Abstract
α–synuclein (α-syn) is a small lipid binding protein implicated in several neurodegenerative diseases, including Parkinson’s disease, whose pathobiology is conserved from yeast to man. There are no therapies targeting these underlying cellular pathologies, or indeed those of any major neurodegenerative disease. Using unbiased phenotypic screens as an alternative to target-based approaches, we discovered an N-aryl benzimidazole (NAB) that strongly and selectively protected diverse cell-types from α-syn toxicity. Three chemical genetic screens in wild-type yeast cells established that NAB promoted endosomal transport events dependent on the E3 ubiquitin ligase, Rsp5/Nedd4. These same steps were perturbed by α–syn itself. Thus, NAB identifies a druggable node in the biology of α-syn that can correct multiple aspects of its underlying pathology, including dysfunctional endosomal and ER-to-Golgi vesicle trafficking.
Phenotypic cell-based drug screens are a powerful yet underutilized approach to identify lead compounds and probe the underlying cellular pathologies that cause human disease (1). Such unbiased screens may be particularly helpful for neurodegenerative diseases (ND), such as Parkinson’s Disease (PD) and Alzheimer’s Disease (AD), where the molecular underpinnings of disease remain unclear. However, establishing robust neuronal phenotypes amenable to high-throughput screening and subsequent target identification remains a challenge. To bridge this gap, we exploit yeast cells that express ND-causing proteins to recapitulate salient cellular pathologies. α-synuclein, for example, causes derangements in vesicle trafficking, metal ion homeostasis and mitochondrial function that are associated with α–synucleinopathies, such as PD (2). The resulting growth inhibition greatly facilitates robust high throughput screening.
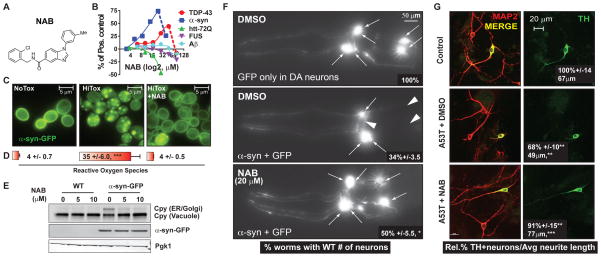
We recently screened ~190,000 compounds for their ability to restore the growth of cells expressing toxic levels of TDP-43 (3), a protein associated with diverse NDs. A weak hit from that screen, an N-Aryl Benzimidazole (NAB; Fig. 1A), proved more potent and effective against α-syn toxicity (Fig. 1B). NAB reversed diverse phenotypes caused by α–syn (4–7), including the accumulation of vesicular α-syn foci (Fig. 1C) (8, 9), the generation of reactive oxygen species (Fig. 1D), the block of ER-Golgi trafficking (Fig. 1E), and the nitration of proteins (6). The toxicity of α–syn is extremely dependent on expression levels (7, 10); however, NAB did not reduce α-syn accumulation (Fig. 1E).
Fig. 1. NAB protects yeast and neurons from α-syn toxicity.
(A) NAB structure. (B) Dose-response curves for NAB in yeast proteinopathy models. (C) α–syn-GFP localization in NoTox, HiTox, and HiTox/10 μM NAB. (D) Percent of ROS-positive cells under same conditions as (C). Values are mean +/− SD, N = 3. (E) Immunoblot of Cpy showing ER-Golgi trafficking defect and α-syn protein levels in WT and HiTox cells treated with NAB. (F) Fluorescence microscopy of the six anterior DA neurons in representative C. elegans expressing either GFP or human α–syn and GFP after treatment with DMSO or NAB. Arrows indicate intact DA neuron cell bodies and arrowheads indicate regions where these cells have degenerated. Inlaid values reflect mean +/− SD, N = 3. (G) Representative images of DA-enriched cultures established from embryonic rat midbrains. ‘Control’, untransduced. ‘A53T’, transduced with A53T α-syn virus. Red, MAP2 (neuronal tubulin). Green, TH-positive neurons. Inlaid values reflect mean with control set to 100% and +/− SD, N = 3 for % TH-positive neurons and mean +/− SEM, N = ~75 neurons for neurite length. For all data: *, P<0.05. **, P<0.01. ***, P<0.001 using one-way ANOVA and a Tukey’s test.
We next asked if the protective activity of NAB was conserved in neurons. First, we tested a nematode model where PD-relevant dopaminergic neurons (DA) degenerate in an age-dependent manner in response to human α-syn expression (5). Second, we tested rat primary neuronal cultures where adenoviral expression of a familial α-syn mutation (A53T) causes selective loss of DA neurons: tyrosine-hydroxylase (TH) positive cells are lost and neuronal processes retract (4, 5). Third, we tested cortical neurons differentiated from PD-patient induced pluripotent stem cells (iPSCs) carrying either the A53T α–syn mutation or an α-syn triplication, both of which caused protein trafficking defects and nitrosative stress (6). In each case, NAB reversed α–syn toxicity or pathology, suggesting the compound’s target and mechanisms of action (MOA) were conserved from yeast to human cells (Fig. 1F, 1G, Fig. S1 (6)).
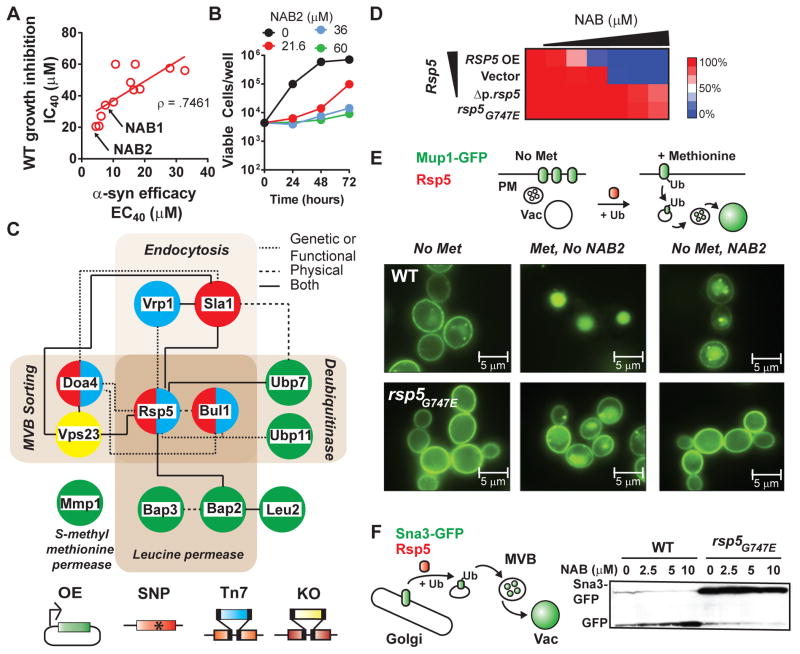
Yeast screens can reveal the target space for small molecules that suppress growth by identifying genetic alterations that restore it (11). At concentrations higher than those that rescued α–syn toxicity, NAB inhibited the growth of WT cells. To test if α-syn rescue and growth inhibition had related MOA, we synthesized twenty-nine NAB analogs (Fig. S2). Compounds inactive in rescuing α-syn did not reverse α–syn foci formation or rescue ER-to-Golgi trafficking (Fig. S3). Compounds that potently rescued α–syn, also more potently inhibited growth in WT cells (Fig. 2A, Fig. S2, supplementary online text). This empowered us to use genetic screens in WT cells to investigate NAB’s MOA. Importantly, though NAB inhibited growth, cells retained full viability (Fig. 2B).
Fig. 2. Chemical genetic screens of NAB2 reveal a network centered on the E3 ligase, Rsp5.
(A) Efficacy (EC40) in α-syn cells versus growth inhibition (IC40) in WT cells for active analogs. NAB1 is the screen hit and NAB2 the most potent analog. (B) Viable cells recovered after prolonged NAB2 treatment. (C) NAB2 interaction network. Node color reflects screen of origin indicated below. Edges are interactions (see legend) according to String database and literature. VPS23 was deleted after identification of other hits. (D) Heat map of RSP5 variant cell growth in response to increasing NAB2 compared to untreated cells. Mutants include rsp5G747E and the hypomorphic allele, Δp.rsp5. (E) (Top) Methionine- and Rsp5-dependent Mup1-GFP endocytosis. (Bottom) Mup1-GFP localization in WT and rsp5G747E strains under indicated conditions. (F) (Left) Schematic of Sna3-GFP endosomal trafficking to the vacuole, where GFP is cleaved. (Right) Immunoblot analysis of Sna3-GFP in WT and rsp5G747E cells treated with NAB2.
Using our most potent analog – NAB2 – we selected for genetic alterations that allowed growth at high concentrations. We used three approaches: (1) a library of over-expression strains covering most genes in the yeast genome (~5,800 genes), (2) a library of ~ 300,000 random transposon-insertions (12), and (3) spontaneous genomic point mutations arising from ~2 million cells (fig. S4). A small number of hits were recovered and formed a highly connected network of functionally related genes (Fig. 2C). These were an E3 ubiquitin ligase that promotes endosomal transport (RSP5), endocytic proteins (SLA1, VRP1), a multivesicular body sorting deubiquitinase (DOA4), an Rsp5 adaptor (BUL1), two proteins that can deubiquitinate Rsp5 substrates (UBP7, UBP11), known and potential Rsp5 substrates (BAP2, BAP3, MMP1) and VPS23, which directs Rsp5 substrates for degradation in the vacuole (Fig. 2C). Analogs ineffective against α–syn did not exhibit dosage sensitivity with NAB network genes (Fig. S5), thus supporting a related MOA between α–syn rescue and growth inhibition of WT cells.
The network topology of screen hits, and the nature of their altered dosage-sensitivity to NAB2 (Fig. S6–S8), suggested that NAB acts on Rsp5 to promote ubiquitin-mediated endosomal transport. With the exception of RSP5, which is essential, every other gene in our network could be deleted. But no deletion (including a double deletion of UBP7 and UBP11) conferred more than partial resistance to NAB2. Thus, while these other proteins are involved in NAB2 activity, they cannot themselves be its target. Indeed, the effects of altering RSP5 gene dosage indicate it is the central node: increased RSP5 dosage increased sensitivity to NAB2 and decreased RSP5 dosage decreased sensitivity to NAB2 (Fig. 2D). Furthermore, in otherwise isogenic cells, a single amino acid substitution in the ~1000 amino acid protein (rsp5G747E) conferred resistance to NAB2 (Fig. 2D, Fig. S7).
Rsp5 is the single yeast member of the highly conserved mammalian family of HECT domain Nedd4 E3 ligases. These proteins catalyze K63 linkages of ubiquitin to diverse membrane proteins and thereby regulate endosomal trafficking, not proteasomal degradation (13, 14). HECT domain ubiquitin ligases contain multiple protein-protein interaction domains that bind diverse adaptor proteins and substrates. Calcium, lipid binding, and autoinhibitory conformations regulate substrate specificity and endosomal transport from either the plasma membrane or Golgi to the vacuole/lysosome. Most aspects of these complex modes of Rsp5 regulation have not been recapitulated in vitro.
Therefore, to further investigate NAB2 activities, we monitored, in WT cells, its effects on three proteins whose trafficking depends on Rsp5: Mup1 (15), Sna3 (16) and Bap2 (17). NAB2 (1) promoted the Rsp5-dependent endocytosis and vacuolar delivery of the methionine permease, Mup1 (Fig. 2E, Fig. S9), (2) promoted the Rsp5-dependent Golgi-to-vacuole trafficking of the adaptor protein, Sna3 (Fig. 2F, Fig. S9), and (3) promoted the Rsp5-dependent degradation of the leucine permease, Bap2 (Fig. S10). (This affected leucine-dependent growth explaining its recovery in our overexpression screen.)
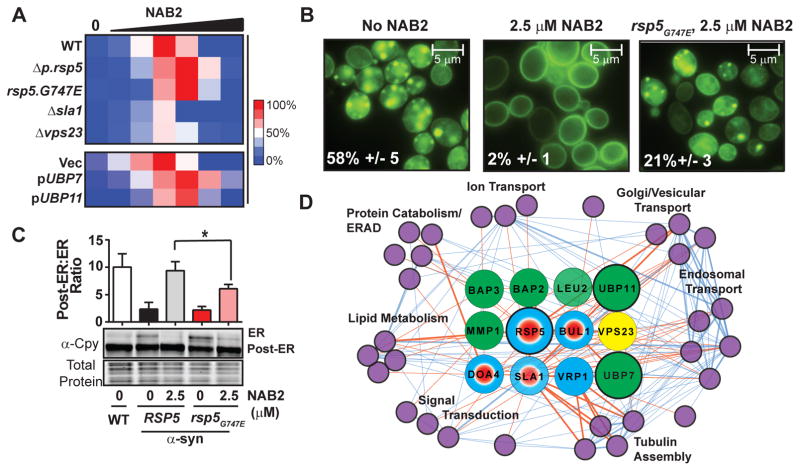
We further established the relevance of the NAB/Rsp5 network by genetically altering screen hits in the context of α–syn. Indeed, most but not all genetic manipulations that antagonized NAB2 activities in WT cells antagonized its activities against α–syn toxicity. For example, deleting Δsla1 and Δvps23, or overexpressing the deubiquitinases UBP7 and UBP11, all partially compromised the ability of NAB2 to rescue α–syn toxicity (Fig. 3A, Fig. S11). Deleting Δubp7 and Δubp11, either singly or in combination, had no effect NAB2’s rescue of α–syn toxicity (Fig. S11).
Fig. 3. NAB/Rsp5 network directly impacts rescue of α–syn toxicity.
(A) Heat map of NAB2 dose-response in WT and modified α–syn strains. Rescue is relative to EC100 for NAB2 in WT α–syn cells. (B) α-syn-GFP localization in WT or rsp5G747E α-syn cells under indicated conditions. Inlaid values indicate “% of cells with large α–syn foci” (mean +/− SD, N = 3). (C) Immunoblot of Cpy trafficking defect α–syn cells with DMSO or NAB2. *,P<0.05 using one-way ANOVA. (D) Interaction network of α-syn and NAB2 genetic modifiers. α-syn nodes, purple; NAB2 nodes are color coded according to screen of origin (Fig. 2C). RSP5, UBP7, and UBP11 are larger because they both suppress NAB2 growth inhibition and enhance α–syn toxicity (4). Edges between nodes depict physical or genetic interactions. Thicker lines indicate both genetic and physical interactions. Red edges link node interactions between α–syn and NAB2 networks. Remaining α-syn edges are blue.
The spontaneous point mutation recovered in our screen, rsp5G747E, which compromised Rsp5 activity (Fig. 2E, 2F, Fig. S7) and conferred resistance to NAB2 in WT cells, shifted the dose of NAB2 required for α-syn rescue in a complementary fashion (Fig. 3A). Consistent with this, rsp5G747E prevented NAB2 from fully reverting the formation of α-syn foci (Fig. 3B) and from restoring ER-to-Golgi trafficking (Fig. 3C) phenotypes. Together, these analyses established Rsp5 as the central node and only potential protein target within the NAB network.
Rsp5’s importance in modifying α-syn toxicity was highlighted by integrating the NAB/Rsp5 and α–syn genetic networks ((4, 18) and Table S3). These interactions connected our NAB network to nearly 30% of the previously established genetic modifiers of α-syn toxicity, including those that function in Golgi/vesicular transport, endosomal transport, lipid metabolism, protein catabolism, and tubulin assembly (Fig. 3D, Fig. S12).
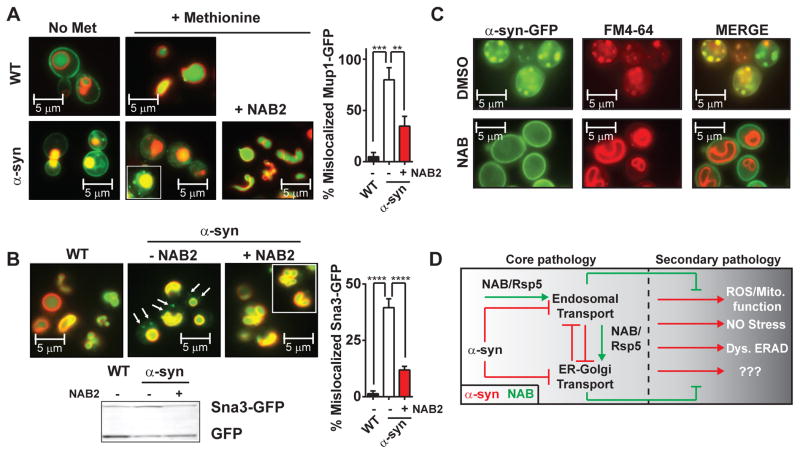
Next we tested α–syn’s effect on Mup1-GFP and Sna3-GFP trafficking. Indeed, α–syn expression impeded both the methionine-induced transport of Mup1-GFP from the plasma membrane to the vacuole (Fig. 4A) and the constitutive trafficking of Sna3-GFP from the Golgi and the vacuole (Fig. 4B). Further, in the presence of α-syn, NAB2 restored trafficking of both substrates (Fig. 4A, 4B).
Fig. 4. NAB2 directly antagonizes α–syn-induced endosomal defects.
(A) Methionine-stimulated Mup1-GFP endocytosis in WT or untagged α–syn strains with DMSO or NAB2. Pulse-labeling cells with FM4-64 during the first hour of α–syn expression marked the vacuole. (B) Effects of α-syn on Sna3-GFP localization. Immunoblot shows Sna3-GFP cleavage in response to α–syn and NAB. FM4-64 labeling as in (B). (C) Pulse-labeling of FM4-64 of α-syn cells after 4 hours of expression in the presence or absence of NAB2. (D) Schematic of NAB2 mechanism in antagonizing core and secondary α–syn pathologies.
In addition to specific substrates, bulk endosomal transport from the plasma membrane to the vacuole was perturbed by α–syn (Fig.4C) (7–9, 19). When FM4-64 was used to pulse-label the endosomal pathway, after prolonged α–syn expression, the dye strongly co-localized with α-syn inclusions and failed to reach the vacuole (Fig. 4C). NAB2 fully restored endocytosis and concomitantly reduced α–syn inclusions (Fig. 4C, bottom panel). Thus, the ability of NAB to promote Rsp5-dependent processes directly restored diverse cellular pathologies caused by α–syn, including both ER-to-Golgi and endosomal trafficking (Fig. 4D, Fig. S8).
Rsp5/Nedd4 can ubiquitinate α–syn and Nedd4 localizes to Lewy Bodies in brain samples from PD patients (20). However, α–syn levels were not altered by NAB2 in vivo (Fig. 1E). And, when tested in vitro, NAB2 did not affect the ubiquitination of α-syn and Sna3 by Rsp5 (Fig. S13). As noted, however, most of the complexities of Rsp5 in vivo activities have yet to be recapitulated in vitro. Thus, NAB2 exemplifies the ability of unbiased in vivo phenotypic screens to uncover chemical probes that cannot be discovered through simple target-based in vitro approaches. Likewise, NAB2 chemical genetics identify a deeply rooted biological node, Rsp5 that had not been identified previous overexpression or deletion screens. Notably, despite their central role in protein homeostasis and several human diseases, to date E3 ubiquitin ligases are virtually untouched by biological probes, let alone therapeutics.
The vesicular trafficking processes perturbed by α–syn and promoted by NAB are fundamental to all eukaryotic cells, yet are particularly important to neurons that rely heavily on efficient synaptic vesicle dynamics and regulated neurotransmitter release. Indeed, dysfunctional endosomal transport is emerging as contributing factor in α-syn pathology in human neurons. Altered cell biology, post-mortem pathology, and human genetic risk factors all implicate altered vesicular trafficking (4, 7–9, 19, 21–25). The ability of NAB to promote endosomal trafficking through Rsp5/Nedd4 and thus ‘reset’ vesicle trafficking homeostasis, in turn, rescued several other, seemingly disparate, α–syn phenotypes. Identifying such deeply rooted pathways that ramify to affect multiple aspects of protein folding pathology may be critical for developing disease-modifying therapies.
Supplementary Material
Acknowledgments
We thank Anuj Kumar for providing the plasmid Tn7 library; B. Wendland/L. Hicke for the Rsp5 antibody; the WIBR Genome Technology Core for Illumina Sequencing; Brenda Schulman (BS and HK funded by NIH grant 5R01GM069530) and Alfred Goldberg for help with in vitro ubiquitination assays; Tom DiCesare for graphics support; and members of the Lindquist Lab for helpful comments on the manuscript. SL is an investigator for the Howard Hughes Medical Institute. DFT was funded by an NRSA Fellowship F32NS061419 and research supported by the JPB Foundation (DFT/SL), the Eleanor Schwartz Charitable Foundation, and an HHMI Collaborative Innovation Award (DFT/VK/CYC/SL, GAC/KAC/MLT, and JCR/MT). NTJ was funded by an NRSA fellowship (F32GM099817) and NTJ/SLB by NIH grant GM58160. HTK was funded by The Bachmann-Strauss Dystonia & Parkinson Foundation. SL, SLB, DFT, and NTJ are inventors on a pending patent application relating to work described in this paper. Genome sequencing data is deposited in NCBI under BioProject accession number PRJNA222476.
References and Notes
- 1.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011 Jul;10:507. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 2.Khurana V, Lindquist S. Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker’s yeast? Nat Rev Neurosci. 2010 Jun;11:436. doi: 10.1038/nrn2809. [DOI] [PubMed] [Google Scholar]
- 3.Tardiff DF, Tucci ML, Caldwell KA, Caldwell GA, Lindquist S. Different 8-hydroxyquinolines protect models of TDP-43 protein, alpha-synuclein, and polyglutamine proteotoxicity through distinct mechanisms. J Biol Chem. 2012 Feb 3;287:4107. doi: 10.1074/jbc.M111.308668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper AA, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006 Jul 21;313:324. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su LJ, et al. Compounds from an unbiased chemical screen reverse both ER-to-Golgi trafficking defects and mitochondrial dysfunction in Parkinson’s disease models. Dis Model Mech. 2010 Mar-Apr;3:194. doi: 10.1242/dmm.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung CY, et al. Identification and Rescue of α-Synuclein Toxicity in Neurons Derived from Parkinson’s Disease Patients co-submission. 2013. [Google Scholar]
- 7.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003 Dec 5;302:1772. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gitler AD, et al. The Parkinson’s disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci U S A. 2008 Jan 8;105:145. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soper JH, Kehm V, Burd CG, Bankaitis VA, Lee VM. Aggregation of alpha-synuclein in S. cerevisiae is associated with defects in endosomal trafficking and phospholipid biosynthesis. J Mol Neurosci. 2011 Mar;43:391. doi: 10.1007/s12031-010-9455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singleton AB, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003 Oct 31;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 11.Smith AM, Ammar R, Nislow C, Giaever G. A survey of yeast genomic assays for drug and target discovery. Pharmacol Ther. 2010 Aug;127:156. doi: 10.1016/j.pharmthera.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A. Multipurpose transposon insertion libraries for large-scale analysis of gene function in yeast. Methods Mol Biol. 2008;416:117. doi: 10.1007/978-1-59745-321-9_8. [DOI] [PubMed] [Google Scholar]
- 13.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009 Jun;10:398. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 14.Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, Andre B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 2010 Apr;20:196. doi: 10.1016/j.tcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Menant A, Barbey R, Thomas D. Substrate-mediated remodeling of methionine transport by multiple ubiquitin-dependent mechanisms in yeast cells. EMBO J. 2006 Oct 4;25:4436. doi: 10.1038/sj.emboj.7601330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacDonald C, Stringer DK, Piper RC. Sna3 is an Rsp5 adaptor protein that relies on ubiquitination for its MVB sorting. Traffic. 2012 Apr;13:586. doi: 10.1111/j.1600-0854.2011.01326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omura F, Kodama Y, Ashikari T. The basal turnover of yeast branched-chain amino acid permease Bap2p requires its C-terminal tail. FEMS Microbiol Lett. 2001 Jan 15;194:207. doi: 10.1111/j.1574-6968.2001.tb09471.x. [DOI] [PubMed] [Google Scholar]
- 18.Yeger-Lotem E, et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nat Genet. 2009 Mar;41:316. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sancenon V, et al. Suppression of alpha-synuclein toxicity and vesicle trafficking defects by phosphorylation at S129 in yeast depends on genetic context. Hum Mol Genet. 2012 Jun 1;21:2432. doi: 10.1093/hmg/dds058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tofaris GK, et al. Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc Natl Acad Sci U S A. 2011 Oct 11;108:17004. doi: 10.1073/pnas.1109356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esposito G, Ana Clara F, Verstreken P. Synaptic vesicle trafficking and Parkinson’s disease. Dev Neurobiol. 2012 Jan;72:134. doi: 10.1002/dneu.20916. [DOI] [PubMed] [Google Scholar]
- 22.Macleod DA, et al. RAB7L1 Interacts with LRRK2 to Modify Intraneuronal Protein Sorting and Parkinson’s Disease. Risk Neuron. 2013 Feb 6;77:425. doi: 10.1016/j.neuron.2012.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vilarino-Guell C, et al. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011 Jul 15;89:162. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimprich A, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 2011 Jul 15;89:168. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zabrocki P, et al. Phosphorylation, lipid raft interaction and traffic of alpha-synuclein in a yeast model for Parkinson. Biochim Biophys Acta. 2008 Oct;1783:1767. doi: 10.1016/j.bbamcr.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Treusch S, et al. Functional links between Abeta toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science. 2011 Dec 2;334:1241. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ju S, et al. A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 2011 Apr;9:e1001052. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, et al. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007 Apr;17:536. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol. 1995;48:3. [PubMed] [Google Scholar]
- 30.Cao S, Gelwix CC, Caldwell KA, Caldwell GA. Torsin-mediated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans. J Neurosci. 2005 Apr 13;25:3801. doi: 10.1523/JNEUROSCI.5157-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F, et al. Methionine sulfoxide reductase A protects dopaminergic cells from Parkinson’s disease-related insults. Free Radic Biol Med. 2008 Aug 1;45:242. doi: 10.1016/j.freeradbiomed.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009 Jul 15;25:1754. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011 May;43:491. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012 Apr 19; doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HC, Steffen AM, Oldham ML, Chen J, Huibregtse JM. Structure and function of a HECT domain ubiquitin-binding site. EMBO Rep. 2011 Apr;12:334. doi: 10.1038/embor.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchiki T, et al. The Ubiquitin-interacting Motif Protein, S5a, Is Ubiquitinated by All Types of Ubiquitin Ligases by a Mechanism Different from Typical Substrate Recognition. J Biol Chem. 2009 May 8;284:12622. doi: 10.1074/jbc.M900556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saeki Y, Isono E, Toh EA. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 2005;399:215. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- 38.Kamadurai HB, et al. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. Elife. 2013;2:e00828. doi: 10.7554/eLife.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mercado G, Valdes P, Hetz C. An ERcentric view of Parkinson’s disease. Trends Mol Med. 2013 Mar;19:165. doi: 10.1016/j.molmed.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Winslow AR, Rubinsztein DC. The Parkinson disease protein alpha-synuclein inhibits autophagy. Autophagy. 2011 Apr;7:429. doi: 10.4161/auto.7.4.14393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lynch-Day MA, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010 Apr 27;107:7811. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duennwald ML, Jagadish S, Giorgini F, Muchowski PJ, Lindquist S. A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci U S A. 2006 Jul 18;103:11051. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gelperin DM, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005 Dec 1;19:2816. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampaio-Marques B, et al. SNCA (alpha-synuclein)-induced toxicity in yeast cells is dependent on sirtuin 2 (Sir2)-mediated mitophagy. Autophagy. 2012 Oct;8:1494. doi: 10.4161/auto.21275. [DOI] [PubMed] [Google Scholar]
- 45.Lee YJ, Wang S, Slone SR, Yacoubian TA, Witt SN. Defects in very long chain fatty acid synthesis enhance alpha-synuclein toxicity in a yeast model of Parkinson’s disease. PLoS One. 2011;6:e15946. doi: 10.1371/journal.pone.0015946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sere YY, Regnacq M, Colas J, Berges T. A Saccharomyces cerevisiae strain unable to store neutral lipids is tolerant to oxidative stress induced by alpha-synuclein. Free Radic Biol Med. 2010 Dec 1;49:1755. doi: 10.1016/j.freeradbiomed.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Kim M, et al. Impairment of microtubule system increases alpha-synuclein aggregation and toxicity. Biochem Biophys Res Commun. 2008 Jan 25;365:628. doi: 10.1016/j.bbrc.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 48.Sharma N, et al. alpha-Synuclein budding yeast model: toxicity enhanced by impaired proteasome and oxidative stress. J Mol Neurosci. 2006;28:161. doi: 10.1385/JMN:28:2:161. [DOI] [PubMed] [Google Scholar]
- 49.Flower TR, Chesnokova LS, Froelich CA, Dixon C, Witt SN. Heat shock prevents alpha-synuclein-induced apoptosis in a yeast model of Parkinson’s disease. J Mol Biol. 2005 Sep 2;351:1081. doi: 10.1016/j.jmb.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 50.Duennwald ML, Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008 Dec 1;22:3308. doi: 10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.