Abstract
Magnetic nanotechnologies have shown significant potential in several areas of nanomedicine such as imaging, therapeutics, and early disease detection. Giant magnetoresistive spin-valve (GMR SV) sensors coupled with magnetic nanotags (MNTs) possess great promise as ultra-sensitive biosensors for diagnostics. We report an integrated sensor interface for an array of 256 GMR SV biosensors designed in 0.18 μm CMOS. Arranged like an imager, each of the 16 column level readout channels contains an analog front- end and a compact ΣΔ modulator (0.054 mm2) with 84 dB of dynamic range and an input referred noise of 49 nT/√Hz. Performance is demonstrated through detection of an ovarian cancer biomarker, secretory leukocyte peptidase inhibitor (SLPI), spiked at concentrations as low as 10 fM. This system is designed as a replacement for optical protein microarrays while also providing real-time kinetics monitoring.
Keywords: biochip, biosensor, magnetic biosensor, GMR spin-valve, proteomics, sigma delta modulator
I. INTRODUCTION
The future of medical diagnostics is predicated on detecting the presence of one or more biomarkers by means of a molecular test. A molecular test examines a patient’s proteome or genome (a snapshot of the protein or gene expression levels) for a biomolecular signature related to a particular disease process. Typically, these disease biomarkers are quantified since ascertaining their concentrations helps in determining the diagnosis, prognosis, or ideal treatment regimen for the disease. Sensitive and early disease diagnosis is essential because the earlier a biomarker signature is detected, the more likely it becomes that the treatment will be successful. Furthermore, utilizing molecular tests is ushering in a new era of therapy known as personalized medicine, where a drug or treatment regime is chosen based on an individual’s genetic or proteomic information [1]. Presently, most molecular tests are probed optically. However, with recent advancements in nanotechnology, which have enabled new transducers on the same size scale as biomolecules, it is now possible to use electronic detection. To this end, novel biosensing platforms are needed to make this transition possible.
Microarrays are a vital tool to perform many molecular tests in parallel, typically for analyzing large-scale gene and protein expression changes in a biological sample. Deoxyribonucleic acid (DNA) microarrays, in particular, have seen prolific adoption and success in genomic research [2]. One of the greatest contributors to this achievement is the existence of an amplification technique, the polymerase chain reaction (PCR), which allows a single copy of DNA to be replicated a million or more times [3]. Unfortunately, protein microarrays have proved to be much more challenging than DNA microarrays, primarily due to the lack of an equivalent amplification technique for proteins. As such, protein microarrays must be significantly more sensitive and require higher dynamic range. For example, the concentration of serum proteins range from as high as 50 mg/mL (in the case of the most abundant protein, albumin) to below 5 pg/mL (in the case of thyroid hormone, free triiodothyronine). There are ten orders of magnitude difference in the relative abundance of these proteins at physiologic levels. Moreover, with early cancer detection and post-surgical tumor marker monitoring, tumor markers may be shed into the blood at concentrations below 1 pg/mL [4]. To date, most protein microarrays have utilized optical or electrochemical tags and are only semi-quantitative (log-fold changes) in their readout. In contrast, we describe a quantitative platform utilizing a magnetic immunoassay (MIA) coupled with an array of magnetic biosensors and an integrated data acquisition system. This platform, which we call MagDAQ 256 (Fig. 1(a)) [5], features both high sensitivity and a large, high-density sensor array, enabling quantitative proteomic analysis. With this platform, we address the need for highly sensitive electronic molecular tests with potential new applications in areas such as the study of protein-protein interactions, clinical diagnostics, monitoring of disease states, and drug discovery.
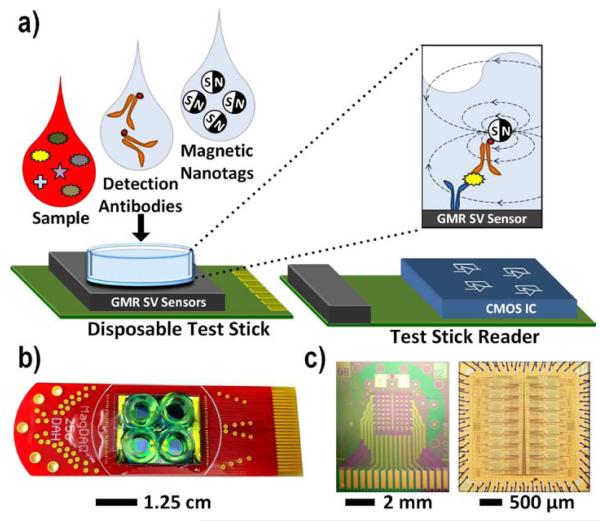
Fig. 1.
a) Illustration of complete system using a disposable test stick and a test stick reader containing the custom designed CMOS chip. Photograph of: b) disposable test stick, c) GMR SV sensor die, and CMOS IC.
This paper is organized as follows: Section II covers magnetic biosensing and the magnetic immunoassay. Section III explains the design of the interface IC containing an array of analog front-ends and ADCs. Sections IV, V and VI contain electrical measurement results, temperature correction measurements, and biological measurement results respectively. Section VII compares this work to previous work on biosensors, and Section VIII contains a conclusion.
II. MAGNETIC BIOSENSING AND THE MAGNETIC IMMUNOASSAY
Biosensors act as a bridge between biology and instrumentation by converting a biological or chemical response into an electrical signal. Biosensors take on many different forms and sensing modalities, but can be broadly classified into two categories: labeled and label-free techniques. Labeled techniques tag a recognition antibody which binds to the biomolecule of interest with an externally observable label such as a fluorophore [6], quantum dot [7], electrochemical tag [8], or magnetic tag [9]–[13]. On the other hand, label-free techniques detect an intrinsic property of the biomolecule, such as the mass [14], charge [15], thermal reactivity [16], size [17], or its optical interaction with a surface [18]. This work concentrates specifically on magnetic labels that offer several key advantages over conventional optical techniques and other competing sensing methods. First, the samples (blood, urine, serum, etc.) naturally lack any detectable magnetic content, providing a sensing platform with a very low background signal. This is a significant and fundamental problem with many optical techniques where one encounters label-bleaching [19] and autofluorescence [20]. Second, the sensors can be arrayed and multiplexed to perform complex protein or nucleic acid analysis in a single assay without resorting to bulky optical scanning. Additionally, the magnetic tags can be manipulated with a magnetic field to potentially speed up the reaction [21] or remove unbound tags [22]. Finally, the sensors are compatible with CMOS IC technology, allowing them to be manufactured with integrated electronic readout, produced in mass quantities (potentially at low cost), and deployed in a one-time use, disposable format for point-of-care testing.
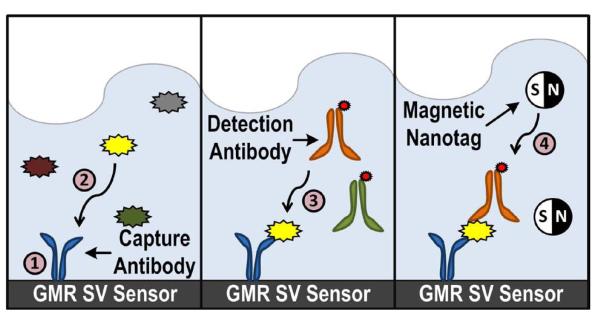
An immunoassay is a biochemical test that measures the presence or concentration of a protein biomarker in a solution. Currently, the enzyme-linked immunosorbent assay (ELISA) is widely regarded as the gold standard for protein detection. In the ELISA, biomarkers are labeled with a colorimetric label, typically an enzyme that reacts with a substrate, that is then imaged by a camera. The MIA (Fig. 2) strongly resembles the ELISA, except that the label has been changed from a colorimetric tag to a magnetic nanotag (MNT) [23]. The MIA is implemented as follows: 1) A capture antibody, highly specific to a particular biomarker, is immobilized on the surface of the sensor through a covalent attachment chemistry [24]. 2) Upon introduction of the sample, which may contain hundreds to thousands of different biomarkers in vastly different concentrations, the capture antibody selectively binds only to the targeted biomarker. The sample is incubated and the unbound biomarkers are washed away. 3) A biotinylated detection antibody is added and binds to a different site on the captured biomarker. After a short incubation, the unbound detection antibodies are washed away. 4) Finally, MNTs are added and attach to the detection antibody through a high affinity biotin-streptavidin bond to complete the MIA. Each MNT is ~50 nm in diameter and composed of several 10 nm superparamagnetic iron oxide cores embedded in a dextran matrix [25]. Multiple biomarkers can be simultaneously detected in the same assay by immobilizing a different capture antibody on each of the individually addressable sensors and adding a cocktail of biomarker specific detection antibodies.
Fig. 2.
Sequence of steps for the magnetic immunoassay.
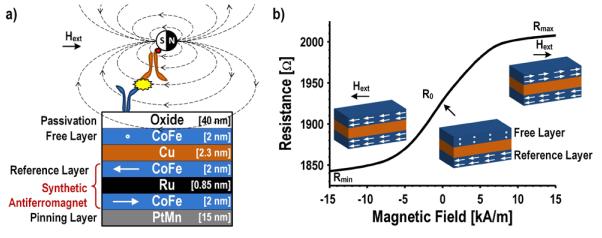
The number of MNTs tethered to the surface is quantified by the underlying giant magnetoresistive spin-valve (GMR SV) sensors, featuring the same type of sensor materials used in hard disk drives in the late 1990’s [26]. GMR SV biosensors are elaborately engineered thin film stacks, typically only a few tens of nanometers thick (Fig. 3(a)) [27]. To explain the operation of this device, it is first necessary to explicate the structure and the magnetization of the different films. The PtMn layer defines the magnetization for the synthetic antiferromagnet (the CoFe/Ru/CoFe tri-layer). The top CoFe layer of the synthetic antiferromagnet is referred to as the reference layer because the magnetization is fixed through indirect exchange coupling. A small conductive copper layer separates the antiferromagnet and the free layer (the upper CoFe layer). The magnetization of the free layer rotates relatively freely to align with the external magnetic field. In the final step, this structure is passivated from the biochemistry with a 40 nm tri-layer oxide (SiO2/Si3N4/SiO2). Minimizing the thickness of this passivation layer is critical to the sensitivity because the detected signal, the stray field from the MNTs, falls off inversely as the distance cubed (~1/d3). It may be possible to increase the sensitivity of these devices further by using a thinner ALD oxide; however, the deposition must be done at a low temperature to avoid degrading the underlying magnetic layers.
Fig. 3.
a) GMR SV structure annotated with film thicknesses and orientation of magnetizations. b) Measured transfer function of a GMR SV sensor.
GMR SV sensors exploit a quantum mechanical effect in which a change in magnetic flux is transduced into a change in electrical resistance through spin-dependent scattering (primarily at the Cu/CoFe interface and the bulk of the CoFe). The resistance of a GMR SV sensor can be empirically written as a function of the angle between the reference layer and the free layer (Eq. 1) where R0 is the resistance without a magnetic field and ΔR is the difference between the maximum and minimum resistances (Rmax and Rmin, respectively).
| (Eq. 1) |
Fig. 3(b) shows a measured transfer curve of a sensor along with the magnetization of the reference and free layers. The sensor has a minimum resistance of 1.84 kΩ when a negative saturating magnetic field is applied. The magnetization of the free layer and the reference layer are in the same direction leading to minimal spin-dependent scattering in this configuration. In this design, when no external field is applied, the orientation of the free layer is set through shape anisotropy, perpendicular to the reference layer. Then, when a positive saturating magnetic field is applied, the reference layer and free layer are antiparallel resulting in significant spin-dependent scattering and thus a high resistance (2.0 kΩ). The magnetoresistance (MR) ratio (Eq. 2), which quantifies the percentage of the resistance that responds to an external magnetic field, is 9.2% for this sensor. The stray field from each MNT attached to a biomarker on the surface of the sensor opposes the external applied magnetic field used to magnetize the superparamagnetic MNTs. This local change in the magnetic field causes the free layer to rotate, slightly reducing the resistance of the sensor. The resistance change is proportional to the number of MNTs and hence the number of captured biomarkers when operated in the linear range of the transfer curve.
| (Eq. 2) |
III. IC DESIGN
A. Challenges
While conceptually straightforward, there are several challenges in designing a sensor interface for GMR SV biosensors. First, each MNT induces only a minute resistance change that is superimposed on a much larger resistance, approximately 3.5 μΩ on a 1.8 kΩ sensor. This low signal to baseline ratio (< 10−8) is due to the small magnetic moment of the MNTs [28] and the limited MR ratio of the sensor. Applying the largest voltage (0.5 Vpk), which avoids dielectric breakdown of the passivation layer, results in a 100 fA current change per MNT. Detection of the MNTs is further complicated by the high 1/f noise of GMR SV sensors that have a flicker noise corner frequency greater than 10 kHz. Furthermore, the sensors operate in a harsh, dynamic sensing environment where sudden temperature changes of up to 30°C are possible during the biochemical fluid removal and addition steps due to the different storage conditions of the reagents. When coupled with the large temperature coefficients of the GMR SV sensors (252.6 ppm°/C and −710.5 ppm/°C for the resistive and magnetoresistive components, respectively), there are large temperature-induced signals that can easily overwhelm the weak signal from the MNTs, particularly at low biomarker concentrations. To monitor the kinetics of the reaction, the sensor interface must rapidly readout (< 10 s) the entire array of sensors. In addition, the sensor interface must be able to tolerate deviations in the mean nominal sensor resistance between 1.5 kΩ and 3.0 kΩ with 5% random process variation within the array (analogous to fixed pattern noise). Taken together, these constraints make the sensor interface challenging.
B. Architecture
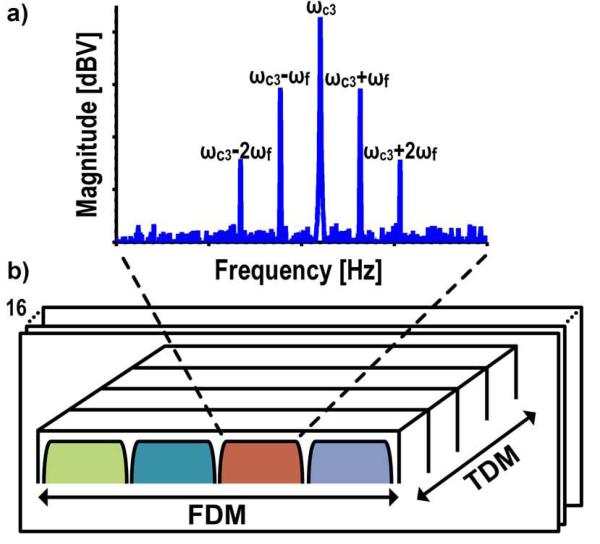
The 1/f noise issue is mitigated by modulating the signal due to the MNTs away from the 1/f noise by applying a time-varying magnetic field (ωf) and a time-varying excitation voltage (ωc) to the sensor. The resulting current (Eq. 3) has three primary tones in the spectrum (Fig. 4(a)) at frequencies ωc and ωc ± ωf. The tone at ωc, referred to as the carrier tone (CT), is due to the non-magnetoresistive portion of the sensor, whereas the side tones (STs) at ωc ± ωf result from the magnetoresistive component. The 1/f noise of the sensor is modulated up with the carrier, but the alternating magnetic field separates the resistive and magnetoresistive components of the sensor, significantly reducing the spot noise at the side tones. Additionally, the signal to baseline ratio is improved by an order of magnitude by the removal of the non-magnetoresistive component. Monitoring the change in the ST amplitude over time, essentially a simplified spectrogram, allows the number of MNTs on the surface of the sensor to be quantified. This multi-domain (magnetic and electronic) modulation scheme has been described previously using a current rather than a voltage excitation [29]–[31]. A voltage excitation facilitates multiplexing the sensors by summing their currents without the need for a dedicated amplifier per sensor. However, there are additional harmonics separated by integer multiples of ωf from ω c from this approach.
Fig. 4.
a) Spectrum showing modulation scheme. b) Readout with FDM and TDM.
| (Eq. 3) |
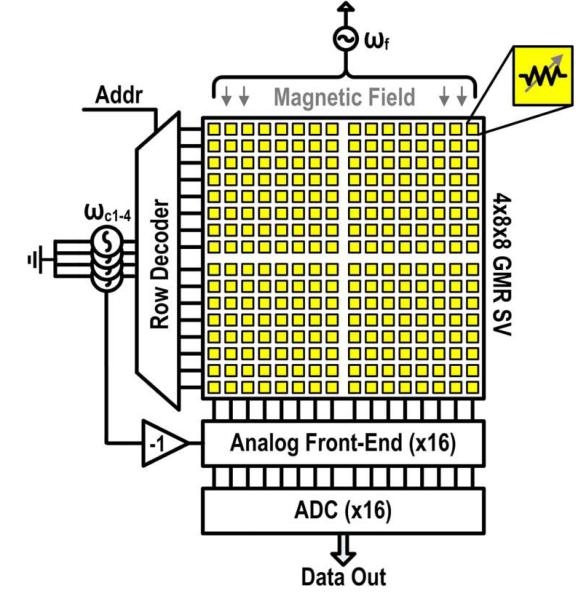
The sensor array is composed of four sub-arrays, each made up of an 8x8 matrix with 100 μm × 100 μm GMR SV sensors on a 300 μm pitch, for a total of 256 individually addressable sensors. Typically with large arrays, time-division multiplexing (TDM) is used to sequentially scan each sensor in a round robin fashion [32]. However, with the signal modulation scheme described previously, this would be prohibitively slow since the noise is coupled to the readout time and requires roughly one second per sensor. We utilize 16 parallel readout channels and frequency-division multiplexing (FDM) to reduce the readout time, which is accomplished by simultaneously exciting one sensor in each of the four sub-arrays with a different carrier frequency (ωc1-4) and summing the resulting currents. All of the sensors share the same magnetic field that is generated by an external Helmholtz coil. The response from each sensor is then isolated after digitization through spectral analysis (Fig. 4(b)).
Previous work has shown that it is possible to manufacture GMR SV sensors and the sensor interface (without an ADC) on the same die [31]. However, the resulting sensors suffered in performance (low MR ratio and low yield) due to complications from the additional fabrication steps negating many of the benefits of integration. In principle, fabricating the sensors on top of the CMOS electronics should not degrade the quality of the sensors, but is difficult in practice without wafer-scale processing. Furthermore, we have not found a method to remove the magnetic tags from the surface of the sensor when using sub-micron magnetic tags relegating the sensors to a single use. Here, we focus on the integrated readout electronics where the sensors are contained on a one-time use test stick (Fig. 1(b)) that plugs into a test stick reader containing a custom designed sensor interface and acquisition IC (Fig. 1(c)). This format introduces an interfacing challenge caused by the large capacitance at the input due to ESD structure, pads, and interconnect that is addressed through circuit design. Architecturally, this system is arranged like an imager (Fig. 5), where a decoder selects a particular row of sensors that are readout in parallel by column level ADCs. The interface IC contains the analog front-ends and ADCs, which are described in the next two sections.
Fig. 5.
Architecture of GMR SV system.
C. Analog Front-End
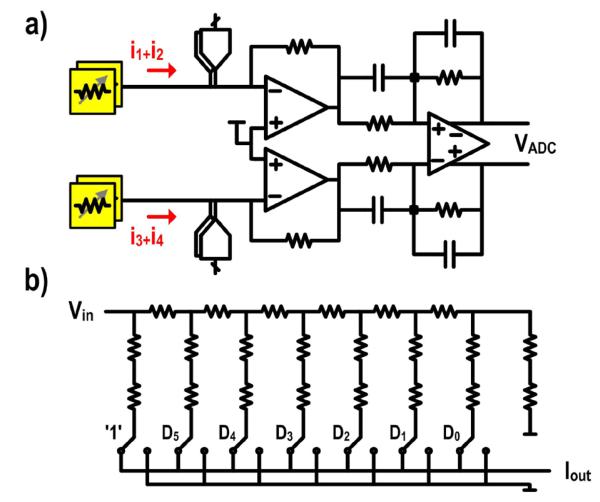
The analog front-end consists of two pseudo-differential transimpedance amplifiers (TIAs) followed by a fully-differential ADC driver (Fig. 6(a)). Each TIA combines currents from two of the four sensors while the ADC driver combines the output of both TIAs and converts the signal from single-ended to fully-differential. This split architecture is chosen over a single amplifier due to the large parasitic input capacitor (> 10 pF) which significantly degrades the feedback factor. It is inefficient to design an amplifier that is highly linear, fast enough to drive the ADC, and has low input referred noise within a reasonable power budget. By partitioning the design, we are able to optimize each amplifier separately: the TIA for low noise and high linearity, and the ADC driver for high speed.
Fig. 6.
a) Schematic of the analog front-end. b) Schematic of the 7-bit R-2R ladder.
Due to the limited MR ratio of the sensor (at most 9.2%, although frequently operated at a fraction of this amount for linearity reasons), the CTs are over an order of magnitude larger than the STs (Fig. 4(a)). To reduce the swing and dynamic range requirements of the signal path, we employ a carrier suppression technique where DACs at the input of each TIA inject signals to suppress the CTs (ωc1-4). In total, there are four DACs corresponding to the four different carrier signals. The carrier tones used for the sensor are inverted and drive each DAC directly. The DAC generates a current that is similar in amplitude, but 180° out of phase to the carrier. Each DAC is implemented by a 7-bit R-2R ladder with the most significant bit tied to a logic one (Fig. 6(b)) permitting a tunable range of 1.5Ωk to 3.0 kΩ. The DAC resolution is dictated by the process spread in the GMR SV sensor array. An extra leg is added as a termination to keep the input resistance constant regardless of the input code. Half-size dummy switches are frequently used in the R branch to accurately match the resistance of the 2R leg. However, the large swing experienced by the dummy switches relative to the actual switches significantly reduces the dynamic linearity. Simulations reveal that the spurious-free dynamic range (SFDR) degrades from 102 dB to less than 65 dB when the dummy switches are added. This is addressed by leaving out the dummy switches and sizing the switches such that the on resistance was negligible compared to the unit element resistance.
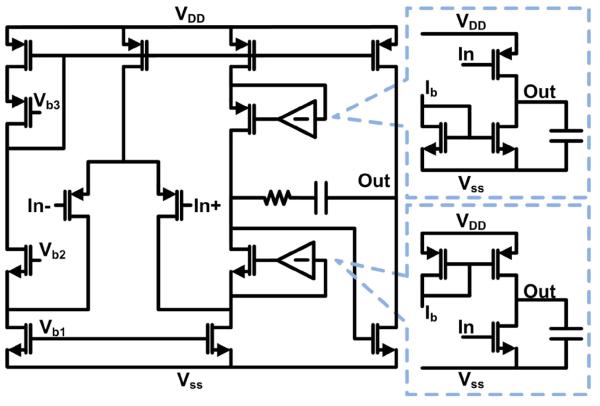
The input architecture is constrained by the requirement to have a fixed common-mode input voltage for the current summing and carrier suppression. In the frequency band of interest (1 kHz to 10 kHz), a fully-differential TIA with a common-mode input control [33] would add too much 1/f noise or require too much area to be an effective solution. This is overcome by using pseudo-differential input stage to provide the virtual ground for the sensor. Each TIA is realized by a two-stage folded-cascode amplifier with gain-boosting and resistive feedback (Fig. 7). The input devices are sized such that the 1/f noise corner of the TIA is less than 1 kHz. The gain-boosters are implemented with common-source amplifiers. Only the output branch of the folded-cascode amplifier is gain-boosted because the other side does not increase the output impedance and thus does not contribute to the overall gain of the amplifier. High loop gain is needed to achieve the necessary linearity and prevent the different sensor frequency components from mixing. The entire signal path gain (84.8 dBΩ) is placed in the first amplifier for noise reasons.
Fig. 7.
Schematic of the TIA and gain-boosters.
D. ADC
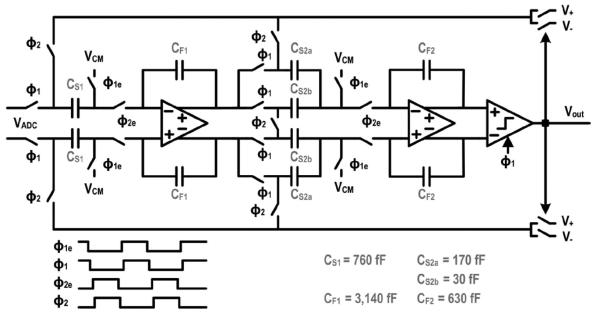
The ADC is implemented with a switched capacitor, 2nd order, single bit, highly oversampled (OSR = 500, fs = 10 MHz) ΣΔ modulator (Fig. 8). Given the low bandwidth (10 kHz) and high resolution (≥ 14 bits) requirements for this application, an oversampling converter allows us to make best use of the high speed transistors by trading resolution in time for resolution in amplitude. The implementation follows the classic Boser-Wooley architecture using a cascade of two delaying integrators [34]. This configuration permits each integrator to settle independently and simplifies the overall timing. The ADC is specifically optimized at the circuit and architectural level to have a small footprint. For example, a shared sampling capacitor (CS1) is chosen over two separate sampling capacitors in the first stage to minimize both the area and the kT/C noise. This approach is also extended to the second integrator where the output of the first integrator is sampled onto a split capacitor consisting of CS2a and CS2b. In the integration phase, CS2a is connected to the reference voltage while the bottom plate of CS2b is shorted with its differential counterpart to implement the necessary transfer function. While the area saving is not substantial for a single channel, it adds up when the modulator is arrayed. Sharing the sampling capacitors, as is done in both integrators, introduces signal dependent loading on the reference voltages requiring low-impedance references, local decoupling capacitors, and budgeting of sufficient settling time to avoid distortion, mixing of high frequency noise into baseband, and crosstalk among the modulators.
Fig. 8.
Schematic of the ΣΔ modulator.
The requirements for the integrators, switches, and comparator were calculated by hand [34], [35] and refined though high level simulation using Simulink due to the highly nonlinear behavior of a single-bit ΣΔ quantizer [36]. The amplifiers in the integrators are implemented using two-stage amplifiers with Miller compensation to allow high output swing and reduce the area of the capacitors. Large PMOS input devices are used to minimize the 1/f noise at the expense of the lower transit frequency in the first integrator. A switched capacitor common-mode feedback (CMFB) circuit controls the common-mode output voltage of the amplifier [37]. The input and output common-mode voltages are both set to mid-supply to minimize the number of voltages that need to be brought onto the chip. The input sampling switch is constructed with a clock boosted NMOS [38] to achieve better than 14-bit linearity. A purely dynamic sense amplifier without preamplification is used for the comparator.
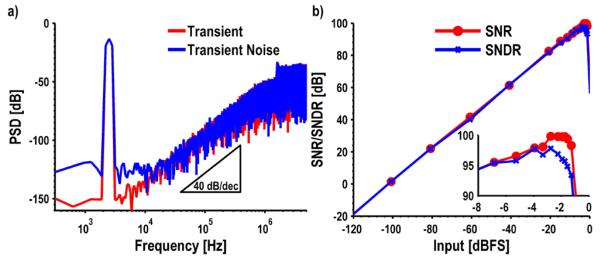
Transient and transient noise simulations of the full modulator are shown in Fig. 9(a). The simulations with a −3.3 dBFS input sinusoid show an SQNR of 115 dB and an SNDR of 97 dB for the transient and transient noise simulations, respectively. The second order 40 dB/decade noise shaping is clearly visible from the spectrum. These simulations are repeated for several different input amplitudes to generate a dynamic range curve (Fig. 9(b)). The designed modulator has a simulated peak SNDR of 98 dB at −1.75 dBFS (1.8 Vpp) and a dynamic range of 99.9 dB. The ΣΔ modulator consumes 910 μW of which 61% is used by the first integrator, 24% by the output buffer, and 12% by the second integrator. The references and comparator consume an almost negligible amount (2% and 1%, respectively).
Fig. 9.
Simulation results for the ΣΔ modulator, a) Transient and transient noise, b) Dynamic range curve.
IV. ELECTRICAL MEASUREMENTS
Owing to the large number of parallel readout channels on the chip, there are no additional pins or area for dedicated test structures. As such, all measurements were taken using the entire signal path. The analog front-end was measured to have an input referred noise of 32.6 pA/√Hz and a 1/f noise corner of 700 Hz. The single tone linearity, characterized by the SFDR, was greater than 80 dB and limited by our measurement setup. However, a two-tone test is a more relevant metric in this application as it more closely mimics the actual usage of the chip with the FDM and modulation scheme. We measured a multi-tone SFDR of 73 dB with the near carrier tones at 78.7 dB. We characterized the static nonlinearity of all 64 DACs on a single chip. The overlaid DNL ranged from −0.32 to 0.31 LSBs and the INL was between −0.23 and 0.18 LSBs. The lack of dummy switches caused noticeable DNL errors at the high codes corresponding to low resistances.
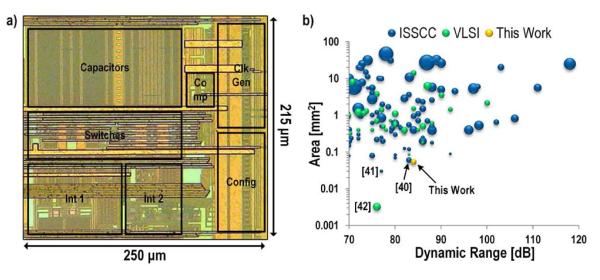
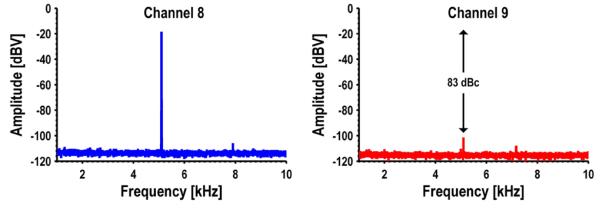
The sensor interface and acquisition system has a measured dynamic range of 84 dB. This is unfortunately slightly lower than the simulated value due to an error in the ADC driver which required the common-mode voltage to be reduced, ultimately reducing the available swing. The layout of the ΣΔ modulator is very compact, only 250 μm × 215 μm (Fig. 10(a)). This is one of the smallest ADCs reported in the literature with a bandwidth of at least 1 kHz and a dynamic range greater than 70 dB, despite being implemented in a relatively old 0.18 μm technology (Fig. 10(b)) [39].The surrounding data points are also ΣΔ modulators. In [40] amplifier sharing was used to reduce the area, and both [41] and [42] use inverter based modulators. However, even though the modulators are compact and in very close proximity, there is still excellent isolation among the different ADCs. Fig. 11 shows the measured spectra from two adjacent channels (8 and 9) when a −9 dBFS tone is input to channel 8. Channel 9 picks up this tone, but attenuated by 83 dB. Channel 7, while not plotted, showed a similar spectral signature as channel 9 whereas channel 10 showed no sign of this input tone confirming that it is proximity based coupling.
Fig. 10.
a) Die micrograph of ΣΔ modulator. b) Survey of ADCs from ISSCC and VLSI. Size of each data point indicates technology node relative to 32 nm.
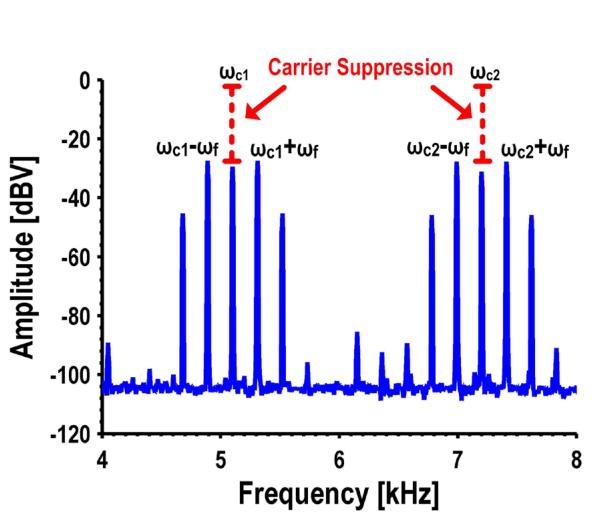
Fig. 12 shows a subset of the spectrum with the GMR SV sensors and the carrier suppression circuitry operating. For clarity, only two of the four tones are shown as the spectrum becomes quite busy looking when all four are shown. The carrier frequencies are 3.1 kHz, 5.1 kHz, 7.2 kHz, and 9.6 kHz with only the 5.1 kHz and 7.2 kHz frequencies shown in the figure. The external Helmholtz coil is driven to 4 kA/mRMS at 215 Hz using a Kepco power amplifier. The figure is annotated to show the unsuppressed amplitude of the carriers. In both cases the carriers have been reduced to below the ST amplitudes (by ~30 dB). The measured input referred noise with all of the sensors connected is 101.5 pA√/Hz, which translates to a sensitivity of 49 nT/√Hz. This sensor interface could theoretically detect as few as 2,000 MNTs with an SNR of 6 dB and a 4 second readout time; however, we have found that the limit of detection (LOD) is often bounded by the stochastic biological variations and non-specific binding rather than the sensor and electronic noise.
V. TEMPERATURE CORRECTION
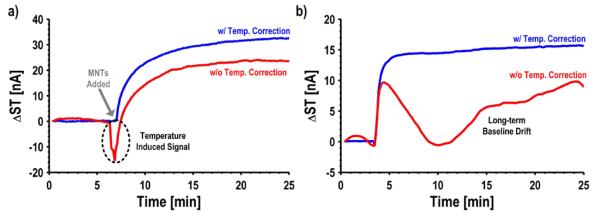
In addition to responding to changes in the local magnetic field, GMR SV sensors also respond to changes in temperature. These temperature-induced signals appear on all of the tones, and from the side tone alone, are indistinguishable from the signal induced by the MNTs. Temperature-induced signals and baseline drift in the binding curves are removed through a reference-less, background temperature correction technique [43]. The crux of our approach relies on using the CT to sense the relative temperature change on a sensor-by-sensor basis and digitally correcting the ST to remove the temperature dependence. This approach is successfully applied to this new sensor interface. To illustrate the effectiveness of this technique, Fig. 13(a) shows a measured trace from a sensor with and without the temperature correction enabled. When the unbound detection antibodies are washed away and the MNTs are added (t = 7 min), the uncorrected sensor exhibits a large temperature-induced signal (−16 nA). As a result, the subsequent response from the sensor underestimates the number of bound MNTs. However, the trace becomes pristine after applying the temperature correction algorithm, showing no signs of temperature-induced signals when the MNTs are added. In addition to the rapid temperature-induced signals, there can also be long-term baseline drift (Fig. 13(b)). This trace has very peculiar behavior and without the temperature correction might have been disregarded as being from a defective sensor. The same temperature correction algorithm also removes the long-term baseline drift components. Although this technique requires a pre-characterization step to determine the ratio of the temperature coefficients, it is preferred over a reference sensor approach due to the matching difficulties for GMR SV sensors. Others have tried to solve the temperature problem by using on-chip heaters to precisely regulate the temperature [10]. The approach used here works even when there are temperature gradients across the chip or the sensors are isolated in different channels via microfluidics because each sensor is measuring the relative temperature change it experiences.
VI. BIOLOGICAL MEASUREMENTS
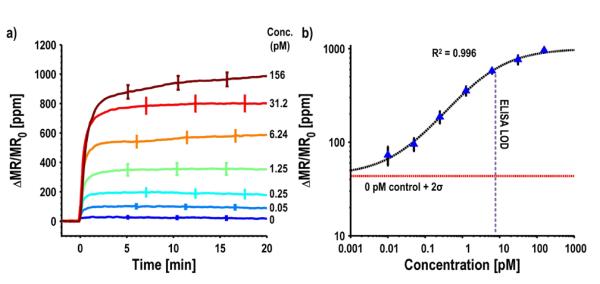
We characterized the biological performance of this system by measuring samples of secretory leukocyte peptidase inhibitor (SLPI) spiked into phosphate buffered solution (PBS). SLPI is being actively investigated as a biomarker indicative of many types of cancers such as gastric and ovarian cancer [44], [45]. We setup the experiment where each of the four sub-arrays is in a separate reaction well and monitors a different concentration of SLPI, so we can observe four different concentrations simultaneously. To monitor the variability within each sub-array, 24 sensors were functionalized with capture antibodies for SLPI. Additionally, there were three groups of four sensors, each coated with either epoxy, bovine serum albumin (BSA), or a noncomplimentary antibody for negative controls. BSA is a high-abundance protein commonly used to monitor nonspecific binding. Fig. 14(a) shows real-time binding curves for seven different concentrations of SLPI overlaid. The curves are presented in units of parts-per-million change in the MR normalized to the initial MR. This unit was devised to normalize out resistance and MR ratio variations across different wafers. Before adding the MNTs, all of the curves remain flat. When the MNTs are added to the assay, they become tethered above the surface of the SLPI functionalized sensors and detected by the underlying GMR SV biosensors. The negative controls all remained flat (< 30 ppm), indicating minimal nonspecific binding while the SLPI functionalized sensors exhibited smooth, clean binding curves. The saturation value for each of these concentrations was compiled into a calibration curve (Fig. 14(b)). A calibration curve allows one to quantify the concentration of an unknown sample. Another experiment (not shown) was run where no SLPI was added to determine the background level. The 0 pM control line represents this value plus two standard deviations. The calibration curve has a sigmoidal shape where the signal varies linearly (on a log-log plot) proportional to the concentration. The LOD for this biomarker was 10 fM with a linear dynamic range of three and a half orders of magnitude. Although 10 fM was clearly discernible above background, the error bars overlapped with the 50 fM data point, indicating a lack of precision in quantifying SLPI in the sub 100 fM regime. This same antibody pair and standard protein were run on ELISA and the LOD was found to be 5 pM, unequivocally demonstrating the superior detection capability of the GMR SV biosensors. This improvement in LOD is due to the lower background and high sensitivity of the GMR SV sensors. A summary of the electrical and biological measurement data is presented in Table 1.
Table 1.
Summary of electrical and biological measurement results.
| Measurement | Value | Units |
|---|---|---|
| Technology | 0.18 (2P/6M) | μm |
| VddA/Vdd/VddD | 2.0/ 2.1/ 1.8 | V |
| Readout Columns | 16 | -- |
| Area (with pads) | 2.7 × 2.7 | mm × mm |
| Power Consumption | 50.4 | mW |
| Gain | 84.9 | dBΩ |
| Sampling Frequency | 10 | MHz |
| Oversampling Ratio | 500 | -- |
| # Sensors | 256 | -- |
| Readout Time | 4 | s |
| Nominal Sensor Resistance | 1.84 | kΩ |
| MR Ratio | 9.2 | % |
| Input Referred Noise | 49 | nT/√Hz |
| DAC DNL | −0.32 to 0.31 | LSBs |
| DAC INL | −0.23 to 0.18 | LSBs |
| SFDR | >80.2 | dB |
| Multitone SFDR | 73.0 | dB |
| Adjacent Channel Crosstalk | 83.0 | dBc |
| Limit of Detection (SLPI) | 10 | fM |
| Linear Dynamic Range (SLPI) | 3.5 | Decades |
This biomarker is part of a panel of eight biomarkers being investigated in an ongoing ovarian cancer study. Most of the biomarkers in this panel are in the femtomolar sensitivity regime with three or more orders of linear dynamic range. The dynamic range is slightly higher than that of many optical setups; however, the sensitivity is improved, typically by 1-3 decades. Furthermore, this large sensor array allows all eight of the biomarkers, in addition to positive and negative controls, to be measured simultaneously. Using multiple biomarkers allows them to be combined using multivariate index analyses to form a diagnostic score for screening or diagnosing patients. Alternatively, the array can be further partitioned and multiple patient samples can be run in parallel.
One of the key advantages of this system over optical-based approaches is the ability to monitor the real-time kinetics of the reaction. Since GMR SV sensors are proximity-based sensors, only sensing the MNTs within ~150 nm of the surface [27], the unbound MNTs do not need to be removed. This is in contrast to an ELISA where the assay cannot be readout until the unreacted tags have been washed away since the imager would detect all of the tags, bound or unbound, distorting the measurement. Here, because the MNTs remain colloidally stable in solution, the sensors detect only the tethered nanotags [46]. One can see that the kinetics of the reaction are different as the concentration is increased (Fig. 14(a)). For low concentrations, the reaction occurs almost instantly whereas the higher concentrations take significantly longer to reach their final saturation value. The real-time binding curves provide the user with visual confirmation that the reaction has reached equilibrium. They also contain valuable information about the kinetics of the reaction between streptavidin (on MNT) and biotin (on the detection antibody captured at the sensor surface), such as the association rate constant (kon) and the disassociation rate constant (koff) of the kinetic reaction of the last step in the sandwich immunoassay [25]. Kinetic information can be used to predict the saturation signal before the reaction has had time to finish. Such modeling is applied to a wash-free magnetic immunoassay where antibody-conjugated MNTs are directly reacting with the surface-captured analytes, reducing the assay time in a point-of-care setting [47]. The potentials of applying magnetoresistive biosensor microarray to simultaneously measure kinetic parameters of many antibodies or antigens [25] make this work even more relevant because a large array of sensors and real-time readout are needed for such applications.
VII. COMPARISON TO OTHER SYSTEMS
This work compares favorably to previously published magnetic biosensors (Table 2). We have shown a scalable circuit architecture for GMR SV sensors, increasing the sensor count from 16 to 256 compared to prior work in our group [31]. Additionally, this work has over an order of magnitude lower readout time per sensor due to the combination of FDM and TDM with parallel readout channels. Most of the prior art on magnetic biosensors, such as in [10], [31], [48], have trended towards smaller superparamagnetic nanotags, likely due to the colloidal stability and increased dynamic range for a given sensor size. We report a theoretical LOD of 2,000 of the 50 nm MNTs. Although 2,000 tags sounds like a lot compared to just one, it is important to recognize that a single 1 μm tag has the same volume as 8,000 nanotags that are 50 nm in diameter. Furthermore, detecting several smaller MNTs leads to more binding events which averages out the stochastic variations inherent in the bioassay and gives more reproducible and reliable measurements.
Table 2.
Comparison to some of the previously published magnetic biosensors.
| [48] | [31] | [10] | [50] | This work | |
|---|---|---|---|---|---|
| Sensor Type | NMR | GMR SV | LC | Hall | GMR SV |
| # Sensors | 1 | 16 | 8 | 10,240 | 256 |
| Sensor Size [μm × μm] | 2,500 × 2,500 | 120 × 120 | N/A | 3 × 4 | 100 × 100 |
| Readout Time/Sensor [ms] | 200 | 250 | N/A | 0.78 | 16 |
| Magnetic Tag Size [nm] | 38 | 50 | 1,000 | 1,000 | 50 |
| Min. Detectable Magnetic Tags | N/A | N/A | 1 | 1 | 2,000 |
| Input Referred Noise [nT/√Hz] | N/A | N/A | N/A | 260 | 49 |
| Biological LOD [pM] | 5,000 (Protein) | 10,000 (DNA) | 1,000 (DNA) | 1,493 (Protein) | 0.01 (Protein) |
| Biological DR [orders] | N/A | N/A | N/A | 2 | 3.5 |
| Temperature Correction | No | Yes | Yes | No | Yes |
Most notably, the work reported here achieved a very low input referred noise of 49 nT√/Hz, which translated into a biological LOD of 10 fM — 100 times lower than most commercial ELISA kits, which have an LOD between 1-5 pM and two decades of linear dynamic range. This LOD is on par with lowest LOD reported for nanowires, microcantilevers, quantum dots, and other biosensors [49]. Furthermore, this work demonstrated biological detection on a relevant biomarker rather than a mock biomarker such as biotynlated albumin (either bovine or human). Often used as a model compound, biotinylated albumin is not a useful biomarker and greatly oversimplifies the assay. In fact, we use biotinylated BSA as a positive control in our assays since it places an absolute limit on the maximum achievable signal, often 2-3 times higher than we can achieve with a sandwich assay. This higher signal is due to the high affinity nature of the biotin-streptavidin bond and the close proximity of the tags to the sensors.
All of the biosensors compared in Table 2 are sensitive to temperature effects and most took design steps to correct the temperature dependence, albeit through a variety of methods. In [10], the authors regulated the temperature using an on-die temperature control loop whereas [31] used a foreground correction technique by modulating a second orthogonal magnetic field. The work described here implemented a digital temperature correction technique that removes the temperature dependence on a sensor by sensor basis in real-time.
VIII. CONCLUSION
This paper presented a sensor interface and acquisition system for a large array of GMR SV biosensors. An imager-like architecture was used with individually addressable GMR SV sensors and a parallel column readout structure. In total there are 16 readout columns each consisting of four DACs for carrier suppression, a pseudo-differential TIA, an ADC driver, and a 2nd order, highly oversampled ΣΔ modulator. A multi-domain modulation scheme reduces the 1/f noise of the sensor and a combination of FDM and TDM reduces the readout time. A background temperature correction algorithm was developed to digitally correct the response on a sensor-by-sensor basis by removing temperature-induced signals and long-term baseline drift. Lastly, we demonstrated state-of-the-art biological protein detection with an LOD of 10 fM while also highlighting the real-time readout capabilities.
Fig. 11.
Measured spectra illustrating adjacent channel isolation.
Fig. 12.
Measured spectrum showing modulation scheme and carrier suppression.
Fig. 13.
Illustrations showing measured binding curves with and without temperature correction enabled.
Fig. 14.
a) Measured real-time curves for various concentrations of SLPI biomarker. Error bars represent ±1σ. b) Calibration curve compiled from (a). Error bars represent ±1σ, background is 0 pM signal plus 2σ.
ACKNOWLEDGMENT
The authors thank the National Semiconductor Corporation for CMOS IC fabrication, funding and mentorship; Sebastian Osterfeld for GMR SV sensor design; NCI grants U54CA119367, U54CA143907, U54CA151459; NSF grant ECCS 0801365; Gates Grand Challenge Exploration Award; and Achievement Rewards for College Scientists (ARCS) for funding. The authors also tk Berkeley Design Automation for the use of the Analog FastSPICE Platform (AFS).
REFERENCES
- 1.Million RP. Impact of genetic diagnostics on drug development strategy. Nature Reviews Drug Discovery. 2006 May;5(6):459–462. doi: 10.1038/nrd2034. [DOI] [PubMed] [Google Scholar]
- 2.Schena M, Shalon D, Davis RW, Brown PO. Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray. Science. 1995 Oct;270(5235):467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 3.Cheng S, Fockler C, Barnes WM, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. PNAS. 1994 Jun;91(12):5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thaxton CS, Elghanian R, Thomas AD, Stoeva SI, Lee J-S, Smith ND, Schaeffer AJ, Klocker H, Horninger W, Bartsch G, Mirkin CA. Nanoparticle-based bio-barcode assay redefines ‘undetectable’ PSA and biochemical recurrence after radical prostatectomy. PNAS. 2009 Nov;106(44):18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall DA, Gaster RS, Osterfeld SJ, Makinwa K, Wang SX, Murmann B. A 256 channel magnetoresistive biosensor microarray for quantitative proteomics; presented at the 2011 Symposium on VLSI Circuits (VLSIC).2011. pp. 174–175. [Google Scholar]
- 6.Curry R, Heitzman H, Riege D, Sweet R, Simonsen M. A systems approach to fluorescent immunoassay: general principles and representative applications. Clin Chem. 1979 Sep;25(9):1591–1595. [PubMed] [Google Scholar]
- 7.Han M, Gao X, Su JZ, Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat Biotech. 2001 Jul;19(7):631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 8.Levine PM, Gong P, Levicky R, Shepard KL. Active CMOS Sensor Array for Electrochemical Biomolecular Detection. IEEE Journal of Solid-State Circuits. 2008 Aug.43(8):1859–1871. [Google Scholar]
- 9.Gaster RS, Hall DA, Nielsen CH, Osterfeld SJ, Yu H, Mach KE, Wilson RJ, Murmann B, Liao JC, Gambhir SS, Wang SX. Matrix-insensitive protein assays push the limits of biosensors in medicine. Nat Med. 2009 Nov;15(11):1327–1332. doi: 10.1038/nm.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Chen Y, Hassibi A, Scherer A, Hajimiri A. A frequency-shift CMOS magnetic biosensor array with single-bead sensitivity and no external magnet; presented at the Solid-State Circuits Conference - Digest of Technical Papers; 2009. pp. 438–439. ISSCC 2009. IEEE International, 2009. 439a. [Google Scholar]
- 11.Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008 Jul;14(8):869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florescu O, Mattmann M, Boser B. Fully integrated detection of single magnetic beads in complementary metal-oxide-semiconductor. J. Appl. Phys. 2008;103(4):046101. [Google Scholar]
- 13.Liu P, Skucha K, Duan Y, Megens M, Kim Jungkyu, Izyumin I, Gambini S, Boser B. Magnetic relaxation detector for microbead labels in biomedical assays; presented at the 2011 Symposium on VLSI Circuits (VLSIC); 2011. pp. 176–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritz J, Baller MK, Lang HP, Rothuizen H, Vettiger P, Meyer E, Güntherodt H-J, Gerber C, Gimzewski JK. Translating Biomolecular Recognition into Nanomechanics. Science. 2000 Apr;288(5464):316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 15.Stern E, F. Klemic J, Routenberg DA, Wyrembak PN, Turner-Evans DB, Hamilton AD, LaVan DA, Fahmy TM, Reed MA. Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature. 2007 Feb;445(7127):519–522. doi: 10.1038/nature05498. [DOI] [PubMed] [Google Scholar]
- 16.Hagleitner C, Hierlemann A, Lange D, Kummer A, Kerness N, Brand O, Baltes H. Smart single- chip gas sensor microsystem. Nature. 2001 Nov;414(6861):293–296. doi: 10.1038/35104535. [DOI] [PubMed] [Google Scholar]
- 17.Clarke J, Wu H-C, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nano. 2009 Apr;4(4):265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 18.Boozer C, Kim G, Cong S, Guan H, Londergan T. Looking towards label-free biomolecular interaction analysis in a high-throughput format: a review of new surface plasmon resonance technologies. Current Opinion in Biotechnology. 2006 Aug;17(4):400–405. doi: 10.1016/j.copbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Giloh H, Sedat J. Fluorescence microscopy: reduced photobleaching of rhodamine and fluorescein protein conjugates by n-propyl gallate. Science. 1982 Sep;217(4566):1252–1255. doi: 10.1126/science.7112126. [DOI] [PubMed] [Google Scholar]
- 20.Aubin JE. Autofluorescence of viable cultured mammalian cells. Journal of Histochemistry & Cytochemistry. 1979 Jan;27(1):36–43. doi: 10.1177/27.1.220325. [DOI] [PubMed] [Google Scholar]
- 21.Bruls DM, Evers TH, Kahlman JAH, van Lankvelt PJW, Ovsyanko M, Pelssers EGM, Schleipen JJHB, de Theije FK, Verschuren CA, van der Wijk T, van Zon JBA, Dittmer WU, Immink AHJ, Nieuwenhuis JH, Prins MWJ. Rapid integrated biosensor for multiplexed immunoassays based on actuated magnetic nanoparticles. Lab Chip. 2009 Oct;9(24):3504–3510. doi: 10.1039/b913960e. [DOI] [PubMed] [Google Scholar]
- 22.Aytur T, Foley J, Anwar M, Boser B, Harris E, Beatty PR. A novel magnetic bead bioassay platform using a microchip-based sensor for infectious disease diagnosis. Journal of Immunological Methods. 2006 Jul.314(1–2):21–29. doi: 10.1016/j.jim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Rife JC, Miller MM, Sheehan PE, Tamanaha CR, Tondra M, Whitman LJ. Design and performance of GMR sensors for the detection of magnetic microbeads in biosensors. Sensors and Actuators A: Physical. 2003 Nov;107(3):209–218. [Google Scholar]
- 24.Yu H, Osterfeld SJ, Xu L, White RL, Pourmand N, Wang SX. Giant magnetoresistive biosensors for molecular diagnosis: surface chemistry and assay development; presented at the Biosensing; San Diego, CA, USA: 2008. pp. 70350E–70350E. 9. [Google Scholar]
- 25.Gaster RS, Xu L, Han S-J, Wilson RJ, Hall DA, Osterfeld SJ, Yu H, Wang SX. Quantification of protein interactions and solution transport using high-density GMR sensor arrays. Nat Nano. 2011 May;6(5):314–320. doi: 10.1038/nnano.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsang CH, Fontana RE, Lin T, Heim DE, Gurney BA, Williams ML. Design, fabrication, and performance of spin-valve read heads for magnetic recording applications. IBM Journal of Research and Development. 1998 Jan;42(1):103–116. [Google Scholar]
- 27.Osterfeld SJ, Yu H, Gaster RS, Caramuta S, Xu L, Han S-J, Hall DA, Wilson RJ, Sun S, White RL, Davis RW, Pourmand N, Wang SX. Multiplex protein assays based on real-time magnetic nanotag sensing. PNAS. 2008 Dec;105(52):20637–20640. doi: 10.1073/pnas.0810822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall DA, Gaster RS, Lin T, Osterfeld SJ, Han S, Murmann B, Wang SX. GMR biosensor arrays: A system perspective. Biosensors and Bioelectronics. 2010 May;25(9):2051–2057. doi: 10.1016/j.bios.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Boer BM, Kahlman JAHM, Jansen TPGH, Duric H, Veen J. An integrated and sensitive detection platform for magneto-resistive biosensors. Biosensors and Bioelectronics. 2007 Apr.22(9–10):2366–2370. doi: 10.1016/j.bios.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 30.Aytur T, Beatty PR, Boser B, Anwar M, Ishikawa T. An immunoassay platform based on CMOS Hall sensors; Proceedings Solid-State and Actuator Workshop; Hilton Head Island, SC. 2002. pp. 126–129. [Google Scholar]
- 31.Han S-J, Yu H, Murmann B, Pourmand N, Wang SX. A High-Density Magnetoresistive Biosensor Array with Drift-Compensation Mechanism; presented at the Solid-State Circuits Conference, 2007. ISSCC 2007. Digest of Technical Papers. IEEE International.2007. pp. 168–594. [Google Scholar]
- 32.Romani A, Manaresi N, Marzocchi L, Medoro G, Leonardi A, Altomare L, Tartagni M, Guerrieri R. Capacitive sensor array for localization of bioparticles in CMOS lab-on-a-chip; presented at the Solid-State Circuits Conference, 2004. Digest of Technical Papers. ISSCC. 2004 IEEE International.2004. pp. 224–225. [Google Scholar]
- 33.Augustyniak M, Paulus C, Brederlow R, Persike N, Hartwich G, Schmitt-Landsiedel D, Thewes R. A 24x16 CMOS-Based Chronocoulometric DNA Microarray; presented at the Solid-State Circuits Conference, 2006. ISSCC 2006. Digest of Technical Papers. IEEE International.2006. pp. 59–68. [Google Scholar]
- 34.Boser BE, Wooley BA. The design of sigma-delta modulation analog-to-digital converters. IEEE Journal of Solid-State Circuits. 1988 Dec;23(6):1298–1308. [Google Scholar]
- 35.Schreier R. Delta Sigma Toolbox. 2000 [Google Scholar]
- 36.Ardalan S, Paulos J. An analysis of nonlinear behavior in delta - sigma modulators. IEEE Transactions on Circuits and Systems. 1987 Jun;34(6):593–603. [Google Scholar]
- 37.Feldman AR, Boser BE, Gray PR. A 13-bit, 1.4-MS/s sigma-delta modulator for RF baseband channel applications. IEEE Journal of Solid-State Circuits. 1998 Oct;33(10):1462–1469. [Google Scholar]
- 38.Dessouky M, Kaiser A. Rail-to-rail operation of very low voltage CMOS switched-capacitor circuits; presented at the Proceedings of the 1999 IEEE International Symposium on Circuits and Systems, 1999. ISCAS ’99; 1999. pp. 144–147. 2. [Google Scholar]
- 39.Murmann B. ADC Performance Survey 1997-2012. [Online]. Available: http://www.stanford.edu/~murmann/adcsurvey.html.
- 40.Goes J, Vaz B, Monteiro R, Paulino N. A 0.9V /spl Delta//spl Sigma/ Modulator with 80dB SNDR and 83dB DR Using a Single-Phase Technique; Solid-State Circuits Conference, 2006. ISSCC 2006. Digest of Technical Papers. IEEE International.2006. pp. 191–200. [Google Scholar]
- 41.van Veldhoven R, Rutten R, Breems LJ. An Inverter-Based Hybrid ΔΣ Modulator; Solid-State Circuits Conference, 2008. ISSCC 2008. Digest of Technical Papers. IEEE International.2008. pp. 492–630. [Google Scholar]
- 42.Chae Youngcheol, Lee Inhee, Han Gunhee. A 0.7V 36μW 85dB-DR Audio ΔΣ Modulator Using Class-C Inverter; Solid-State Circuits Conference, 2008. ISSCC 2008. Digest of Technical Papers. IEEE International.2008. pp. 490–630. [Google Scholar]
- 43.Hall DA, Gaster RS, Osterfeld SJ, Murmann B, Wang SX. GMR biosensor arrays: Correction techniques for reproducibility and enhanced sensitivity. Biosensors and Bioelectronics. 2010 May;25(9):2177–2181. doi: 10.1016/j.bios.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng W-L, Wang C-S, Huang Y-H, Liang Y, Lin PY, Hsueh C, Wu Y-C, Chen W-J, Yu C-J, Lin S-R, Lin K-H. Overexpression of a secretory leukocyte protease inhibitor in human gastric cancer. Int. J. Cancer. 2008 Oct;123(8):1787–1796. doi: 10.1002/ijc.23746. [DOI] [PubMed] [Google Scholar]
- 45.Tsukishiro S, Suzumori N, Nishikawa H, Arakawa A, Suzumori K. Use of serum secretory leukocyte protease inhibitor levels in patients to improve specificity of ovarian cancer diagnosis. Gynecologic Oncology. 2005 Feb;96(2):516–519. doi: 10.1016/j.ygyno.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 46.Gaster RS, Hall DA, Wang SX. Autoassembly Protein Arrays for Analyzing Antibody Cross- Reactivity. Nano Letters. 2011 Jul;11(7):2579–2583. doi: 10.1021/nl1026056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaster RS, Hall DA, Wang SX. nanoLAB: An ultraportable, handheld diagnostic laboratory for global health. Lab Chip. 2011 Jan.11(5):950. doi: 10.1039/c0lc00534g. [DOI] [PubMed] [Google Scholar]
- 48.Sun Nan, Yoon Tae-Jong, Lee Hakho, Andress W, Weissleder R, Ham Donhee. Palm NMR and 1- Chip NMR. IEEE Journal of Solid-State Circuits. 2011 Jan;46(1):342–352. [Google Scholar]
- 49.Arlett JL, Myers EB, Roukes ML. Comparative advantages of mechanical biosensors. Nature Nanotechnology. 2011 Mar;6(4):203–215. doi: 10.1038/nnano.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gambini S, Skucha K, Liu P, Kim J, Krigel R, Mathies R, Boser B. A CMOS 10kpixel Baseline-Free Magnetic Bead Detector with Column-Parallel Readout for Miniturized Immunoassays; presented at the Solid-State Circuits Conference, 2012. Digest of Technical Papers. ISSCC. 2012 IEEE International.2012. pp. 126–127. [Google Scholar]