Abstract
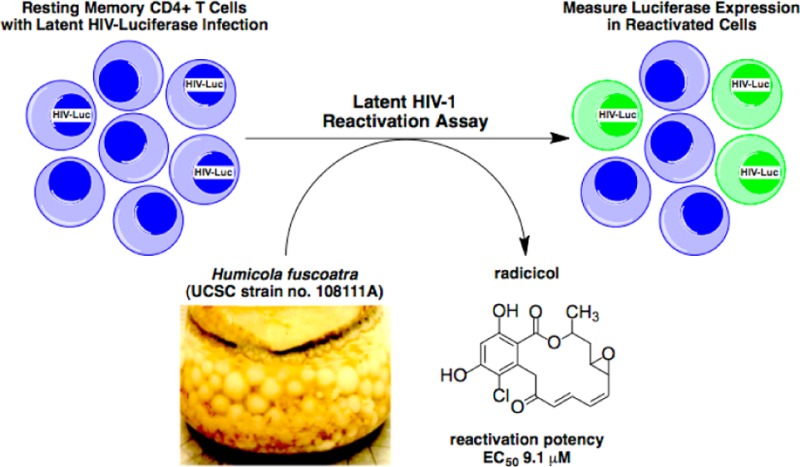
An extract of Humicola fuscoatra (UCSC strain no. 108111A) was shown to reactivate latent HIV-1 expression in an in vitro model of central memory CD4+ T cells. We report the bioassay-guided isolation and structure determination of several resorcyclic acid lactones, including four known compounds, radicicol (1, aka. monorden) and pochonins B (2), C (3), and N (4), and three new analogues, radicicols B–D (5–7). Compounds 1–3 and 5 showed moderate activities in the memory T cell model of HIV-1 latency. Radicicol (1) displayed lower potency in reactivating latent HIV-1 (EC50 = 9.1 μM) relative to the HDAC inhibitors apicidin (EC50 = 0.3 μM), romidepsin (EC50 = 0.003 μM), and SAHA (EC50 = 0.6 μM); however, it achieved equivalent maximum efficacy relative to the positive control compounds (98% of SAHA and romidepsin).
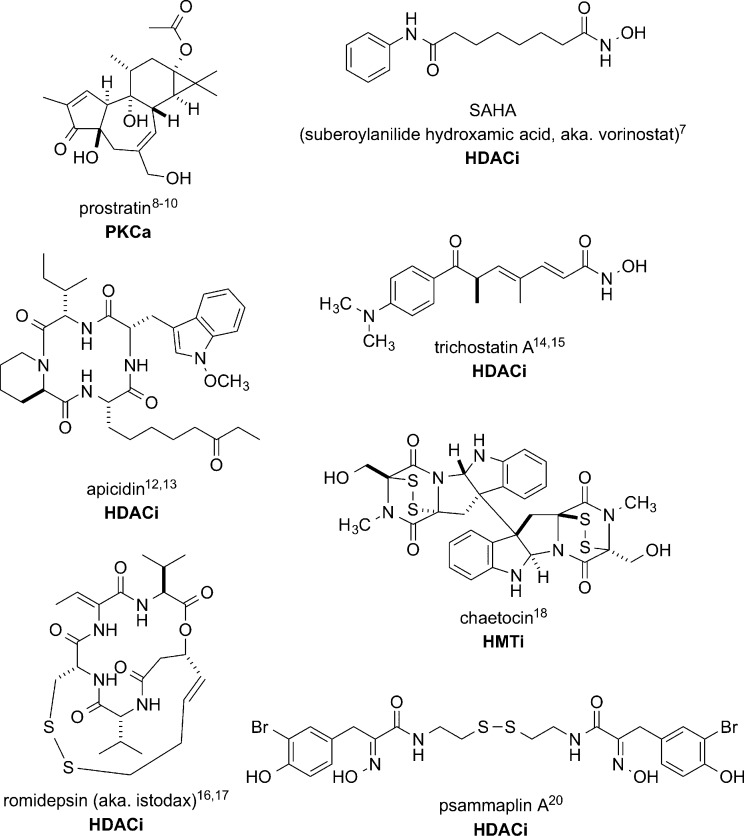
Human immunodeficiency virus type-1 infection (HIV-1) depletes CD4+ T cells and causes acquired immunodeficiency syndrome (AIDS). Development of combination antiretroviral therapy (cART), which targets multiple stages of the viral life cycle, has transformed this lethal disease to a manageable condition by reducing plasma viremia to clinically undetectable levels1 and restoring normal CD4+ T cell counts in HIV-1 patients.2,3 However, residual provirus harbored in cellular reservoirs quickly rebounds when treatment is interrupted.4 One of the major obstacles to HIV-1 eradication is the latent viral reservoir in long-lived resting CD4+ central memory T cells, which harbor transcriptionally silent proviral DNA.5 With a half-life of 44 weeks, it is estimated that 60 years, or a lifetime of cART, would be required to achieve a cure.6 One promising strategy to expunge HIV-1 infection is to pharmacologically reactivate the latent viral reservoir in CD4+ T cells, thus inducing cell surface expression of viral antigen to facilitate clearance by either host cytolytic effector mechanisms or targeted cytopathic therapies. The mechanisms of HIV-1 latency maintenance and reversal are complex, and effective small-molecule agents that reactivate latent provirus (Figure 1), such as the histone deacetylase inhibitor SAHA (suberoylanilide hydroxamic acid, aka. vorinostat; HDACi)7 and the protein kinase C activator prostratin (PKCa),8−10 have dose-limiting toxicity. Therefore, safe and effective latency-reversing agents are critically needed.
Figure 1.
Small-molecule HIV-1 latency-reversing agents with known modes of action: protein kinase C activation (PKCa), histone deacetylase inhibition (HDACi), and histone methytransferase inhibition (HMTi).
Three years ago, we began a collaborative screening program to discover small-molecule marine natural products that could be tools for reactivation of the latent HIV-1 reservoir. Recently, a summary of small-molecule latency-reversing agents was published and included 32 molecular structures.11 Some entries that regulate epigenetic mechanisms, shown in Figure 1, have been especially useful as potential therapeutics, e.g., SAHA,7 or as tool compounds, e.g., apicidin (a natural product cyclic peptide);12,13 both are histone deacetylase inhibitors (HDACi). Other epigenetic natural product latency-reversing agents include trichostatin A (HDACi),14,15 romidepsin (aka. istodax; HDACi),16,17 and chaetocin (histone methyl transferase inhibitor, HMTi).18 Our team screened a marine natural product library with a sensitive and scalable in vitro memory T cell model of HIV-1 latency,16,17 modified from that of Bosque and Planelles.19 Two positive controls consisted of SAHA (EC50 = 0.6 μM) and romidepsin (EC50 = 0.003 μM), which behaved in a dose-dependent manner to increase HIV-1 expression in the model. Small molecules that include both epigenetic and nonepigenetic hits were identified and are the subject of results described herein.
Results and Discussion
We screened a collection of sponge- and marine fungi-derived extracts (n = 2500) and pure compounds (n = 400) using a primary cell-based in vitro model of HIV-1 latency that is re-engineered for optimal sensitivity and miniaturized for high-throughput screening.16,17 Two hits were initially given high priority for further investigation: (1) the extract of a sponge putatively identified as Pseudoceratina purpurea (UCSC Coll. No. 06135), based on morphological similarity to previously identified sponges, and (2) the extract of a marine-derived fungus that was not taxonomically identified (UCSC strain no. 108107B). Briefly, the known compounds psammaplin A20 and apicidin,12 shown in Figure 1, were isolated from the sponge and fungal extracts, respectively, and retested to show significant potencies (EC50 = 0.2 μM and EC50 = 0.3 μM, respectively). A critical first step in these outcomes was the application of our recently published method for natural product peak library fractionation,21 whereby HPLC coupled with MS and evaporative light scattering detection (ELSD) maps the distribution of metabolites into biological screening wells as a function of retention time (see Experimental Section and Figure S1, Supporting Information). Subsequent operations involved scale-up isolation via peak libraries annotated with m/z MS data and bioassay hit information. The activity of psammaplin A likely stems from its known HDACi properties. Apicidin has previously been shown to function as an HIV-1 reactivator via an HDACi mechanism.13
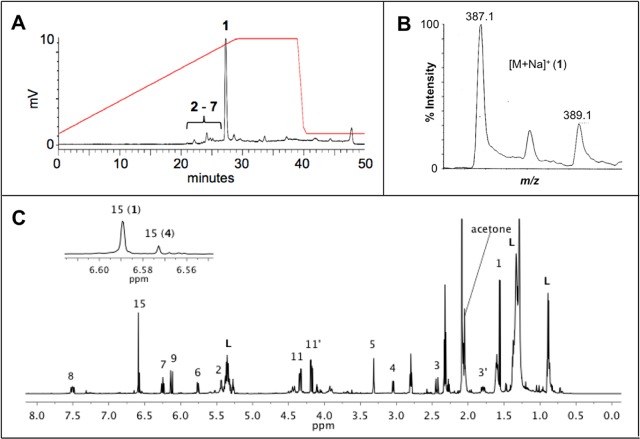
The successful workflow described above was employed to investigate the active extract of a marine-derived Humicola fuscoatra (UCSC strain no. 108111A), whose peak library ELSD profile is shown in Figure 2, panel A. The major component (tR = 27–28 min) showed a positive response in the latent HIV-1 reactivation assay. The mass spectrum (ESITOF+) of this analyte displayed a chlorinated ion at m/z = 387.1:389.1 (3:1 intensity, Figure 2, panel B), which we duly assigned as the sodiated molecular ion [M + Na]+ in conjunction with an ancillary [M + H]+ signal observed at m/z 365.1. Isolation of this active material (EC50 = 9.1 μM) was accomplished by retrieving a pure sample from an archived library plate, and an accurate mass was measured (ESITOF+) as 387.0637 ([M + Na]+), consistent with the molecular formula of C18H17ClO6, UN = 10. Conducting a literature search based upon taxonomy and this molecular formula yielded radicicol (1),22 a well-known fungal metabolite and potent in vitro HSP90 inhibitor,23 as a solitary dereplication hit. Indeed, the 1H NMR spectrum of the active extract (Figure 2, panel C) was dominated by the resonances of this metabolite. Additionally, the specific rotation of +203 for our sample from H. fuscoatra matched the literature value for 1 (+216);22 therefore, we assigned the absolute configuration of the three asymmetric carbon atoms in 1 as all R, based on a previously reported single-crystal XRD assignment.29
Figure 2.
Analytical data for the active extract of Humicola fuscoatra (UCSC strain no. 108111A). (A) Evaporative light scattering chromatogram collected during automated peak library fractionation. Annotations indicate the distribution of compounds 1–7 (see Experimental Section for elution conditions). (B) ESITOF(+) m/z data collected during peak library fractionation. Expansion of [M + Na]+ ion for radicicol (1), at tR = 27 min (see panel A). (C) 600 MHz 1H NMR spectrum in acetone-d6. Numbered annotations are those of the major component, radicicol (1). “L” denotes lipids. The expanded region shows the aromatic singlet of the most abundant minor congener, compound 4.
Chart 1.
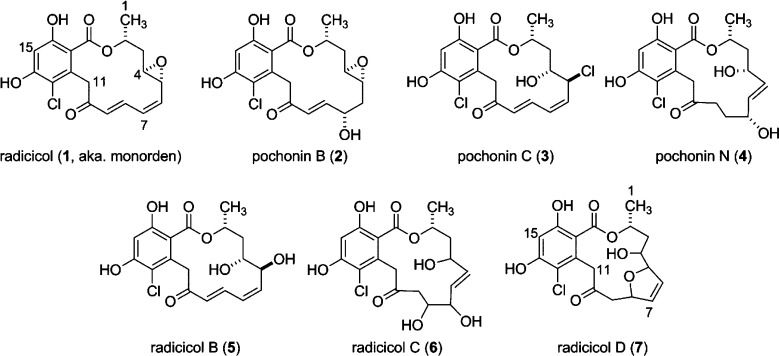
We were inspired to further mine the extract for radicicol congeners by two pieces of information: (1) the 1H NMR spectrum shown in Figure 2, panel C, displayed a characteristic aromatic singlet of considerably less intensity than that of the major metabolite, and (2) the mass chromatogram generated during the peak library fractionation was rich with chlorinated ions in the region of 20–26 min (Figure 2, panel A). Successive rounds of MS and NMR-guided HPLC purification yielded minor quantities of several additional radicicol congeners: three known compounds, pochonins B (2), C18H19ClO7,24 C (3), C18H18Cl2O6,24 and N (4), C18H21ClO7,25 and three new natural products, radicicols B–D (5–7). The molecular formulas and NMR properties of the known compounds 1–4 provided important baseline data to characterize the new compounds 5–7.
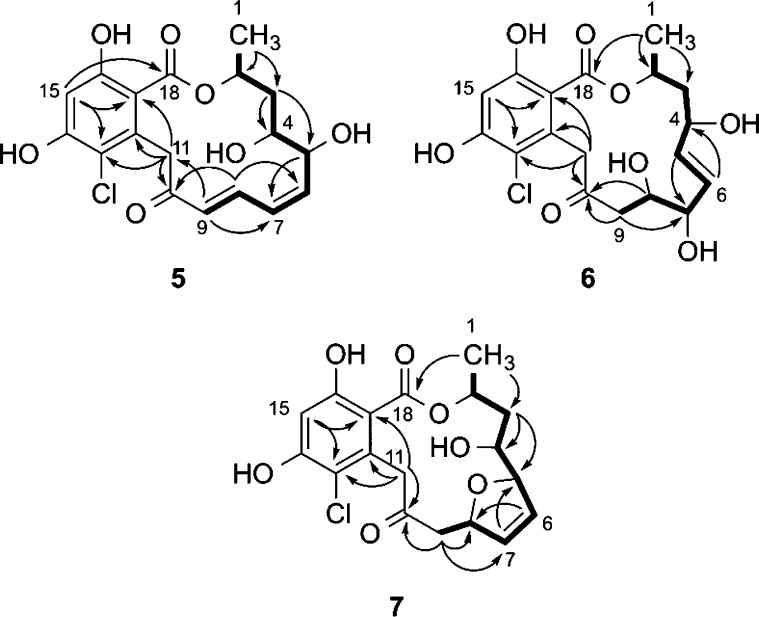
Radicicol B (5, C18H19ClO7, UN = 9) contains an additional molecule of H2O and one less degree of unsaturation relative to radicicol (1). The 1H and 13C NMR data of 5 closely matched those of 1 with the following exceptions: (i) the H-4 and H-5 proton resonances of 1 were transposed downfield from δH 3.05 and 3.32 to δH 3.62 and 4.50, respectively, and (ii) the C-4 and C-5 carbon resonances of 1 were transposed downfield from δC 56.1 and 55.8 to δC 71.9 and 69.4, respectively. Taken together with the gCOSY and gHMBC correlations shown in Figure 3, the data suggest the epoxide of 1 is a 1,2-diol in 5. The planar structure of 5 is identical to semisynthetic epoxide-opened diasteromers of natural radicicol described in both patent26 and peer-reviewed literature.27,28 The configuration of position C-2 is proposed to be analogous to that of radicicol (1), and the anti relative configuration for the 1,2-diol of 5 is assigned based on a vicinal proton coupling constant of J4,5 = 5.4 Hz, which matched the reported value of the synthetic anti diastereomer (lit.27J4,5 = 6.1 Hz).
Figure 3.
Selected gCOSY (bold) and gHMBC (arrow) correlations for radicicols B–D (5–7).
The molecular formula of radicicol C (6, C18H21ClO8, UN = 8) contains two additional H2O molecules in comparison to 1. The 13C NMR data (Table 1) matched those of 1 for all positions, apart from C-3 to C-10, and suggested the two equivalents of H2O were positioned in the macrocyclic chain. With the aromatic ring functionality and macrocyclic ester linkage deemed intact, the two remaining degrees of unsaturation were assigned to resonances of a ketone (δC 204.1) and a trans double bond (δC 135.7 and 131.7; δH 5.47 and 5.39, 3JHH = 15.6 Hz). Additionally, three secondary alcohols were detected at δH 3.60, 3.93, and 4.27 (δC 78.7, 69.1, and 70.2). A single extended spin system in the gCOSY spectrum was delineated from H3-1 to H2-9, and long-range gHMBC correlations finalized the planar structure of 6 as shown in Figure 3. The stereochemistry of position C-2 is proposed to be analogous to that of radicicol (1); however due to a lack of both epoxide and α,β,γ,δ-unsaturated ketone functionalities, radicicol C (6) is too flexible to assign the relative configurations of the three secondary alcohols by NOESY correlation spectroscopy.
Table 1. 1H (600 MHz) and 13C (150 MHz) NMR Data for Radicicols B–D (5–7) in Acetone-d6.
|
5 |
6 |
7 |
||||
|---|---|---|---|---|---|---|
| position | δC | δH, mult (J in Hz) | δC | δH, mult (J in Hz) | δC | δH, mult (J in Hz) |
| 1 | 21.5 | 1.46, d (6.3) | 20.7 | 1.36, d (6.5) | 19.1 | 1.48, d (6.6) |
| 2 | 70.6 | 5.66, m | 71.6 | 5.56, m | 73.5 | 5.35, m |
| 3 | 38.5 | 2.09, m | 43.0 | 2.10, m | 40.3 | 2.33, ddd (3.0, 3.3, 15.1) |
| 1.88, ddd (2.3, 9.3, 14.9) | 1.78, ddd (4.7, 7.1, 15.1) | |||||
| 4 | 71.9 | 3.62, ddd (2.3, 5.4, 8.7) | 70.2 | 4.27, m | 72.3 | 3.50, ddd (3.0, 7.1, 9.6) |
| 5 | 69.4 | 4.50, ddd (1.1, 5.4, 8.7) | 135.7 | 5.47, dd (8.5, 15.6) | 89.7 | 4.36, m |
| 6 | 143.4 | 5.87, dd (8.7, 10.8) | 131.7 | 5.39, dd (8.5, 15.6) | 129.9 | 5.98, ddd (1.5, 2.0, 6.2) |
| 7 | 127.2 | 6.16, t (10.8) | 78.7 | 3.60, t (8.7) | 131.6 | 6.09, ddd (1.5, 2.0, 6.2) |
| 8 | 139.7 | 7.43, dd (10.8, 16.2) | 69.1 | 3.93, ddd (2.5, 7.1, 8.9) | 84.1 | 5.15, m |
| 9 | 131.3 | 5.95, d (16.2) | 46.8 | 2.56, m | 47.3 | 2.87, dd (4.6, 14.6) |
| 2.56, dd (6.8, 14.6) | ||||||
| 10 | 196.8 | 204.3 | 203.7 | |||
| 11 | 44.6 | 4.46, d (15.5) | 47.6 | 4.23, d (17.5) | 50.2 | 4.35, d (15.5) |
| 4.08, d (15.5) | 4.00, d (17.5) | 4.24, d (15.5) | ||||
| 12 | 136.8 | 137.7 | 136.9 | |||
| 13 | 116.2 | 116.1a | 115.5a | |||
| 14 | 158.2 | 159.7b | 158.3a | |||
| 15 | 103.9 | 6.59, s | 104.1 | 6.60, s | 103.5 | 6.55, s |
| 16 | 159.6 | 159.7c | 161.2a | |||
| 17 | 111.2 | 107.1a | 110.3a | |||
| 18 | 168.2 | 168.3a | 171.3a | |||
Assigned from gHMBC data.
Assigned from gHMQC.
Exchangeable.
Radicicol D (7) was assigned the same molecular formula as 2 and 5 based on accurate mass measurement and required nine degrees of unsaturation. Comparison of 13C chemical shifts to those of 1 indicated the aromatic ring functionality and macrocyclic ester linkage remained intact, and positions C-4 to C-10 were modified. Two of the three remaining degrees of unsaturation were assigned to a ketone detected at δC 203.7 and a cis double bond at δC 129.9 and 131.6 (δH 5.98 and 6.09, 3JHH = 6.2 Hz). Additionally, three oxygenated carbons were detected at δC 72.3, 84.1, and 89.7. The connectivity from positions 1 through 9 was established from the gCOSY and gHMBC data (Figure 3), and on the basis of the vicinal coupling constant of 6.2 Hz between olefinic protons H-6 and H-7, the final unit of unsaturation was assigned to a 2,5-disubstituted-2,5-dihydrofuran ring system. The stereochemistry of position C-2 in radicicol D (7) is proposed to be analogous to that of radicicol (1); the relative configuration of positions 4, 5, and 8 could not be assigned using data sets collected in this study.
The possibility that radicicols B–D (5–7) are artifacts produced during the isolation process was ruled out, as they were detected in the LCMS analysis of the original EtOAc extract. Furthermore, compounds 5–7 were not detected by LCMS or 1H NMR after prolonged exposure of both natural (1) and commercial (Sigma) radicicol samples to the isolation conditions.
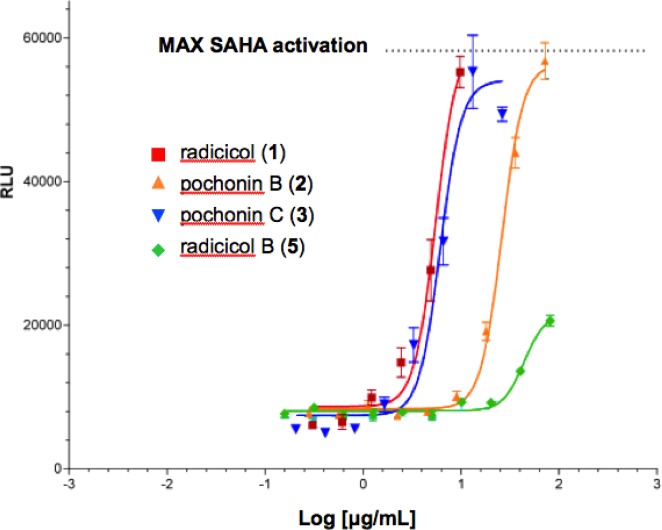
The screening data obtained in this study are summarized in Table 2, and some of the patterns deserve additional discussion. Radicicol (1) was isolated as the most abundant metabolite of H. fuscoatra and exhibited 98% reactivation efficiency of latent HIV-1 relative to the tool compound SAHA (Figure 4) and an EC50 of 9.1 μM. Radicicol activation of HIV-1 was confirmed by qPCR of HIV-1 RNA in the same latency model (see Supplementary Experimental Procedures and Figure S15, Supporting Information). Of the additional congeners 2–7, isolated from the fungal extract, pochonins B (2) and C (3) and radicicol B (5) were reactivators (Figure 4), while pochonin N (4) and radicicols C (6) and D (9) were found to be inactive. Comparison of the structures of the active compounds suggests the epoxide functionality is not required for activation in our assay; however, the active compounds all contain a Michael acceptor functionality, something the inactive compounds 4, 6, and 7 do not possess. While the potencies in reactivating latent HIV-1 for the active radicicols and pochonins are lower than that of romidepsin, it appears that their mechanism of action is unique and remains to be elucidated. Indeed, radicicol was previously reported to lack in vitro activity against histone deacetylase.30 Although radicicol is a potent in vitro HSP90 inhibitor (IC50 = 19 nM),23 a structurally unrelated HSP90 inhibitor, geldanamycin, is inactive in our assay, indicating that the mechanism of HIV-1 reactivation by 1 is unlikely to involve HSP90 inhibition. In addition, the most potent analogue, pochonin C (3), is unlikely to act as an HSP90 inhibitor based on the conformational hypothesis advanced by Winssinger: that HSP90 inhibition by radicicol-type resorcylides is closely tied to a specific bioactive molecular topology attenuated by modifications at the epoxide moiety.31,32 Finally, radicicol did not induce detectable NFkB reporter gene activation in Jurkat cells (see Supplementary Experimental Procedures and Figure S15, Supporting Information), indicating it reactivates latent HIV-1 by a PKC-independent mechanism.
Table 2. Latent HIV-1 Reactivation Activities of Compounds 1–7, Psammaplin A, Apicidin, Romidepsin, and SAHAa.
| latent
HIV-1 reactivation assay |
||
|---|---|---|
| compound | % reactivation/SAHA | EC50 (μM) |
| radicicol (1) | 98 | 9.1 |
| pochonin B (2) | 98 | 39.6 |
| pochonin C (3) | 92 | 6.3 |
| pochonin N (4) | not active | not active |
| radicicol B (5) | 25 | 24.9 |
| radicicol C (6) | not active | not active |
| radicicol D (7) | not active | not active |
| psammaplin A | 100 | 0.2 |
| apicidin | 100 | 0.3 |
| romidepsin | 100 | 0.003 |
| SAHA | 0.6 | |
Reported values include % reactivation relative to SAHA and potency (EC50 in μM).
Figure 4.
In vitro latent HIV-1 reactivation assay: dose–response profiles of radicicol (1), pochonin B (2) pochonin C (3), and radicicol B (5) relative to the maximum activation of the positive control SAHA.
There are several significant outcomes associated with our study. A screening paradigm has been created consisting of a sensitive and high-throughput primary cell-based HIV-1 latency assay that was well suited to investigate diverse marine natural product collections for novel reactivators of latent HIV-1. The assay was robust with highly reproducible EC50 values for reference compound SAHA in all donors used in this screen. Starting from an extract of the marine-derived fungus H. fuscoatra and followed by successive rounds of purification, dereplication, and bioassay, we isolated seven compounds in the radicicol/pochonin class, of which four display latent HIV-1 reactivation activity, providing a preliminary understanding of the structure–activity relationship of these compounds. At this stage we believe that the new HIV-1 reactivators summarized in Table 2 can be either the seeds for further scaffold modification or useful in combination with agents possessing a distinct mechanism of action, such as romidepsin, to achieve a higher magnitude of latent HIV-1 reactivation.
Experimental Section
General Experimental Procedures
Optical rotations were measured using a JASCO P-2000 polarimeter. Standard pulse sequences were used for all NMR experiments, which were run on either a Varian UNITY INOVA spectrometer (600 and 150 MHz for 1H and 13C, respectively) equipped with a 5 mm triple resonance (HCN) cold probe or a Varian spectrometer (500 and 125 MHz for 1H and 13C, respectively) equipped with an inverse detection probe. Residual solvent shifts for acetone-d6 were referenced to δH 2.05 and δC 29.84, respectively. Accurate mass measurements were obtained on a Mariner ESITOF instrument for molecular formula determinations. All chromatographic work was done in reversed-phase and utilized HPLC grade CH3CN (solvent A) and Milli-Q H2O (solvent B), both adjusted to contain 0.1% formic acid. The analytical LCMS system was composed of Waters HPLC components (i.e., solvent pumps and autosampler) and controlled by Empower software. A 150 × 4.60 mm 5 μm Luna C18 column (Phenomenex) was utilized, and the system operated at a flow rate of 1 mL/min. The postcolumn flow of the eluent was first through a photodiode array (Waters) and then split (1:1) between an evaporative light-scattering detector (SEDEX model 55) and an ESITOF mass spectrometer (Applied Biosystems Mariner). The semipreparative HPLC system was composed of Waters HPLC components (i.e., solvent pumps and gradient controller) and equipped with a 250 × 10 mm 5 μm Luna C18 column (Phenomenex) and tunable UV-absorbance detector (Waters).
Biological Material
The Humicola fuscoatra strain (UCSC coll. no. 108111A) was isolated from sediment collected at <33 m depth by scuba in Tutuila, American Samoa (S 14°16.600, W 170°36.719), in 2010. The strain was taxonomically identified by molecular (100% by ITS and D1/D2 regions of rDNA) and morphological methods at the University of Texas Fungus Testing Laboratory. It is maintained as a cryopreserved glycerol stock at UCSC.
Culture Conditions
The 125 mL and 5 L cultures of 108111A were grown in media that consisted of the following: cornmeal (4.5 g/150 mL) and 5xASW (200 mL/L) with the pH adjusted to 7.4. The stock solution of 5xASW contained the following: NaCl (135 g/L), MgSO4·7H2O (35 g/L), Tris (10 g/L), KCl (3 g/L), CaCl2 (1.5 g/L). After autoclave sterilization, the cultures were inoculated and shaken at 75 rpm for 21 days at ambient temperature.
Extraction and Isolation
At the time of harvest, the fungal cultures (5 × 1 L) were homogenized, and the liquid was extracted three times with equal volumes of EtOAc. The combined organic extracts were concentrated in vacuo to yield 2 g of oil. A portion of the 108111A CMa extract (194.9 mg) was separated into 15 fractions (fraction codes H1–H15, Figure S2) using the semipreparative HPLC system utilizing a flow rate of 3 mL/min, 310 nm detection, and the following elution conditions: 15 min isocratic 25:75, 25 min gradient from 25:75 to 70:30, 3 min gradient from 70:30 to 100:0, 10 min isocratic 100:0, and 3 min gradient from 100:0 to 25:75. Fraction H5 was pochonin N (4, 5.0 mg), fraction H9 was pochonin B (2,1.8 mg), and fraction H9 was pochonin C (3, 1.9 mg). Fraction H12 was semipure radicicol, and an additional run under identical HPLC conditions yielded pure radicicol (1, 11.2 mg) and three new analogues: radicicol B (5, 1.2 mg), radicicol C (6, 2.2 mg), and radicicol D (7, 1.0 mg).
Automated Peak Library Fractionation
The instrumentation setup for our natural product peak library generation has been recently published.21 The HPLC stage utilized two Waters 510 pumps and a Waters 717plus autosampler, both controlled with Empower 2 software. Separation was performed on a 250 × 10 mm 5 μm Luna C18 column (Phenomenex). Spectra from three detectors were acquired during peak library fractionation: Waters 996 photo diode array, SEDEX 55 ELSD, and Mariner 5054 ESI-TOF-MS. The mobile phase parameters are CH3CN (A) and H2O (B) with a flow rate of 2 mL/min and the following elution conditions: 30 min gradient from 10:90, 10 min isocratic 100:0, 1 min gradient from 100:0 to 10:90, 9 min isocratic 10:90. The injection amount was 15 mg/150 μL. Sample collection was performed using a Gilson 215 liquid handler controlled with Gilson Unipoint LC software. Samples were collected into BD Biosciences 96-deep-well plates, with a working volume of 2 mL (part number: 353966). Fractions were collected every minute. After the LC-MS-UV-ELSD library is collected, a duplicate archive plate is generated for analytical reference using a 12-channel pipet, creating an exact copy and counter balance for centrifugal drying. Plates were dried and concentrated using a Savant AES2010 SpeedVac.
Radicicol (1):
[α]20D +203 (c 1.0, CHCl3), lit.22 [α]D +216 (c 1.00, chloroform); UV, 1H NMR, 13C NMR, and HRESITOFMS data were in accordance with reported values.22
Pochonin B (2):
[α]20D +9 (c 0.8, acetone), lit.25 [α]D +20 (c 1.0, acetone); UV, 1H NMR, 13C NMR, and HRESITOFMS data were in accordance with reported values.24
Pochonin C (3):
[α]20D −111 (c 1.1, acetone), lit. (−)-2R,4S,5R31 [α]D −68.3 (c 0.06, chloroform); UV, 1H NMR, 13C NMR, and HRESITOFMS data were in accordance with reported values.24
Pochonin N (4):
[α]20D −73 (c 0.5, MeOH), lit.25 [α]D −77.3 (c 1.00, methanol); UV, 1H NMR, 13C NMR, and HRESITOFMS data were in accordance with reported values.25
Radicicol B (5):
[α]20D +32 (c 0.8, methanol); UV (LC eluent H2O/CH3CN/formic acid, 1:1:0.1) λmax (AU) 219 (0.87), 276 (0.60) nm; 1H and 13C NMR data, see Table 1; HRESITOFMS m/z 405.0739 [M + Na]+ (calcd for C18H19ClO7Na, 405.0712).
Radicicol C (6):
[α]20D −69 (c 1.0, methanol); UV (LC eluent H2O/CH3CN/formic acid, 1:1:0.1) λmax (AU) 219 (0.94) 262 (0.36) 309 (0.20) nm; 1H and 13C NMR data, see Table 1; HRESITOFMS m/z 423.0826 [M + Na]+ (calcd for C18H21ClO8Na, 423.0817).
Radicicol D (7):
[α]20D −53 (c 0.4, methanol); UV (LC eluent H2O/CH3CN/formic acid, 1:1:0.1) λmax (AU) 217 (0.48) 257 (0.12) 297 (0.08) nm; 1H and 13C NMR data, see Table 1; HRESITOFMS m/z 405.0668 [M + Na]+ (calcd for C18H19ClO7Na, 405.0712).
HIV-1-Luc Construct
An NL4-3-based vector with frame-shift mutations in vpr and env genes was engineered with a codon-optimized firefly luciferase gene in place of nef to generate the resulting construct pKS13. The NL4-3-Luc virus was generated by co-transfection of HEK-293T cells with pKS13 and a plasmid containing the HIV-1 env gene using Lipofectamine 2000 (Life Technologies).
In Vitro Model of HIV-1 Latency
Generation of latent infected cells was performed as described.16,17,19 Briefly, naive CD4+ T cells were purified by negative selection using PBMCs from healthy HIV-1 negative donors. Purified naive CD4+ T cells were activated by incubation with anti-CD3/CD28 magnetic Dynabeads (1 bead:2 cells ratio, Life Technologies), 1 mg/mL anti-IL-4 (R&D Systems), 2 mg/mL anti-IL-12p70 antibodies (R&D Systems), and 10 ng/mL TGF-β (R&D Systems) for 3 days. Cells were maintained in 30 U/mL IL-2 (Life Technologies) for 2 days followed by infection with NL4-3-Luc. Seven days postinfection, 20 μL of latently infected cells at 10 000 cells/well was dispensed into 384-well plates containing 350 nL of compound solutions in 50 μL of culture medium. After a 48 h incubation, 40 μL/well BriteGlo (Promega) was added, and luminescence measured using the Envision plate reader (Perkin-Elmer).
Acknowledgments
This work was supported by an NIH grant (R01 CA 047135) and NMR equipment grants NSF CHE 0342912 and NIH S10-RR19918, and the IMSD (MBRS) program (NIH/NIGMS research grant R25GM058903).
Supporting Information Available
Supplemental experimental procedures and Figures S1–S15. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Dedication
Dedicated to Prof. Dr. Otto Sticher, of ETH-Zurich, Zurich, Switzerland, for his pioneering work in pharmacognosy and phytochemistry.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Blankson J. N.; Persaud D.; Siliciano R. F. Annu. Rev. Med. 2002, 53, 557–593. [DOI] [PubMed] [Google Scholar]
- Barreiro P.; Soriano V.; Casas E.; González-lahoz J. AIDS 2002, 16, 245–249. [DOI] [PubMed] [Google Scholar]
- Sempowski G.; Haynes B. Annu. Rev. Med. 2002, 53, 269–284. [DOI] [PubMed] [Google Scholar]
- Wong J. K.; Hezareh M.; Günthard H. F.; Havlir D. V.; Ignacio C. C.; Spina C. A.; Richman D. D. Science 1997, 278, 1291–1295. [DOI] [PubMed] [Google Scholar]
- Palmer S.; Maldarelli F.; Wiegand A.; Bernstein B.; Hanna G. J.; Brun S. C.; Kempf D. J.; Mellors J. W.; Coffin J. M.; King M. S. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 3879–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano J. D.; Kajdas J.; Finzi D.; Quinn T. C.; Chadwick K.; Morgolick J. B.; Kovacs C.; Gange S. J.; Siliciano R. F. Nat. Med. 2003, 9, 727–728. [DOI] [PubMed] [Google Scholar]
- Archin N. M.; Liberty A. L.; Kashuba A. D.; Choudhary S. K.; Kuruc J. D.; Crooks A. M.; Parker D. C.; Anderson E. M.; Kearney M. F.; Strain M. C.; Richman D. D.; Hudgens M. G.; Bosch R. J.; Coffin J. M.; Eron J. J.; Hazuda D. J.; Margolis D. M. Nature 2012, 487, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miana G. A.; Bashir M.; Evans F. J. Planta Med. 1985, 4, 353–354. [DOI] [PubMed] [Google Scholar]
- Kulkosky J.; Culnan D. M.; Roman J.; Dornadula G.; Schnell M.; Boyd M. R.; Pomerantz R. J. Blood 2001, 98, 3006–3015. [DOI] [PubMed] [Google Scholar]
- Korin Y. D.; Brooks D. G.; Brown S.; Korotzer A.; Zack J. A. J. Virol. 2002, 76, 8118–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S.; Sliciano R. F. Drug Discovery Today 2013, 18, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-S.; Lee K.-R.; Kim J.-C.; Lim S.-H.; Seo J.-A.; Lee Y.-W. Appl. Eviron. Microbiol. 1999, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Zhang Y.; Zhu H. Curr. HIV Res. 2011, 9, 202–208. [DOI] [PubMed] [Google Scholar]
- Tsuji N.; Kobayashi M.; Nagashima K.; Wakisaka Y.; Koizumi K. J. Antibiot. 1976, 24, 1–6. [DOI] [PubMed] [Google Scholar]
- Huber K.; Doyon G.; Plaks J.; Fyne E.; Mellors J. W.; Sluis-Cremer N. J. Biol. Chem. 2011, 286, 22211–22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Barnes T.; Stepan G.; Balakrishnan M.; Stray K.; Callebaut C.; Wei G.; Geleziunas R.; Pagratis N.; Cihlar T.. Sensitive and Scalable In Vitro Memory T-Cell Model of HIV-1 Latency for High Throughput Screening and Identification of Agents That Activate HIV, 20th Conference on Retroviruses and Opportunistic Infections, Atlanta, GA, March 3–6, 2013.
- Wei G., Chiang V., Fyne E., Balakrishnan M., Barnes T., Graupe M., Hesselgesser J., Stepan G., Stray K., Tsai A., Yu H., McMahon D., Lalezari J., Sloan D., John Mellors J., Geleziunas R., Cihlar T.. Histone Deacetylase Inhibitor Romidepsin Induces HIV Expression in CD4 T Cells from Patients on Suppressive Antiretroviral Therapy at Concentrations Achieved by Clinical Dosing. Submitted. [DOI] [PMC free article] [PubMed]
- Bernhard W.; Barreto K.; Saunders A.; Dahabieh M. S.; Johnson P.; Sadowski I. FEBS Lett. 2011, 585, 3549–3554. [DOI] [PubMed] [Google Scholar]
- Bosque A.; Planelles V. Blood 2009, 113, 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña I. C.; Gautschi J. T.; Wang G-Y-S.; Sanders M. L.; Schmitz F. J.; France D.; Cornell-Kennon S.; Sambucetti L. C.; Remiszewski S. W.; Perez L. B.; Bair K. W.; Crews P. J. Org. Chem. 2003, 68, 3866–3873. [DOI] [PubMed] [Google Scholar]
- Johnson T. A.; Sohn J.; Inman W. D.; Estee S. A.; Loveridge S. T.; Vervoort H. C.; Tenney K.; Liu J.; Ang K. K-H.; Ratnam J.; Bray W. M.; Gassner N. C.; Shen Y. Y.; Lokey R. S.; McKerrow J. H.; Boundy-Mills K.; Nukanto A.; Kanti A.; Julistiono H.; Kardono L. B. S.; Bjeldanes L. F.; Crews P. J. Nat. Prod. 2011, 74, 2545–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirrington R. N.; Ritchie E.; Shoppee C. W.; Taylor W. C.; Sternhell S. Tetrahedron Lett. 1964, 7, 365–370. [Google Scholar]
- Roe S. M.; Prodromou C.; O’Brien R.; Ladbury J. E.; Piper P. W.; Pearl L. H. J. Med. Chem. 1999, 42, 260–266. [DOI] [PubMed] [Google Scholar]
- Hellwig V.; Mayer-Bartschmid A.; Muller H.; Greif G.; Kleymann G.; Zitzmann W.; Tichy H.-V.; Stadler M. J. Nat. Prod. 2003, 66, 829–837. [DOI] [PubMed] [Google Scholar]
- Shinonaga H.; Kawamura Y.; Ikeda A.; Aoki M.; Sakai N.; Fujimoto N.; Kawashima A. Tetrahedron 2009, 65, 3446–3453. [Google Scholar]
- Ikeda A.; Shinonaga H.; Fujimoto N.; Kasai Y. (Taisho Pharmaceutical Co., Ltd., Japan) PCT Int. Appl., 2003. [Google Scholar]
- Shinonaga H.; Noguchi T.; Ikeda A.; Aoki M.; Fujimoto N.; Kawashima A. Bioorg. Med. Chem. 2009, 4622–4635. [DOI] [PubMed] [Google Scholar]
- Agatsuma T.; Ogawa H.; Akasaka K.; Asai A.; Yamashita Y.; Mizukami T.; Akinaga S.; Saitoh Y. Bioorg. Med. Chem. 2002, 10, 3445–3454. [DOI] [PubMed] [Google Scholar]
- Cutler H. G.; Arrendale R. F.; Springer J. P.; Cole P. D.; Roberts R. G.; Hanlin R. T. Agric. Biol. Chem. 1987, 3331–3338. [Google Scholar]
- Kwon H. J.; Owa T.; Hassig C. A.; Shimada J.; Schreiber S. L. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 3356–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barluenga S.; Moulin E.; Lopez P.; Winssinger N. Chem.—Eur. J. 2005, 11, 4935–4952. [DOI] [PubMed] [Google Scholar]
- Moulin E.; Zoete V.; Barluenga S.; Karplus M.; Winssinger N. J. Am. Chem. Soc. 2005, 127, 6999–7004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.