of the cellular uptake and metabolic conversion of acetylated N-acetylmannosamine (ManNAc) analogues to sialic acids. Biotechnol. Bioeng. 85, 394–405. [DOI] [PubMed] [Google Scholar]
- Tian L.; Yang Y.; Wysocki L. M.; Arnold A. C.; Hu A.; Ravichandran B.; Sternson S. M.; Looger L. L.; Lavis L. D. (2012) Selective esterase-ester pair for targeting small molecules with cellular specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 4756–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetke C.; Brossmer R.; Mantey L. R.; Hinderlich S.; Isecke R.; Reutter W.; Keppler O. T.; Pawlita M. (2002) Versatile biosynthetic engineering of sialic acid in living cells using synthetic sialic acid analogues. J. Biol. Chem. 277, 6688–95. [DOI] [PubMed] [Google Scholar]
- North S. J.; Huang H. H.; Sundaram S.; Jang-Lee J.; Etienne A. T.; Trollope A.; Chalabi S.; Dell A.; Stanley P.; Haslam S. M. (2010) Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J. Biol. Chem. 285, 5759–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Hu T.; Frantom P. A.; Zheng T.; Gerwe B.; Del Amo D. S.; Garret S.; Seidel R. D. 3rd; Wu P. (2009) Chemoenzymatic synthesis of GDP-L-fucose and the Lewis X glycan derivatives. Proc. Natl. Acad. Sci. U.S.A. 106, 16096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T.; Ge X.; Lindeman R.; Nair S.; Runke G.; Mullins M. C.; Pelegri F. (2009) The maternal-effect gene cellular island encodes aurora B kinase and is essential for furrow formation in the early zebrafish embryo. PLoS Genet. 5, e1000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.