Abstract
Purpose of review
This review highlights recent findings about the known DNA repair machinery, its impact on chromosomal translocation mechanisms and their relevance to leukemia in the clinic.
Recent findings
Chromosomal translocations regulate the behavior of leukemia. They not only predict outcome but they define therapy. There is a great deal of knowledge on the products of leukemic translocations, yet little is known about the mechanism by which those translocations occur. Given the large number of DNA double-strand breaks that occur during normal progression through the cell cycle, especially from V(D)J recombination, stalled replication forks or failed decatenation, it is surprising that leukemogenic translocations do not occur more frequently. Fortunately, hematopoietic cells have sophisticated repair mechanisms to suppress such translocations. When these defenses fail leukemia becomes far more common, as seen in inherited deficiencies of DNA repair. Analyzing translocation sequences in cellular and animal models, and in human leukemias, has yielded new insights into the mechanisms of leukemogenic translocations.
Summary
New data from animal models suggest a two hit origin of leukemic translocations, where there must be both a defect in DNA double-strand break repair and a subsequent failure of cell cycle arrest for leukemogenesis.
Keywords: chromosomal translocations, DNA double-strand break repair, nonhomologous end-joining, V(D)J recombination
Introduction
The recognition that specific balanced chromosomal translocations not only were diagnostic of acute leukemia but also defined the future behavior of the disease was a seminal advance. Understanding how these translocations generated acute leukemias consumed much of the attention of those working in this area. The paradigm has been that acute leukemia required two genetic lesions, one which blocks differentiation of hematopoietic progenitors, usually a translocation that predates the formation of leukemia, and one that occurs later and stimulates proliferation, usually a point mutation [1]. However, the question of how such translocations are generated in the first place has received less attention. The mechanisms by which translocations originally occur are just beginning to be understood, as the pathways that repair DNA double-strand breaks (DSBs) are defined.
Requirements for chromosomal translocations
Chromosomal translocations require at least two simultaneous DSBs in separate chromosomes. A reciprocal translocation occurs when the free double-strand ends (DSEs) from one chromosome are ligated to the DSEs from the other. Thus, chromosomal translocations result from misrepair of DSBs. DSBs can be repaired through two major pathways, homologous recombination and nonhomologous end joining (NHEJ), both described below.
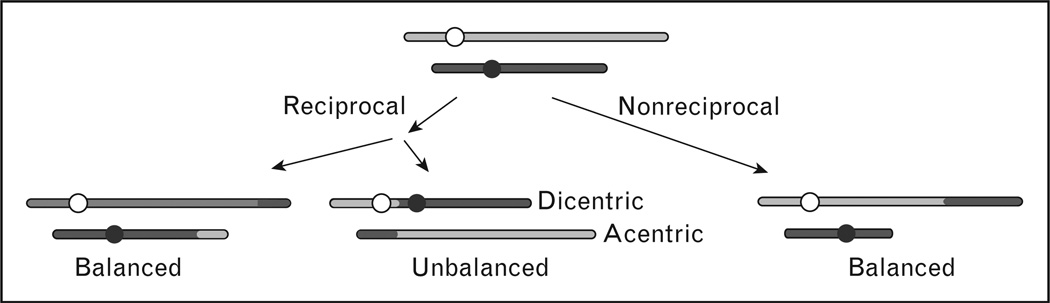
Chromosomal translocations reflect exchanges of chromosome arms and can theoretically occur between any loci on any two chromosomes. However, half of random exchanges produce unbalanced translocations that result in dicentric and acentric chromosomes (Fig. 1). Dicentric chromosomes typically cause lethal DSBs during mitosis, and acentric fragments are mitotically unstable and usually lost. Cells that manage to survive after forming a dicentric chromosome will usually show dramatic chromosomal instability as a result of breakage–bridge–fusion cycles, which are also a common outcome of telomere loss [2,3]. Translocations can also be reciprocal, with arms from different chromosomes switching places, or nonreciprocal if only one arm transfers to the broken end of another chromosome. Most leukemia translocations are reciprocal translocations. Although reciprocal translocations can arise by crossing over during homologous recombination, most translocations appear to arise by NHEJ [4,5].
Figure 1. Types of chromosomal translocations.
Nonhomologous chromosomes are diagrammed in gray or black, and circles indicate centromeres. Translocation junctions are often imperfect with small deletions or insertions created during NHEJ. Perfect junctions may be present in rare translocations that result from crossovers during homologous recombination. NHEJ, nonhomologous end joining.
Because translocation involves joining fragments from different chromosomes, the basic requirements are two simultaneous DSBs, juxtaposition of the involved chromosomes, and abnormal DSB repair. There has been some debate about whether chromosomal translocations that are commonly seen in cancer reflect biases in DSB formation and chromosome juxtaposition or tumorigenic selection pressure. Even when it is clear that translocations result from defined DSBs, the requirement for rapid and uncontrolled growth imposes a strong selection for oncogene activation or tumor suppressor inactivation, and this selection pressure adequately explains translocation patterns in many cancers.
Translocations are greatly enhanced when DSBs occur simultaneously in two chromosomes [5]. For translocations to occur, the two broken ends at a DSB must first dissociate from each other and associate with broken ends from a different chromosome. Chromosomes occupy territories within the nucleus [6], suggesting another reason why DSBs at a certain locus may lead to a limited set of translocation products. For example, three-dimensional fluorescent in-situ hybridization (3D-FISH) analysis showed that mixed lineage leukemia (MLL) and its translocation partners, Eleven–nineteen leukaemia protein (ENL) and AML fused gene from chromosome 4 (AF4) are adjacently located in interphase nuclei [7]. Two factors that may regulate chromosome DSB end-coordination during NHEJ and homologous recombination are the Ku70/Ku80 heterodimer, which has end-binding, end-protection, and self-association activities, and the RAD50 subunit of the MRE11/RAD50/NBS1 (MRN) complex.
RAD50 is a member of the structural maintenance of chromosomes (SMC) protein family that may prevent translocations and other types of aberrant DSB repair by tethering broken ends [8]. Because translocations predominantly arise via NHEJ, it was surprising that the Ku70/Ku80 complex suppressed translocations. By monitoring broken ends cytologically, it was shown that in the presence of Ku70/Ku80, broken ends remain near each other for long periods of time, thus promoting their rejoining, whereas in cells lacking Ku70, ends migrate to different positions in the nuclease and repair is thus more likely to produce a translocation [9••,10••]. This result also indicates that these translocations are mediated by an alternative NHEJ pathway, independent of Ku70/Ku80.
Sources of endogenous double-strand breaks
DSBs arise spontaneously during normal DNA metabolism, including DNA replication and repair, and during programmed genome rearrangements, such as V(D)J recombination. Many DNA lesions block DNA polymerase, causing replication fork stalling. Stalled forks are stabilized by many factors including the checkpoint kinases ataxia telangiectasia mutated (ATM) and ataxia-telangiectasia and Rad3-related protein kinase (ATR), the DNA repair protein BLM, and the multifunctional single-strand DNA (ssDNA) binding protein replication protein A (RPA) [11]. However, if a stalled fork does not restart in timely manner, it may collapse, producing a DSE. Because a DSE has no second end with which to rejoin, conservative repair of collapsed replication forks is thought to involve homologous recombination-mediated strand invasion to reform the replication fork [12]. Alternatively, if multiple forks collapse, DSEs from different chromosomes may join and produce translocations.
In addition, topoisomerase IIα (TopoIIα) creates DSBs and passes double-stranded DNA through the break, decatenating tangled chromosomes before mitosis. If decatenation fails, DSBs may form when catenated chromosomes are pulled toward opposite spindle poles [13•].
Endogenous DSBs are also normally formed during lymphoid development. In B cells for example, V(D)J recombination is initiated by specific DSBs introduced into recombination signal sequences by the RAG1/2 endonuclease. These DSBs are repaired by an error-prone, NHEJ-mediated deletion mechanism that creates novel V(D)J junctions in antibody coding sequences, a process that is instrumental in generating antibody diversity in mammals [14].
After B cells undergo the V(D)J maturation process, these antibody-producing cells can undergo a second round of genome rearrangement that switches a Cµ constant heavy chain region for a different region (Cα, Cγ, or Cε) that converts an immunoglobulin M(IgM) producing cell to one that produces immunoglobulin A (IgA), immunoglobulin G (IgG), or immunoglobulin E (IgE). This process is called class switch recombination (CSR), which is also stimulated by DSBs, but unlike V(D)J recombination, DSBs that catalyze CSR are not produced by direct nuclease cleavage. Rather, they arise indirectly following the deamination of cytidine residues in transcriptionally active, repetitive heavy chain switch regions by activation-induced cytidine deaminase. Deaminated cytidine residues are then processed by base excision repair enzymes to yield DSBs. These DSBs are repaired by a deletional NHEJ-mediated mechanism [15••].
During meiosis, endogenous DSBs are created at many loci by SPO11 endonuclease. Although SPO11 does not appear to recognize specific DNA sequences, most DSBs are created in gene promoter regions, although even within these hotspots, DSBs occur at varied locations. Meiotic DSBs are repaired by homologous recombination and typically involve interactions between homologous chromosomes [16]. As with other programmed DSBs, improper processing of meiotic DSBs can result in translocations [17].
Sources of exogenous DNA double-strand breaks
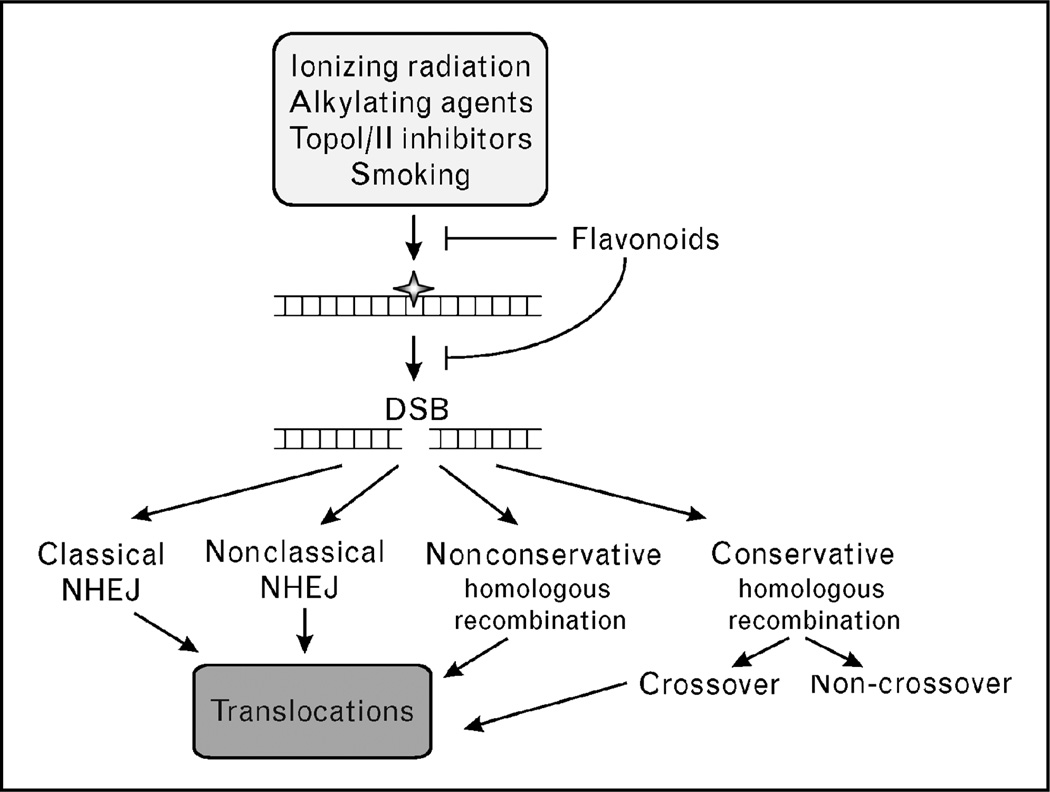
DSBs are produced by a wide variety of exogenous DNA damaging agents. Ionizing radiation, including X-rays, γ-rays, β-particles, and α-particles, can cause many types of DNA damage (Fig. 2). These include base damage (e.g. broken rings), single-strand breaks, DSBs, and inter and intra-strand crosslinks. Of these lesions, the vast majority of the cytotoxicity associated with ionizing radiation is due to DSBs. DSBs are also caused by radiomimetic drugs such as bleomycin, and by topoisomerase inhibitors.
Figure 2. Genotoxic agents produce DNA lesions that are converted to double-strand breaks and processed by several repair pathways, most of which carry a risk of inducing chromosomal translocations.
DSB, double-strand break; NHEJ, nonhomologous end joining.
Topoisomerase I (TopoI) is inhibited by the camptothecins, and TopoIIα is inhibited by the anthracyclines and etoposide, all of which are used in chemotherapy [18]. TopoI and TopoII inhibitors induce ATM Ser-1981 phosphorylation and phosphorylation of histone H2AX (γH2AX), both hallmarks of DSB damage [19]. Translocation breakpoints cluster in regions that are enriched in TopoIIα cleavage sites, matrix attachment regions, and nuclease hypersensitivity sites [20]. Importantly, TopoIIα inhibitors have been implicated in therapy-induced leukemias carrying translocations with breakpoints that map to TopoII cleavage sites, and display NHEJ-type junctions [20–22].
Finally, tobacco smoke is a known carcinogen that induces mutations and DSBs [23]. This DSB damage is mediated through free radicals and is dose dependent [24].
Mechanisms of double-strand break repair
In mammalian cells, DSBs are repaired by NHEJ and homologous recombination. Homologous recombination can occur by conservative or nonconservative pathways. Conservative homologous recombination occurs in a stepwise manner that begins with 5′–3′ end resection regulated by the MRN complex, producing 3′ ssDNA tails of a kb or longer. ssDNA is coated with RPA, which is subsequently replaced by RAD51 in a reaction promoted by mediator proteins including five RAD51 paralogs (XRCC2, XRCC3, RAD51B, RAD51C, and RAD51D), RAD52, and breast cancer type 2 susceptibility protein (BRCA2). The RAD51–ssDNA filament searches for, and invades homologous sequences. The 3′-end of the invading strand is then extended by DNA polymerase past the original site of the DSB, and may then dissociate and anneal with the other resected end. Alternatively, the second can invade producing a double Holliday junction intermediate that can be resolved in two ways to produce crossover or noncrossover products.
One nonconservative form of homologous recombination is called single-strand annealing (SSA). SSA can repair a DSB present between or within linked repeats, or between repeats on different chromosomes if DSBs occur within or near each repeat. With linked repeats, end-resection uncovers complementary single strands in the two repeats that anneal to form a single repeat. This deletes one repeat and sequences between the repeats. Translocations result when SSA occurs between unlinked repeats, but this is rare because of the requirement for DSBs to occur essentially contemporaneously in or near both repeats [25]. In the absence of BRCA2, DSBs are repaired by nonconservative (SSA) homologous recombination subpathways that presumably lead to chromosomal instability [26].
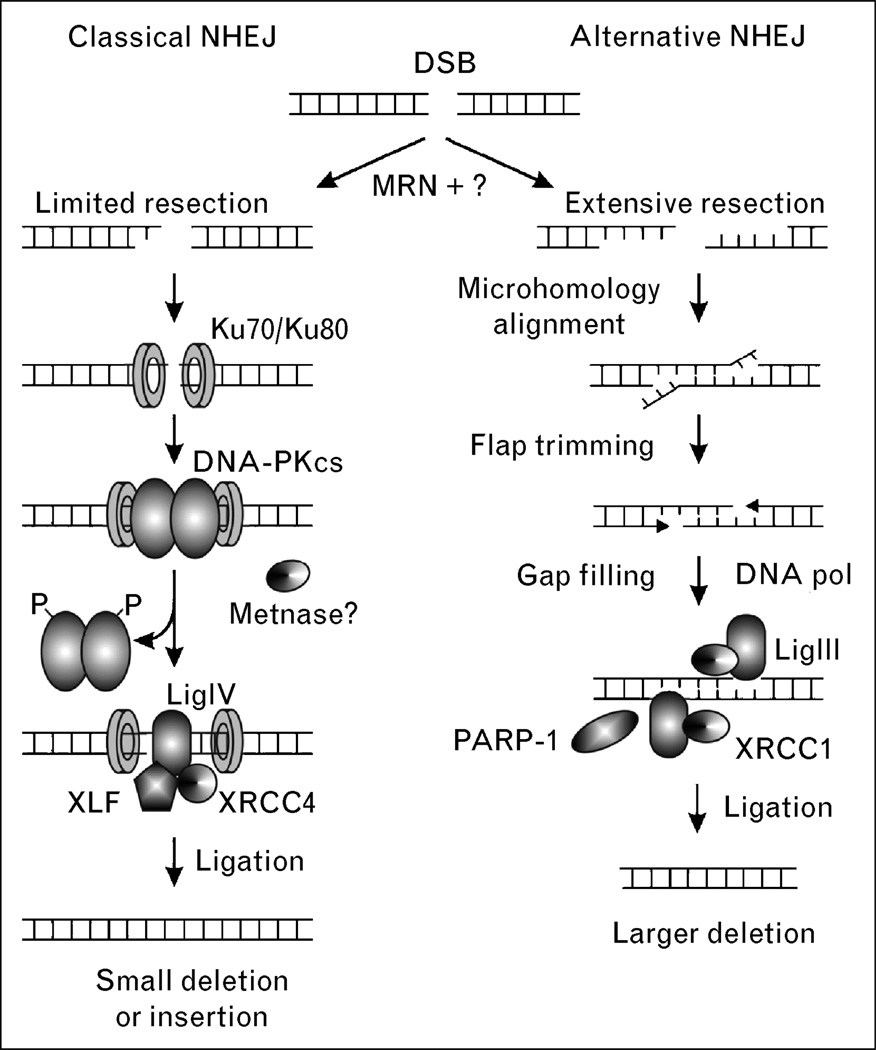
There are two NHEJ pathways, including an efficient and well defined classical pathway and a less efficient alternative pathway (Fig. 3). Classical NHEJ is mediated by Ku complex (Ku70/Ku80), the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), XRCC4, and LigIV. The first step in NHEJ is the signaling of a DSB by the MRN complex, and then binding of Ku to broken ends, and recruitment of DNA-PKcs to form the active DNA-PK complex. DNA-PK is a member of the PI3 kinase family that is activated upon binding to DNA ends. In-vivo substrates of DNA-PK are unclear; however, there are several clusters of phosphorylation sites that are either autophosphorylated, or phosphorylated by ATM, or both. DNA-PK probably recruits the LigIV/XRCC4 complex to the break site, which catalyzes the rejoining step [27]. Depending on the particular type of DSB, other NHEJ accessory factors may be involved, including MRN (important for processing the dirty ends created by ionizing radiation) and the Artemis nuclease which is important for processing hairpins formed during V(D)J recombination [28]. Cernunos (also called XLF) is another accessory protein that interacts with the LigIV/XRCC4 complex, promotes classical NHEJ [29], and has been shown to be mutated in human immunodeficiency [30]. We recently described another accessory NHEJ component termed Metnase that methylates histones to open chromatin and processes DNA ends [31]. The suppressor of variegation-enhancer of zeste-trithorax (SET) domain protein Metnase promotes foreign DNA integration and links integration to NHEJ repair [31,32•].
Figure 3. Double-strand breaks can be repaired by two NHEJ pathways.
The classical pathway may be activated via the MRN complex, followed by end resection and binding of the Ku complex. DNA-PKcs is recruited next, along with other proteins. Finally, the ends are re-ligated by XRCC4/LigIV. In the nonclassical pathway, there is more extensive resection of the DNA ends and more frequent end alignment through microhomology. DNA flaps are trimmed and LigIII/XRCC1 ligate the ends. In the nonclassical pathway there is a greater chance of errors, such as large deletions, due to the extensive DNA end resection, microhomology alignment, and flap trimming. The second pathway is more likely to lead to translocations. DNA-PKcs, DNA-dependent protein kinase; DSB, double-strand break; MRN, MRE11/RAD50/NBS1; NHEJ, nonhomologous end joining; PARP-1, poly ADP ribose polymerase-1; XRCC1, X-ray repair complementing defective repair in Chinese hamster cells 1; XRCC4, X-ray repair complementing defective repair in Chinese hamster cells 4.
Alternative NHEJ (also called backup or nonclassical NHEJ) is defined as NHEJ occurring in the absence of one or more classical NHEJ factors [14,15••,33]. Alternative NHEJ is less efficient than classical NHEJ, in part because of competition from the classical NHEJ factor DNA-PK [34]. At least in some contexts alternative NHEJ appears to be mediated by LigIII, and also involves poly(ADP)ribose polymerase-1 (PARP-1), a factor with roles in single-strand break repair [35]. In addition to its low efficiency, alternative NHEJ is characterized by a strong dependence on microhomology, and thus is more error prone than classical NHEJ. Both classical and nonclassical NHEJ are thought to mediate misrepair of DSBs leading to chromosomal translocations [15••,36].
Inherited diseases related to double-strand break repair and associated malignancies
Inherited mutations in DSB repair components in humans generate a predisposition to leukemia and other cancers. These include Bloom’s syndrome, Werner’s syndrome, ataxia telangiectasia, Nijmegen breakage syndrome, and Fanconi’s anemia (Table 1).
Table 1.
Syndromes associated with DNA repair deficiencies, increased incidence of translocations, leukemia and other cancers
| DNA repair disorder | Affected protein(s) | Clinical manifestations | Cellular function |
|---|---|---|---|
| Bloom’s syndrome | BLM | Narrowing of the face, prominent nose, a butterfly shaped rash from sun exposure, a high-pitched voice, abnormal skin pigmentation, learning disabilities, mental retardation, immune deficiency, diabetes, anemia, male infertility, irregular female menstruation and cancer predisposition | Sensitive to replication fork challenges [hydroxyurea, ultraviolet (UV)], involved in S-phase checkpoint control, TopoIII interaction and Holliday junction resolvase activity |
| Werner’s syndrome | WRN | Extreme early aging including a severe early onset cancer predisposition | 3′–5′ Exonuclease activity, Holliday junction migration activity, telomere maintenance |
| Ataxia telangiectasia | ATM (AT-mutated) | Neurodegeneration, radiosensitivity, sterility, immunodeficiency and a predisposition to lymphomas and leukemias | Kinase involved in the G1/S, S, and G2/M checkpoints and DNA repair through both homologous recombination and NHEJ |
| Nijmegen break syndrome | NBS-1 | Radiosensitivity, immunodeficiency, cancer predisposition, microcephaly and nuerodegeneration | Identification and signalling of DNA DSBs and the ensuing repair |
| Fanconi’s anemia | FANCA, B, C, D1, D2, E, F, G, I, J, L, or M | Bone marrow failure, developmental abnormalities, and severe cancer predisposition | Replication fork re-start |
ATM, ataxia telangiactasia mutated; DSBs, double-strand breaks; FANCA, Fanconi anemia-complementation group A; NHEJ, nonhomologous end joining.
Bloom’s and Werner’s syndromes result from mutations in genes that code for proteins related to the E. coli 3′–5′ RecQ helicase [37]. Bloom’s syndrome cells in culture are markedly sensitive to hydroxyurea and ultraviolet light, but not ionizing radiation [38,39]. These studies firmly implicate the Bloom’s syndrome protein BLM in replication-based DSB repair. BLM has recently been demonstrated to play a role in the processing of Holliday junctions resulting from stalled and collapsed replication forks. In this model, BLM associates with topoisomerase III (TopoIII) and functions as a Holliday junction resolvase [40,41••].
Mutations that cause Werner’s syndrome affect the WRN protein, another RecQ family helicase that also possesses an ATP dependent 3′–5′ exonuclease motif [42–46]. Constantinou et al. [47] showed that WRN has the ability to branch migrate Holliday junctions during homologous recombination. WRN has also been shown to be important in telomere maintenance and cellular senescence [48•].
Ataxia telangiectasia has been shown to be the result of mutations in a specific gene named ATM. ATM is a serine/threonine kinase activated by DNA DSBs [49•]. It is a homodimer in undamaged cells and undergoes autophosphorylation in the presence of DNA DSBs [50]. Once phosphorylated on serine 1981, ATM is activated and it phosphorylates a number of important DNA repair factors including p53, CHK2, BRCA1, RPAp34, H2AX, SMC1, Fanconi anemia-complementation group D2 (FANCD2), RAD17, Artemis and NBS1 [49•]. ATM functions as an upstream regulator of both NHEJ and homologous recombination.
Nijmegen breakage syndrome is an autosomal recessive disorder shown to be the result of mutations in the NBS1 protein. It is part of the MRN complex that is conserved from yeast to mammals and functions in the identification and signaling of DNA DSBs [51]. The MRN complex is important in the activation of ATM and the initiation of a proper DNA damage response.
Fanconi’s anemia is an inherited syndrome that presents as bone marrow failure, developmental abnormalities, and a severe cancer predisposition. Fanconi’s anemia consists of 13 complementation groups [Fanconi anemia-complementation group A (FANCA), B, C, D1, D2, E, F, G, I, J, L, and M], each of which represent a specific gene that has been mutated or deleted [52•]. Fanconi’s anemia can be autosomal recessive or sex-linked in inheritance, depending on the gene mutated. Fanconi’s anemia cells are extremely sensitive to DNA crosslinking drugs such as mitomycin C and show phenotypes consisting of abnormal cell cycle regulation (extended G2), hypersensitivity to oxygen, increased apoptosis and accelerated telomere shortening [53].
All known Fanconi’s anemia proteins function in a DNA repair pathway that is involved in the re-start of stalled replication forks. The majority of these protein products have been shown to form a complex that functions as theE3 specificity enzyme inmonoubiquitination of FANCD2 after recognition of specific DNA structures. Ubiquitinated FANCD2 performs multiple tasks including the recruitment of BRCA2, enhancement of homologous recombination, and possibly the promotion of translesion DNA synthesis [52•].
Lessons learned from models of translocation
Franco et al. [54] have generated a number of animal models that examine the role of NHEJ components in translocations and oncogenesis, and the role of DNA breaks in genomic instability and tumorigenesis [55,56]. Mice deficient for NHEJ components develop either embryonic lethality (seen in LigIV and XRCC4 deficiency) or severe combined immunodeficiency (seen with Ku70, Ku80, DNA-PK and Artemis deficiency). Occasionally these immunodeficient mice develop lymphoid neoplasia as they age. Interestingly, the embryonic lethality can be rescued by deleting p53, indicating that apoptosis after decreased repair of endogenous DSBs is the culprit for the lethality. When cells lacking both LigIV and p53 were exposed to radiation, large numbers of translocations were seen, and these mice developed lymphoma at a high rate. In contrast, wild-type or p53-deficient cells had few genomic abnormalities and a low incidence of lymphoma (LigIV-deficient but p53 +/+ cells arrested after radiation and could not be tested). These studies revealed several important concepts. First, the relative lack of translocations after the same radiation dose in wild-type cells compared with the LigIV−/− and p53 −/− cells indicates that the NHEJ pathway is less promiscuous than once thought. Second, translocations occur when a damaged cell with deficient NHEJ is forced to progress through the cell cycle, such as when p53 is deleted. This same phenomenon has been seen in p53 −/− mice also deficient inKu80,DNA-PKcs, orXRCC4.These mice also develop lymphomas at a high rate.
Specific mechanisms of translocation in human leukemia
Etoposide has been associated with chromosomal translocations that result in acute leukemia, most commonly through translocations involving the MLL gene [57,58], or the runt-related transcription factor 1 (RUNX1) gene [59]. Balanced translocations involving chromosome bands 11q23 and 21q22 are highly characteristic of myelodysplasia and leukemia following therapy with etoposide [60–62]. The MLL translocations occur in a defined breakpoint cluster region (BCR) that spans 8.5 kbp between exons 5 and 11. This MLL BCR contains known nuclear matrix attachment regions, DNase hypersensitive sites, and TopoIIα cleavage sites. Treatment of cells with etoposide results in DSBs within this BCR [63–65].
Many breakpoints from both MLL and RUNX1 translocations have been sequenced. Virtually none of these breakpoint junctions have any sequence homology between the partner genes, indicating that it is unlikely that homologus recombination played any role in the aberrant ligation of the translocation partners [20]. The junction sequences, however, do reveal classic signatures of NHEJ repair, such as microdeletions, duplications, microhomologies, and nontemplate insertions [20].
Leukemic rearrangements of MLL after DNA damage can also take the form of partial tandem duplications (PTDs) as opposed to reciprocal translocations. PTDs at the MLL locus are probably mediated by inter or intra-chromosomal SSA between Alu sequences within the MLL locus [66]. In seven of nine MLL PTD junction sequences analyzed, recombination took place between Alu elements within the MLL gene locus [67]. This is consistent with SSA repair. Although most breakpoint junction sequences do not show evidence for such Alu-mediated recombination [20], as described above, when DSBs occur in Alu sequences, translocations can be generated by the SSA pathway [68].
In lymphoid cells endogenous DSBs are generated during V(D)J recombination or CSR. There are sequences elsewhere in the genome that resemble the heptamer and nonamer V(D)J recombination signal sequences [69]. These sites can become targets of RAG-mediated DSB generation and subsequent leukemogenic translocations [70] and can mediate LMO2, Ttg1, SCL/TAL1, and SIL translocations in acute T-cell leukemia [70].
Class switching from the µ heavy chain locus to the α, β, δ, or ε loci in the maturation of the immunoglobulin response can also generate DSBs that can be targets of translocations. For example, in the t(8;14) seen in Burkitt’s lymphoma and some B-cell acute lymphoblastic leukemia (ALL), c-myc can be translocated to the switch region of the µ heavy chain gene [71].
Conclusion
It is clear that NHEJDNA repair components are critical for preventing leukemogenic translocations. The finding that leukemia is a well described complication of inherited disorders of NHEJ repair as opposed to inherited disorders of homologus recombination such as BRCA1/2 deficiency is clinical evidence for this postulate. The role of subtle decreases in NHEJ activity in genomic instability in leukemia is not well explored. The most fascinating question is whether such subtle genomic instability predates the leukemogenic translocation, or whether it is a result of the leukemogenic transformation. There are several important clinical implications of defects in DNA repair predating the original translocation. First, such defects could be used to screen for patients at risk for leukemia. Second, reversing such defects would be an important therapeutic target. Such therapy could be used especially in environmental situations where exposure to DNA damaging agents could lead to leukemia, such as an accidental or purposeful radiation exposure.
There has been considerable discussion about leukemia being the result of two genetic lesions, one that stimulates proliferation, usually a point mutation, and one that blocks differentiation, usually a translocation. However, these genetic lesions are probably the result of a preexisting genomic instability. This preexisting genomic instability may be the result of small defects in NHEJ repair and a coincidental defect in DNA damage cell cycle arrest. The coincidental existence of these two defects could underlie much of leukemia, and produce the translocations that have defined our efforts in this disease thus far. It is possible that these defects become obscured by the resultant transformation of the hematopoietic progenitor after translocation. Perhaps we should be focusing on the conditions that predate the translocation, as this may be an easier place to intervene. One could envision the time when our efforts in leukemia prevention exceed those in leukemia therapy.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 425–426).
- 1.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 2.Murnane JP. Telomeres and chromosome instability. DNA Repair. 2006;5:1082–1092. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 3.Vukovic B, Beheshti B, Park P, et al. Correlating breakage–fusion–bridge events with the overall chromosomal instability and in vitro karyotype evolution in prostate cancer. Cytogenet Genome Res. 2007;116:1–11. doi: 10.1159/000097411. [DOI] [PubMed] [Google Scholar]
- 4.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by inter-chromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 6.Cremer T, Cremer M, Dietzel S, et al. Chromosome territories–a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Gue M, Sun JS, Boudier T. Simultaneous localization of MLL, AF4 and ENL genes in interphase nuclei by 3D-FISH: MLL translocation revisited. BMC Cancer. 2006;6:20. doi: 10.1186/1471-2407-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jager M, van Noort J, van Gent DC, et al. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 9. Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. The major repair pathway mediating translocations is NHEJ. However, the Ku complex, a major component of classical NHEJ, is not required indicating a significant role for nonclassical NHEJ in translocations.
- 10. Soutoglou E, Dorn JF, Sengupta K, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. Broken ends show spatial stability and do not migrate around the cell. This stability is a key factor in preventing translocations.
- 11.Andreassen PR, Ho GP, D’Andrea AD. DNA damage responses and their many interactions with the replication fork. Carcinogenesis. 2006;27:883–892. doi: 10.1093/carcin/bgi319. [DOI] [PubMed] [Google Scholar]
- 12.Shen Z, Nickoloff JA. Mammalian homologous recombination repair and cancer intervention. In: Wei Q, Li L, Chen DJ, editors. DNA repair, genetic instability, and cancer. Singapore: World Scientific Publishing Co.; 2007. pp. 119–156. [Google Scholar]
- 13. Damelin M, Bestor TH. The decatenation checkpoint. Br J Cancer. 2007;96:201–205. doi: 10.1038/sj.bjc.6603537. A complete review of chromosome decatenation, highlighting the role it plays in chromosomal stability.
- 14.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair. 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 15. Yan CT, Boboila C, Souza EK, et al. IgH class switching and translocations use a robust nonclassical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. Shows that translocations associated with CSR are often mediated by nonclassical NHEJ.
- 16.Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans. 2006;34(Pt 4):523–525. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- 17.Bandyopadhyay R, Heller A, Knox-DuBois C, et al. Parental origin and timing of de novo Robertsonian translocation formation. Am J Hum Genet. 2002;71:1456–1462. doi: 10.1086/344662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang CH, Treat J. New advances in lung cancer chemotherapy: topotecan and the role of topoisomerase I inhibitors. Oncology. 2001;61(Suppl 1):14–24. doi: 10.1159/000055387. [DOI] [PubMed] [Google Scholar]
- 19.Kurose A, Tanaka T, Huang X, et al. Assessment of ATM phosphorylation on Ser-1981 induced by DNA topoisomerase I and II inhibitors in relation to Ser-139-histone H2AX phosphorylation, cell cycle phase, and apoptosis. Cytometry A. 2005;68:1–9. doi: 10.1002/cyto.a.20186. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair. 2006;5:1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 21.Aplan PD. Causes of oncogenic chromosomal translocation. Trends Genet. 2006;22:46–55. doi: 10.1016/j.tig.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felix CA, Kolaris CP, Osheroff N. Topoisomerase II and the etiology of chromosomal translocations. DNA Repair. 2006;5:1093–1108. doi: 10.1016/j.dnarep.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 23.Kim H, Liu X, Kobayashi T, et al. Reversible cigarette smoke extract-induced DNA damage in human lung fibroblasts. Am J Respir Cell Mol Biol. 2004;31:483–490. doi: 10.1165/rcmb.2002-0300OC. [DOI] [PubMed] [Google Scholar]
- 24.Collier AC, Dandge SD, Woodrow JE, Pritsos CA. Differences in DNA-damage in nonsmoking men and women exposed to environmental tobacco smoke (ETS) Toxicol Lett. 2005;158:10–19. doi: 10.1016/j.toxlet.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Weinstock DM, Richardson CA, Elliott B, Jasin M. Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair. 2006;5:1065–1074. doi: 10.1016/j.dnarep.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 26.Tutt A, Bertwistle D, Valentine J, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobrich M, Jeggo PA. Harmonising the response to DSBs: a new string in the ATM bow. DNA Repair. 2005;4:749–759. doi: 10.1016/j.dnarep.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 29.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4–DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 30.Buck D, Malivert L, de Chasseval R, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 31.Lee SH, Oshige M, Durant ST, et al. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci U S A. 2005;102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Roman Y, Oshige M, Lee YJ, et al. Biochemical characterization of a SET and transposase fusion protein. Metnase: its DNA binding and DNA cleavage activity. Biochemistry. 2007;46:11369–11376. doi: 10.1021/bi7005477. Emphasizes the importance of nuclease activities in NHEJ repair.
- 33.Wang H, Rosidi B, Perrault R, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 34.Perrault R, Wang H, Wang M, et al. Backup pathways of NHEJ are suppressed by DNA-PK. J Cell Biochem. 2004;92:781–794. doi: 10.1002/jcb.20104. [DOI] [PubMed] [Google Scholar]
- 35.Wang M, Wu W, Wu W, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Povirk LF. Biochemical mechanisms of chromosomal translocations resulting from DNA double-strand breaks. DNA Repair. 2006;5:1199–1212. doi: 10.1016/j.dnarep.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 37.Umezu K, Nakayama K, Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci U S A. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W, Seki M, Otsuki M, et al. The absence of a functional relationship between ATM and BLM, the components of BASC, in DT40 cells. Biochim Biophys Acta. 2004;1688:137–144. doi: 10.1016/j.bbadis.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Seki M, Narita Y, et al. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 2000;19:3428–3435. doi: 10.1093/emboj/19.13.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. First report linking the Bloom’s syndrome protein to repair of replication fork damage.
- 42.Shen JC, Gray MD, Oshima J, et al. Werner syndrome protein. I. DNA helicase and DNA exonuclease reside on the same polypeptide. J Biol Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 43.Huang S, Li B, Gray MD, et al. The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease. Nature Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S, Beresten S, Li B, et al. Characterization of the human and mouse WRN 3′→5′ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamath-Loeb AS, Shen JC, Loeb LA, Fry M. Werner syndrome protein. II. Characterization of the integral 3′→5′ DNA exonuclease. J Biol Chem. 1998;273:34145–34150. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki N, Shimamoto A, Imamura O, et al. DNA helicase activity in Werner’s syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res. 1997;25:2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Constantinou A, Tarsounas M, Karow JK, et al. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. Comprehensive review of the human DNA damage clinical syndromes, Bloom’s and Werner’s, that result from mutations of proteins related to RecQ.
- 49. Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. Comprehensive review of ATM function.
- 50.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 51.Demuth I, Digweed M. The clinical manifestation of a defective response to DNA double-strand breaks as exemplified by Nijmegen breakage syndrome. Oncogene. 2007;26:7792–7798. doi: 10.1038/sj.onc.1210876. [DOI] [PubMed] [Google Scholar]
- 52. Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. Description of the role of the Fanconi’s anemia complex in DNA repair.
- 53.Dokal I. Fanconi’s anaemia and related bone marrow failure syndromes. Br Med Bull. 2006;77–78:37–53. doi: 10.1093/bmb/ldl007. [DOI] [PubMed] [Google Scholar]
- 54.Franco S, Alt FW, Manis JP. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair. 2006;5:1030–1041. doi: 10.1016/j.dnarep.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 55.Ferguson DO, Alt FW. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene. 2001;20:5572–5579. doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- 56.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 57.Libura J, Slater DJ, Felix CA, Richardson C. Therapy-related acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion. Blood. 2005;105:2124–2131. doi: 10.1182/blood-2004-07-2683. [DOI] [PubMed] [Google Scholar]
- 58.Felix CA. Leukemias related to treatment with DNA topoisomerase II inhibitors. Medical and pediatric oncology. 2001;36:525–535. doi: 10.1002/mpo.1125. [DOI] [PubMed] [Google Scholar]
- 59.Larson RA, Le Beau MM, Ratain MJ, Rowley JD. Balanced translocations involving chromosome bands 11q23 and 21q22 in therapy-related leukemia. Blood. 1992;79:1892–1893. [PubMed] [Google Scholar]
- 60.Pedersen-Bjergaard J, Philip P. Two different classes of therapy-related and de-novo acute myeloid leukemia? Cancer Genet Cytogenet. 1991;55:119–124. doi: 10.1016/0165-4608(91)90246-q. [DOI] [PubMed] [Google Scholar]
- 61.Izumi T, Ohtsuki T, Ohya K, et al. Therapy-related leukemia with a novel 21q22 rearrangement. Cancer Genet Cytogenet. 1996;90:45–48. doi: 10.1016/0165-4608(96)00044-1. [DOI] [PubMed] [Google Scholar]
- 62.Slovak ML, Bedell V, Popplewell L, et al. 21q22 balanced chromosome aberrations in therapy-related hematopoietic disorders: report from an international workshop. Genes Chromosomes Cancer. 2002;33:379–394. doi: 10.1002/gcc.10042. [DOI] [PubMed] [Google Scholar]
- 63.Strissel PL, Strick R, Rowley JD, Zeleznik-Le NJ. An in vivo topoisomerase II cleavage site and a DNase I hypersensitive site colocalize near exon 9 in the MLL breakpoint cluster region. Blood. 1998;92:3793–3803. [PubMed] [Google Scholar]
- 64.Domer PH, Head DR, Renganathan N, et al. Molecular analysis of 13 cases of MLL/11q23 secondary acute leukemia and identification of topoisomerase II consensus-binding sequences near the chromosomal breakpoint of a secondary leukemia with the t(4;11) Leukemia. 1995;9:1305–1312. [PubMed] [Google Scholar]
- 65.Stanulla M, Schunemann HJ, Thandla S, et al. Pseudo-rearrangement of the MLL gene at chromosome 11q23: a cautionary note on genotype analysis of leukaemia patients. Mol Pathol. 1998;51:85–89. doi: 10.1136/mp.51.2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aplan PD. Chromosomal translocations involving the MLL gene: molecular mechanisms. DNA Repair. 2006;5:1265–1272. doi: 10.1016/j.dnarep.2006.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strout MP, Marcucci G, Bloomfield CD, Caligiuri MA. The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia. Proc Natl Acad Sci U S A. 1998;95:2390–2395. doi: 10.1073/pnas.95.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elliott B, Richardson C, Jasin M. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell. 2005;17:885–894. doi: 10.1016/j.molcel.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 69.Lewis SM, Agard E, Suh S, Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol Cell Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raghavan SC, Kirsch IR, Lieber MR. Analysis of the V(D)J recombination efficiency at lymphoid chromosomal translocation breakpoints. J Biol Chem. 2001;276:29126–29133. doi: 10.1074/jbc.M103797200. [DOI] [PubMed] [Google Scholar]
- 71.Unniraman S, Schatz DG. AID and Igh switch region-Myc chromosomal translocations. DNA Repair. 2006;5:1259–1264. doi: 10.1016/j.dnarep.2006.05.019. [DOI] [PubMed] [Google Scholar]